Abstract
Most cancers arise from oncogenic changes in the genomes of somatic cells, and while the cells may migrate by metastasis, they remain within that single individual. Natural transmission of cancer cells from one individual to another has been observed in two distinctive cases in mammals (Tasmanian devils1 and dogs2,3), but these are generally considered to be rare exceptions in nature. The discovery of transmissible cancer in soft-shell clams (Mya arenaria)4 suggested that this phenomenon might be more widespread. Here we analyzed disseminated neoplasia in mussels (Mytilus trossulus), cockles (Cerastoderma edule), and golden carpet shell clams (Polititapes aureus) and found that neoplasias in all three species are attributable to independent transmissible cancer lineages. In mussels and cockles, the cancer lineages are derived from their respective host species, but unexpectedly, cancer cells in P. aureus are all derived from Venerupis corrugata, a different species living in the same geographic area. No cases of disseminated neoplasia have thus far been found in V. corrugata from the same region. These findings show that transmission of cancer cells in the marine environment is common in multiple species, that it has originated many times, and that while most transmissible cancers were found spreading within the species of origin, cross-species transmission of cancer cells can occur.
Disseminated neoplasia, or hemic neoplasia, a leukemia-like disease, occurs with high prevalence in multiple bivalve species5,6. Here we investigated the possibility that cancers in three species could be attributed to transmissible cancer cells, and whether these cancers are restricted to the species of origin or can undergo cross-species transmission.
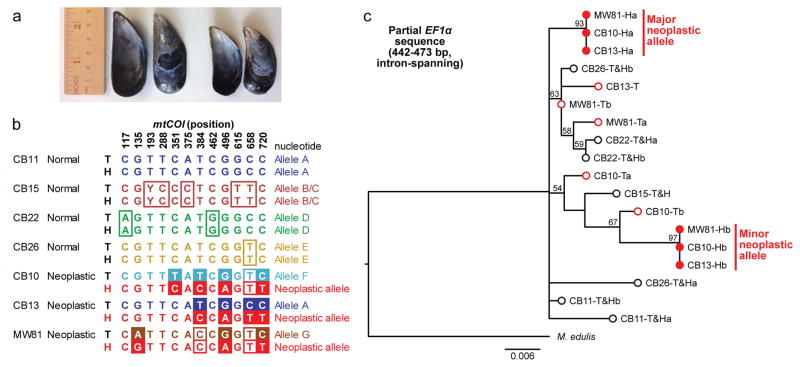
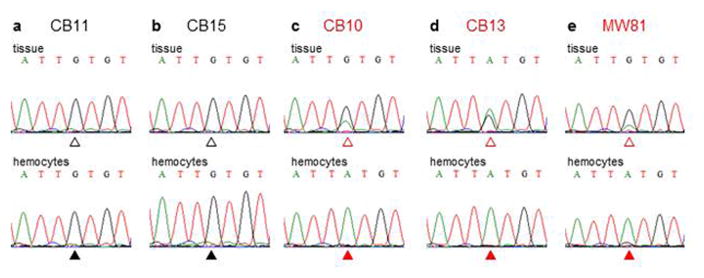
Mussels (Mytilus trossulus, Fig. 1a) are subject to disseminated neoplasia in the Pacific Northwest Coast7,8, and evidence of common polymorphisms in neoplasias suggested that these might represent a transmissible cancer9. 28 Mussels (M. trossulus) collected from West Vancouver were screened for neoplasia by drawing hemolymph and analyzing hemocytes for the rounded, non-adherent morphology of neoplastic cells. Two were identified with high levels of neoplastic cells. We sequenced part of the mitochondrial cytochrome c oxidase I (mtCOI) gene in host tissue and neoplastic hemocytes from the two diseased animals and four normal animals to test whether the neoplastic cells were derived from the host individuals or exhibited a distinct genotype, the hallmark of transmissible neoplasia. While solid tissue and hemocyte genotypes within each normal animal were always identical, solid tissue and neoplastic hemocyte genotypes of the diseased animals were discordant (Fig. 1b, Extended Data Fig. 1). Moreover, the same single nucleotide polymorphisms (SNPs) were found in neoplastic cells of the two diseased individuals, indicating that the cancer cells were not of host origin and suggesting that they arose from a single clonal origin. EF1α gene sequences also revealed that the genotypes of the host cells and neoplastic hemocytes were discordant, and that the genotypes of the neoplastic cells of the two different animals were again identical to each other (Fig. 1c).
Figure 1. Analysis of tissue and hemocyte genotypes of normal mussels (M. trossulus) and mussels with disseminated neoplasia using mitochondrial and nuclear DNA markers.
a, Representative M. trossulus, with ruler for scale. b, Sequencing of mtCOI (bases 99–759) was conducted for both host solid tissue (T) and hemocytes (H) of normal (N=4) and neoplastic mussels (N=3). Open boxes mark SNPs which differ from allele A. Filled boxes mark discordance between neoplastic hemocyte and tissue genotypes. Sequences are numbered using AY823625.1. c, Maximum likelihood phylogenetic tree based on sequences of an intron-containing region of EF1α, with bootstrap values over 50 shown and a scale bar showing genetic distance. M. edulis (614 bp, EU684203.1), was used as an outgroup. In normal animals, the sequences of tissue and hemocyte DNA were identical and are presented together (T&H; black circles). For neoplastic animals, the tissue (T; open red circles) and hemocyte (H; filled red circles) alleles differed. Letters a and b denote multiple alleles in heterozygous individuals, and major and minor alleles from neoplastic alleles are marked. See Extended Data Fig 1 for further details.
To determine if this transmissible cell line was widespread in the M. trossulus population, 250 were collected from Vancouver Island, and seven potentially diseased individuals were analyzed. In one neoplastic sample (MW81), the hemocyte and tissue genotypes did not match, and the hemocytes contained the same mtCOI allele and the same EF1α major and minor alleles found in the other samples (Fig. 1b, c, Extended Data Fig. 1). These genotypes strongly indicate the existence of a M. trossulus-derived transmissible cancer lineage circulating in the wild population.
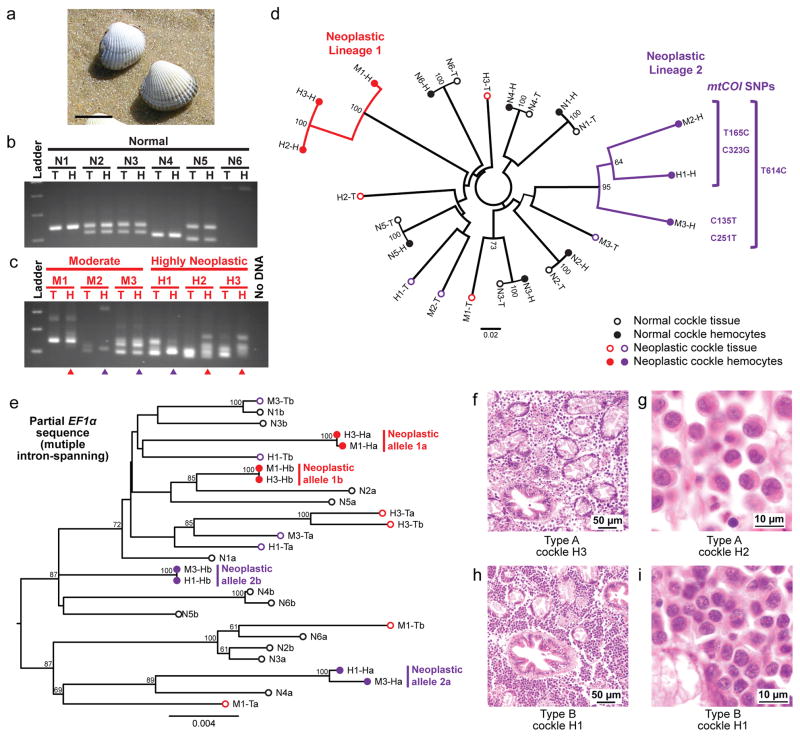
High prevalence of disseminated neoplasia has been observed in two species of bivalves on the Galician Coast, cockles (Cerastoderma edule)10,11 and golden carpet shell clams (Polititapes aureus, previously named Venerupis aurea)12. The disease in cockles (C. edule) exhibits one of two distinct morphologies, termed types A and B13,14. We collected about 150 cockles (C. edule), and examined the genotypes of solid tissue and hemocytes of six normal individuals and six (Fig. 2a) with high (>75%) or moderate (15–75%) amounts of neoplastic cells in the hemolymph. Nine polymorphic microsatellite loci15 were amplified from normal animals, and allele sizes from tissue and hemocytes of each normal animal all matched, but allele sizes in hemocytes and tissue of diseased animals were discordant (Fig. 2b, c). In a phylogenetic tree based on microsatellite alleles16,17 the neoplastic hemocyte genotypes did not group with the host tissue genotypes, consistent with transmissible cancer, and instead clustered into two distinct branches, suggesting two independent cancer lineages (Fig. 2d). We sequenced the mtCOI gene and identified several SNPs that were present only in lineage 2, and not in any of the normal animals. No unique SNPs were identified in the mtCOI region sequenced in lineage 1.
Figure 2. Analysis of lineages of transmissible neoplasia in cockles (C. edule).
a, Representative C. edule (scale = 20 mm). b–c, Microsatellite loci were amplified from solid tissue (T) and hemocyte (H) DNA from normal (samples N1-6) and diseased (M1-3 & H1-3) cockles. Products of a representative locus (CeATC1-5) are shown. Triangles mark samples corresponding to lineages 1 (red) and 2 (purple). d, Neighbor-joining phylogenetic tree based on nine microsatellite loci (See Source Data). Bootstrap values over 50 are shown and scale bar is based on comparison across all 104 observed alleles. mtCOI SNPs unique to lineage 2 are marked. e, Maximum likelihood tree of EF1α (2725–4249 bp, spanning four introns), rooted at the midpoint, with bootstrap values above 50 and scale bar showing genetic distance. Letters a and b denote multiple alleles from heterozygous samples, and alleles from neoplastic lineages 1 and 2 are marked. f–g, Histology of cockles H3 and H2, representative of Type A neoplastic cells. h–i, Histology of cockle H1, representative of Type B neoplastic cells.
The microsatellite alleles and mtCOI SNPs suggest two independent cancer lineages, but these data are also consistent two subgroups which have diverged from a single transmissible cancer lineage. To investigate this question, we sequenced an approximately 3 kb intron-spanning region in EF1α from six normal individuals and two diseased individuals from each lineage (Fig. 2e). Both neoplastic hemocyte alleles were different from the tissue alleles in all diseased individuals, and the two alleles of the neoplasm in each diseased individual were nearly identical to the two alleles of the other individual in the lineage. However, both alleles in lineage 1 cells were different from those in lineage 2. Moreover, the neoplastic alleles were more closely related to some normal alleles than they were to the alleles of the alternate lineage. These data strongly suggest an independent origin of these two lineages.
Histological and morphometric examination showed that all three samples in lineage 1 were type A (characterized by a looser arrangement of neoplastic cells in the connective tissue, with pleomorphic nuclei) and all three samples in lineage 2 were type B (tighter arrangement, with smaller cells than type A and rounded, smaller nuclei, Fig. 2f–i, Extended Data Table 1). Altogether, these results argue that two distinct lineages of transmissible cancer, with distinct morphologies and genotypes, arose independently in cockles and are circulating in this species.
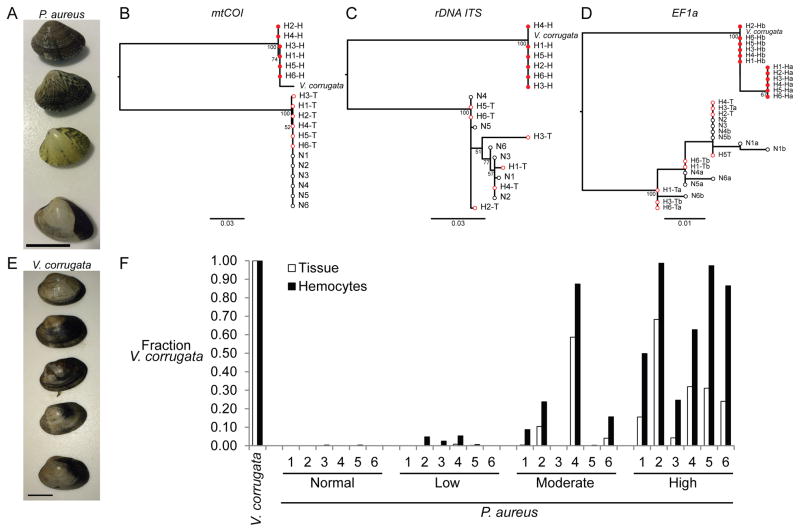
We also examined golden carpet shell clams (P. aureus, Fig. 3a), which are present in the same habitat as several other bivalves, including the closely related pullet shell clam (Venerupis corrugata). Of 74 P. aureus individuals tested, nine had high levels of disease, and 22 had low-to-medium disease. We sequenced regions of mtCOI, the ribosomal DNA Internal Transcribed Spacer (rDNA ITS), and EF1α from tissue and hemocyte DNA of six highly diseased and six normal P. aureus individuals, and again found that the genotypes of neoplastic cells were nearly identical and did not match those of their hosts (Fig. 3b–d). In contrast to the transmissible cancers in other species, however, the sequences of the neoplastic cells were highly dissimilar from normal host sequence (only 78.4–78.5% identical in the mtCOI locus and 89.3–93.2% identical at nuclear loci, ignoring insertions and deletions). The neoplastic genotypes were instead near-perfect matches to the sequences of V. corrugata (98.6–7% and 99.3–100% identical, respectively).
Figure 3. Phylogenetic analysis of neoplastic cells in golden carpet shell clams (P. aureus) and quantification of V. corrugata cell engraftment in normal and diseased animals.
a, Representative P. aureus clams (scale = 20 mm). b–d, Maximum likelihood trees of sequences from solid tissue (T) and hemocyte (H) DNA from normal (N1-6) and highly diseased (H1-6) P. aureus and normal V. corrugata, based on (b) mtCOI (658 bp), (c) rDNA ITS (454–373 bp), and (d) EF1α (148 bp, letters a and b denote multiple alleles). Open black circles, tissue and hemocytes of normal P. aureus; open red circles, tissue of diseased P. aureus; filled red circles, hemocytes of diseased P. aureus; scale bars show genetic distance. e, Representative V. corrugata clams (scale = 20 mm). f, Quantification of cancer cell engraftment using species-specific amplification of EF1a from hemocyte and tissue DNA of normal V. corrugata, normal P. aureus, and P. aureus diagnosed as having low, moderate, or high levels of disseminated neoplasia.
To confirm that detection of V. corrugata (Fig. 3e) sequence reflected the neoplastic cells observed morphologically, we used species-specific EF1α qPCR to quantify the fraction of cells that were derived from each species in the tissue and hemocyte samples (Fig. 3f). As expected, no V. corrugata sequences were detected in normal P. aureus animals, but high frequencies of V. corrugata DNA were detected in all six individuals diagnosed with high levels of neoplasia and lower amounts in most individuals diagnosed with medium and low levels. The small number of individuals diagnosed with low and medium disease in which V. corrugata DNA was not detected could have primary host-derived neoplasia, a different transmissible cancer lineage, too low an abundance of tumor DNA to detect, or even a small amount of normal cells with an unusually rounded morphology. In diseased animals, the highest levels of V. corrugata sequence were detected in hemocytes, and lower levels were present in tissues, likely due to infiltration of cancer cells via the circulation. We conclude that the cancer spreading in the P. aureus population resulted from cross-species transmission of cancer cells of V. corrugata origin. It is noteworthy that no cases of disseminated neoplasia have thus far been found in V. corrugata from the same region, despite analysis of hundreds of clams, suggesting that the V. corrugata cancer cells do not initiate disease in the species of origin and were only observed to colonize P. aureus animals.
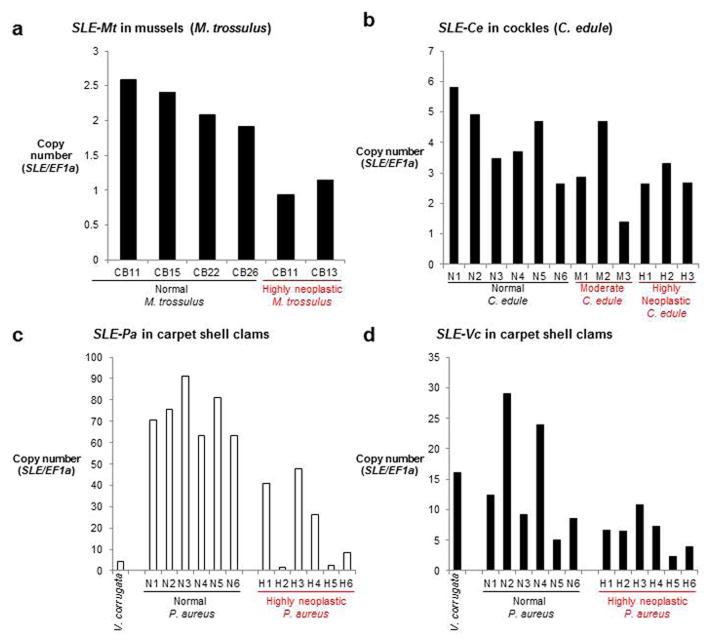
We previously identified an LTR-retrotransposon, Steamer, which was amplified in the transmissible neoplasia lineage in the soft-shell clam (M. arenaria)18. Using degenerate primers we identified at least one Steamer-like element (SLE) in each of the three species studied here. In each case, copy numbers were variable among individuals, but no massive amplification of these particular retrotransposons was observed in any of the transmissible cancers assayed (Extended Data Fig. 2). This suggests that Steamer-like retrotransposon amplification is not essential in development of transmissible clones.
Our results indicate that transmission of contagious cancer cells is a widespread phenomenon in the marine environment, with multiple independent lineages developing in multiple species in four bivalve families. Along with the recent identification of a second independent lineage of transmissible cancer in Tasmanian devils19, these findings confirm that under suitable conditions, the development of transmissible cancer can occur multiple times, and suggests that some species may be more susceptible to development of transmissible neoplasia than others.
Spontaneous hemic neoplasia must occur at some frequency in order for transmissible lineages to arise, but cases of transmissible cancer appear to outnumber spontaneous disease, at least in the species investigated so far (i.e. all neoplastic cases tested in cockles, golden carpet shell clams, and three of nine samples in mussels, in addition to all samples in the previous report of soft-shell clams4).
Transmissible cancers appear largely restricted to the species of origin in nature. While canine transmissible venereal tumor has been experimentally transplanted to coyotes20, jackals21, and foxes22–24, no examples of natural transmission beyond dogs have been reported25. Disseminated neoplasias of molluscs have only been transplanted to members of the same species (soft-shell clams, mussels, and others5,6). Attempts to transfer M. arenaria neoplasia through water exposure to both M. arenaria and M. trossulus only resulted in transfer of disease to M. arenaria26, and injection of M. trossulus cancer cells into multiple bivalve species only resulted in engraftment in M. trossulus27. Our finding that multiple cancer lineages are most often found to spread within the original host species is consistent with these previous experiments, and suggests that there may be species-specific restriction factors that prevent engraftment into divergent hosts.
In this context, our observation of cross-species transmission, a cancer from one species spreading through another, is particularly striking. They both belong to the Veneridae family (some studies suggest that P. aureus should belong to the Venerupis genus28), and coexist in the same beds. This close relationship may have aided the transmission. It is notable that despite ongoing surveillance of the bivalves of the Galician coast since the 1990s, only one V. corrugata has been identified as harboring disseminated neoplasia (personal communication, Susana Darriba, at INTECMAR), while the prevalence of disease in P. aureus is quite high12 (12% of the individuals collected for the current study had high levels of neoplasia and 42% had some detectable level of disease). This would be explained if the cancer originated in V. corrugata, led to selective loss of susceptible animals, and left the current population of V. corrugata resistant to engraftment and disease. The spread of transmissible cancers and the evolutionary pressure to defend against them may be a strong and underappreciated selective force in evolution of multicellular organisms.
In summary, our findings show that transmission of cancers between individuals within a species may be more common than previously assumed, and that transmission can even occur between species. We now know of eight transmissible cancers in nature: one lineage in dogs, two lineages in devils, and five lineages circulating in four species of molluscs, including one example of cross-species transmission. Further studies may well reveal additional examples. The recent report of a cancer composed of transformed tapeworm cells in an immunocompromised AIDS patient29 highlights the possibility that transmissible tumors could arise in humans. These transmissible cancers constitute a distinct class of infectious agent and show the remarkable ability of tumors to acquire new phenotypes that promote their own survival and propagation.
METHODS
Collection and Diagnosis of M. trossulus
Mussel (M. trossulus) specimens 5 to 6 cm long were collected from the intertidal zone at low tide (noon) on April 18, 2015 at Copper Beach (N 49° 22′41″ W 123° 16′ 44″, West Vancouver, B.C., Canada). They were transported to the laboratory in aerated seawater from Copper Beach. In the laboratory, 0.5 – 1.0 mL hemolymph was removed from the posterior adductor muscle. For each individual, one drop of hemolymph was placed on a poly-L-lysine-coated slide and let sit for 10–15 min to allow the cells to spread and attach to the slide. Thereafter, the slide was viewed under a Zeiss Axiostar light microscope at 40× magnification. Normal (non-neoplastic) specimens were those with greater than 90% cells with normal appearance, i.e. agranular or granular hemocytes with spread pseudopodia30. Fully leukemic (diseased) specimens were those with prolific amounts (> 90%) of round, non-adherent cells. Hemolymph was added to 1.5 mL Eppendorf tubes and spun at 900 g for 3 min. After the supernatant was withdrawn, the cells were re-suspended in absolute ethanol before shipping. Excised tissues (mantle, foot, and gills) were preserved in absolute ethanol before shipping. Of 28 individuals collected from West Vancouver, two had high levels of neoplastic disease.
A second set of M. trossulus samples were collected from Esquimalt, Vancouver Island, B.C., Canada. Hemolymph was extracted and cell morphology was used for diagnosis as above. Hemocyte and solid tissue samples were fixed in ethanol before DNA extraction. Of 250 individuals collected from Esquimalt, nine were scored as potentially moderately or highly diseased, with seven samples available for analysis.
Collection and Diagnosis of C. edule and P. aureus
Cockles (C. edule) were collected from an intertidal bed named O Sarrido (42° 30′ N, 8° 49′ W) and golden carpet shell clams (P. aureus) and pullet shell clams (V. corrugata) were collected from a subtidal bed named O Bohído (42 ° 32′ N, 8° 51′ W); both shellfish beds are located in the ria of Arousa (Galicia, NW Spain). Once in the laboratory of CIMA, each cockle and clam was notched through the shell margin close to the posterior adductor muscle and hemolymph (as much as possible) was collected from the posterior adductor muscle using a 2-ml syringe with a 21 gauge needle. A small quantity (ca. 100 μl) of hemolymph was used to produce a cell monolayer onto a slide by cytocentrifugation (92 × g, 5 min, 4°C), which was fixed, stained with Hemacolor (Merck) kit and examined with light microscopy for diagnosis of disseminated neoplasia; the remaining hemolymph was preserved in absolute ethanol for molecular analysis. After hemolymph collection, molluscs were shucked and a small piece of mantle (ca. 5 × 5 mm square) was removed and preserved in absolute ethanol for molecular analysis; additionally, an approximately 5 mm thick section of meat, containing gills, visceral mass, foot and mantle lobes, was fixed in Davidson’s solution and embedded in paraffin. Sections of 5 μm thickness were stained with Harris-hematoxylin and eosin31. Histological sections were examined under light microscopy for histopathological analysis.
Morphometric analysis was conducted on histological sections of cockle (C. edule) samples. Neoplastic cells (both A and B types) had a unique morphology, and were clearly distinguished from normal cells, with much larger overall size, and much larger nucleus. The longest diameters of the cell and the nucleus of at least 10 neoplastic cells for each individual were measured by direct examination of histological sections with light microscopy using a reticle. Two-tailed T-tests were used for comparisons of morphometric data from different individuals and types.
Cockles (C. edule) and golden carpet shell clams (P. aureus) were ranked according to a scale of disease severity based on hemolymph diagnosis: non-affected (N0, or N); low severity (N1, or L), when individuals showed proportions of neoplastic cells lower than 15% in the hemolymph cell monolayers; moderate severity (N2, or M), when the proportion ranged from 15% to 75%; and high severity (N3, or H), when the proportion was higher than 75%32. 74 golden carpet shell clams (P. aureus) were collected with 12 diagnosed with light, 10 with moderate, and 9 with heavy neoplasia. About 150 cockles (C. edule) were collected and a subset was analyzed for both disease and morphological type of neoplastic cells. Of the 30 in this subset, two had type A neoplastic cells (one light and the other moderate) and one had low levels of type B neoplastic cells. We have also collected and analyzed hundreds of pullet clams (V. corrugata) from multiple beds throughout Galicia from 1988 to the present time, including about 100 from the same bed in which the samples of P. aureus were collected for this study (O Bohído). To this date, we have not observed any V. corrugata samples with neoplastic disease. One V. corrugata individual from a different location has been identified by a different researcher as harboring disseminated neoplasia, with a different morphology than that observed in P. aureus neoplasia (personal communication, Susana Darriba, at INTECMAR).
DNA extraction
DNA was extracted from ethanol-fixed hemocytes using DNeasy Blood and Tissue Kit (Qiagen). DNA extraction of tissues used the same kit, but included an additional step in order to reduce the amount of PCR-inhibiting polysaccharides. After tissue lysis, 63 μl of buffer P3 was added to lysate and allowed to precipitate for 5 min. Lysate was spun 10 minutes at full speed at 4°C, and the resulting supernatant was mixed with ethanol and added to the column, continuing with the standard protocol.
mtCOI, EF1α, ITS, Steamer-like PCR
Primers and annealing temperatures used are listed in Extended Data Table 2. PfuUltra II Fusion HS DNA Polymerase (Agilent) was used to amplify 10 ng of genomic DNA for 35 cycles. Extension was at 72°C for 15 sec for mtCOI for all bivalves33. For P. aureus, ITS was amplified (using primers modified from34,35) with extension for 30 sec. Despite multiple copies of ITS in genomic DNA, a single sequence was observed for each normal P. aureus and V. corrugata, and a single pair of host/neoplasm sequences was observed in each diseased individual. PCR for EF1α followed the same program, with extension for 20 sec in mussels (M. trossulus) and clams (P. aureus and V. corrugata) and 1 min 30 sec in cockles (C. edule). For amplification of Steamer-like elements, a degenerate primer pair was designed to match conserved regions in reverse transcriptase and integrase of the LTR-retrotransposon, Steamer, and 10 ng of DNA was amplified for 35 cycles, annealing at 55°C for 20 sec and extending for 1 min at 72°C. PCR products were directly sequenced. When multiple alleles could not be resolved by direct sequencing, PCR products were cloned using the Zero Blunt TOPO Kit (Invitrogen), and at least 6 colonies were sequenced. Sequences were aligned with Clustal W, with some manual adjustment. Primer-binding regions were excluded from analysis and are not included in the sizes listed as sequenced. Maximum likelihood phylogenetic trees were generated using PhyML 3.036, using the HKY85 substitution model, with 100 bootstrap replicates, treating gaps in the alignment as missing data. Trees were visualized using FigTree v1.4, with addition of markers at the branch termini. Each phylogenetic tree based on alignment of sequence at a single locus (Figure 1c, 2e, 3b–d) includes a scale bar which shows genetic distance (0–1) based on the frequency of nucleotide divergence. All sequences are available in GenBank (accession numbers: KX018521-KX018605).
Extended Data Table 2.
Primers used in PCR and qPCR
| Target | Forward Primer | Reverse Primer | Temp | ||
|---|---|---|---|---|---|
| mtCOI | |||||
| LCO1490 | GGTCAACAAATCATAAAGATATTGG | HC02198 | TAAACTTCAGGGTGACCAAAAAATCA | 50°C | |
| ITS | |||||
| its-3d | gcgtcgatgaagagcgca | its-4r | agttttttttcctccgctta | 55°C | |
| EF1α | |||||
| Mussel (M. trossulus) | consEF1-F1 | ACCATTGATATTGCTYTNTGGAA | MtEF1-R2 | ACAATCAAAATGGCACAATC | 50°C |
| Cockle (C. edule) | CeEF1-F2 | TGGTATCACCATCGATATTGC | CeEF1-R3 | CCGTTTCGGATCTCTACAGG | 55°C |
| P. aureus & V. corrugata | consVEF1F3b | AGGAACCTCTCAAGCTGAYTG | consVEF1R3b | AGCAATTACCTCGCTGTANGG | 55°C |
| Cockle (C. edule) Microsatellite Loci | |||||
| CeATC1-5F-6FAM | cgttctaccggcatatgtcac | CeATC1-5R | caccttccaccactagaagaaaa | 60°C | |
| CeATC1-22F-VIC | caaacctgaccgggtttatt | CeATC1-22R | tgactccactttttcagttcca | 60°C | |
| CeATC1-36F-NED | gacatgacaaacaggcctca | CeATC1-36R | tggcctggtcttatttccac | 60°C | |
| CeATC1-52F-PET | aatctgattttgccacctct | CeATC1-52R | agctcataggagttgtatacgtaag | 55°C | |
| CeATC1-54F-6FAM | tacaaggccgagaaactgct | CeATC1-54R | caatgactgccaaatgagga | 55°C | |
| CeATC2-4F-VIC | tggaaatgcattcattgagc | CeATC2-4R | ccgattgcgttctttgatct | 55°C | |
| CeATC2-11F-NED | tggtgtgcaattagatgcttg | CeATC2-11R | taggctcgcagaaagatggt | 60°C | |
| CeATC2-34F-6FAM | gccatagaggccaccctatt | CeATC2-34R | gggctgacaagatttgacatt | 60°C | |
| CeATC2-46F-NED | accaaggcagatatcgatcc | CeATC2-46R | tccagttttaaacgcactctga | 55°C | |
|
Cloning qPCR Standards
| |||||
| EF1α | |||||
| Mussel (M. trossulus) | consEF1F1 | ACCATTGATATTGCTYTNTGGAA | consEF1R2 | ACGTTGAAACCAACRTTRTC | 50°C |
| Cockle (C. edule) | CeEF1-F1 | TGGAACAAACTGAAGGCCGA | CeEF1-R3 | CCGTTTCGGATCTCTACAGG | 50°C |
| P. aureus & V. corrugata | consVEF1F3b | AGGAACCTCTCAAGCTGAYTG | consVEF1R3b | AGCAATTACCTCGCTGTANGG | 55°C |
| SLE | |||||
| Mussel (M. trossulus) | DHKPL-F1 | TTGAAAGCGACCACAARCCNYT | PXRPW-R1 | TTCCGACTTTGGCCCANGGNC | 55°C |
| Cockle (C. edule) | LVW-F1 | GGTATCCAGCCTAGTNGTNGT | PXRPW-R1 | TTCCGACTTTGGCCCANGGNC | 55°C |
| P. aureus & V. corrugata | DHKPL-F1 | TTGAAAGCGACCACAARCCNYT | PXRPW-VaR1 | CTGCAATTTTCTGCCANGGNC | 55°C |
| qPCR | Forward Primer | Reverse Primer | Control Plasmids | ||
|
| |||||
| EF1α | |||||
| Mussel (M. trossulus) | MtEF1qF2 | TAGGTATTGGAACAGTGCCAGT | MtEF1qR1 | AGACTCGTGGTGCATTTCTAC | pMtEF1-SLE4 |
| Cockle (C. edule) | CeEF1qF2 | CCAGTGGGCCGAGTTGAGAC | CeEF1qR2 | CTTCAGTGGTCACGTTGGCAG | pCockleSLE-EF1 |
| P. aureus | VaEF1qF1 | AAAGAATGGACAGACCAGAGAG | VaEF1qR1 | TGCTTCACACCCAATGTGAAA | pVaN2-EF1SLE |
| V. corrugata | VcEF1qF1 | CAAGAATGGACAGACAAGAGAA | VcEF1qR1 | TGTTTCACACCCAAGGTGAAG | pVaH2-EF1SLE |
| SLE | |||||
| SLE-Mt | MtSLEqF2 | TGGACTACTTTACAAAAGCCAACG | MtSLEqRI | GCTTCGTGCAATTTCACTAACAT | pMtEF1-SLE4 |
| SLE-Ce | cockleRT-F2 | GCAGTGTGCAATGTCCCAGT | cockleRT-R2 | GACCTGTGTCCGAGGCATCA | pCockleSLE-EF1 |
| SLE-Pa | VaNSLEqF1 | TAGAGACGAATTGTCCGTGGG | VaNSLEqR4 | CGTGCTCGCTGTAAGCATTT | pVaN2-EF1SLE |
| SLE-Vc | VaHSLEqF1 | TAGAGATGAACTCTCTTTTGA | VaHSLEqRI | TGCACGGCTTAATGTTTTGGA | pVaH2-EF1SLE |
Analysis of microsatellite loci from cockles (C. edule)
Microsatellites were amplified using primers for 12 loci, reported previously to be polymorphic in cockle (C. edule) populations15. Of the 12 primer pairs, 9 pairs amplified products in all cockle DNA samples, with 1–2 alleles observed in normal samples and 1–4 neoplastic alleles observed. These were used in all further analyses, with fluorescent modifications on the 5′ end of the forward primers as listed (Extended Data Table 2). KOD polymerase (Millipore) was used to prevent ambiguity of untemplated addition of A residues by Taq polymerase. Products were run on an agarose gel for visualization. Allele sizes were identified to single-base precision by fragment analysis using florescent primers (6-FAM, PET, NED, and VIC) using a 3730xl Genetic Analyzer with the LIZ-500 size standard, (Applied Biosystems, operated by Genewiz). Peaks were called using Peak Scanner 2.0 (Applied Biosystems), rounding sizes to whole bases, with some manual adjustment to keep alleles of the same size together.
We used the R package Poppr16 to generate distance matrices and phylogenetic trees using the Bruvo’s method with the infinite alleles model. The value of c (repeat size) was set to 0.00001 in order to calculate distances using the band sharing model, in which alleles are either identical sizes (distance=0) or non-identical (distance=1). Each allele was coded as a dominant marker17, with a single variable for each allele observed at each position (104 total markers from 9 loci). For each allele (a single size at a single locus), a sample is observed to be either present, absent, or the information is missing. For example, a sample with sizes 195 and 199 at locus A would be marked as “present” for marker “A-195” and “A-199,” but “absent” for all other sizes at that locus (like “A-203” and “A-207”). This analysis method ignores the uncertainty in the copy number of each locus in aneuploid cancer cells and allows for ambiguity at particular alleles. In cases where the hemocyte and tissue genotypes could be clearly differentiated, all alleles were analyzed (H1 and H3). In some cases (M1, M2, M3, and H2), cancer-specific alleles could be detected in the hemocyte samples, but represented less than 50% of the total hemocyte DNA, and the host tissue alleles obscured one or two positions (depending on whether the normal genotype was homozygous or heterozygous at that locus). In these cases, since the tissue allele obscured a potential neoplastic allele, that size was coded as “missing” for the hemocyte genotype. The total distance between any two samples is calculated as the average of the pairwise differences at all allele sizes observed (104 pairwise comparisons if no data is missing). In pairwise comparisons for each allele, two samples either have a distance of 0 (meaning that the particular allele size is either present in both samples or absent in both samples) or a distance of 1 (meaning that the particular allele size is present in one sample but absent in the other). If data are missing for one or both then that specific comparison is dropped with no contribution to the total distance between the two samples. The genetic distances calculated are based on the alleles observed at the 9 loci, so the absolute value of the scale itself is therefore dependent on the number of observed alleles that are included in the analysis. Source Data for the generation of the phylogenetic tree lists all observed alleles coded as present (1), absent (2), or missing (0) for each sample.
As an alternate analysis method, the data were analyzed using Bruvo’s method as polyploid data with 9 loci, using only the individuals which could be confidently identified. In this alternate analysis, the topology of the resulting tree was not different—all nodes with bootstrap values above 50 were maintained. In this analysis, the genetic distance between the representatives of lineage 1 (H3) and lineage 2 (H1) was 0.676, and the distances to the closest normal sample were 0.595 (H3 to N5) and 0.648 (H1 to N1 and N2).
qPCR
pecies-specific qPCR of the EF1α locus in P. aureus and V. corrugata was done using FastStart Universal SYBR Green Master Mix (Roche). Primers were designed to amplify the same region, with the 3′ end of both forward and reverse primers on sites which differ between the two species (Extended Data Table 2). Standard curves for each primer set were generated using two control plasmids (one containing the P. aureus EF1α fragment and P. aureus SLE fragment and the other containing the fragments from V. corrugata). EF1α fragments were amplified using conserved primers (Extended Data Table 2). Fragments were cloned with the Zero Blunt TOPO Kit (Invitrogen), and plasmids were linearized with NotI before qPCR. Standard curve samples (104 through 102 copies per reaction) and experimental samples (2.5 ng per reaction) were done in triplicate. No amplification was detected with the P. aureus primers on the V. corrugata plasmid or with the V. corrugata primers on P. aureus plasmid (using up to 107 copies per reaction). The fraction of V. corrugata in each sample was calculated as the copy number using V. corrugata-specific primers divided by copy number of P. aureus-plus V. corrugata-specific amplification.
Quantification of Steamer-like elements in the three species was performed by the same method, using primers in the RT-IN region of the retrotransposon (based on sequence obtained through amplification using degenerate primers) and control primers amplifying a region in EF1α. All primers used to amplify SLEs and EF1α from genomic DNA for cloning into standards and primers used for qPCR are listed in Extended Data Table 2. For each species, a single control plasmid containing both the EF1α and the SLE fragment was used.
Extended Data
Extended Data Figure 1. Analysis of mtCOI amplified from tissue and hemocyte DNA of normal and diseased mussels (M. trossulus).
A partial region of the mtCOI gene was amplified from genomic DNA of solid tissue and hemocytes from mussels (M. trossulus) and directly sequenced (Figure 1b). Trace images show a region flanking a representative SNP marked with an open triangle (tissue) or closed triangle (hemocytes). a–b, In normal mussels, tissue and hemocyte alleles match (with G496 at all positions). c–e, In mussels with disseminated neoplasia, the tissue and hemocyte alleles are different. Neoplastic hemocytes have A at position 496 and G in tissue, with some A observable in tissue, likely due to infiltration of neoplastic hemocytes.
Extended Data Figure 2. Quantification of Steamer-like element genomic copy number in mussels (M. trossulus), cockles (C. edule), and golden carpet shell clams (P. aureus).
a–d, Fragments from the SLE reverse transcriptase region and EF1α genes were cloned from each species. Haploid copy numbers of Steamer-like elements (SLE) were quantified by determining the ratio of SLE/EF1a in genomic DNA from hemocytes. Single species-specifc SLEs were analyzed in (a) mussels (M. trossulus) and (b) cockles (C. edule). (c, d) In golden carpet shell clams, one SLE (SLE-Pa) was cloned from a normal P. aureus (clam N2) and a different one (SLE-Vc) was cloned from neoplastic cells (clam H2). Both SLEs could be found in both species, and qPCR analysis confirmed that SLE-Pa is more highly amplified in P. aureus and has fewer copies in V. corrugata and in the neoplastic cells derived from V. corrugata.
Extended Data Table 1.
Morphometric analysis of type A and type B cockle (C. edule) neoplasia
| Cockle ID | Type | Cells counted (N) | Cell diameter ± SD (μm)a | Nuclear diameter ± SD(μm)b | Nuclear/cell ratio ± SDc |
|---|---|---|---|---|---|
| M1 | A | 15 | 9.0 ± 1.2 | 6.7 ± 1.0 | 0.75 ± 0.09 |
| H2 | A | 15 | 8.7 ± 0.9 | 6.6 ± 0.8 | 0.76 ± 0.06 |
| H3 | A | 10 | 8.5 ± 1.1 | 6.5 ± 1.0 | 0.77 ± 0.07 |
| total | A | 40 | 8.8 ± 1.0 | 6.6 ± 0.9 | 0.76 ± 0.07 |
| M2 | B | 10 | 7.7 ± 0.7 | 5.5 ± 0.5 | 0.72 ± 0.07 |
| M3 | B | 15 | 6.7 ± 1.2 | 4.5 ± 0.6 | 0.70 ± 0.15 |
| H1 | B | 15 | 7.3 ± 1.0 | 5.4 ± 0.8 | 0.75 ± 0.09 |
| total | B | 40 | 7.2 ± 1.1 | 5.1 ± 0.8 | 0.72 ± 0.11 |
Cell diameter is statistically different between types A and B (p < 0.0001). All tests are two-tailed T-tests.
Nuclear diameter is statistically different between types A and B (p < 0.0001); for all pairwise comparisons between type A and type B individuals p < 0.05.
Ratio of nuclear diameter to cell diameter is not significantly different between types.
Acknowledgments
MJM and SPG were supported by the Howard Hughes Medical Institute and Training Grant T32 CA009503. DI, MJC, and AV were supported by the Consellería do Mar da Xunta de Galicia, through the project PGIDIT-CIMA 13/03.
Footnotes
Contributions
MJM and SPG wrote the manuscript. MJM conducted molecular analyses. AFM and SAB collected and diagnosed M. trossulus from West Vancouver. JS and CR collected and diagnosed M. trossulus from Vancouver Island. DI, MJC, and AV collected and diagnosed C. edule and P. aureus. MJCD produced micrographs of C. edule neoplastic hemocytes, and DI conducted morphometric analysis.
Sequences generated in this work have been deposited in Genbank under accession numbers KX018521-KX018605.
The authors declare no competing financial interests.
Contributor Information
Michael J. Metzger, Email: mm4184@cumc.columbia.edu.
Antonio Villalba, Email: antonio.villalba.garcia@xunta.es.
María J. Carballal, Email: maria.jesus.carballal.duran@xunta.es.
David Iglesias, Email: david.iglesias.estepa@xunta.es.
James Sherry, Email: Jim.Sherry@ec.gc.ca.
Carol Reinisch, Email: creinisc@mbl.edu.
Annette F. Muttray, Email: amuttray@gmail.com.
Susan A. Baldwin, Email: sbaldwin@mail.ubc.ca.
Stephen P. Goff, Email: spg1@cumc.columbia.edu.
References
- 1.Pearse AM, Swift K. Allograft theory: transmission of devil facial-tumour disease. Nature. 2006;439:549. doi: 10.1038/439549a. [DOI] [PubMed] [Google Scholar]
- 2.Murgia C, Pritchard JK, Kim SY, Fassati A, Weiss RA. Clonal origin and evolution of a transmissible cancer. Cell. 2006;126:477–487. doi: 10.1016/j.cell.2006.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rebbeck CA, Thomas R, Breen M, Leroi AM, Burt A. Origins and evolution of a transmissible cancer. Evolution. 2009;63:2340–2349. doi: 10.1111/j.1558-5646.2009.00724.x. [DOI] [PubMed] [Google Scholar]
- 4.Metzger MJ, Reinisch C, Sherry J, Goff SP. Horizontal transmission of clonal cancer cells causes leukemia in soft-shell clams. Cell. 2015;161:255–263. doi: 10.1016/j.cell.2015.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barber BJ. Neoplastic diseases of commercially important marine bivalves. Aquatic Living Resources. 2004;17:449–466. doi: 10.1051/alr:2004052. [DOI] [Google Scholar]
- 6.Carballal MJ, Barber BJ, Iglesias D, Villalba A. Neoplastic diseases of marine bivalves. J Invertebr Pathol. 2015;131:83–106. doi: 10.1016/j.jip.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 7.Moore JD, Elston RA, Drum AS, Wilkinson MT. Alternate pathogenesis of systemic neoplasia in the bivalve mollusc Mytilus. J Invertebr Pathol. 1991;58:231–243. doi: 10.1016/0022-2011(91)90067-z. [DOI] [PubMed] [Google Scholar]
- 8.Vassilenko E, Baldwin SA. Using flow cytometry to detect haemic neoplasia in mussels (Mytilus trossulus) from the Pacific Coast of Southern British Columbia, Canada. J Invertebr Pathol. 2014;117:68–72. doi: 10.1016/j.jip.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 9.Vassilenko EI, Muttray AF, Schulte PM, Baldwin SA. Variations in p53-like cDNA sequence are correlated with mussel haemic neoplasia: A potential molecular-level tool for biomonitoring. Mutat Res. 2010;701:145–152. doi: 10.1016/j.mrgentox.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 10.Villalba A, Carballal MJ, Lopez C. Disseminated neoplasia and large foci indicating heavy haemocytic infiltration in cockles Cerastoderma edule from Galicia (NW Spain) Dis Aquat Organ. 2001;46:213–216. doi: 10.3354/dao046213. [DOI] [PubMed] [Google Scholar]
- 11.Diaz S, Iglesias D, Villalba A, Carballal MJ. Long-term epidemiological study of disseminated neoplasia of cockles in Galicia (NW Spain): temporal patterns at individual and population levels, influence of environmental and cockle-based factors and lethality. J Fish Dis. 2016 doi: 10.1111/jfd.12436. [DOI] [PubMed] [Google Scholar]
- 12.Carballal MJ, Iglesias D, Diaz S, Villalba A. Disseminated neoplasia in clams Venerupis aurea from Galicia (NW Spain): histopathology, ultrastructure and ploidy of the neoplastic cells, and comparison of diagnostic procedures. J Invertebr Pathol. 2013;112:16–19. doi: 10.1016/j.jip.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 13.Carballal MJ, Iglesias D, Santamarina J, Ferro-Soto B, Villalba A. Parasites and pathologic conditions of the cockle Cerastoderma edule populations of the coast of Galicia (NW Spain) J Invertebr Pathol. 2001;78:87–97. doi: 10.1006/jipa.2001.5049. [DOI] [PubMed] [Google Scholar]
- 14.Iglesias D. PhD thesis. University of Santiago do Compostela; 2006. Estudio patológico de las poblaciones de berberecho Cerastoderma edule (L.) de Galicia. [Google Scholar]
- 15.Martinez L, Arias A, Mendez J, Insua A, Freire R. Development of twelve polymorphic microsatellite markers in the edible cockle Cerastoderma edule (Bivalvia: Cardiidae) Conserv Genet Resour. 2009;1:107–109. doi: 10.1007/s12686-009-9026-7. [DOI] [Google Scholar]
- 16.Kamvar ZN, Tabima JF, Grunwald NJ. Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ. 2014;2:e281. doi: 10.7717/peerj.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodzen JA, May B. Inheritance of microsatellite loci in the white sturgeon (Acipenser transmontanus) Genome. 2002;45:1064–1076. doi: 10.1139/g02-083. [DOI] [PubMed] [Google Scholar]
- 18.Arriagada G, et al. Activation of transcription and retrotransposition of a novel retroelement, Steamer, in neoplastic hemocytes of the mollusk Mya arenaria. Proc Natl Acad Sci U S A. 2014;111:14175–14180. doi: 10.1073/pnas.1409945111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pye RJ, et al. A second transmissible cancer in Tasmanian devils. Proc Natl Acad Sci U S A. 2016;113:374–379. doi: 10.1073/pnas.1519691113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cockrill JM, Beasley JN. Transmission of transmissible venereal tumor of the dog to the coyote. Am J Vet Res. 1979;40:409–410. [PubMed] [Google Scholar]
- 21.Samso A. Docteur es Sciences Naturalles thesis. University de Paris; 1965. Recherches experimentales sur le Sarcome de Sticker. [Google Scholar]
- 22.Stickler A. Transplantables Rundzellensarkom des Hundes. Zeitschrift für Krebsforschung. 1906;4:227–314. [Google Scholar]
- 23.Wade H. An experimental investigation of infective sarcoma of the dog, with a consideration of its relationship to cancer. J Pathol Bacteriol. 1908;12:384–425. doi: 10.1002/path.1700120221. [DOI] [Google Scholar]
- 24.Dungern V. Zur Biologie des Rundzellensarkoms des Hundes. Muenchener Medizinische Wochenschrift. 1912:238–239. [Google Scholar]
- 25.Strakova A, Murchison EP. The changing global distribution and prevalence of canine transmissible venereal tumour. BMC Vet Res. 2014;10:168. doi: 10.1186/s12917-014-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mateo DR, MacCallum GS, Davidson J. Field and laboratory transmission studies of haemic neoplasia in the soft-shell clam, Mya arenaria, from Atlantic Canada. J Fish Dis. 2015 doi: 10.1111/jfd.12426. [DOI] [PubMed] [Google Scholar]
- 27.Kent ML, Wilkinson MT, Drum AS, Elston RA. Failure of transmission of hemic neoplasia of bay mussels, Mytilus trossulus, to other bivalve species. J Invertebr Pathol. 1991;57:435–436. doi: 10.1016/0022-2011(91)90148-j. [DOI] [PubMed] [Google Scholar]
- 28.Canapa A, Schiaparelli S, Marota I, Barucca M. Molecular data from the 16S rRNA gene for the phylogeny of Veneridae (Mollusca : Bivalvia) Mar Biol. 2003;142:1125–1130. doi: 10.1007/s00227-003-1048-1. [DOI] [Google Scholar]
- 29.Muehlenbachs A, et al. Malignant Transformation of Hymenolepis nana in a Human Host. N Engl J Med. 2015;373:1845–1852. doi: 10.1056/NEJMoa1505892. [DOI] [PubMed] [Google Scholar]
- 30.Muttray AF, Schulte PM, Baldwin SA. Invertebrate p53-like mRNA isoforms are differentially expressed in mussel haemic neoplasia. Mar Environ Res. 2008;66:412–421. doi: 10.1016/j.marenvres.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 31.Howard D, Lewis E, Keller B, Smith C. NOAA Technical Memorandum NOS NCCOS. Vol. 5. NOAA/National Centers for Coastal Ocean Science; Oxford, MD: 2004. [Google Scholar]
- 32.Diaz S, Cao A, Villalba A, Carballal MJ. Expression of mutant protein p53 and Hsp70 and Hsp90 chaperones in cockles Cerastoderma edule affected by neoplasia. Dis Aquat Organ. 2010;90:215–222. doi: 10.3354/dao02231. [DOI] [PubMed] [Google Scholar]
- 33.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- 34.Oliverio M, Mariottini P. Contrasting morphological and molecular variation in Coralliophila meyendorffii (Muricidae, Coralliophilinae) J Mollus Stud. 2001;67:243–245. doi: 10.1093/mollus/67.2.243. [DOI] [Google Scholar]
- 35.Salvi D, Mariottini P. Molecular phylogenetics in 2D: ITS2 rRNA evolution and sequence-structure barcode from Veneridae to Bivalvia. Mol Phylogenet Evol. 2012;65:792–798. doi: 10.1016/j.ympev.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 36.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]