Abstract
Introduction
Lipidomics is the large-scale profiling and characterization of lipid species in a biological system using mass spectrometry. The skin barrier is mainly comprised of corneocytes and a lipid-enriched extracellular matrix. The major skin lipids are ceramides, cholesterol and free fatty acids. Lipid compositions are altered in inflammatory skin disorders with disrupted skin barrier such as atopic dermatitis (AD).
Areas covered
Here we discuss some of the recent applications of lipidomics in human skin biology and in inflammatory skin diseases such as AD, psoriasis and Netherton syndrome. We also review applications of lipidomics in human skin equivalent and in pre-clinical animal models of skin diseases to gain insight into the pathogenesis of the skin disease.
Expert commentary
Skin lipidomics analysis could be a fast, reliable and noninvasive tool to characterize the skin lipid profile and to monitor the progression of inflammatory skin diseases such as AD.
Keywords: skin, barrier, corneocytes, lipidomics, mass spectrometry, atopic dermatitis, Netherton syndrome, psoriasis
1. Introduction
A lipidome is defined as the complete spectrum of lipids in a tissue, organelle or membrane. Lipidomics is the large-scale profiling and characterization of a set of lipid species in a biological system[1,2]. Lipidomics was first introduced by Han and Gross in 2003 and has experienced unprecedented progress, mainly because of continuous advances in mass spectrometric techniques over the years[1]. Currently, the development of MS-based lipidomic approaches can be classified into two broad directions, including unbiased global (non-targeted) lipidomics and targeted lipidomics[3]. Global lipidomic analysis refers to identification and quantification of hundreds to thousands of cellular lipid species via a high-throughput process, while targeted lipidomic analysis identifies one or a few lipid classes of interest, and is typically performed via liquid chromatography coupled to mass spectrometry (LC-MS) and liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) based methods[4]. As lipids play essential roles in the maintenance of body health, lipidomics has been introduced for unraveling the role of lipids in life science. One objective of lipidomics studies is to profile the lipids in a biological system and to determine the fundamental biological processes of lipid metabolism. Another objective of lipidomics studies is to find changes in lipid profiles of patients with a certain disease compared to healthy controls and to examine the role of lipids in the progression of the disease and to understand the possible mechanisms of this disease. The applications of lipidomics in various diseases, such as cancers, diabetes, circulation and cardiovascular diseases (e.g. atherosclerosis), neurodegenerative disease (e.g. Alzheimer’s disease) and infection disease, have been well summarized in other reviews[3,5–7]. In the past few years, the application of lipidomics to studies of skin biology and inflammatory skin diseases has drawn particular attention. Several reported studies demonstrate that alteration of lipid composition may play an important role in an impaired skin barrier, which is a major cause of several inflammatory skin diseases[8–10]. In this review, we will focus on discussing some of the recent applications of lipidomics in skin biology and skin disease research.
2. Skin and epidermal barrier
Skin is the largest organ serving multiple functions in the mammalian body, mainly providing a barrier that protects the body against external irritation and infections as well as preventing water loss[11,12]. The skin is composed of the epidermis, dermis and subcutis. The epidermis, the exterior of the skin, mainly consists of four distinctive cell layers: the stratum basale (SB), the stratum spinosum (SS), the stratum granulosum (SG) and the stratum corneum (SC)[13]. The outmost layer of the epidermis, the stratum corneum (SC), is comprised of large, flattened, enucleated corneocytes and a lipid-enriched extracellular matrix[14–16]. The SC is often referred to as a “bricks in mortar” structure, in which corneocytes act as the bricks and the lipids act as the mortar, and it plays a critical role in skin permeability barrier formation and maintenance[17,18].
The presence of intercellular lipids is essential for the maintenance of a moist, pliable and healthy skin barrier. Alteration of SC lipids composition leads to disrupted or impaired skin barrier functions and results in an increase in transepidermal water loss (TEWL)[11,19]. Increased TEWL has been observed in many skin diseases, such as atopic dermatitis (AD) and psoriasis[20–22]. The major classes of lipids found in the SC include ceramides (CERs), cholesterol (Chol), and free fatty acids (FFAs)[17,23,24].
3. Lipidomics for the identification of skin lipid composition in healthy individuals
CERs are the most abundant lipid constituent in human SC, comprising approximately 50% of the intercellular lipid content by mass [25]. Lipidomics has revealed the wide diversity of CERs found in skin SC in humans. CERs consist of a sphingoid base linked via an amide bond to a fatty acid and both variation in the fatty acid carbon chain and the sphingoid base architecture result in a large number of CER subclasses with a wide variation in chain length distribution[26,27]. As early as 1978, Gary and White used quantitative thin layer chromatography (TLC) to identify several CER subclasses in human SC[28]. In 2003, Ponec et al. first reported a reasonably complete description of the human epidermal CERs by a using a combination of high performance thin layer chromatography (HPTLC) and nuclear magnetic resonance spectroscopy (NMR)[29]. Nine CER subclasses were identified: (1) CEREOS] containing ester-linked ω-hydroxy fatty acids and sphingosines, (2) CEREOP] containing ester-linked ω-hydroxy fatty acids and phytosphingosines, (3) CEREOH] containing ester-linked ω-hydroxy fatty acids and 6-hydroxysphingosines, (4) CERNS] containing nonhydroxy fatty acids and sphingosines, (5) CERNP] containing nonhydroxy fatty acids and phytosphingosines, (6) CERNH] containing nonhydroxy fatty acids and 6-hydroxysphingosines, (7) CERAS] containing α-hydroxy fatty acids and sphingosines, (8) CERAP] containing α-hydroxy fatty acids and phytosphingosines, and (9) CERAH] containing α-hydroxy fatty acids and 6-hydroxysphingosines[29]. Due to the rapid advances in mass spectrometric techniques, Masukawa et al. (2008) developed an improved method for the comprehensive quantification of CER species in human SC by using simultaneous selected ion monitoring measurement of as many as 182 molecular-related ions derived from overall CER species in a normal-phase liquid chromatography-electrospray ionization-mass spectrometry (NPLC-ESI-MS) technique combined with a calculation procedure using relative responses of respective authentic CERs[27]. In addition to the previous nine classes, this analysis identified two more CER subclasses: CERNDS] contains nonhydroxy fatty acids and dihydrosphingosines and CERADS] contains ω-hydroxy fatty acids and dihydrosphingosines[27,29]. More recently, Smeden et al. reported a total of 12 CER subclasses (Table 1) in human SC harvested from tape strips including a new CER subclass, CEREODS], containing ester-linked ω-hydroxy fatty acids and dihydrosphinosines, and all 11 previously known subclasses by using liquid chromatography-tandem mass spectrometry (LC-MS/MS) with an ion trap (IT) system, a Fourier transform-ion cyclotron resonance system and a triple quadrupole system[26]. The combined LC-MS platform provides both information on lipid subclasses as well as the lipid chain length distribution in each of the subclasses[24]. Moreover, four additional CER subclasses, based on four different sphingoid bases and each consisting of non-esterified ω-hydroxy fatty acids [CER (OS/OH/OP/OdS], have been reported only in healthy human SC [30,31].
Table 1.
Twelve subclasses of ceramide in human SC. (Modified from Janessens et. al, 2012, J. Lipid Res)
| Fatty acids | Non-hydroxy fatty acid [N] |
α-hydroxy fatty acid [A] |
Esterified ω-hydroxy fatty acid [EO] |
|---|---|---|---|
| Sphingoid bases | |||
| Dihydrosphingosine [dS] | [NDS] | [Ads] | [EOdS] |
| Sphingosine [S] | [NS] | [AS] | [EOS] |
| Phytosphingosine [P] | [NP] | [AP] | [EOP] |
| 6-hydroxy sphingosine [H] | [NH] | [AH] | [EOH] |
Fatty acids have been identified as another major constituent of the SC and are crucial for epidermal barrier structure and function, such as creating an acidic environment at the surface of the SC[23,32]. As early as 1970, Ansari et al. performed gas/liquid chromatography (GLC) to analyze the fatty acid composition of the human epidermis. The presence of saturated fatty acids, mono-unsaturated fatty acids (MUFAs), poly-unsaturated fatty acids (PUFAs) and hydroxy-free fatty acids were detected in the human epidermis[33]. In 1998, a gas-liquid chromatography coupled to high-performance liquid chromatography (HPLC)/light scattering detection method (LSD) was developed by Norlen et al. to quantitative free fatty acids in the SC[34]. Very long chain (C32-C36) saturated and monounsaturated FFAs were detected in human SC and C24:0 and C26:0 were the most abundant FFAs[34]. Recently, an LC-MS-based lipidomics analysis, a newly developed method with enhanced sensitivity, confirmed the previous observations about the FFAs composition in the human SC[35].
4. Lipidomics in skin diseases
Lipidomics analysis is not only applied to identify the lipid composition in normal skin, but is also applied to investigate the changes in lipid profiles of skin with impaired barrier functions associated with diseases such as atopic dermatitis, Netherton syndrome (NTS) and psoriasis.
4.1. Atopic dermatitis
Atopic dermatitis is a common chronic inflammatory skin disease characterized by disrupted epidermal barrier functions, which can be indicated by increased TEWL[36,37]. About 15%–20% of children and 1–3% of adults are affected by AD[11,38]. The role of lipids, especially CERs, in skin permeability barrier function in AD has been widely studied and reviewed[11,22,39]. Briefly, the abnormalities of SC lipids in AD were first reported by Melnik et al. in 1988[40]. In 1991, Imokawa et al. showed for the first time that the CEREOS] level had been reduced in AD [41]. Since then, several articles reported the SC lipid abnormalities in AD, especially CERs [42–44]. The most recent applications of MS-based lipidomics analysis in AD will be discussed in detail here. In 2010, Ishikawa et al. performed lipidomics analysis using normal-phase liquid chromatography coupled with electrospray ionization-mass spectrometry to compare the SC CERs profile between AD patients and healthy individuals within small groups[45]. Their study showed that the level of total CERs as well as those of the CERNH], CERNP], CEREOS], CEREOH], and CEREOP] classes were significantly lower in AD lesional skin compared with controlled healthy non-lesional skin[45]. In addition, the levels of certain CER species with short chain lengths (e.g. CERNS], CERNDS], CERAS]) were lower in AD patients and certain CER species with long chain lengths (e.g. CERNS], CERNDS], CERNH], CERAS], CERAH] were significantly higher in AD subjects[45]. Later in 2012, a more comprehensive comparison of CER composition in the SC of AD and control subjects was performed by Janssens et al using LC-MS[8]. Consistent with a previous report, they observed that the CER chain length is altered in AD subjects by significantly elevated C34 CERs levels (e.g CERNS], CERAS] and CERAH]) and decreased ω-esters of (ω)-O-acyl-CER levels. The changes in CER chain length were strongly correlated with aberrant lipid organization and decreased SC permeability barrier functions (as measured by TEWL). In addition, it was demonstrated that altered CER composition and abnormal lipid organization significantly correlated with disease severity and levels of natural moisturizing factors derived from filaggrin in skin [8].
4.2. Netherton syndrome
Netherton syndrome (NTS) is a severe genetic skin disease that is caused by autosomal recessive mutations in the SPINK5 gene encoding the serine protease inhibitor, lympho-epithelial Kazal-type 5 inhibitor (LEKT1). The mutations in the SPINK5 gene lead to increased activity of several kallikrein-related peptidases, thereby compromising formation of the skin barrier[46,47]. The disease is characterized by chronic inflammation, hair shaft defects and epidermal hyperplasia with a drastically impaired cutaneous barrier function[47]. However, little is known about the SC lipid composition and whether SC lipids are important in NTS patients. Recently, a comprehensive LC-MS based lipidomics analysis by Smeden et al. has revealed drastic alterations of the SC lipid profile in NTS patients[10]. In this study, decreased FFA chain lengths and increased levels of monounsaturated fatty acids (MUFAs) have been seen in the SC of NTS patients compared with healthy individuals. Furthermore, the study showed the total amount of CERs is reduced, whereas the level of short-chain CERs was increased, and unsaturated CERs of different chain length and subclasses were observed in NTS patients compared to healthy individuals. In addition, it was also found that the amount of acyl-CERs is reduced in a subgroup of NTS patients compared to the controls. Finally, they elucidated that the changes in lipid profiles (FFAs and CERs) in NTS patients corresponded to altered expression of enzymes involved in SC lipid processing, including stearoyl-CoA desaturase-1 (involved in converting FFAs into MUFAs), elongase (ELOVL)-1 and 6 (responsible for elongation of the FFAs), beta-glucocerebrosidase [48] and acid sphingomeylinase (aSMase) (responsible for conversion of CER precursors into their specific CER subclass). Thus, these results indicated the importance of the SC lipids in maintenance of a healthy and functional skin barrier, the alteration of which may lead to diseased skin, such as NTS[10].
4.3. Psoriasis
Psoriasis is a common incurable chronic inflammatory skin disease that is characterized by disrupted epidermal homeostasis and decreased skin barrier function[49]. Altered ceramide distribution and content has been previously reported by a number of studies. As early as 1993, Motta et al. reported the changes in the CERs profile of psoriasis patients, including reduction of CEREOS], [NP], [AP] and induction of CERAS] and [NS], using a TLC-based method. Recently, Tawada and colleagues published an important article in which they analyze the CER profiles of the SC in psoriasis patients by using matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF-MS)[50]. Significant differences in the fatty acid composition profiles of certain CER subclasses of the SC between psoriasis patients and normal controls (e.g CER [ADS], [NP], CERNH] and CERAP]) were observed. The proportion of CERs with long-chain fatty acids was significantly lower in psoriasis patients than in healthy controls[50]. However, the three classes of ω-hydroxy CERs (CEREOS], CEREOH] and CEREOP]) were unable to be analyzed in their studies [30]. Not only the ceramide profiles but also the levels of sphingoid bases were changed in the psoriatic epidermis. Sung and colleagues reported that levels of sphingosine and sphinganine were increased in the psoriatic epidermis and increased expression of ceramidase was positively correlated with the clinical severity of psoriasis[9]. These studies suggest that the alteration of SC lipids could, at least partially, be linked to the decreased skin barrier function in the skin of psoriasis patients. More comprehensive lipidomics analysis is needed to characterize the SC lipid profile in psoriasis patients and to further illustrate the role of SC lipids in skin barrier maintenance in these patients.
5. Lipidomics in pre-clinical animal models
Pre-clinical animal models of human disease are commonly utilized to gain insight into the pathogenesis of specific diseases[48]. In this review, we also briefly summarize some applications of lipidomics in pre-clinical mouse models of certain skin diseases (e.g. AD). To elucidate potential mechanisms underlying depletion of long-chain CERs in the SC of AD, Park et al. used a hapten-induced AD mice model[51]. First, they conducted a lipidomics study by using LC-MS/MS-electrospray ionization (ESI) technique to investigate the lipid profile in AD-like skin. A reduction in CER chain length was observed in AD-like skin, which was in agreement with previous observations in human SC. Next, they further examined the expression of the genes involved in the synthesis of long-chain CERs and found the level of certain fatty acid elongases, such as ELOVL-1 and 4, was significantly reduced at both the RNA and protein level. These results suggested that the alteration in the profile of fatty acid lengths of CERs might due to the changes in expression of elongase genes. Recently, our lab performed a lipidomics analysis in a pre-clinical eczema model in which the mice were deficient in a transcription factor, COUP-TF-Interacting protein 2 (CTIP2), by using an LC-MS/MS technique[52]. Mice lacking Ctip2 (Ctip2−/− mice) exhibit epidermal permeability barrier defects (e.g increased TEWL), and selective ablation of Ctip2 in epidermal keratinocytes leads to the development of an atopic dermatitis-like phenotype in skin[53,54]. The lipidomics study showed an altered composition of major epidermal lipids, including a decreased CERNS] level and an increased level of sphingomyelins in Ctip2 knockout mice. Interestingly, the changes in the sphingolipid (ceramide and sphingomyelin) profile in Ctip2-null mice were accompanied by differences in the expression of genes involved in sphingolipid biosynthesis and metabolism, including ceramide synthases (Lass1-,2,-3 and 6), serine palmitoyltransferases (Sptlc1 and -3), sphingomyelinases (Smpd1 and -2), sphingomyelin synthase 2 (Sgm2), acid β-glucosidase (Gba2), glucosylceramide synthase (Ugcg), N-acylsphingosine amidohydrolases (Asah2), sphingosine kinases (Sphk1 and 2), and alkaline ceramidases (Acer1)[52]. The results from this lipidomics study indicated a crucial role of CTIP2 in regulating epidermal lipid metabolism. A comparative bioinformatics analysis between the top human and mouse skin-associated genes has recently been reported. This study identified about 200 commonly shared genes between mice and humans, and the majority of those encode proteins that participate in structural and barrier functions[55], highlighting the importance of the epidermal barrier in these species to maintain skin homeostasis and prevent onset of AD. The utilization of lipidomics analysis in a mouse model will lead to the identification of specific genes (e.g. key epidermal lipid-regulating transcription factors) involved in skin lipid biosynthesis and metabolism with respect to skin diseases and will contribute to the understanding of possible mechanisms involved in lipid metabolism in certain diseases, such as AD.
6. Lipidomics in human skin equivalents
The most commonly used skin substitute, the 3-dimensional (3D) tissue engineered human skin equivalent, which is generated from keratinocytes and fibroblasts, is an alternative to animal studies. Drongelen and colleagues used a filaggrin (FLG) knock-down human skin equivalent to investigate the effect of FLG on SC lipid properties. They found that a knock-down of FLG did not alter the lipid composition and organization in the human skin equivalents, which is in line with previous studies showing that changes in SC lipid properties were independent of FLG mutations in AD patients[8,47,56]. In another study, it was shown that Th2 cytokines and tumor necrosis factor alpha (TNF-α reduced the level of long chain FFAs and acyl-CERs, consequently resulted in altered lipid organization in human skin equivalents, indicating the influence of cytokines on AD pathogenesis[57]. These studies provide information that human skin equivalents can be a useful skin substitute for future studies. However, there are still noticeable differences in lipid (ceramide and FFA) profiles between human skin equivalents and human skin SC obtained from tape strips [24]. Development of improved types of human skin equivalent that can more closely mimic human skin will be important in the future.
7. Expert commentary & five-year view
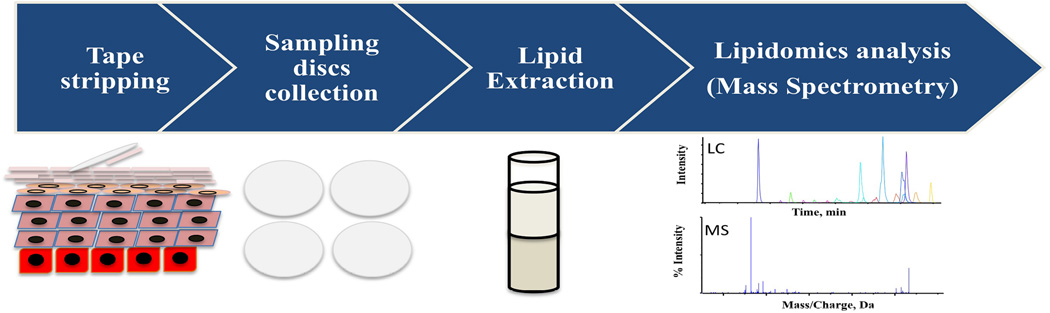
A typical lipidomics analysis workflow for a human SC sample is briefly summarized below (Figure 1): 1) A human SC sample is collected by tape stripping; 2) The SC sample is then subjected to lipid extraction using an extraction protocol optimized according to specific lipids of interest [e.g modified Bligh and Dyer method[58]]; 3) The extracted lipid is subjected to MS analysis after chromatographic separation; 4) Data is collected for identification and quantification of specific lipids of interest; 5) The levels of the specific lipid of interest are compared between samples under different conditions (e.g. healthy skin vs. AD skin) as represented in Figure 2.
Figure 1.
A typical lipidomics analysis workflow on a human SC sample.
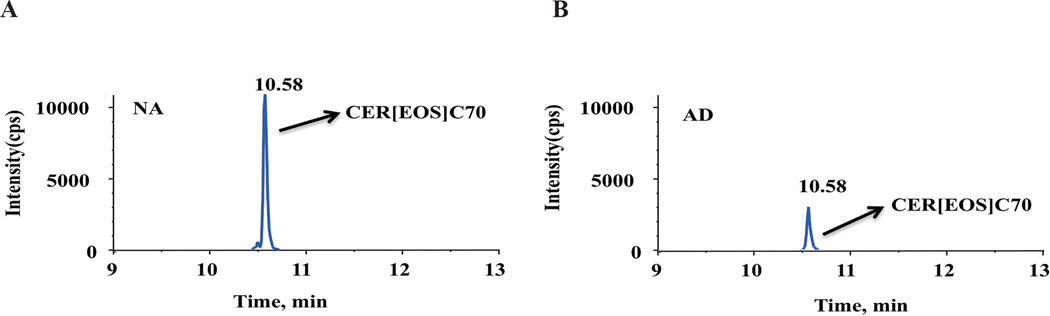
Figure 2.
A representative example of an extracted-ion chromatogram in the positive ion mode from LC-MS analysis of human skin: (A) normal healthy skin and (B) atopic dermatitis skin. NA: non-atopic; AD: atopic dermatitis. cps: counts per second.
The applications of lipidomics in studies of skin biology and skin disorders have revealed and enhanced the knowledge of SC lipids under different physiological or pathological conditions. Particularly, lipidomics analysis can be a fast, reliable, and powerful tool to characterize lipid profiles in diseased skin and to estimate the barrier status in inflammatory skin diseases. Although much profiling of lipids in diseased skin has been accomplished, the exact relationship between the alteration of the lipid composition and the onset of inflammatory skin disease is still unclear. Ultimately, a combination of lipidomics, genomics and proteomics will be needed to investigate the possible mechanism(s) underlying the pathogenesis of certain skin diseases, but lipidomic analysis should be extremely helpful for exploring new treatment options for skin diseases such as AD.
Key issues.
Lipidomics is the large-scale profiling and characterization of a set of lipid species in a biological system.
The major lipid constituents of the stratum corneum are ceramides, cholesterol and free fatty acids.
Lipid compositions are altered in certain inflammatory skin disorders, such as atopic dermatitis, Netherton syndrome and psoriasis.
Applications of mass spectrometry-based lipidomics to study skin biology and skin disorders will further our knowledge of skin lipid functions.
Skin lipidomics analysis could be a fast, reliable and non-invasive tool for estimating barrier status and for monitoring the progression of inflammatory skin diseases.
Acknowledgments
The authors thank Dr. Jaewoo Choi for helping with figure preparation and Dr. Phil Proteau for critical reading of the manuscript. The authors also thank Drs. Mark Zabriskie, Theresa Filtz and Mark Leid of the OSU-OHSU College of Pharmacy for continuous support and encouragement.
The studies on Ctip2/Bcl11b were supported by grant AR056008 from NIAMS. This project has been funded in whole or in part with funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract/grant no: HHSN272201000020C, HHSN272201000017C, UM2AI117870, U19AI117673-01.
Footnotes
Declaration of interest
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
References
Papers of special note have been highlighted as:
* of interest
** of considerable interest
- 1.Han X, Gross RW. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: a bridge to lipidomics. J Lipid Res. 2003;44:1071–1079. doi: 10.1194/jlr.R300004-JLR200. [DOI] [PubMed] [Google Scholar]
- 2.Spener F, Lagarde M, Geloen A, Record M. What is lipidomics? Eur. J. Lipid Sci. Technol. 2003;105:481–482. [Google Scholar]
- 3.Lam SM, Shui G. Lipidomics as a principal tool for advancing biomedical research. J Genet Genomics. 2013;40:375–390. doi: 10.1016/j.jgg.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 4.Murphy SA, Nicolaou A. Lipidomics applications in health, disease and nutrition research. Mol Nutr Food Res. 2013;57:1336–1346. doi: 10.1002/mnfr.201200863. [DOI] [PubMed] [Google Scholar]
- 5.Checa A, Bedia C, Jaumot J. Lipidomic data analysis: tutorial, practical guidelines and applications. Anal Chim Acta. 2015;885:1–16. doi: 10.1016/j.aca.2015.02.068. [DOI] [PubMed] [Google Scholar]
- 6.Dehairs J, Derua R, Rueda-Rincon N, Swinnen JV. Lipidomics in drug development. Drug Discov Today Technol. 2015;13:33–38. doi: 10.1016/j.ddtec.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 7.Yang L, Li M, Shan Y, Shen S, et al. Recent advances in lipidomics for disease research. J Sep Sci. 2016;39:38–50. doi: 10.1002/jssc.201500899. [DOI] [PubMed] [Google Scholar]
- 8.Janssens M, van Smeden J, Gooris GS, Bars W, et al. Increase in short-chain ceramides correlates with an altered lipid organization and decreased barrier function in atopic eczema patients. J Lipid Res. 2012;53:2755–2766. doi: 10.1194/jlr.P030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moon SH, Kim JY, Song EH, Shin MK, et al. Altered levels of sphingosine and sphinganine in psoriatic epidermis. Ann Dermatol. 2013;25:321–326. doi: 10.5021/ad.2013.25.3.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Smeden J, Janssens M, Boiten WA, van Drongelen V, et al. Intercellular skin barrier lipid composition and organization in Netherton syndrome patients. J Invest Dermatol. 2014;134:1238–1245. doi: 10.1038/jid.2013.517. [DOI] [PubMed] [Google Scholar]
- 11.van Smeden J, Janssens M, Gooris GS, Bouwstra JA. The important role of stratum corneum lipids for the cutaneous barrier function. Biochim Biophys Acta. 2014;1841:295–313. doi: 10.1016/j.bbalip.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 12.Elias PM, Friend DS. The permeability barrier in mammalian epidermis. J Cell Biol. 1975;65:180–191. doi: 10.1083/jcb.65.1.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsui T, Amagai M. Dissecting the formation, structure and barrier function of the stratum corneum. Int Immunol. 2015;27:269–280. doi: 10.1093/intimm/dxv013. [DOI] [PubMed] [Google Scholar]
- 14.Elias PM. The Epidermal Permeability Barrier: From the Early Days at Harvard to Emerging Concepts. J Invest Dermatol. 2004;122:xxxvi–xxxix. doi: 10.1046/j.0022-202X.2004.22233.x. [DOI] [PubMed] [Google Scholar]
- 15.Candi E, Schmidt R, Melino G. The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol. 2005;6:328–340. doi: 10.1038/nrm1619. [DOI] [PubMed] [Google Scholar]
- 16.Hachem JP, Houben E, Crumrine D, Man MO, et al. Serine protease signaling of epidermal permeability barrier homeostasis. J Invest Dermatol. 2006;126:2074–2086. doi: 10.1038/sj.jid.5700351. [DOI] [PubMed] [Google Scholar]
- 17.Elias PM, Menon GK. Structural and lipid biochemical correlates of the epidermal permeability barrier. Adv Lipid Res. 1991;24:1–26. doi: 10.1016/b978-0-12-024924-4.50005-5. [DOI] [PubMed] [Google Scholar]
- 18.Elias PM. Epidermal lipids, barrier function, and desquammation. J Invest Dermatol. 1983;80:44s–49s. [PubMed] [Google Scholar]
- 19.Feingold KR, Elias PM. Role of lipids in the formation and maintenance of the cutaneous permeability barrier. Biochim Biophys Acta. 2014;1841:280–294. doi: 10.1016/j.bbalip.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 20.Takahashi H, Tsuji H, Minami-Hori M, Miyauchi Y, et al. Defective barrier function accompanied by structural changes of psoriatic stratum corneum. J Dermatol. 2014;41:144–148. doi: 10.1111/1346-8138.12393. [DOI] [PubMed] [Google Scholar]
- 21.Lee CH, Chuang HY, Shih CC, Jong SB, et al. Transepidermal water loss, serum IgE and beta-endorphin as important and independent biological markers for development of itch intensity in atopic dermatitis. Br J Dermatol. 2006;154:1100–1107. doi: 10.1111/j.1365-2133.2006.07191.x. [DOI] [PubMed] [Google Scholar]
- 22.Borodzicz S, Rudnicka L, Mirowska-Guzel D, Cudnoch-Jedrzejewska A. The role of epidermal sphingolipids in dermatologic diseases. Lipids Health Dis. 2016;15:13. doi: 10.1186/s12944-016-0178-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feingold KR. The outer frontier: the importance of lipid metabolism in the skin. J Lipid Res. 2009;50:S417–S422. doi: 10.1194/jlr.R800039-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. van Smeden J, Boiten WA, Hankemeier T, Rissmann R, et al. Combined LC/MS-platform for analysis of all major stratum corneum lipids, and the profiling of skin substitutes. Biochim Biophys Acta. 2014;1841:70–79. doi: 10.1016/j.bbalip.2013.10.002. * This study describes a semi-quantitative method to analzye all main lipid classes present in human SC and did a comparative analysis of lipid profile between human skin subsitutes and human SC.
- 25.Meckfessel MH, Brandt S. The structure, function, and importance of ceramides in skin and their use as therapeutic agents in skin-care products. J Am Acad Dermatol. 2014;71:177–184. doi: 10.1016/j.jaad.2014.01.891. [DOI] [PubMed] [Google Scholar]
- 26. van Smeden J, Hoppel L, van der Heijden R, Hankemeier T, et al. LC/MS analysis of stratum corneum lipids: ceramide profiling and discovery. J Lipid Res. 2011;52:1211–1221. doi: 10.1194/jlr.M014456. **They developed a LC-MS based method to identify all 12 SC CER subclasses.
- 27. Masukawa Y, Narita H, Sato H, Naoe A, et al. Comprehensive quantification of ceramide species in human stratum corneum. J Lipid Res. 2009;50(8):1708–1719. doi: 10.1194/jlr.D800055-JLR200. ** This is the first literature to develop a method for comprehensive quantification of CER species in human stratum corneum.
- 28.Gray GM, White RJ. Glycosphingolipids and ceramides in human and pig epidermis. J Invest Dermatol. 1978;70:336–341. doi: 10.1111/1523-1747.ep12543527. [DOI] [PubMed] [Google Scholar]
- 29.Ponec M, Weerheim A, Lankhorst P, Wertz P. New acylceramide in native and reconstructed epidermis. J Invest Dermatol. 2003;120:581–588. doi: 10.1046/j.1523-1747.2003.12103.x. [DOI] [PubMed] [Google Scholar]
- 30.Shin JH, Shon JC, Lee K, Kim S, et al. A lipidomic platform establishment for structural identification of skin ceramides with non-hydroxyacyl chains. Anal Bioanal Chem. 2014;406:1917–1932. doi: 10.1007/s00216-013-7601-y. [DOI] [PubMed] [Google Scholar]
- 31.van Smeden J, Bouwstra JA. Stratum Corneum Lipids: Their Role for the Skin Barrier Function in Healthy Subjects and Atopic Dermatitis Patients. Curr Probl Dermatol. 2016;49:8–26. doi: 10.1159/000441540. [DOI] [PubMed] [Google Scholar]
- 32.Grubauer G, Feingold KR, Harris RM, Elias PM. Lipid content and lipid type as determinants of the epidermal permeability barrier. J Lipid Res. 1989;30:89–96. [PubMed] [Google Scholar]
- 33.Ansari MN, Nicolaides N, Fu HC. Fatty acid composition of the living layer and stratum corneum lipids of human sole skin epidermis. Lipids. 1970;5:838–845. doi: 10.1007/BF02531977. [DOI] [PubMed] [Google Scholar]
- 34.Norlén L, Nicander I, Lundsjö A, Cronholm T, et al. A new HPLC-based method for the quantitative analysis of inner stratum corneum lipids with special reference to the free fatty acid fraction. Arch Dermatol Res. 1998;290:508–516. doi: 10.1007/s004030050344. [DOI] [PubMed] [Google Scholar]
- 35.van Smeden J, Janssens M, Kaye EC, Caspers PJ, et al. The importance of free fatty acid chain length for the skin barrier function in atopic eczema patients. Exp Dermatol. 2014;23:45–52. doi: 10.1111/exd.12293. [DOI] [PubMed] [Google Scholar]
- 36.Leung DY. New insights into atopic dermatitis: role of skin barrier and immune dysregulation. Allergol Int. 2013;62:151–161. doi: 10.2332/allergolint.13-RAI-0564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leung DYM, Boguniewicz M, Howell MD, Nomura I, et al. New insights into atopic dermatitis. Journal of Clinical Investigation. 2004;113:651–657. doi: 10.1172/JCI21060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nutten S. Atopic dermatitis: global epidemiology and risk factors. Ann Nutr Metab. 2015;66:8–16. doi: 10.1159/000370220. [DOI] [PubMed] [Google Scholar]
- 39.Breiden B, Sandhoff K. The role of sphingolipid metabolism in cutaneous permeability barrier formation. Biochim Biophys Acta. 2014;1841:441–452. doi: 10.1016/j.bbalip.2013.08.010. [DOI] [PubMed] [Google Scholar]
- 40.Melnik B, Hollmann J, Plewig G. Decreased stratum corneum ceramides in atopic individuals--a pathobiochemical factor in xerosis? Br J Dermatol. 1988;119:547–549. doi: 10.1111/j.1365-2133.1988.tb03262.x. [DOI] [PubMed] [Google Scholar]
- 41.Imokawa G, Abe A, Jin K, Higaki Y, et al. A. Decreased Level of Ceramides in Stratum Corneum of Atopic Dermatitis: An Etiologic Factor in Atopic Dry Skin? Journal of Investigative Dermatology. 1991;96:523–526. doi: 10.1111/1523-1747.ep12470233. [DOI] [PubMed] [Google Scholar]
- 42.Yamamot A, Serizawa S, Ito M, Sato Y. stratum corneum lipid abnormalities in atopic dermatitis. Arch Dermatol Res. 1991;284:219–223. doi: 10.1007/BF01106105. [DOI] [PubMed] [Google Scholar]
- 43.Di Nardo A, Wertz P, Giannetti A, Seidenari S. Ceramide and cholesterol composition of the skin of patients with atopic dermatitis. Acta Derm Venereol. 1998;78:27–30. doi: 10.1080/00015559850135788. [DOI] [PubMed] [Google Scholar]
- 44.Matsumoto M, Umemoto N, Sugiura H, Uehara M. Difference in ceramide composition between "dry" and "normal" skin in patients with atopic dermatitis. Acta Derm Venereol. 1999;79:246–247. doi: 10.1080/000155599750011183. [DOI] [PubMed] [Google Scholar]
- 45.Ishikawa J, Narita H, Kondo N, Hotta M, et al. Changes in the ceramide profile of atopic dermatitis patients. J Invest Dermatol. 2010;130:2511–2514. doi: 10.1038/jid.2010.161. [DOI] [PubMed] [Google Scholar]
- 46.Chavanas S, Bodemer C, Rochat A, Hamel-Teillac D, et al. Mutations in SPINK5, encoding a serine protease inhibitor, cause Netherton syndrome. Nat Genet. 2000;25:141–142. doi: 10.1038/75977. [DOI] [PubMed] [Google Scholar]
- 47.Raghunath M, Tontsidou L, Oji V, Aufenvenne K, et al. SPINK5 and Netherton syndrome: novel mutations, demonstration of missing LEKTI, and differential expression of transglutaminases. J Invest Dermatol. 2004;123:474–483. doi: 10.1111/j.0022-202X.2004.23220.x. [DOI] [PubMed] [Google Scholar]
- 48.Morgan SJ, Elangbam CS, Berens S, Janovitz E, et al. Use of animal models of human disease for nonclinical safety assessment of novel pharmaceuticals. Toxicol Pathol. 2013;41:508–518. doi: 10.1177/0192623312457273. [DOI] [PubMed] [Google Scholar]
- 49.Roberson ED, Bowcock AM. Psoriasis genetics: breaking the barrier. Trends Genet. 2010;26:415–423. doi: 10.1016/j.tig.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tawada C, Kanoh H, Nakamura M, Mizutani Y, et al. Interferon-gamma decreases ceramides with long-chain fatty acids: possible involvement in atopic dermatitis and psoriasis. J Invest Dermatol. 2014;134:712–718. doi: 10.1038/jid.2013.364. * They showed changes in the fatty acid compostion profiles of SC CERs in psoriasis patients.
- 51.Park YH, Jang WH, Seo JA, Park M, et al. Decrease of ceramides with very long-chain fatty acids and downregulation of elongases in a murine atopic dermatitis model. J Invest Dermatol. 2012;132:476–479. doi: 10.1038/jid.2011.333. [DOI] [PubMed] [Google Scholar]
- 52. Wang Z, Kirkwood JS, Taylor AW, Stevens JF, et al. Transcription factor Ctip2 controls epidermal lipid metabolism and regulates expression of genes involved in sphingolipid biosynthesis during skin development. J Invest Dermatol. 2013;133:668–676. doi: 10.1038/jid.2012.358. ** This study showed for the first time the correlation between skin lipid compostion and lipid metabolism genes in a mouse model.
- 53.Golonzhka O, Liang X, Messaddeq N, Bornert JM, et al. Dual role of COUP-TF-interacting protein 2 in epidermal homeostasis and permeability barrier formation. J Invest Dermatol. 2009;129:1459–1470. doi: 10.1038/jid.2008.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Z, Zhang LJ, Guha G, Li S, et al. Selective ablation of Ctip2/Bcl11b in epidermal keratinocytes triggers atopic dermatitis-like skin inflammatory responses in adult mice. PLoS One. 2012;7:e51262. doi: 10.1371/journal.pone.0051262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gerber PA, Buhren BA, Schrumpf H, Homey B, et al. The top skin-associated genes: a comparative analysis of human and mouse skin transcriptomes. Biol Chem. 2014;395:577–591. doi: 10.1515/hsz-2013-0279. [DOI] [PubMed] [Google Scholar]
- 56.van Drongelen V, Alloul-Ramdhani M, Danso MO, Mieremet A, et al. Knock-down of filaggrin does not affect lipid organization and composition in stratum corneum of reconstructed human skin equivalents. Exp Dermatol. 2013;22:807–812. doi: 10.1111/exd.12271. [DOI] [PubMed] [Google Scholar]
- 57.Danso MO, van Drongelen V, Mulder A, van Esch J, et al. TNF-alpha and Th2 cytokines induce atopic dermatitis-like features on epidermal differentiation proteins and stratum corneum lipids in human skin equivalents. J Invest Dermatol. 2014;134:1941–1950. doi: 10.1038/jid.2014.83. [DOI] [PubMed] [Google Scholar]
- 58. Bligh E, Dyer W. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. **This study described for the first time isolation and purification of total skin lipids.