Abstract
Spodoptera frugiperda (Sf) cell lines are used to produce several biologicals for human and veterinary use. Recently, it was discovered that all tested Sf cell lines are persistently infected with Sf-rhabdovirus, a novel rhabdovirus. As part of an effort to search for other adventitious viruses, we searched the Sf cell genome and transcriptome for sequences related to Sf-rhabdovirus. To our surprise, we found intact Sf-rhabdovirus N- and P-like ORFs, and partial Sf-rhabdovirus G- and L-like ORFs. The transcribed and genomic sequences matched, indicating the transcripts were derived from the genomic sequences. These appear to be endogenous viral elements (EVEs), which result from the integration of partial viral genetic material into the host cell genome. It is theoretically impossible for the Sf-rhabdovirus-like EVEs to produce infectious virus particles as 1) they are disseminated across 4 genomic loci, 2) the G and L ORFs are incomplete, and 3) the M ORF is missing. Our finding of transcribed virus-like sequences in Sf cells underscores that MPS-based searches for adventitious viruses in cell substrates used to manufacture biologics should take into account both genomic and transcribed sequences to facilitate the identification of transcribed EVE's, and to avoid false positive detection of replication-competent adventitious viruses.
Keywords: Adventitious virus, Endogenous viral element, Baculovirus insect cell system, Massively parallel sequencing, Sf-rhabdovirus, Spodoptera frugiperda cells
1. Introduction
Insect cell lines are commonly used with the baculovirus-insect cell system (BICS) to produce recombinant proteins, including several biologicals for human and veterinary use (see Table 1). Presently, most BICS-produced biologicals are manufactured using cells derived from the fall armyworm, Spodoptera frugiperda (Sf). All Sf cell lines currently used to produce biologicals are derived from a single progenitor cell line, IPLB-Sf-21, also known as Sf-21[1]. These derivatives include the most widely used cell line, Sf9[2], and several others isolated by various biological manufacturers, such as Super9[3] (GlaxoSmithKline) and Sf900+[4] (marketed as expresSF+, Protein Sciences Corp.).
Table 1.
Human and veterinary biologicals produced in insect cells
| Drug name | Protein | Application | Company | Cell line | Approval (year) / trial stage* |
|---|---|---|---|---|---|
| Human biological | |||||
| Cervarix | HPV L1 protein | HPV Vaccine | GlaxoSmithKline Biologicals | High5 | FDA (2009) EMA (2007) |
| Provenge | PAP-GM-CSF fusion | Prostate cancer therapy | Dendreon Corp. | Sf21 | FDA (2010) EMA† (2013) |
| Flublok | Influenza HA | Seasonal influenza vaccine | Protein Sciences Corp. | expresSF+ | FDA approved (2013) |
| n/a | H7N9 HA, NA, M | H7N9 influenza vaccine | Novavax | Sf9 | Phase II (completed) |
| n/a | H5N1 HA, NA, M | H5N1 influenza vaccine | Novavax | Sf9 | Phase II (completed) |
| n/a | Influenza HA, NA, M | Seasonal influenza vaccine | Novavax | Sf9 | Phase II (completed) |
| n/a | Ebola G | Ebola vaccine | Novavax | Sf9 | Phase I (completed) |
| n/a | RSV F | RSV vaccine | Novavax | Sf9 | Phase III (ongoing) |
| Panblok | Influenza HA | H5N1 influenza vaccine | Protein Sciences Corp. | expresSF+ | Phase II (completed) |
| Panblok | Influenza HA | H7N9 influenza vaccine | Protein Sciences Corp. | expresSF+ | Phase II (completed) |
| Diamyd | Human GAD65 | Diabetes autoantigen therapy | Diamyd Therapeutics AB | expresSF+ | Phase III (terminated) |
| Glybera | rAAV vector | LPL deficiency gene therapy | uniQure | expresSF+ | Phase III (completed) |
| rHEV | 56 kDa ORF2 | Hepatitis E vaccine | GlaxoSmithKline Biologicals | High5 | Phase II (completed) |
| n/a | Parvovirus VP1 / 2 | Parvovirus B19 vaccine | Meridian Life Science Inc. | Sf9 | Phase II (terminated) |
| n/a | Norovirus capsid | Norovirus vaccine | Ligocyte (Takeda) | Sf9 | Phase II (completed) |
| n/a | FGF-1 | Traumatic Spinal Cord Injury | BioArctic Neuroscience AB | expresSF+ | Phase I/II (recruiting) |
| Veterinary biologicals | |||||
| Porcilis Pesti | CSFV E2 | CSFV Vaccine | MSD Animal Health | Sf-21-CB | EMA (2000) |
| Bayovac CSF E2 / Advasure | CSFV E2 | CSFV Vaccine | Bayer / Pfizer Animal Health | Sf21 | EMA† (2001) |
| Circumvent PCV | PCV2 ORF2 protein | PCV2 Vaccine | Merck Animal Health | Sf9 | USDA (2007) EMA (2009) |
| Ingelvac CircoFLEX | PCV2 ORF2 protein | PCV2 Vaccine | Boehringer Ingelheim | expresSF+ | USDA (2006) EMA (2008) |
| Porcilis PCV | PCV2 ORF2 protein | PCV2 Vaccine | MSD Animal Health | Sf-21-CB | EMA (2009) |
| Virbagen Omega | Feline Interferon ω | Immunostimulant | VirBac SA | Bm-N | EMA (2001) |
Only US / EU approval or US clinical trial stage shown. Several products in the list have been approved for clinical use outside the USA / EU or are in trials outside the USA.
License has been withdrawn since
Recently, it was found that all Sf cell lines tested, including those used for biological manufacturing, are contaminated with Sf-rhabdovirus, a previously unknown adventitious virus[5, 6]. At GlycoBac, we found that all Sf cell lines in our labs, including Sf-21, Sf9, and expresSF+ are also contaminated with this virus. Considering that the ancestral Sf-21 cell line is contaminated, Sf-rhabdovirus infection has likely been passed on to Sf-21-derived cell lines by vertical transmission.
While the FDA has previously approved biologicals produced using Sf-rhabdovirus-contaminated cell lines, best practices will demand an Sf-rhabdovirus-negative cell line for new biological license applications[7]. We recently resolved this issue by establishing Sf-RVN, a new, Sf-rhabdovirus-negative Sf cell line that is not contaminated with Sf-rhabdovirus [8].
To search for other potential adventitious viruses that might be present in Sf cells, we searched the Sf cell genome and transcriptome for viral or virus-like sequences using the BLAST algorithm. As part of this effort, we searched the Sf cell genome and transcriptome using the Sf-rhabdovirus proteome as the query in a TBLASTN search. To our surprise, we found additional Sf-rhabdovirus-like sequences that were clearly distinct from, but closely related to Sf-rhabdovirus in both the Sf cell genome and transcriptome. Here, we describe these novel sequences and their relationships to their Sf-rhabdovirus homologues, and discuss the implications of our findings for adventitious virus searches using massively parallel sequencing combined with bioinformatics.
2. Materials and Methods
2.1. Bioinformatics
Bioinformatics searches of the Sf cell genome and transcriptome were conducted using the publicly accessible NCBI TBLASTN interface (blast.ncbi.nlm.nih.gov/blast/Blast.cgi). The Sf cell genome (Whole Genome Shotgun, WGS) and transcriptome (Transcribed Sequence Assembly, TSA) were queried with the concatenated predicted Sf-rhabdovirus proteome using the default settings except the low complexity region filter was disabled and word size was 3, and the E-value cutoff was raised to 100. Bioinformatics searches against the silkworm (Bombyx mori) genome were conducted using the publicly accessible silkdb TBLASTN interface (http://www.silkdb.org/silksoft/blast2.html) using the same settings. Ka/Ks calculations were performed using a publicly accessible interface at the Norwegian Bioinformatics Platform (http://services.cbu.uib.no/tools/kaks) using the default settings, with homologous proteins encoded by the Sanxia water strider virus, a related insect rhabdovirus, as the comparator [9].
2.2. Cells and cell culture
Sf-RVN cells [8] and Sf9 cells [10] are previously described Spodoptera frugiperda cell lines. Sf9 and Sf-RVN cells were routinely grown in 50 mL ESF-921 medium (96-001, Expression Systems) in 125 mL deLong flasks at 28°C at 125 RPM as described previously [8].
2.3. DNA and RNA extraction, reverse transcription
DNA was extracted from 2×107 exponential-phase Sf9 or Sf-RVN cells as described previously[11], and dissolved in 500 μL TE buffer. RNA was extracted from 1×106 exponential-phase Sf9 or Sf-RVN cells with the E.Z.N.A. HP Total RNA Kit (R6812, Omega Biotek) and treated with RNAse-free DNAse (E1091, Omega Biotek) according to the manufacturer's instructions. RNA was reverse-transcribed with the ProtoScript® II RT kit (M0368, New England Biolabs) using random hexamers according to the manufacturer's instructions.
2.4. PCR amplification, gel electrophoresis and sequencing
PCRs were routinely performed in 50 μL reactions containing 1 μM of each primer, 1 μL dNTP mix (N0447, New England Biolabs), and 1 U Phusion DNA polymerase (M0530, New England Biolabs) in the manufacturer's 1X HF buffer with 1 μL DNA or reverse transcribed RNA as the template in a Biometra Tprofessional thermal cycler. Negative control reactions contained no template or mock reverse-transcribed RNA from reactions where the reverse transcriptase was omitted. PCR reaction products were separated on 1% agarose gels in Tris acetate EDTA buffer. DNA was extracted from excised bands using the E.Z.N.A. Gel Extraction Kit (D2501, Omega Biotek) according to the manufacturer's instructions, and sequenced by a commercial service provider (Genewiz). Sequences of oligonucleotides used for the PCRs and sequencing reactions are shown in Supplemental Table 1.
2.5. Sequence comparison
Sequences were assembled and compared using Vector NTI 10.3.1 (Invitrogen), and alignments were performed using ClustalX 2.1. Phylogenetic trees were drawn using the Phylip 3.695 package.
3. Results
3.1. Identification of the Sf-rhabdovirus genome in the assembled Sf cell transcriptome, comparison to published sequences
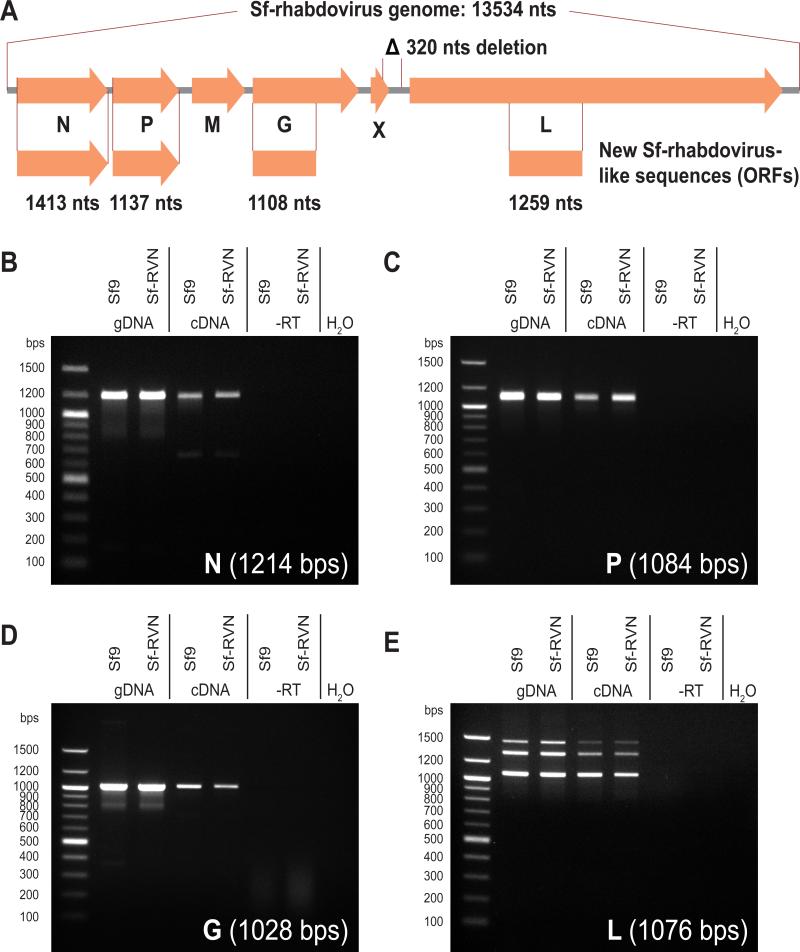
In an effort to search Sf cells for adventitious rhabdoviruses other than Sf-rhabdovirus [5], we searched the published Sf21 cell genome and transcriptome [12, 13] for sequences putatively encoding Sf-rhabdovirus-like proteins using the TBLASTN algorithm with the Sf-rhabdovirus proteome as the query. As expected, the assembled Sf-rhabdovirus genome was readily found in the Sf cell transcriptome. This assembled sequence (Genbank accession number GCTM01002581) mostly matched the previously published Sf-rhabdovirus genomic sequences. Surprisingly, in comparison with the sequence published by Ma et al. [5], a 320 nt deletion was found, spanning the last 120 nts of the putative X gene ORF and the first 200 nts of the intergenic region between the X and L ORFs (shown schematically in Figure 1 A). The same deletion was present in the sequence published by Takeda Vaccines, Inc [6], and in Sf-rhabdovirus RNA from our in-house Sf9 cells (data not shown). The presence of a 320 bps deletion in these Sf-rhabdovirus sequences suggests that the X gene product is not required for persistence in cultured Sf cells. Small, non-conserved, putative accessory genes such as the Sf-rhabdovirus X gene are common in genomes of Rhabdoviruses and other ssRNA viruses, and have been suggested to potentially function as viral porins [14].
Figure 1.
Sf-rhabdovirus-like sequences are present in and transcribed from Sf cell genomes. A) The relative sizes and positions of the newly found N-, P-, G-, and L-like ORFs as compared to their homologs in the Sf-rhabdovirus genome, and the position of a 320 nt deletion found in some Sf-rhabdovirus sequences. PCR demonstrating presence and transcription of Sf-rhabdovirus N- (B), P- (C), G- (D) and L-like (E) sequences in Sf9 and Sf-RVN cells. Expected PCR amplimer sizes are indicated. Templates were genomic DNA (gDNA), reverse-transcribed total cellular RNA (cDNA), mock reverse-transcribed total cellular RNA (−RT) or no template (H2O).
Apart from the deletion in X, there were differences between the leader and trailer sequences of the three Sf-rhabdovirus genomes, which can likely be ascribed to incorrect or partial coverage of one or more sequences. Furthermore, several SNPs could be identified where the TSA-derived Sf-rhabdovirus sequence was different from the sequences published either by either Takeda Vaccines, Inc [6], or Ma et al. [5]. Such sequence variants can readily be explained by the poor replication fidelity provided of rhabdoviral RNA directed RNA polymerases[15, 16].
3.2. Sf-rhabdovirus N-, P-, G-, and L-like sequences are present in and transcribed from both the Sf9 and Sf-RVN cell genome
Surprisingly, our TBLASTN searches also produced four additional significant hits for the N, P, G, and L genes in both the Sf21 cell genome and transcriptome. The GenBank accession numbers of these sequences and their positions are listed in Table 2. Curated and annotated versions of these sequences are also included in Genbank format as Supplementary material. The new Sf-rhabdovirus-like sequences found in the transcriptome and genome largely overlapped and were identical in the overlapping regions, providing a first hint that the Sf-rhabdovirus-like sequences found in the transcriptome were derived from the Sf cell genome.
Table 2.
Accession numbers and positions of the Sf-rhabdovirus N-, P-, G-, and L-like ORFs found in the Sf21 cell genome and transcriptome
| ORF | WGS Genbank accession number | Position (strand) | TSA Genbank accession number | Position (strand) |
|---|---|---|---|---|
| N | JQCY02013702.1 | 2041 – 3543 (−) | GCTM01018210.1 | 6 – 1277 (−) |
| P | JQCY02009702.1 | 9618 – 10700 (−) | GCTM01015401.1 | 1216 – 2352 (−) |
| G | JQCY02005970.1 | 100 – 1186 (−) | GCTM01008235.1 | 98 – 1165 (+) |
| L | JQCY02007162.1 | 12405 – 13664 (−) | GCTM01018396.1 | 1 – 438 (+) |
The newly found Sf-rhabdovirus N- and P-like sequences comprised intact ORF's, and the N-like ORF had a polyadenyl tract immediately following the stop codon. The newly identified Sf-rhabdovirus G- and L-like sequences comprised partial ORFs, with the G gene ORF missing approximately 248 C-terminal codons, and the L gene ORF missing approximately 570 N-terminal, and 1150 C-terminal codons. The relative sizes and positions of the newly found ORFs as compared to their homologous ORFs in the Sf-rhabdovirus genome are shown schematically in Figure 1 A. The DNA sequences of the newly found N-, P-, G-, and L-like ORFs, and their predicted translated amino acid sequences are shown with their Sf-rhabdovirus homologs in Supplementary Figures 1, 2, 3, and 4, respectively.
In order to determine if these newly found Sf-rhabdovirus N-, P-, G-, and L-like ORFs were present in the genome of Sf9 and Sf-RVN cells, we next attempted to PCR amplify them using genomic DNA from those cell lines as the template. In order to determine whether they are transcribed into RNA, we also performed PCRs with cDNA as the template. To rule out false positive results due to contamination with genomic DNA, RNA was DNAse-treated, and control PCRs with mock reverse-transcribed RNA or water as the template were included. The results of these PCRs are shown in Figure 1 B-E. In all cases strong bands of the expected size were obtained from reactions containing genomic DNA or cDNA from both Sf9 and Sf-RVN cells. The amplimers were directly sequenced, and corresponded exactly to the expected sequences, indicating that the amplification products were specific, and that the Sf-rhabdovirus N-, P-, G-, and L-like sequences were present in and expressed from the genome of both cell lines
We also performed PCRs to determine if the predicted, abrupt ends of the partial G-and L-like ORFs were correct. The results of those PCRs showed that the partials ORFs ended where indicated by the genomic sequences (data not shown).
3.3. Comparison of the Sf-rhabdovirus N-, P-, G-, and L-like sequences to their Sf-rhabdovirus homologs
The similarity of the newly found Sf-rhabdovirus N-, P-, G-, and L-like sequences to their Sf-rhabdovirus homologs is evident from the alignments in Supplementary Figures 1 - 4. Of interest is that the third codon positions are most highly variable, and largely randomized. However, the alignments also show that the overall levels of similarity are distinct for the 4 fossils. The similarity between the Sf-rhabdovirus N-, P-, G-, and L-like sequences and their Sf-rhabdovirus homologs is quantified in Table 3.
Table 3.
Similarity between the Sf-rhabdovirus N-, P-, G-, and L-like EVE's and their Sf-rhabdovirus homologs
| Gene | N | P | G | L |
|---|---|---|---|---|
| % amino acid identity | 35% | 79% | 82% | 92% |
| % amino acid similarity | 52% | 86% | 91% | 96% |
| BLASTP amino acids aligned | all | all | all | all |
| BLASTP E value | 1e-92 | 0.0 | 0.0 | 0.0 |
| BLASTN nucleotides aligned | 37 / 1506 | 1133 / 1137 | 1059 / 1085 | 1259 / 1259 |
| BLASTN E value | 3e-07 | 2e-134 | 0.0 | 0.0 |
| Ka / Ks ratio | 0.5648 | 0.1305 | 0.1121 | 0.055 |
As can be seen, the overall level of similarity at both the nucleic acid levels as well as the amino acid level is N<P<G<L. Further TBLASTN searches of the newly found Sf-rhabdovirus-like sequences against the Genbank non-redundant (nr) database confirmed that they are most similar to Sf-rhabdovirus, and less similar to other related mononegaviruses.
3.4. Rhabdovirus-like sequences in the silkworm genome are distinct from Sf-rhabdovirus
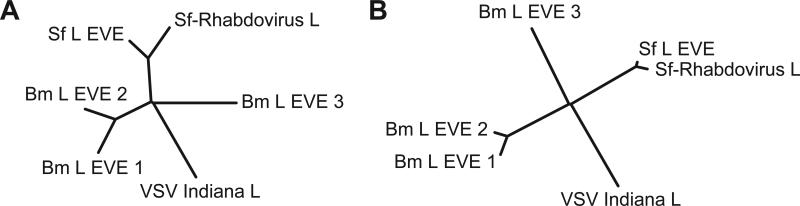
The Sf-rhabdovirus infection in Sf cell lines could conceivably have originated from the S. frugiperda pupa originally used as a source of ovarian tissue or the silkworm hemolymph used to supplement the culture medium in the initial stages of cell culture [1]. The presence of Sf-rhabdovirus-like sequences in the Sf cell genome suggests that Sf-rhabdovirus is associated with S. frugiperda, and not B. mori. This conclusion would be strengthened if rhabdovirus-like sequences in the B. mori genome are less similar to Sf-rhabdovirus than the rhabdovirus-like sequences in the S. frugiperda genome. Thus, we searched the B. mori genome for Sf-rhabdovirus-like sequences. We found three previously unidentified rhabdovirus L-like sequences in the B. mori genome. The GenBank accession numbers of these sequences and their positions are listed in Table 4. Curated and annotated versions of these sequences are also included in Genbank format as Supplementary material. A comparison of these rhabdovirus-like sequences from B. mori to the Sf-rhabdovirus-like sequences from the Sf cell genome and Sf-rhabdovirus revealed that the B. mori sequences were distinct from the latter, and clustered separately (Figure 2 A). A comparison of their translations similarly showed that the B. mori rhabdovirus-like sequences clustered separately from Sf-rhabdovirus and the Sf-rhabdovirus-like sequences encoded by the S. frugiperda genome (Figure 2 B).
Table 4.
Accession numbers and positions of the rhabdovirus L-like sequences found in the B. mori genome
| Genbank accession number (original) | Position (strand) | Length (nts) | Internal stop codons | |
|---|---|---|---|---|
| Bm L EVE 1 | NW_004581722.1 | 831136 – 836836 (−) | 5661 | 0 |
| Bm L EVE 2 | NW_004581840.1 | 67242 – 69297 (+) | 2055 | 1 |
| Bm L EVE 3 | NW_004581726.1 | 806500 – 807791 (+) | 1323 | 6 |
Figure 2.
Phylogenetic trees of Sf cell and B. mori rhabdovirus L-like EVE's and rhabdovirus L gene ORFs (A) and predicted gene products (B). The alignment of the predicted gene products is shown in Supplementary Figure 5.
4. Discussion
4.1. The newly found Sf-rhabdovirus like sequences are endogenous viral elements (EVE's)
Endogenous viral elements (EVE's) are viral genes or genomes that have become integrated into host germline chromosomes and are inherited as host alleles [17]. Since their initial discovery in 2004 in two Aedes mosquito species and cell lines [18], EVE's derived from RNA viruses that lack a DNA stage in their replication cycle have been found to be ubiquitous throughout the animal kingdom [17, 19-22].
In particular, the presence of rhabdovirus-like sequences has previously been found to be common in many insect genomes[17, 23]. Similar to the rhabdovirus EVE's in those reports, our newly found Sf-rhabdovirus-like sequences comprise long, uninterrupted ORFs, they are most similar to extant rhabdoviruses (in our case, to Sf-rhabdovirus), and they have high levels of similarity to extant rhabdoviruses as demonstrated by low BLAST E values. Similar to those previously reported EVE's, our N-like sequence comprised an apparently full-length ORF, and our G- and L-like sequences only comprised partial sequences. Unlike previous reports, we did find a P-like sequence, indicating that the P ORF can also fossilize into the insect cell genome. Only our N-like sequence had a polyadenlyl tract following the ORF, however, for unknown reasons, this is common only for N-like EVE's, not for EVE's of other mononegaviral genes [17]. Considering these similarities between our newly found Sf-rhabdovirus-like sequences and previously reported mononegaviral EVE's, we conclude the newly found Sf-rhabdovirus-like sequences comprise EVE's derived from ancient infection(s) with a Sf-rhabdovirus-like virus.
4.2. Sf-rhabdovirus-like EVE's might be exapted for antiviral defense
The Sf-rhabdovirus-like EVE's comprise long continuous ORFs without internal stop codons (1413, 1137, 1108, and 1259 nts, for the N, P, G, and L-like EVE's, respectively), indicating these sequences are subject to purifying selection, as stop codons would have been randomly inserted under conditions of neutral selection [17]. The Sf-rhabdovirus-like EVE's encode predicted proteins with strong similarity to proteins encoded by the extant Sf-rhabdovirus. However, the Sf-rhabdovirus-like EVE's have diverged substantially at the nucleic acid level from their Sf-rhabdovirus homologs, especially at the third codon positions, also indicating purifying selection. Ka/Ks ratios for the predicted N, P, G, and L protein (fragments) encoded by the EVE's as compared to their Sf-rhabdovirus orthologs were estimated at 0.56, 0.13, 0.11, and 0.05, respectively. Each of these values is <1, again indicating purifying selection. The Sf-rhabdovirus N, P, G, and L-like EVE's have accumulated large numbers of mutations as compared to their Sf-rhabdovirus homologs and are only 49%, 69%, 73% and 75% identical, respectively, suggesting the EVE's must have persisted over relatively long periods of time to allow for the accumulation of these mutations. Combined with our demonstration that the Sf-rhabdovirus-like EVE's are expressed, at least at the RNA level (Fig. 1), evidence for purifying selection and long persistence suggests that the Sf-rhabdovirus-like EVE's are not pseudogenes, but represent functional protein-coding genes.
Thus, Sf-rhabdovirus-like EVEs are likely exapted viral genes that have acquired new, cellular functions. Host cell exaptation has been demonstrated for various genes that originated from viruses, and often results in a reversal of function by co-opting virus-derived genes in antiviral defense (reviewed by [24, 25]). For example, a bornavirus N-like EVE has been demonstrated to inhibit bornaviral replication, presumably by functioning as a dominant-negative inhibitor by incorporation into viral ribonucleoprotein complexes [26]. The Sf-rhabdovirus N-like EVE could conceivably function similarly. While there is not yet any experimental evidence indicating EVE's derived from other mononegaviral genes have been similarly exapted for antiviral defenses, this seems likely.
The ability of Sf-rhabdovirus to persistently infect Sf cells, without cytopathic or other apparent ill effects [8], suggests that Sf cells have mechanisms to control Sf-rhabdovirus, and the Sf-rhabdovirus-like EVE's would be plausible candidates for such mechanisms. However, experimental confirmation of the function of the Sf-rhabdovirus-like EVE's is beyond the scope of this study.
4.3. Sf-rhabdovirus is likely associated with S. frugiperda in nature
Sf-rhabdovirus could already have been present in the S. frugiperda pupae from which Sf cell lines were derived, or was transmitted from silkworm hemolymph used as a tissue culture supplement. We found four Sf-rhabdovirus-like EVE's in the Sf cell genome, each of which was highly similar to Sf-rhabdovirus (Table 3 and Figure 2). Although we also found three rhabdoviral L-like EVE's in the silkworm genome, these sequences were substantially more different from the Sf-rhabdovirus L gene than the Sf-rhabdovirus L-like EVE (Figure 2). As the S. frugiperda, but not the B. mori genome comprises EVE's that are very similar to Sf-rhabdovirus, we conclude that in nature, Sf-rhabdovirus is likely associated with S. frugiperda, not B. mori, and that the Sf-rhabdovirus infection in Sf cell lines is likely derived from the S. frugiperda pupa originally used as a source of ovarian tissue. Conversely, our finding of three rhabdovirus L-like EVE's that are distinct from Sf-rhabdovirus in the B. mori genome suggests that an as of yet unidentified B. mori-specific rhabdovirus with sequence similarity to these EVE's infects B. mori populations in nature. Considering we found four Sf-rhabdovirus-like EVE's in the Sf cell genome, it appears that Sf-rhabdovirus has been a pathogen of S. frugiperda for an appreciable time.
4.4. Implications for adventitious virus searches using MPS
Cell lines used for biological production should lack any adventitious agents in order to ensure that biologicals produced therein are safe for human and veterinary use. Until recently, adventitious virus searches in cell substrates used to produce biologicals relied mainly on detecting the effects of viral infections. One method used to probe for infectious viral particles comprises looking for cytopathic effects in cell cultures exposed to potentially contaminated material. Another method comprises exposing hypersensitive animals, such as suckling mice or athymic nude mice, to potentially contaminated material. However, both methods suffer from two fundamental limitations: the cells or animals used in the test must be susceptible to infection with the adventitious virus of interest, and infection must result in observable effects. These limitations are not theoretical: SV40 in polio vaccines initially eluded detection because SV40 does not produce cytopathic effects in rhesus monkey kidney cells, which were originally used to probe for adventitious viruses[27]. Similarly, Sf-rhabdovirus eluded detection as Sf-rhabdovirus infection produces cytopathic effects neither in Sf or other insect cell types [5, 6], nor in mammalian cells, suckling mice or embryonated eggs [4].
Recent developments in massively parallel sequencing (MPS) technology have made detection of (unknown) adventitious viruses less dependent on the availability of susceptible cell substrates or animals [28-31]. MPS has made detection of adventitious viruses much more sensitive, as very low amounts of genetic material can be detected. MPS also has made detection of adventitious viruses more specific, as the exact sequence, and therefore virus species and strain can be identified. Facilitated by the very large number or known viral sequences, MPS data can be searched for unknown viruses similar to known viruses. Finding viral sequences in expressed sequence databases would logically imply that the cells are infected with a virus that gives rise to those sequences. However, our findings demonstrate this is not necessarily so, and that expressed viral-like sequences can in fact be derived entirely from genomically integrated EVE's. In our case, we found 4 expressed sequences in Sf cells that are clearly related to 4 out of 5 viral genes encoded by Sf-rhabdovirus. Such high coverage could easily be mistaken for an active infection, especially in cases such as presented in this study, where the previously unidentified virus-like sequences are very similar to a virus known to infect the cell line of interest.
If the Sf-rhabdovirus-like EVE's were in fact derived from a novel replication-competent adventitious virus, they would not have been present in the Sf cell genome, as Rhabdoviridae are ssRNA+ viruses, which do not encode a reverse transcriptase, and do not have a DNA stage in their lifecycle. Only a direct comparison of the transcribed Sf-rhabdovirus-like sequences with the Sf cell genome revealed their origin in the cellular genome. This finding exemplifies the importance in verifying whether transcripts that appear to be of viral origin are derived from a replication competent virus with an intact genome, or from (partial) viral sequences integrated into the cellular genome. Thus, MPS-based searches for adventitious viruses in cell substrates used to produce biologicals should compare both genomic and transcribed sequences to facilitate the identification of transcribed EVE's, and to avoid false positive detection of replication-competent adventitious viruses.
Supplementary Material
We searched the Sf cell genome and transcriptome for adventitious virus sequences
We found rhabdovirus-like sequences transcribed from the Sf cell genome
The rhabdovirus-like sequences are closely related to Sf-rhabdovirus
Transcribed endogenous viral elements may result in false positive adventitious virus detection
Sequencing and comparing both the genome and transcriptome is critical to avoid false positives
Acknowledgements
We thank Dr. J.C. Gatlin (University of Wyoming) for constructive comments. This work was supported by Awards R43 GM102982 and R43 AI112118 from the National Institutes of Health, Institutes of General Medical Sciences and Allergy and Infectious Diseases, respectively. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, National Institute of General Medical Sciences or National Institute of Allergy and Infectious Diseases.
Abbreviations
- BICS
Baculovirus insect cell system
- EVE
Endogenous viral element
- ORF
Open reading frame
- Sf
Spodoptera frugiperda
- TSA
Transcriptome Sequence Assembly
- WGS
Whole Genome Shotgun
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
8. Supplementary figure legends
Supplementary Figure 1: Alignment of Sf-rhabdovirus N-like EVE and Sf-rhabdovirus N gene (A) ORFs and (B) predicted gene products. Grey shading indicates conserved nucleotides or amino acids.
Supplementary Figure 2: Alignment of Sf-rhabdovirus P-like EVE and Sf-rhabdovirus P gene (A) ORFs and (B) predicted gene products. Grey shading indicates conserved nucleotides or amino acids.
Supplementary Figure 3: Alignment of Sf-rhabdovirus G-like EVE and Sf-rhabdovirus G gene (A) ORFs and (B) predicted gene products. Grey shading indicates conserved nucleotides or amino acids.
Supplementary Figure 4: Alignment of Sf-rhabdovirus L-like EVE and Sf-rhabdovirus L gene (A) ORFs and (B) predicted gene products. Grey shading indicates conserved nucleotides or amino acids.
Supplementary Figure 5: Alignment of predicted gene products of two rhabdovirus L genes and rhabdovirus L-like EVE's from Sf cell lines and B. mori. Shading corresponds to the degree of conservation, with darker shades indicating more highly conserved amino acid residues.
References
- 1.Vaughn JL, Goodwin RH, Tompkins GJ, McCawley P. The establishment of two cell lines from the insect Spodoptera frugiperda (Lepidoptera; Noctuidae). In Vitro. 1977;13:213–7. doi: 10.1007/BF02615077. [DOI] [PubMed] [Google Scholar]
- 2.Summers MD, Smith GE. Texas Agricultural Experiment Station Bulletin. Texas Agricultural Experiment Station; College Station, TX: 1987. A manual of methods for baculovirus vectors and insect cell culture procedures. [Google Scholar]
- 3.Marino JPJ, McAtee JJ, Washburn DG. In: SEH and 11 β-HSD1 inhibitors and their use. PCT, editor. SmithKline Beecham Corp; 2009-06-04. [Google Scholar]
- 4.Smith GE, Foellmer HG, Knell J, DeBartolomeis J, Voznesensky AI. In: Spodoptera frugiperda single cell suspension cell line in serum-free media, methods of producing and using. USPTO, editor. Protein Sciences Corporation; USA: 2000. [Google Scholar]
- 5.Ma H, Galvin TA, Glasner DR, Shaheduzzaman S, Khan AS. Identification of a novel rhabdovirus in Spodoptera frugiperda cell lines. J Virol. 2014;88:6576–85. doi: 10.1128/JVI.00780-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haynes J. In: Methods of detection and removal of rhabdoviruses from cell lines. Organization WIP, editor. Takeda Vaccines, Inc.; 2014. [Google Scholar]
- 7.Research CfBEa, editor. Guidance for Industry - Characterization and Qualification of Cell Substrates and Other Biological Materials Used in the Production of Viral Vaccines for Infectious Disease Indications. Office of Communication, Outreach and Development; Rockville, MD: 2010. [Google Scholar]
- 8.Maghodia AB, Geisler C, Jarvis DL. Characterization of an Sf-rhabdovirus-negative Spodoptera frugiperda cell line as an alternative host for recombinant protein production in the baculovirus-insect cell system. Protein Expr Purif. 2016;122:45–55. doi: 10.1016/j.pep.2016.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li C-X, Shi M, Tian J-H, Lin X-D, Kang Y-J, Chen L-J, et al. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. eLife. 2015;4:e05378. doi: 10.7554/eLife.05378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith GE, Summers MD, Fraser MJ. Production of human beta interferon in insect cells infected with a baculovirus expression vector. Mol Cell Biol. 1983;3:2156–65. doi: 10.1128/mcb.3.12.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laird PW, Zijderveld A, Linders K, Rudnicki MA, Jaenisch R, Berns A. Simplified mammalian DNA isolation procedure. Nucleic Acids Res. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kakumani PK, Malhotra P, Mukherjee SK, Bhatnagar RK. A draft genome assembly of the army worm, Spodoptera frugiperda. Genomics. 2014;104:134–43. doi: 10.1016/j.ygeno.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 13.Kakumani P, Shukla R, Todur V, Malhotra P, Mukherjee S, Bhatnagar R. De novo transcriptome assembly and analysis of Sf21 cells using illumina paired end sequencing. Biol Direct. 2015;10:1–7. doi: 10.1186/s13062-015-0072-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walker PJ, Firth C, Widen SG, Blasdell KR, Guzman H, Wood TG, et al. Evolution of Genome Size and Complexity in the Rhabdoviridae. PLoS Pathog. 2015;11:e1004664. doi: 10.1371/journal.ppat.1004664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steinhauer DA, Domingo E, Holland JJ. Lack of evidence for proofreading mechanisms associated with an RNA virus polymerase. Gene. 1992;122:281–8. doi: 10.1016/0378-1119(92)90216-c. [DOI] [PubMed] [Google Scholar]
- 16.Drake JW. The distribution of rates of spontaneous mutation over viruses, prokaryotes, and eukaryotes. Ann N Y Acad Sci. 1999;870:100–7. doi: 10.1111/j.1749-6632.1999.tb08870.x. [DOI] [PubMed] [Google Scholar]
- 17.Katzourakis A, Gifford RJ. Endogenous viral elements in animal genomes. PLoS Genet. 2010:6. doi: 10.1371/journal.pgen.1001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crochu S, Cook S, Attoui H, Charrel RN, De Chesse R, Belhouchet M, et al. Sequences of flavivirus-related RNA viruses persist in DNA form integrated in the genome of Aedes spp. mosquitoes. J Gen Virol. 2004;85:1971–80. doi: 10.1099/vir.0.79850-0. [DOI] [PubMed] [Google Scholar]
- 19.Belyi VA, Levine AJ, Skalka AM. Unexpected inheritance: multiple integrations of ancient bornavirus and ebolavirus/marburgvirus sequences in vertebrate genomes. PLoS Pathog. 2010;6:e1001030. doi: 10.1371/journal.ppat.1001030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horie M, Honda T, Suzuki Y, Kobayashi Y, Daito T, Oshida T, et al. Endogenous non-retroviral RNA virus elements in mammalian genomes. Nature. 2010;463:84–7. doi: 10.1038/nature08695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu H, Fu Y, Jiang D, Li G, Xie J, Cheng J, et al. Widespread Horizontal Gene Transfer from Double-Stranded RNA Viruses to Eukaryotic Nuclear Genomes. J Virol. 2010;84:11876–87. doi: 10.1128/JVI.00955-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor DJ, Leach RW, Bruenn J. Filoviruses are ancient and integrated into mammalian genomes. BMC Evol Biol. 2010;10:1–10. doi: 10.1186/1471-2148-10-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fort P, Albertini A, Van-Hua A, Berthomieu A, Roche S, Delsuc F, et al. Fossil rhabdoviral sequences integrated into arthropod genomes: ontogeny, evolution, and potential functionality. Mol Biol Evol. 2012;29:381–90. doi: 10.1093/molbev/msr226. [DOI] [PubMed] [Google Scholar]
- 24.Aswad A, Katzourakis A. Paleovirology and virally derived immunity. Trends Ecol Evol. 2012;27:627–36. doi: 10.1016/j.tree.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 25.Malfavon-Borja R, Feschotte C. Fighting Fire with Fire: Endogenous Retrovirus Envelopes as Restriction Factors. J Virol. 2015;89:4047–50. doi: 10.1128/JVI.03653-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujino K, Horie M, Honda T, Merriman DK, Tomonaga K. Inhibition of Borna disease virus replication by an endogenous bornavirus-like element in the ground squirrel genome. Proc Natl Acad Sci U S A. 2014;111:13175–80. doi: 10.1073/pnas.1407046111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petricciani J, Sheets R, Griffiths E, Knezevic I. Adventitious agents in viral vaccines: Lessons learned from 4 case studies. Biologicals. 2014;42:223–36. doi: 10.1016/j.biologicals.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 28.Onions D, Kolman J. Massively parallel sequencing, a new method for detecting adventitious agents. Biologicals. 2010;38:377–80. doi: 10.1016/j.biologicals.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 29.Onions D, Cote C, Love B, Toms B, Koduri S, Armstrong A, et al. Ensuring the safety of vaccine cell substrates by massively parallel sequencing of the transcriptome. Vaccine. 2011;29:7117–21. doi: 10.1016/j.vaccine.2011.05.071. [DOI] [PubMed] [Google Scholar]
- 30.McClenahan SD, Uhlenhaut C, Krause PR. Evaluation of cells and biological reagents for adventitious agents using degenerate primer PCR and massively parallel sequencing. Vaccine. 2014;32:7115–21. doi: 10.1016/j.vaccine.2014.10.022. [DOI] [PubMed] [Google Scholar]
- 31.Onions D, Côté C, Love B, Kolman J. Deep Sequencing Applications for Vaccine Development and Safety. In: Nunnally BK, Turula VE, Sitrin RD, editors. Vaccine Analysis: Strategies, Principles, and Control. Springer; Berlin Heidelberg: 2015. pp. 445–77. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.