Abstract
Objectives
Genome-wide association studies (GWAS) of lung cancer have identified regions of common genetic variation with lung cancer risk in Europeans who smoke and never-smoking Asian women. This study aimed to conduct a GWAS in African Americans, who have higher rates of lung cancer despite smoking fewer cigarettes per day when compared with Caucasians. This population provides a different genetic architecture based on underlying African ancestry allowing the identification of new regions and exploration of known regions for finer mapping.
Materials and Methods
We genotyped 1,024,001 SNPs in 1737 cases and 3602 controls in stage 1, followed by a replication phase of 20 SNPs (p<1.51×10−5) in an independent set of 866 cases and 796 controls in stage 2.
Results and Conclusion
In the combined analysis, we confirmed two loci to be associated with lung cancer that achieved the threshold of genome-wide significance: 15q25.1 marked by rs2036527 (p = 1.3 × 10−9; OR = 1.32; 95% CI = 1.20–1.44) near CHRNA5, and 5p15.33 marked by rs2853677 (p = 2.8 × 10−9; OR = 1.28; 95% CI = 1.18–1.39) near TERT. The association with rs2853677 is driven by the adenocarcinoma subtype of lung cancer (p = 1.3 × 10−8; OR = 1.37; 95% CI = 1.23–1.54). No SNPs reached genome-wide significance for either of the main effect models examining smoking - cigarettes per day and current or former smoker. Our study was powered to identify strong risk loci for lung cancer in African Americans; we confirmed results previously reported in African Americans and other populations for two loci near plausible candidate genes, CHRNA5 and TERT, on 15q25.1 and 5p15.33 respectively, are associated with lung cancer. Additional work is required to map and understand the biological underpinnings of the strong association of these loci with lung cancer risk in African Americans.
Keywords: Genome-wide Association Study, Lung Neoplasms, Smoking, African Americans, Telomerase, Receptors, Cholinergic
1. Introduction
Lung cancer is the second most common cancer in US men and women and has the highest death rate [1]. There is a substantial disparity in African-American men, who have higher age-adjusted incidence rates (99.9 per 100,000) when compared with Caucasian and Asian men (76.4 and 52.2 per 100,000, respectively), as well as higher age-adjusted death rates (African American 82.6 per 100,000; Caucasian 65.3 per 100,000; Asian 35.9 per 100,000) [2]. Smoking represents the major risk factor for lung cancer [3–5]. Known differences in smoking-related exposures for African Americans could contribute to the observed differences in incidence, including age of smoking initiation, intensity of smoking, and quitting and relapse behaviors [6]. It has been estimated that approximately eighty-five percent of lung cancer risk is attributed to cigarette smoking; however, the higher incidence seen in African Americans occurs even though they smoke fewer cigarettes per day than their Caucasian counterparts [7]. Family and twin studies, as well as lung cancer genome-wide association studies (GWAS) have confirmed an underlying genetic contribution to lung cancer risk as well. To date, GWAS performed in populations of European and Asian descent have identified over 15 lung cancer risk loci [8–21] (Supplementary Table 1). Interestingly, other than the hTERT region on chromosome 5p15, the regions identified in non-smoking Asian women do not overlap with the regions identified in Europeans who smoke, suggesting distinct genetic contributions to primary lung carcinogenesis and smoking-related carcinogenesis. Although African Americans have higher lung cancer incidence and poorer lung cancer survival when compared with other racial and ethnic populations in the United States, no GWAS has been conducted in this population. In addition, African admixture in African Americans could provide an opportunity to identify new lung cancer risk alleles [22]. Smoking behavior also has a significant genetic component, as reported in family, twin and GWAS studies [12, 23–26]. Given differences in smoking exposures and genetic architecture in African Americans as compared to Europeans and Asians, studies of genetic variation contributing to risk of lung cancer in African Americans might help explain the racial difference observed in lung cancer incidence and survival, and identify novel markers of lung cancer susceptibility.
Studies have been conducted to further interrogate chromosomal regions 2q31.1, 5p15.33, 6p22.1-p21.31, and 15q25.1 in African Americans, including fine mapping studies [27–31]. The 15q25.1 region harbors three nicotinic acetylcholine receptor subunit genes (CHRNA3, CHRNA4, and CHRNA5), which are associated with lung cancer [9, 11, 12], smoking behavior [32], nicotine addiction [33], and increased exposure to specific nicotine metabolites [34]. These studies have identified SNPs potentially associated with lung cancer risk in African Americans, either weakly associated with or independent of smoking behavior [28, 29]. Additionally, variants on 5p15.33 and 6p21.33 were observed to have histology-specific associations with lung cancer risk in African Americans [31, 35]. Specifically, variants on 5p15.33 appear to have a stronger association with adenocarcinoma, whereas variants on 6p21.33 had a specific association with squamous cell carcinoma.
In light of the current evidence suggesting that a genetic contribution to lung cancer in African Americans, we conducted a two-stage GWAS comprised of 1737 cases and 3602 controls (stage 1) and followed by replication of the most significantly associated genotypes in an independent sample set of 866 cases and 796 controls (stage 2).
2. Materials and Methods
2.1 Study populations
A two-stage case-control study was designed to evaluate the association between common genetic variants and the risk of lung cancer. Detailed study descriptions of the eight studies included in the discovery phase (stage 1) GWAS and seven studies in the replication phase (stage 2) can found in Supplementary Information. The stage 1 studies included 1737 cases and 3602 controls from the following studies: MD Anderson Lung Cancer Epidemiology Study, The Multiethnic Cohort Study (MEC), NCI-MD Lung Cancer-Case Control Study, Northern California Lung Cancer Study, Project CHURCH (Creating a Higher Understanding of Cancer Research & Community Health), Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial (PLCO), Southern Community Cohort Study (SCCS), and the Karmanos Cancer Institute at Wayne State University (KCI/WSU). The Project CHURCH dataset contained controls only; therefore, these data were combined with the MD Anderson Lung Cancer Epidemiology Study to generate one analytical unit for the analyses (designated as MDA). The stage 2 studies included an independent set of 866 cases and 796 controls from the following studies: The Black Women’s Health Study (BWHS), The Harvard-MGH Lung Cancer Susceptibility Study (HLCS), MD Anderson Lung Cancer Epidemiology Study, MD Anderson/LBJ Hospital Biorepository, NCI-MD Lung Cancer Case-Control Study, Northern California Lung Cancer Study, Philadelphia Lung Cancer Study on Gene Environment Interactions (Plus-Gene), Southern Community Cohort Study (SCCS), and KCI/WSU. The MD Anderson Lung Cancer Epidemiology Study and MD Anderson/LBJ Hospital Biorepository were provided as one dataset from the investigators, so were analyzed as one analytical unit (designated as MDA).
Each participating study obtained informed consent from study participants and approval of the study from its IRB; studies also obtained IRB certification permitting data sharing in accordance with the US National Institutes of Health (NIH) Policy for Sharing of Data Obtained in NIH-Supported or -Conducted Genome-Wide Association Studies (GWAS).
2.2 Genome-wide SNP genotyping
Genome-wide SNP genotyping of lung cancer cases from MD Anderson Lung Cancer Epidemiology Study, NCI-MD Lung Cancer Case-Control Study, Northern California Lung Cancer Study, Project CHURCH, PLCO, SCCS, and KCI/WSU (Supplementary Table 2a) was conducted using the Illumina HumanHap 1M Duo chip at the NCI Cancer Genomics Research Laboratory (CGR) in the Division of Cancer Epidemiology and Genetics (DCEG) at the National Cancer Institute. SNPs from the Multi-ethnic Cohort (MEC) were genotyped using the same chip at University of Southern California and the primary iDat files were shared for the combined genotype calling.
2.3 Quality control assessment
SNPs with less than a 90% completion rate were excluded from further analysis. Samples were excluded on the basis of (i) completion rates lower than 94% (n = 556 samples); (ii) abnormal heterozygosity values of less than 24% or greater than 32% (n = 54); (iii) discordant expected duplicates (n=10); (iv) abnormal X-chromosome heterozygosity including gender discordant samples (n = 52); and (v) phenotype exclusion (due to ineligibility or incomplete information) (n = 114).
Using the high quality data based on the sample level filters above, we performed an assay concordance analysis and identified all 164 expected duplicates with average concordance rate > 99.9%. Genotypes for all subject pairs were also examined for close relationships (presence of first- and second-degree relatives) using the Genotyping Library and Utilities (GLU) version 1.0 qc.ibds module with an IBD0 threshold of 0.70, and 122 first-degree relatives were identified and excluded from analysis.
Using a set of population informative SNPs [36] and data from HapMap 27, we excluded 27 subjects that displayed either Asian admixture coefficients >20% or European admixture coefficients >90% [37], as determined on the basis of the GLU strct.admix module with the HapMap I+II CEU, YRI, ASA (JPT +CHB) samples used as reference populations [38] (Supplementary Figure 1). The final association analysis included 1737 cases and 3602 controls of African American ancestry. After quality control filtering, data from 1,024,001 SNPs were available.
2.4 Replication and TaqMan genotyping
A total of 20 SNPs with p < 1.51 × 10−5 based on the adjusted trend model was advanced for replication in stage 2. TaqMan genotyping assays (ABI) were optimized in the CGR pipeline stage 2 genotyping, which included BWHS, HLCS, MD Anderson Lung Cancer Epidemiology Study, MD Anderson/LBJ Hospital Biorepository, NCI-MD Lung Cancer Case-Control Study, Plus-Gene, KCI/WSU, and the Northern California Lung Cancer Study (Supplementary Table 2b). SCCS genotyped their samples at Vanderbilt using the Sequenom platform for 21 assays where rs17568263 was substituted with rs2128611 [a perfect linkage disequilibrium (LD) surrogate] and rs55781567 was genotyped as a redundant assay because it is in LD with rs2036527. rs2036527 was not genotyped in Northern California Lung Cancer Study. We analyzed a total of 20 assays in 866 cases and 796 controls drawn from 7 studies, including additional samples from 5 of the stage 1 studies and 355 controls pooled from an African American renal cancer GWAS [39].
2.5 Statistical analysis
Associations between SNPs and risk of lung cancer (overall, adenocarcinoma only, and squamous cell carcinoma only) were estimated using multivariate logistic regression assuming a trend genetic effect (i.e. linear increase in risk per affected allele) and adjusting for study, age, gender, smoking status (never/former/current), and admixture coefficient for YRI (stage 1); and study, age, gender, smoking status (never/former/current) (stage 2). When included in the null model (baseline model and covariates only), none of the top 10 eigenvectors is significantly (P < 0.05) associated with the case/control outcome, and therefore were they were not included in the final main effect model as adjustment for population substructure. For smoking status, we ran a logistic regression model for current versus former smokers assuming linear trend for each SNP and adjusting for study, age, gender, admixture coefficient for YRI, one significant eigenvector, and lung cancer case/control status. We also ran a linear regression model for log transformation of cigarette smoked per day (CPD) and assuming a trend effect for SNP and adjusting for study, age, gender, admixture coefficient for YRI and lung cancer case/control status. To examine the association between smoking behavior and lung cancer, we performed linear regression analysis for CPD and pack years as the outcomes, adjusting for study, age, gender, coefficient for YRI, current smokers, and eigenvectors.
2.6 Data analysis
Data analysis and management were performed with GLU (https://code.google.com/p/glu-genetics/).
3. Results
The stage 1 analysis included 1737 cases and 3602 controls among African Americans drawn from eight studies of lung cancer (Supplementary Table 2a). We observed no evidence of systematic genomic inflation of the test statistic (λ = 1.003) for the 1,024,001 genotyped SNPs analyzed in stage 1 and had no excess of small p values beyond what was expected (Supplementary Figure 2).
Three SNPs were observed to be associated with lung cancer below the threshold of genome-wide significance (p > 5 × 10−8) in stage 1 (rs55781567, rs3019885, and rs6580649) (Supplementary Figure 3). The most significant association was observed in rs55781567 (p = 7.1 × 10−9; OR=1.34; 95% CI=1.21–1.48; MAF = 0.25 in cases and 0.308 in controls), located in the 5′-untranslated region (UTR) of the gene CHRNA5 (cholinergic receptor, nicotinic, alpha 5) on chromosome 15q25. Twenty promising SNPs (p < 1.51 × 10−5) from stage 1 were genotyped in stage 2 in 866 cases and 796 controls from 7 studies, including additional samples from 5 of the stage 1 studies and 355 controls pooled from an African American renal cancer GWAS [39] (Supplementary Table 2b).
We selected the 20 SNPs for replication in stage 2 (Supplementary Table 3). SNP rs55781567 failed the assay design; however, rs2036527, also located in the 5′-untranslated region (UTR) of the gene CHRNA5 on chromosome 15q25, is a surrogate (r2=1 and 0.48 in 1000 Genomes Project CEU and YRI data, respectively) for rs55781567 and was genotyped in the replication phase. The surrogate selection was based on rs2036527 being the next most significant SNP observed in our scan in the same region. Among the 20 SNPs evaluated, the stage 2 ORs were similar to or somewhat attenuated from the stage 1 ORs for 11, but in different directions for 9.
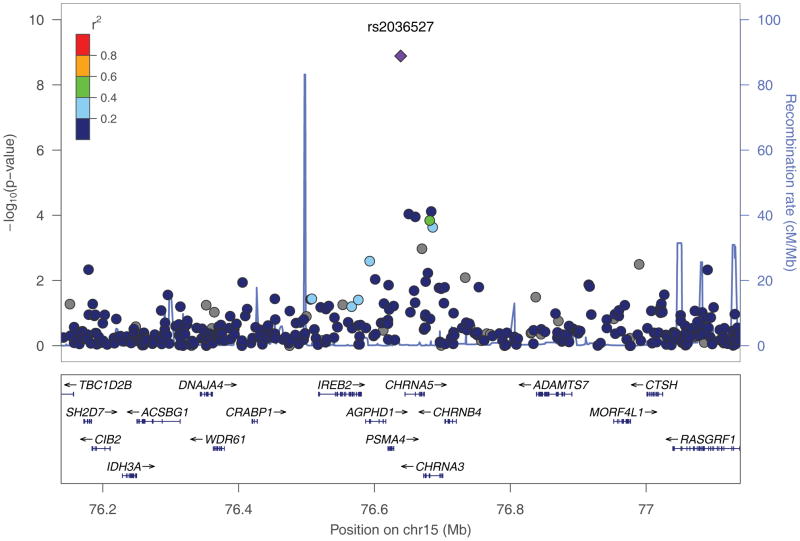
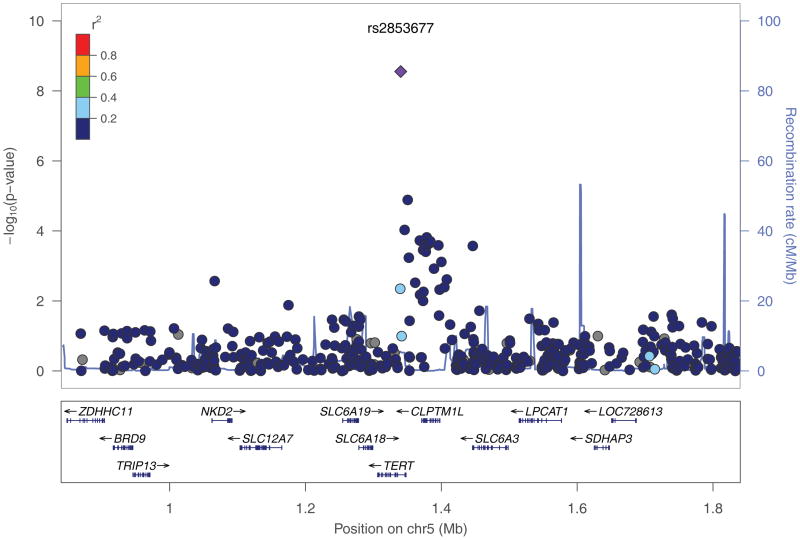
Combining results from stages 1 and 2, two SNPs were significant at the genome-wide significance level. rs2036527 (p = 1.3 × 10−9; OR = 1.32; 95% CI = 1.20–1.44) and rs2853677 (p = 2.8 × 10−9; OR=1.28; 95% CI = 1.18–1.39), which are located 511 base pairs from the initiation sequence of CHRNA5 and in the first intron of telomerase gene (TERT), respectively (Figure 1, Table 1, Supplementary Table 3). SNPs rs3019885 and rs6580649 did not replicate. When stratified by histology, we observed an association with adenocarcinoma below the threshold for genome-wide significance (p = 1.3 × 10−8; OR=1.37; 95% CI = 1.23–1.53), but not squamous cell carcinoma (p = 0.42; OR=1.06; 95% CI = 0.92–1.23). Notably, four loci that failed to replicate displayed substantial differences in allele frequencies between stage1 (GWAS scan) and stage 2 (TaqMan or Sequenom together). Moreover, the allele frequencies of stage 1 were quite different from those estimates from HapMap samples (CEU or YRI); whereas the allele frequencies based on stage 2 genotypes for all four loci were very similar (Supplementary Table 3). In this regard, we consider that the Illumina genotype assays may be problematic for these four SNPs and thus not reliable [40].
Fig. 1.
Regional plots of association results, recombination rates, and linkage disequilibrium for the (a) 15q25.1 marked by rs2036527; (b) 5p15.33 marked by rs2853677. Association results from a log-additive genetic model in −log10 P-values of the SNPs were shown according to their chromosomal positions (hg18) within 1Mb region centering around the index SNP. P values were based on the stage 1 only except for both rs2036527 and rs2853677 where the combined P values were shown. Linkage Disequilibrium (LD) r2 values were estimated on 1000 Genomes Project data June 2010 YRI population (hg18). The colors of the dots reflect the LD with the index SNP as shown in the legend box. The figures were generated using locusZoom (http://csg.sph.umich.edu/locuszoom/).
Table 1.
Two genome-wide significant SNPs for lung cancer risk after combining stage 1 and 2
| Locus | Genes | SNP (reference, effect) | Model | Stage | Controls | Cases | EAF in controlc | EAF in casesc | OR | 95%CI | P | Phet |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 15q25.1 | CHRNA5 | rs2036527 (C,T)a | Overall | Stage1 | 3601 | 1736 | 0.212 | 0.255 | 1.33 | (1.20–1.48) | 1.03E-07 | |
| Stage2 | 1062 | 762 | 0.222 | 0.282 | 1.28 | (1.09–1.51) | 2.92E-03 | |||||
| Combinedb | 4663 | 2498 | 0.214 | 0.263 | 1.32 | (1.20–1.44) | 1.30E-09 | 0.70432 | ||||
| Adeno only | Stage1 | 3601 | 739 | 0.212 | 0.254 | 1.28 | (1.12–1.47) | 4.06E-04 | ||||
| Stage2 | 1062 | 284 | 0.222 | 0.282 | 1.31 | (1.05–1.64) | 1.83E-02 | |||||
| Combinedb | 4663 | 1023 | 0.214 | 0.262 | 1.29 | (1.15–1.45) | 2.28E-05 | 0.87615 | ||||
| Squamous only | Stage1 | 3601 | 399 | 0.212 | 0.237 | 1.25 | (1.04–1.51) | 1.91E-02 | ||||
| Stage2 | 1062 | 176 | 0.222 | 0.259 | 1.24 | (0.93–1.66) | 1.36E-01 | |||||
| Combinedb | 4663 | 575 | 0.214 | 0.244 | 1.25 | (1.07–1.46) | 5.59E-03 | 0.96628 | ||||
|
| ||||||||||||
| 5p15.33 | TERT | rs2853677 (T,C) | Overall | Stage1 | 3600 | 1736 | 0.277 | 0.320 | 1.26 | (1.14–1.38) | 4.28E-06 | |
| Stage2 | 1137 | 851 | 0.261 | 0.321 | 1.35 | (1.16–1.56) | 1.15E-04 | |||||
| Combinedb | 4737 | 2587 | 0.273 | 0.320 | 1.28 | (1.18–1.39) | 2.80E-09 | 0.45653 | ||||
| Adeno only | Stage1 | 3600 | 739 | 0.277 | 0.341 | 1.34 | (1.18–1.52) | 4.44E-06 | ||||
| Stage2 | 1137 | 311 | 0.261 | 0.339 | 1.45 | (1.17–1.78) | 5.55E-04 | |||||
| Combinedb | 4737 | 1050 | 0.273 | 0.340 | 1.37 | (1.23–1.53) | 1.27E-08 | 0.55297 | ||||
| Squamous only | Stage1 | 3600 | 400 | 0.277 | 0.278 | 1.04 | (0.87–1.25) | 6.34E-01 | ||||
| Stage2 | 1137 | 192 | 0.261 | 0.276 | 1.11 | (0.85–1.46) | 4.44E-01 | |||||
| Combinedb | 4737 | 592 | 0.273 | 0.277 | 1.06 | (0.92–1.23) | 4.16E-01 | 0.69993 | ||||
Study design failed; rs2036527 was used as surrogate for the original top SNP rs55781567 in stage 1.
For the combined stage, the odds ratio and p values were generated using a fixed-effects meta-analysis approach. Heterogeneity was assessed using p-values for Cochran’s Q statistic.
Effective allele frequency in controls or cases
We examined whether SNPs were associated with smoking, including cigarettes smoked per day and smoking status (former vs. current smokers). No SNPs reached genome-wide significance for either of the main effect models examining smoking behavior. When comparing current with former smokers, the most significant association was observed with rs1293936 (p = 5.41 × 10−7; OR = 1.28, 95% CI = 1.16 – 1.41), which is located in ESR1 (estrogen receptor 1) on chromosome 6q25.1 (data not shown). When examining cigarettes smoked per day (log transformed) in a linear regression model, the most significant association was observed with rs1372626 (p = 2.5 × 10−6), which is located in the intron of Deleted in Colorectal Cancer (DCC) encoding netrin 1 receptor (data not shown).
Additionally, we evaluated the association between smoking behavior (CPD and pack years) and lung cancer case-control status. We observed that lung cancer is strongly associated with both CPD and pack years in this population (Supplementary Tables 4a and 4b).
4. Discussion
Herein, we report the first GWAS of common genetic variation for lung cancer and smoking phenotypes in 1737 African American cases and 3602 controls, followed by a replication phase including an independent set of 866 cases and 796 controls. Two loci, previously reported in GWAS in other populations, were significant at the genome-wide significance level when combining the discovery and replication phases (Table 1); 15q25.1 marked by rs2036527 near CHRNA5, and 5p15.33 marked by rs2853677 near TERT. The first SNP marker, rs2036527 on 15q25.1 is located 511 base pairs from the initiation sequence for CHRNA5, in the gene cluster CHRNA5-CHRNA3-CHRNA4 that codes for the α5, α3, β5 subunits of the nicotinic acetylcholine receptor (nAChR). Nicotinic acetylcholine receptors initiate the primary brain and peripheral responses to smoking. A number of studies have implicated the nAChR gene cluster in smoking-related phenotypes, including cigarettes smoked per day [12, 32, 41–49], smoking 100 cigarettes or more in a lifetime [42], persistent smoking [50], heavy smoking [32], quitting attempts [42], age at smoking initiation [33, 51, 52], pleasurable early smoking experience [53], reported physical effects after smoking first cigarette [54] and serum cotinine levels [44]. These observations have been reported primarily in populations of European ancestry. Some of the above-mentioned studies include African Americans [45, 52, 53, 55]; yet, small sample size was an issue for some [52, 53]. However, rs2036527 was observed to be associated with smoking quantity in African Americans through a GWAS meta-analysis of 32,389 subjects from the Study of Tobacco in Minority Populations Genetics Consortium [56]. This SNP has also been associated with nicotine dependence in the Finnish Twin Cohort Study [57] and cigarettes smoked per day [43]. However, we did not see a similar association in our data when examining current versus former smokers (p=0.29), and only nominally significant (p=0.038) if modeling cigarettes smoked per day, perhaps due to the small sample size and that African Americans on average smoke cigarettes differently than Caucasians. It has been shown that CPD is less predictive of smoke intake in African-American smokers when compared with Caucasian smokers [58]. When examining carbon monoxide (CO), a biomarker of cigarette smoke, Bloom et al. observed that the correlation between CO and CPD is significantly lower in African Americans when compared with European Americans [59]. Additionally, we recognize that comparing current versus former smokers or examining CPD is not an ideal surrogate for nicotine dependency; however, in the assembled dataset, these are the best measures available. Furthermore, the relationship between rs2036527 and smoking behavior is not likely to be mediated solely through CPD.
rs16969968, a nonsynonomous SNP in CHRNA5 that is in LD with rs2036527 (r2 = 0.88, 1000 Genomes Project CEU data [60]) in populations of European descent [61], replaces an aspartic acid with an asparagine at codon 398 of the protein. This SNP has been associated with smoking behavior in European populations and been shown to have functional consequences, including partial loss of the function of the protein [50, 62]. rs16969968 has been reported to be associated with smoking behavior and nicotine dependence in several small studies with African Americans subjects [32, 61, 63]. In 2012, a large meta-analysis in African American smokers showed a significant association between rs2036527 and self-reported CPD, but not between rs16969968 CPD [56]. However, a more recent meta-analysis showed rs16969968 is associated with increased risk for nicotine dependence in European and African Americans and provided evidence that the A allele increases risk for heavy smoking [64]. In another study, rs16969968 was not found to be consistently associated with smoking abstinence in African Americans; whereas rs2036527 was associated with active pharmacotherapy [65]. Additionally, rs588765 in high LD with rs2036527, and was observed to be associated with CHRNA5 mRNA expression levels, using a diploytpe analysis [66]. Therefore, it is possible that rs16969968 and/or rs588765 are the functional variants in CHRNA5 underlying the associations we have observed with rs2036527. Although these SNPs may be functional variants, they only partially capture variation in nicotine dependency and are one driver of heavy smoking. For example, recent report observed that rs2273500-C, a splice site acceptor variant in CHRNA4 on chromosome 20 accounts for some nicotine dependency [67].
The SNP marker, rs2036527 has been previously shown to be associated with lung cancer in African Americans in a fine mapping study of the nAChR region [28]. It has been shown that rs2036527 is associated with tumor DNA methylation levels in the promoter of CHRNB4, suggesting that genotype-specific methylation may be associated with susceptibility to lung cancer [68]. Because this SNP has been associated with smoking habits, it is not clear whether smoking may also play a role in the epigenetic deregulation. Further investigations into the functional implications of this relationship are warranted.
The second genome-wide significant SNP observed in our study, rs2853677, is located in the first intron of TERT on chromosome 5p15.33. In a case-control study of Caucasian and African-American women, rs2853677 was associated with non-small cell lung cancer in both populations when the data were stratified by race [69]. Furthermore, rs2853677 has been shown to be associated with lung cancer susceptibility in a Japanese GWAS [20]. In a recent report, the A allele of rs2853677 displayed pleiotropy between different cancer types. The same allele is positively associated with testicular germ cell tumor and pancreatic cancer, but negatively associated with glioma and lung cancers, which includes some of the African-American individuals in this study [70]. Additionally, concordant with earlier work, we observed that rs2853677 had a histology-specific association with risk of adenocarcinoma. Variants on 5p15.33 detected upstream of, and within, intron 1 of TERT (rs2735940 and rs2736100) have been observed to be associated with risk of adenocarcinoma [31, 35]. rs2853677 is in moderate linkage disequilibrium (LD) with rs2735940 and rs2736100 in 1000 Genome Project CEU data (r2 = 0.42 and 0.56, respectively) as compared with moderate to low LD (r2 = 0.34 and 0.04, respectively) in 1000 Genomes Project YRI data [60]. This suggests that using an African-American population can refine the association signals in this complex region. We have further confirmed its association with lung cancer in African Americans at a genome-wide significance level by replicating the finding in additional samples of African Americans. Further functional interrogation is warranted.
5. Conclusions
Our study was powered to identify strong risk loci for lung cancer in African Americans; we confirmed data previously reported in African Americans and other populations for two loci near plausible candidate genes, CHRNA5 and TERT, rs2036527 and rs2853677 on 15q25.1 and 5p15.33 respectively, are associated with lung cancer. Additional work is required to map and understand the biological underpinnings of the strong association of these loci with lung cancer risk in African Americans. Further studies of African-American smokers and never-smokers are warranted to investigate the underlying genetic contribution of common variants to lung cancer.
Supplementary Material
Highlights.
Confirmed previous reports that two loci on 15q25.1 and 5p15.33 are associated with lung cancer.
Two loci associated with lung cancer are near plausible candidate genes, CHRNA5 and TERT.
No SNPs reached genome-wide significance for the main effect model examining cigarettes per day.
Acknowledgments
Funding Acknowledgments:
The Black Women’s Health Study (BWHS): NIH grants R01CA098663 and R01CA05842
The Harvard-MGH Lung Cancer Susceptibility Study (HLCS): National Cancer Institute (R01CA074386, R01CA092824, and P20CA090578)
The Multiethnic Cohort Study (MEC): The MEC study, blood collection, DNA extraction and genotyping was supported by National Institutes of Health grants CA63464, CA54281, CA1326792, CA164973, CA148085, CA138338, CA148127 and HG004726 and a Department of Defense Breast Cancer Research Program Era of Hope Scholar Award to CAH (W81XWH-08-1-0383).
MD Anderson Lung Cancer Epidemiology Study: The University of Texas MD Anderson Cancer Center Duncan Family Institute for Cancer Prevention and Risk Assessment; R01CA111646, P50CA070907, R01CA55769, R01CA121197S2, U19CA148127.
NCI-MD Lung Cancer Case-Control Study: Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research
Northern California Lung Cancer Study: National Institute of Environmental Health Sciences (R01ES06717); and the National Cancer Institute (R01CA52689)
Philadelphia Lung Cancer Study on Gene Environment Interactions (Plus-Gene): This work was funded by grant PA4100038714 from the Pennsylvania Department of Health, NCI R21CA156087, and NIH-NIEHS grant P30ES013508 for the Center of Excellence in Environmental Toxicology.
Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial (PLCO): The NCI African American lung cancer GWAS was supported by the intramural research program of the National Institutes of Health, National Cancer Institute. This project has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Project CHURCH (Creating a Higher Understanding of Cancer Research & Community Health): University Cancer Foundation, the Duncan Family Institute through the Center for Community-Engaged Translational Research, the Ms. Regina J. Rogers Gift: Health Disparities Research Program, the Cullen Trust for Health Care Endowed Chair Funds for Health Disparities Research, the Morgan Foundation Funds for Health Disparities Research and Educational Programs, and the National Institutes of Health through MD Anderson’s Cancer Center Support Grant (CA016672)
The Southern Community Cohort Study (SCCS): Funding for the SCCS research was provided by NIH grant R01CA092447. DNA Sample preparation was conducted at the Survey and Biospecimen Shared Resources, which is supported in part by Vanderbilt-Ingram Cancer Center (P30CA068485), and Dr. Melinda Aldrich’s effort was supported by NIH/NCI grant K07CA172294.
Karmanos Cancer Institute at Wayne State University: NIH grants R01CA060691, R01CA87895, contracts N01-PC35145 and P30CA22453, Department of Health and Human Services contract HHSN261201000028C, and the Herrick Foundation.
Additional Acknowledgments:
NCI-MD Lung Cancer Case-Control Study: We would like to thank Glenwood Trivers for managing the contract; Dean Mann, Donna Perlmutter, and Leoni Leondaridis for study management; and the Surgery and Pathology Departments from the participating Hospitals.
Project CHURCH (Creating a Higher Understanding of Cancer Research & Community Health): We would like to acknowledge the research staff at The University of Texas MD Anderson Cancer Center who assisted with implementation of the original project. We especially want to thank the church leadership and participants, whose efforts made this study possible.
The Southern Community Cohort Study (SCCS): We would like to thank Ms. Regina Courtney for her excellent laboratory assistance.
Footnotes
Conflict of Interest Statement:
We wish to confirm that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Krista A. Zanetti, Email: zanettik@mail.nih.gov.
Zhaoming Wang, Email: wangzha@mail.nih.gov.
Melinda Aldrich, Email: melinda.aldrich@Vanderbilt.Edu.
Christopher I. Amos, Email: Christopher.I.Amos@Dartmouth.edu.
William J. Blot, Email: blotw@iei.us.
Elise D. Bowman, Email: bowmane@intra.nci.nih.gov.
Laurie Burdette, Email: burdettl@mail.nih.gov.
Quiyin Cai, Email: qiuyin.cai@Vanderbilt.Edu.
Neil Caporaso, Email: caporasn@mail.nih.gov.
Charles C. Chung, Email: chungcc@mail.nih.gov.
Elizabeth M. Gillanders, Email: lgilland@mail.nih.gov.
Christopher A. Haiman, Email: haiman@usc.edu.
Helen M. Hansen, Email: helen.hansen@ucsf.edu.
Laurence N. Kolonel, Email: lkolonel@crch.hawaii.edu.
Loic Le Marchand, Email: Loic@cc.hawaii.edu.
Shengchao Li, Email: shengchao.li@nih.gov.
Lorna Haughton McNeill, Email: lmcneill@mdanderson.org.
Bríd M. Ryan, Email: ryanb@mail.nih.gov.
Ann G. Schwartz, Email: schwarta@karmanos.org.
Jennette D. Sison, Email: jennette.sison@ucsf.edu.
Margaret R. Spitz, Email: spitz@bcm.edu.
Margaret Tucker, Email: tuckerp@mail.nih.gov.
Angela S. Wenzlaff, Email: wenzlaff@karmanos.org.
John K. Wiencke, Email: john.wiencke@ucsf.edu.
Lynne Wilkens, Email: lynne@cc.hawaii.edu.
Margaret R. Wrensch, Email: Margaret.Wrensch@ucsf.edu.
Xifeng Wu, Email: xwu@mdanderson.org.
Wei Zheng, Email: wei.zheng@vanderbilt.edu.
Weiyin Zhou, Email: zhouw@mail.nih.gov.
David Christiani, Email: dchris@hsph.harvard.edu.
Julie R. Palmer, Email: jpalmer@bu.edu.
Trevor M. Penning, Email: PENNING@UPENN.EDU.
Alyssa G. Rieber, Email: arieber@mdanderson.org.
Lynn Rosenberg, Email: lrosenbe@bu.edu.
Edward A. Ruiz-Narvaez, Email: eruiznar@bu.edu.
Li Su, Email: lisu@hsph.harvard.edu.
Anil Vachani, Email: avachani@mail.med.upenn.edu.
Yongyue Wei, Email: ywei@hsph.harvard.edu.
Alexander S. Whitehead, Email: aswhiteh@mail.med.upenn.edu.
Stephen J. Chanock, Email: chanocks@mail.nih.gov.
Curtis C. Harris, Email: harrisc@mail.nih.gov.
References
- 1.American Cancer Society; American Cancer Society, editor. Cancer Facts & Figures 2015. Atlanta: 2015. [Google Scholar]
- 2.NCI Surveillance Research Program. Fast Stats: An interactive tool for access to SEER cancer statistics. 2013. [Google Scholar]
- 3.Doll R, Hill AB. A study of the aetiology of carcinoma of the lung. Br Med J. 1952;2:1271–1286. doi: 10.1136/bmj.2.4797.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doll R, Peto R. Cigarette smoking and bronchial carcinoma: dose and time relationships among regular smokers and lifelong non-smokers. J Epidemiol Community Health. 1978;32:303–313. doi: 10.1136/jech.32.4.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spitz MR, Wu X, Wilkinson A, Wei Q. Cancer of the Lung. In: Scottenfeld D, Fraumeni JF Jr, editors. Cancer Epidemiology adn Prevention. New York, NY: Oxford Univ Press; 2006. pp. 638–658. [Google Scholar]
- 6.Fagan P, Moolchan ET, Lawrence D, Fernander A, Ponder PK. Identifying health disparities across the tobacco continuum. Addiction. 2007;102(Suppl 2):5–29. doi: 10.1111/j.1360-0443.2007.01952.x. [DOI] [PubMed] [Google Scholar]
- 7.Haiman CA, Stram DO, Wilkens LR, Pike MC, Kolonel LN, Henderson BE, Le Marchand L. Ethnic and racial differences in the smoking-related risk of lung cancer. N Engl J Med. 2006;354:333–342. doi: 10.1056/NEJMoa033250. [DOI] [PubMed] [Google Scholar]
- 8.Broderick P, Wang Y, Vijayakrishnan J, Matakidou A, Spitz MR, Eisen T, Amos CI, Houlston RS. Deciphering the impact of common genetic variation on lung cancer risk: a genome-wide association study. Cancer Res. 2009;69:6633–6641. doi: 10.1158/0008-5472.CAN-09-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D, Mukeria A, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, Fabianova E, Mates D, Bencko V, Foretova L, Janout V, Chen C, Goodman G, Field JK, Liloglou T, Xinarianos G, Cassidy A, McLaughlin J, Liu G, Narod S, Krokan HE, Skorpen F, Elvestad MB, Hveem K, Vatten L, Linseisen J, Clavel-Chapelon F, Vineis P, Bueno-de-Mesquita HB, Lund E, Martinez C, Bingham S, Rasmuson T, Hainaut P, Riboli E, Ahrens W, Benhamou S, Lagiou P, Trichopoulos D, Holcatova I, Merletti F, Kjaerheim K, Agudo A, Macfarlane G, Talamini R, Simonato L, Lowry R, Conway DI, Znaor A, Healy C, Zelenika D, Boland A, Delepine M, Foglio M, Lechner D, Matsuda F, Blanche H, Gut I, Heath S, Lathrop M, Brennan P. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 10.Hsiung CA, Lan Q, Hong YC, Chen CJ, Hosgood HD, Chang IS, Chatterjee N, Brennan P, Wu C, Zheng W, Chang GC, Wu T, Park JY, Hsiao CF, Kim YH, Shen H, Seow A, Yeager M, Tsai YH, Kim YT, Chow WH, Guo H, Wang WC, Sung SW, Hu Z, Chen KY, Kim JH, Chen Y, Huang L, Lee KM, Lo YL, Gao YT, Liu L, Huang MS, Jung TH, Jin G, Caporaso N, Yu D, Kim CH, Su WC, Shu XO, Xu P, Kim IS, Chen YM, Ma H, Shen M, Cha SI, Tan W, Chang CH, Sung JS, Zhang M, Yang TY, Park KH, Yuenger J, Wang CL, Ryu JS, Xiang Y, Deng Q, Hutchinson A, Kim JS, Cai Q, Landi MT, Yu CJ, Tucker M, Hung JY, Lin CC, Perng RP, Boffetta P, Chen CY, Chen KC, Yang SY, Hu CY, Chang CK, Fraumeni JF, Jr, Chanock S, Yang PC, Rothman N, Lin D. The 5p15.33 locus is associated with risk of lung adenocarcinoma in never-smoking females in Asia. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, Dong Q, Zhang Q, Gu X, Vijayakrishnan J, Sullivan K, Matakidou A, Wang Y, Mills G, Doheny K, Tsai YY, Chen WV, Shete S, Spitz MR, Houlston RS. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40:616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, Manolescu A, Thorleifsson G, Stefansson H, Ingason A, Stacey SN, Bergthorsson JT, Thorlacius S, Gudmundsson J, Jonsson T, Jakobsdottir M, Saemundsdottir J, Olafsdottir O, Gudmundsson LJ, Bjornsdottir G, Kristjansson K, Skuladottir H, Isaksson HJ, Gudbjartsson T, Jones GT, Mueller T, Gottsater A, Flex A, Aben KK, de Vegt F, Mulders PF, Isla D, Vidal MJ, Asin L, Saez B, Murillo L, Blondal T, Kolbeinsson H, Stefansson JG, Hansdottir I, Runarsdottir V, Pola R, Lindblad B, van Rij AM, Dieplinger B, Haltmayer M, Mayordomo JI, Kiemeney LA, Matthiasson SE, Oskarsson H, Tyrfingsson T, Gudbjartsson DF, Gulcher JR, Jonsson S, Thorsteinsdottir U, Kong A, Stefansson K. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Broderick P, Webb E, Wu X, Vijayakrishnan J, Matakidou A, Qureshi M, Dong Q, Gu X, Chen WV, Spitz MR, Eisen T, Amos CI, Houlston RS. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat Genet. 2008;40:1407–1409. doi: 10.1038/ng.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKay JD, Hung RJ, Gaborieau V, Boffetta P, Chabrier A, Byrnes G, Zaridze D, Mukeria A, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, Fabianova E, Mates D, Bencko V, Foretova L, Janout V, McLaughlin J, Shepherd F, Montpetit A, Narod S, Krokan HE, Skorpen F, Elvestad MB, Vatten L, Njolstad I, Axelsson T, Chen C, Goodman G, Barnett M, Loomis MM, Lubinski J, Matyjasik J, Lener M, Oszutowska D, Field J, Liloglou T, Xinarianos G, Cassidy A, Vineis P, Clavel-Chapelon F, Palli D, Tumino R, Krogh V, Panico S, Gonzalez CA, Ramon Quiros J, Martinez C, Navarro C, Ardanaz E, Larranaga N, Kham KT, Key T, Bueno-de-Mesquita HB, Peeters PH, Trichopoulou A, Linseisen J, Boeing H, Hallmans G, Overvad K, Tjonneland A, Kumle M, Riboli E, Zelenika D, Boland A, Delepine M, Foglio M, Lechner D, Matsuda F, Blanche H, Gut I, Heath S, Lathrop M, Brennan P. Lung cancer susceptibility locus at 5p15.33. Nat Genet. 2008;40:1404–1406. doi: 10.1038/ng.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong J, Jin G, Wu C, Guo H, Zhou B, Lv J, Lu D, Shi Y, Shu Y, Xu L, Chu M, Wang C, Zhang R, Dai J, Jiang Y, Yu D, Ma H, Zhao X, Yin Z, Yang L, Li Z, Deng Q, Cao S, Qin Z, Gong J, Sun C, Wang J, Wu W, Zhou G, Chen H, Guan P, Chen Y, Liu X, Liu L, Xu P, Han B, Bai C, Zhao Y, Zhang H, Yan Y, Liu J, Amos CI, Chen F, Tan W, Jin L, Wu T, Hu Z, Lin D, Shen H. Genome-wide association study identifies a novel susceptibility locus at 12q23.1 for lung squamous cell carcinoma in han chinese. PLoS Genet. 2013;9:e1003190. doi: 10.1371/journal.pgen.1003190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahn MJ, Won HH, Lee J, Lee ST, Sun JM, Park YH, Ahn JS, Kwon OJ, Kim H, Shim YM, Kim J, Kim K, Kim YH, Park JY, Kim JW, Park K. The 18p11.22 locus is associated with never smoker non-small cell lung cancer susceptibility in Korean populations. Hum Genet. 2012;131:365–372. doi: 10.1007/s00439-011-1080-z. [DOI] [PubMed] [Google Scholar]
- 17.Hu Z, Wu C, Shi Y, Guo H, Zhao X, Yin Z, Yang L, Dai J, Hu L, Tan W, Li Z, Deng Q, Wang J, Wu W, Jin G, Jiang Y, Yu D, Zhou G, Chen H, Guan P, Chen Y, Shu Y, Xu L, Liu X, Liu L, Xu P, Han B, Bai C, Zhao Y, Zhang H, Yan Y, Ma H, Chen J, Chu M, Lu F, Zhang Z, Chen F, Wang X, Jin L, Lu J, Zhou B, Lu D, Wu T, Lin D, Shen H. A genome-wide association study identifies two new lung cancer susceptibility loci at 13q12.12 and 22q12.2 in Han Chinese. Nat Genet. 2011;43:792–796. doi: 10.1038/ng.875. [DOI] [PubMed] [Google Scholar]
- 18.Lan Q, Hsiung CA, Matsuo K, Hong YC, Seow A, Wang Z, Hosgood HD, 3rd, Chen K, Wang JC, Chatterjee N, Hu W, Wong MP, Zheng W, Caporaso N, Park JY, Chen CJ, Kim YH, Kim YT, Landi MT, Shen H, Lawrence C, Burdett L, Yeager M, Yuenger J, Jacobs KB, Chang IS, Mitsudomi T, Kim HN, Chang GC, Bassig BA, Tucker M, Wei F, Yin Z, Wu C, An SJ, Qian B, Lee VH, Lu D, Liu J, Jeon HS, Hsiao CF, Sung JS, Kim JH, Gao YT, Tsai YH, Jung YJ, Guo H, Hu Z, Hutchinson A, Wang WC, Klein R, Chung CC, Oh IJ, Chen KY, Berndt SI, He X, Wu W, Chang J, Zhang XC, Huang MS, Zheng H, Wang J, Zhao X, Li Y, Choi JE, Su WC, Park KH, Sung SW, Shu XO, Chen YM, Liu L, Kang CH, Hu L, Chen CH, Pao W, Kim YC, Yang TY, Xu J, Guan P, Tan W, Su J, Wang CL, Li H, Sihoe AD, Zhao Z, Chen Y, Choi YY, Hung JY, Kim JS, Yoon HI, Cai Q, Lin CC, Park IK, Xu P, Dong J, Kim C, He Q, Perng RP, Kohno T, Kweon SS, et al. Genome-wide association analysis identifies new lung cancer susceptibility loci in never-smoking women in Asia. Nat Genet. 2012;44:1330–1335. doi: 10.1038/ng.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miki D, Kubo M, Takahashi A, Yoon KA, Kim J, Lee GK, Zo JI, Lee JS, Hosono N, Morizono T, Tsunoda T, Kamatani N, Chayama K, Takahashi T, Inazawa J, Nakamura Y, Daigo Y. Variation in TP63 is associated with lung adenocarcinoma susceptibility in Japanese and Korean populations. Nat Genet. 2010;42:893–896. doi: 10.1038/ng.667. [DOI] [PubMed] [Google Scholar]
- 20.Shiraishi K, Kunitoh H, Daigo Y, Takahashi A, Goto K, Sakamoto H, Ohnami S, Shimada Y, Ashikawa K, Saito A, Watanabe S, Tsuta K, Kamatani N, Yoshida T, Nakamura Y, Yokota J, Kubo M, Kohno T. A genome-wide association study identifies two new susceptibility loci for lung adenocarcinoma in the Japanese population. Nat Genet. 2012;44:900–903. doi: 10.1038/ng.2353. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, McKay JD, Rafnar T, Wang Z, Timofeeva MN, Broderick P, Zong X, Laplana M, Wei Y, Han Y, Lloyd A, Delahaye-Sourdeix M, Chubb D, Gaborieau V, Wheeler W, Chatterjee N, Thorleifsson G, Sulem P, Liu G, Kaaks R, Henrion M, Kinnersley B, Vallee M, LeCalvez-Kelm F, Stevens VL, Gapstur SM, Chen WV, Zaridze D, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, Fabianova E, Mates D, Bencko V, Foretova L, Janout V, Krokan HE, Gabrielsen ME, Skorpen F, Vatten L, Njolstad I, Chen C, Goodman G, Benhamou S, Vooder T, Valk K, Nelis M, Metspalu A, Lener M, Lubinski J, Johansson M, Vineis P, Agudo A, Clavel-Chapelon F, Bueno-de-Mesquita HB, Trichopoulos D, Khaw KT, Weiderpass E, Tjonneland A, Riboli E, Lathrop M, Scelo G, Albanes D, Caporaso NE, Ye Y, Gu J, Wu X, Spitz MR, Dienemann H, Rosenberger A, Su L, Matakidou A, Eisen T, Stefansson K, Risch A, Chanock SJ, Christiani DC, Hung RJ, Brennan P, Landi MT, Houlston RS, Amos CI. Rare variants of large effect in BRCA2 and CHEK2 affect risk of lung cancer. Nat Genet. 2014;46:736–741. doi: 10.1038/ng.3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwartz AG, Wenzlaff AS, Bock CH, Ruterbusch JJ, Chen W, Cote ML, Artis AS, Van Dyke AL, Land SJ, Harris CC, Pine SR, Spitz MR, Amos CI, Levin AM, McKeigue PM. Admixture mapping of lung cancer in 1812 African-Americans. Carcinogenesis. 2011;32:312–317. doi: 10.1093/carcin/bgq252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vink JM, Willemsen G, Boomsma DI. Heritability of smoking initiation and nicotine dependence. Behav Genet. 2005;35:397–406. doi: 10.1007/s10519-004-1327-8. [DOI] [PubMed] [Google Scholar]
- 24.Li MD, Cheng R, Ma JZ, Swan GE. A meta-analysis of estimated genetic and environmental effects on smoking behavior in male and female adult twins. Addiction. 2003;98:23–31. doi: 10.1046/j.1360-0443.2003.00295.x. [DOI] [PubMed] [Google Scholar]
- 25.Vink JM, Beem AL, Posthuma D, Neale MC, Willemsen G, Kendler KS, Slagboom PE, Boomsma DI. Linkage analysis of smoking initiation and quantity in Dutch sibling pairs. Pharmacogenomics J. 2004;4:274–282. doi: 10.1038/sj.tpj.6500255. [DOI] [PubMed] [Google Scholar]
- 26.Niu T, Chen C, Ni J, Wang B, Fang Z, Shao H, Xu X. Nicotine dependence and its familial aggregation in Chinese. Int J Epidemiol. 2000;29:248–252. doi: 10.1093/ije/29.2.248. [DOI] [PubMed] [Google Scholar]
- 27.Schwartz AG, Cote ML, Wenzlaff AS, Land S, Amos CI. Racial differences in the association between SNPs on 15q25.1, smoking behavior, and risk of non-small cell lung cancer. J Thorac Oncol. 2009;4:1195–1201. doi: 10.1097/JTO.0b013e3181b244ef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amos CI, Gorlov IP, Dong Q, Wu X, Zhang H, Lu EY, Scheet P, Greisinger AJ, Mills GB, Spitz MR. Nicotinic acetylcholine receptor region on chromosome 15q25 and lung cancer risk among African Americans: a case-control study. J Natl Cancer Inst. 2010;102:1199–1205. doi: 10.1093/jnci/djq232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hansen HM, Xiao Y, Rice T, Bracci PM, Wrensch MR, Sison JD, Chang JS, Smirnov IV, Patoka J, Seldin MF, Quesenberry CP, Kelsey KT, Wiencke JK. Fine mapping of chromosome 15q25.1 lung cancer susceptibility in African-Americans. Hum Mol Genet. 2010;19:3652–3661. doi: 10.1093/hmg/ddq268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walsh KM, Amos CI, Wenzlaff AS, Gorlov IP, Sison JD, Wu X, Spitz MR, Hansen HM, Lu EY, Wei C, Zhang H, Chen W, Lloyd SM, Frazier ML, Bracci PM, Seldin MF, Wrensch MR, Schwartz AG, Wiencke JK. Association study of nicotinic acetylcholine receptor genes identifies a novel lung cancer susceptibility locus near CHRNA1 in African-Americans. Oncotarget. 2012;3:1428–1438. doi: 10.18632/oncotarget.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walsh KM, Gorlov IP, Hansen HM, Wu X, Spitz MR, Zhang H, Lu EY, Wenzlaff AS, Sison JD, Wei C, Lloyd SM, Chen W, Frazier ML, Seldin MF, Bierut LJ, Bracci PM, Wrensch MR, Schwartz AG, Wiencke JK, Amos CI. Fine-mapping of the 5p15.33, 6p22.1-p21.31, and 15q25.1 regions identifies functional and histology-specific lung cancer susceptibility loci in African-Americans. Cancer Epidemiol Biomarkers Prev. 2013;22:251–260. doi: 10.1158/1055-9965.EPI-12-1007-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stevens VL, Bierut LJ, Talbot JT, Wang JC, Sun J, Hinrichs AL, Thun MJ, Goate A, Calle EE. Nicotinic receptor gene variants influence susceptibility to heavy smoking. Cancer Epidemiol Biomarkers Prev. 2008;17:3517–3525. doi: 10.1158/1055-9965.EPI-08-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weiss RB, Baker TB, Cannon DS, von Niederhausern A, Dunn DM, Matsunami N, Singh NA, Baird L, Coon H, McMahon WM, Piper ME, Fiore MC, Scholand MB, Connett JE, Kanner RE, Gahring LC, Rogers SW, Hoidal JR, Leppert MF. A candidate gene approach identifies the CHRNA5-A3-B4 region as a risk factor for age-dependent nicotine addiction. PLoS Genet. 2008;4:e1000125. doi: 10.1371/journal.pgen.1000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le Marchand L, Derby KS, Murphy SE, Hecht SS, Hatsukami D, Carmella SG, Tiirikainen M, Wang H. Smokers with the CHRNA lung cancer-associated variants are exposed to higher levels of nicotine equivalents and a carcinogenic tobacco-specific nitrosamine. Cancer Res. 2008;68:9137–9140. doi: 10.1158/0008-5472.CAN-08-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Landi MT, Chatterjee N, Yu K, Goldin LR, Goldstein AM, Rotunno M, Mirabello L, Jacobs K, Wheeler W, Yeager M, Bergen AW, Li Q, Consonni D, Pesatori AC, Wacholder S, Thun M, Diver R, Oken M, Virtamo J, Albanes D, Wang Z, Burdette L, Doheny KF, Pugh EW, Laurie C, Brennan P, Hung R, Gaborieau V, McKay JD, Lathrop M, McLaughlin J, Wang Y, Tsao MS, Spitz MR, Wang Y, Krokan H, Vatten L, Skorpen F, Arnesen E, Benhamou S, Bouchard C, Metspalu A, Vooder T, Nelis M, Valk K, Field JK, Chen C, Goodman G, Sulem P, Thorleifsson G, Rafnar T, Eisen T, Sauter W, Rosenberger A, Bickeboller H, Risch A, Chang-Claude J, Wichmann HE, Stefansson K, Houlston R, Amos CI, Fraumeni JF, Jr, Savage SA, Bertazzi PA, Tucker MA, Chanock S, Caporaso NE. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet. 2009;85:679–691. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu K, Wang Z, Li Q, Wacholder S, Hunter DJ, Hoover RN, Chanock S, Thomas G. Population substructure and control selection in genome-wide association studies. PLoS One. 2008;3:e2551. doi: 10.1371/journal.pone.0002551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bryc K, Durand EY, Macpherson JM, Reich D, Mountain JL. The genetic ancestry of African Americans, Latinos, and European Americans across the United States. Am J Hum Genet. 2015;96:37–53. doi: 10.1016/j.ajhg.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.International HapMap Consortium. Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, Pasternak S, Wheeler DA, Willis TD, Yu F, Yang H, Zeng C, Gao Y, Hu H, Hu W, Li C, Lin W, Liu S, Pan H, Tang X, Wang J, Wang W, Yu J, Zhang B, Zhang Q, Zhao H, Zhao H, Zhou J, Gabriel SB, Barry R, Blumenstiel B, Camargo A, Defelice M, Faggart M, Goyette M, Gupta S, Moore J, Nguyen H, Onofrio RC, Parkin M, Roy J, Stahl E, Winchester E, Ziaugra L, Altshuler D, Shen Y, Yao Z, Huang W, Chu X, He Y, Jin L, Liu Y, Shen Y, Sun W, Wang H, Wang Y, Wang Y, Xiong X, Xu L, Waye MM, Tsui SK, Xue H, Wong JT, Galver LM, Fan JB, Gunderson K, Murray SS, Oliphant AR, Chee MS, Montpetit A, Chagnon F, Ferretti V, Leboeuf M, Olivier JF, Phillips MS, Roumy S, Sallee C, Verner A, Hudson TJ, Kwok PY, Cai D, Koboldt DC, Miller RD, Pawlikowska L, Taillon-Miller P, Xiao M, Tsui LC, Mak W, Song YQ, Tam PK, Nakamura Y, Kawaguchi T, Kitamoto T, Morizono T, Nagashima A, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Purdue MP, Ye Y, Wang Z, Colt JS, Schwartz KL, Davis FG, Rothman N, Chow WH, Wu X, Chanock SJ. A genome-wide association study of renal cell carcinoma among African Americans. Cancer Epidemiol Biomarkers Prev. 2014;23:209–214. doi: 10.1158/1055-9965.EPI-13-0818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mitry D, Campbell H, Charteris DG, Fleck BW, Tenesa A, Dunlop MG, Hayward C, Wright AF, Vitart V. SNP mistyping in genotyping arrays--an important cause of spurious association in case-control studies. Genet Epidemiol. 2011;35:423–426. doi: 10.1002/gepi.20559. [DOI] [PubMed] [Google Scholar]
- 41.Berrettini W, Yuan X, Tozzi F, Song K, Francks C, Chilcoat H, Waterworth D, Muglia P, Mooser V. Alpha-5/alpha-3 nicotinic receptor subunit alleles increase risk for heavy smoking. Mol Psychiatry. 2008;13:368–373. doi: 10.1038/sj.mp.4002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Erlich PM, Hoffman SN, Rukstalis M, Han JJ, Chu X, Linda Kao WH, Gerhard GS, Stewart WF, Boscarino JA. Nicotinic acetylcholine receptor genes on chromosome 15q25.1 are associated with nicotine and opioid dependence severity. Hum Genet. 2010;128:491–499. doi: 10.1007/s00439-010-0876-6. [DOI] [PubMed] [Google Scholar]
- 43.Caporaso N, Gu F, Chatterjee N, Sheng-Chih J, Yu K, Yeager M, Chen C, Jacobs K, Wheeler W, Landi MT, Ziegler RG, Hunter DJ, Chanock S, Hankinson S, Kraft P, Bergen AW. Genome-wide and candidate gene association study of cigarette smoking behaviors. PLoS One. 2009;4:e4653. doi: 10.1371/journal.pone.0004653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keskitalo K, Broms U, Heliovaara M, Ripatti S, Surakka I, Perola M, Pitkaniemi J, Peltonen L, Aromaa A, Kaprio J. Association of serum cotinine level with a cluster of three nicotinic acetylcholine receptor genes (CHRNA3/CHRNA5/CHRNB4) on chromosome 15. Hum Mol Genet. 2009;18:4007–4012. doi: 10.1093/hmg/ddp322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li MD, Xu Q, Lou XY, Payne TJ, Niu T, Ma JZ. Association and interaction analysis of variants in CHRNA5/CHRNA3/CHRNB4 gene cluster with nicotine dependence in African and European Americans. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:745–756. doi: 10.1002/ajmg.b.31043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu JZ, Tozzi F, Waterworth DM, Pillai SG, Muglia P, Middleton L, Berrettini W, Knouff CW, Yuan X, Waeber G, Vollenweider P, Preisig M, Wareham NJ, Zhao JH, Loos RJ, Barroso I, Khaw KT, Grundy S, Barter P, Mahley R, Kesaniemi A, McPherson R, Vincent JB, Strauss J, Kennedy JL, Farmer A, McGuffin P, Day R, Matthews K, Bakke P, Gulsvik A, Lucae S, Ising M, Brueckl T, Horstmann S, Wichmann HE, Rawal R, Dahmen N, Lamina C, Polasek O, Zgaga L, Huffman J, Campbell S, Kooner J, Chambers JC, Burnett MS, Devaney JM, Pichard AD, Kent KM, Satler L, Lindsay JM, Waksman R, Epstein S, Wilson JF, Wild SH, Campbell H, Vitart V, Reilly MP, Li M, Qu L, Wilensky R, Matthai W, Hakonarson HH, Rader DJ, Franke A, Wittig M, Schafer A, Uda M, Terracciano A, Xiao X, Busonero F, Scheet P, Schlessinger D, St Clair D, Rujescu D, Abecasis GR, Grabe HJ, Teumer A, Volzke H, Petersmann A, John U, Rudan I, Hayward C, Wright AF, Kolcic I, Wright BJ, Thompson JR, Balmforth AJ, Hall AS, Samani NJ, Anderson CA, Ahmad T, Mathew CG, Parkes M, Satsangi J, Caulfield M, Munroe PB, Farrall M, Dominiczak A, Worthington J, et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet. 2010;42:436–440. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saccone NL, Culverhouse RC, Schwantes-An TH, Cannon DS, Chen X, Cichon S, Giegling I, Han S, Han Y, Keskitalo-Vuokko K, Kong X, Landi MT, Ma JZ, Short SE, Stephens SH, Stevens VL, Sun L, Wang Y, Wenzlaff AS, Aggen SH, Breslau N, Broderick P, Chatterjee N, Chen J, Heath AC, Heliovaara M, Hoft NR, Hunter DJ, Jensen MK, Martin NG, Montgomery GW, Niu T, Payne TJ, Peltonen L, Pergadia ML, Rice JP, Sherva R, Spitz MR, Sun J, Wang JC, Weiss RB, Wheeler W, Witt SH, Yang BZ, Caporaso NE, Ehringer MA, Eisen T, Gapstur SM, Gelernter J, Houlston R, Kaprio J, Kendler KS, Kraft P, Leppert MF, Li MD, Madden PA, Nothen MM, Pillai S, Rietschel M, Rujescu D, Schwartz A, Amos CI, Bierut LJ. Multiple independent loci at chromosome 15q25.1 affect smoking quantity: a meta-analysis and comparison with lung cancer and COPD. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spitz MR, Amos CI, Dong Q, Lin J, Wu X. The CHRNA5-A3 region on chromosome 15q24-25.1 is a risk factor both for nicotine dependence and for lung cancer. J Natl Cancer Inst. 2008;100:1552–1556. doi: 10.1093/jnci/djn363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F, Sulem P, Rafnar T, Esko T, Walter S, Gieger C, Rawal R, Mangino M, Prokopenko I, Magi R, Keskitalo K, Gudjonsdottir IH, Gretarsdottir S, Stefansson H, Thompson JR, Aulchenko YS, Nelis M, Aben KK, den Heijer M, Dirksen A, Ashraf H, Soranzo N, Valdes AM, Steves C, Uitterlinden AG, Hofman A, Tonjes A, Kovacs P, Hottenga JJ, Willemsen G, Vogelzangs N, Doring A, Dahmen N, Nitz B, Pergadia ML, Saez B, De Diego V, Lezcano V, Garcia-Prats MD, Ripatti S, Perola M, Kettunen J, Hartikainen AL, Pouta A, Laitinen J, Isohanni M, Huei-Yi S, Allen M, Krestyaninova M, Hall AS, Jones GT, van Rij AM, Mueller T, Dieplinger B, Haltmayer M, Jonsson S, Matthiasson SE, Oskarsson H, Tyrfingsson T, Kiemeney LA, Mayordomo JI, Lindholt JS, Pedersen JH, Franklin WA, Wolf H, Montgomery GW, Heath AC, Martin NG, Madden PA, Giegling I, Rujescu D, Jarvelin MR, Salomaa V, Stumvoll M, Spector TD, Wichmann HE, Metspalu A, Samani NJ, Penninx BW, Oostra BA, Boomsma DI, Tiemeier H, van Duijn CM, Kaprio J, Gulcher JR, McCarthy MI, Peltonen L, Thorsteinsdottir U, Stefansson K. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet. 2010;42:448–453. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bierut LJ, Stitzel JA, Wang JC, Hinrichs AL, Grucza RA, Xuei X, Saccone NL, Saccone SF, Bertelsen S, Fox L, Horton WJ, Breslau N, Budde J, Cloninger CR, Dick DM, Foroud T, Hatsukami D, Hesselbrock V, Johnson EO, Kramer J, Kuperman S, Madden PA, Mayo K, Nurnberger J, Jr, Pomerleau O, Porjesz B, Reyes O, Schuckit M, Swan G, Tischfield JA, Edenberg HJ, Rice JP, Goate AM. Variants in nicotinic receptors and risk for nicotine dependence. Am J Psychiatry. 2008;165:1163–1171. doi: 10.1176/appi.ajp.2008.07111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grucza RA, Johnson EO, Krueger RF, Breslau N, Saccone NL, Chen LS, Derringer J, Agrawal A, Lynskey M, Bierut LJ. Incorporating age at onset of smoking into genetic models for nicotine dependence: evidence for interaction with multiple genes. Addict Biol. 2010;15:346–357. doi: 10.1111/j.1369-1600.2010.00220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schlaepfer IR, Hoft NR, Collins AC, Corley RP, Hewitt JK, Hopfer CJ, Lessem JM, McQueen MB, Rhee SH, Ehringer MA. The CHRNA5/A3/B4 gene cluster variability as an important determinant of early alcohol and tobacco initiation in young adults. Biol Psychiatry. 2008;63:1039–1046. doi: 10.1016/j.biopsych.2007.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sherva R, Wilhelmsen K, Pomerleau CS, Chasse SA, Rice JP, Snedecor SM, Bierut LJ, Neuman RJ, Pomerleau OF. Association of a single nucleotide polymorphism in neuronal acetylcholine receptor subunit alpha 5 (CHRNA5) with smoking status and with ‘pleasurable buzz’ during early experimentation with smoking. Addiction. 2008;103:1544–1552. doi: 10.1111/j.1360-0443.2008.02279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoft NR, Stitzel JA, Hutchison KE, Ehringer MA. CHRNB2 promoter region: association with subjective effects to nicotine and gene expression differences. Genes Brain Behav. 2011;10:176–185. doi: 10.1111/j.1601-183X.2010.00650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sherva R, Kranzler HR, Yu Y, Logue MW, Poling J, Arias AJ, Anton RF, Oslin D, Farrer LA, Gelernter J. Variation in nicotinic acetylcholine receptor genes is associated with multiple substance dependence phenotypes. Neuropsychopharmacology. 2010;35:1921–1931. doi: 10.1038/npp.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.David SP, Hamidovic A, Chen GK, Bergen AW, Wessel J, Kasberger JL, Brown WM, Petruzella S, Thacker EL, Kim Y, Nalls MA, Tranah GJ, Sung YJ, Ambrosone CB, Arnett D, Bandera EV, Becker DM, Becker L, Berndt SI, Bernstein L, Blot WJ, Broeckel U, Buxbaum SG, Caporaso N, Casey G, Chanock SJ, Deming SL, Diver WR, Eaton CB, Evans DS, Evans MK, Fornage M, Franceschini N, Harris TB, Henderson BE, Hernandez DG, Hitsman B, Hu JJ, Hunt SC, Ingles SA, John EM, Kittles R, Kolb S, Kolonel LN, Le Marchand L, Liu Y, Lohman KK, McKnight B, Millikan RC, Murphy A, Neslund-Dudas C, Nyante S, Press M, Psaty BM, Rao DC, Redline S, Rodriguez-Gil JL, Rybicki BA, Signorello LB, Singleton AB, Smoller J, Snively B, Spring B, Stanford JL, Strom SS, Swan GE, Taylor KD, Thun MJ, Wilson AF, Witte JS, Yamamura Y, Yanek LR, Yu K, Zheng W, Ziegler RG, Zonderman AB, Jorgenson E, Haiman CA, Furberg H. Genome-wide meta-analyses of smoking behaviors in African Americans. Transl Psychiatry. 2012;2:e119. doi: 10.1038/tp.2012.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Broms U, Wedenoja J, Largeau MR, Korhonen T, Pitkaniemi J, Keskitalo-Vuokko K, Happola A, Heikkila KH, Heikkila K, Ripatti S, Sarin AP, Salminen O, Paunio T, Pergadia ML, Madden PA, Kaprio J, Loukola A. Analysis of detailed phenotype profiles reveals CHRNA5-CHRNA3-CHRNB4 gene cluster association with several nicotine dependence traits. Nicotine Tob Res. 2012;14:720–733. doi: 10.1093/ntr/ntr283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Benowitz NL, Dains KM, Dempsey D, Wilson M, Jacob P. Racial differences in the relationship between number of cigarettes smoked and nicotine and carcinogen exposure. Nicotine Tob Res. 2011;13:772–783. doi: 10.1093/ntr/ntr072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bloom AJ, Hartz SM, Baker TB, Chen LS, Piper ME, Fox L, Martinez M, Hatsukami D, Johnson EO, Laurie CC, Saccone NL, Goate A, Bierut LJ. Beyond cigarettes per day. A genome-wide association study of the biomarker carbon monoxide. Ann Am Thorac Soc. 2014;11:1003–1010. doi: 10.1513/AnnalsATS.201401-010OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Genomes Project Consortium. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saccone NL, Wang JC, Breslau N, Johnson EO, Hatsukami D, Saccone SF, Grucza RA, Sun L, Duan W, Budde J, Culverhouse RC, Fox L, Hinrichs AL, Steinbach JH, Wu M, Rice JP, Goate AM, Bierut LJ. The CHRNA5-CHRNA3-CHRNB4 nicotinic receptor subunit gene cluster affects risk for nicotine dependence in African-Americans and in European-Americans. Cancer Res. 2009;69:6848–6856. doi: 10.1158/0008-5472.CAN-09-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morel C, Fattore L, Pons S, Hay YA, Marti F, Lambolez B, De Biasi M, Lathrop M, Fratta W, Maskos U, Faure P. Nicotine consumption is regulated by a human polymorphism in dopamine neurons. Mol Psychiatry. 2014;19:930–936. doi: 10.1038/mp.2013.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen LS, Saccone NL, Culverhouse RC, Bracci PM, Chen CH, Dueker N, Han Y, Huang H, Jin G, Kohno T, Ma JZ, Przybeck TR, Sanders AR, Smith JA, Sung YJ, Wenzlaff AS, Wu C, Yoon D, Chen YT, Cheng YC, Cho YS, David SP, Duan J, Eaton CB, Furberg H, Goate AM, Gu D, Hansen HM, Hartz S, Hu Z, Kim YJ, Kittner SJ, Levinson DF, Mosley TH, Payne TJ, Rao DC, Rice JP, Rice TK, Schwantes-An TH, Shete SS, Shi J, Spitz MR, Sun YV, Tsai FJ, Wang JC, Wrensch MR, Xian H, Gejman PV, He J, Hunt SC, Kardia SL, Li MD, Lin D, Mitchell BD, Park T, Schwartz AG, Shen H, Wiencke JK, Wu JY, Yokota J, Amos CI, Bierut LJ. Smoking and genetic risk variation across populations of European, Asian, and African American ancestry--a meta-analysis of chromosome 15q25. Genet Epidemiol. 2012;36:340–351. doi: 10.1002/gepi.21627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Olfson E, Saccone NL, Johnson EO, Chen LS, Culverhouse R, Doheny K, Foltz SM, Fox L, Gogarten SM, Hartz S, Hetrick K, Laurie CC, Marosy B, Amin N, Arnett D, Barr RG, Bartz TM, Bertelsen S, Borecki IB, Brown MR, Chasman DI, van Duijn CM, Feitosa MF, Fox ER, Franceschini N, Franco OH, Grove ML, Guo X, Hofman A, Kardia SL, Morrison AC, Musani SK, Psaty BM, Rao DC, Reiner AP, Rice K, Ridker PM, Rose LM, Schick UM, Schwander K, Uitterlinden AG, Vojinovic D, Wang JC, Ware EB, Wilson G, Yao J, Zhao W, Breslau N, Hatsukami D, Stitzel JA, Rice J, Goate A, Bierut LJ. Rare, low frequency and common coding variants in CHRNA5 and their contribution to nicotine dependence in European and African Americans. Mol Psychiatry. 2015 doi: 10.1038/mp.2015.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu AZ, Zhou Q, Cox LS, David SP, Ahluwalia JS, Benowitz NL, Tyndale RF. Association of CHRNA5-A3-B4 SNP rs2036527 with smoking cessation therapy response in African-American smokers. Clin Pharmacol Ther. 2014;96:256–265. doi: 10.1038/clpt.2014.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang JC, Bierut LJ, Goate AM. Variants weakly correlated with CHRNA5 D398N polymorphism should be considered in transcriptional deregulation at the 15q25 locus associated with lung cancer risk. Clin Cancer Res. 2009;15:5599. doi: 10.1158/1078-0432.CCR-09-1108. author reply 5599. [DOI] [PubMed] [Google Scholar]
- 67.Hancock DB, Reginsson GW, Gaddis NC, Chen X, Saccone NL, Lutz SM, Qaiser B, Sherva R, Steinberg S, Zink F, Stacey SN, Glasheen C, Chen J, Gu F, Frederiksen BN, Loukola A, Gudbjartsson DF, Bruske I, Landi MT, Bickeboller H, Madden P, Farrer L, Kaprio J, Kranzler HR, Gelernter J, Baker TB, Kraft P, Amos CI, Caporaso NE, Hokanson JE, Bierut LJ, Thorgeirsson TE, Johnson EO, Stefansson K. Genome-wide meta-analysis reveals common splice site acceptor variant in CHRNA4 associated with nicotine dependence. Transl Psychiatry. 2015;5:e651. doi: 10.1038/tp.2015.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Scherf DB, Sarkisyan N, Jacobsson H, Claus R, Bermejo JL, Peil B, Gu L, Muley T, Meister M, Dienemann H, Plass C, Risch A. Epigenetic screen identifies genotype-specific promoter DNA methylation and oncogenic potential of CHRNB4. Oncogene. 2013;32:3329–3338. doi: 10.1038/onc.2012.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Van Dyke AL, Cote ML, Wenzlaff AS, Abrams J, Land S, Iyer P, Schwartz AG. Chromosome 5p Region SNPs Are Associated with Risk of NSCLC among Women. J Cancer Epidemiol. 2009;2009:242151. doi: 10.1155/2009/242151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang Z, Zhu B, Zhang M, Parikh H, Jia J, Chung CC, Sampson JN, Hoskins JW, Hutchinson A, Burdette L, Ibrahim A, Hautman C, Raj PS, Abnet CC, Adjei AA, Ahlbom A, Albanes D, Allen NE, Ambrosone CB, Aldrich M, Amiano P, Amos C, Andersson U, Andriole G, Jr, Andrulis IL, Arici C, Arslan AA, Austin MA, Baris D, Barkauskas DA, Bassig BA, Beane Freeman LE, Berg CD, Berndt SI, Bertazzi PA, Biritwum RB, Black A, Blot W, Boeing H, Boffetta P, Bolton K, Boutron-Ruault MC, Bracci PM, Brennan P, Brinton LA, Brotzman M, Bueno-de-Mesquita HB, Buring JE, Butler MA, Cai Q, Cancel-Tassin G, Canzian F, Cao G, Caporaso NE, Carrato A, Carreon T, Carta A, Chang GC, Chang IS, Chang-Claude J, Che X, Chen CJ, Chen CY, Chen CH, Chen C, Chen KY, Chen YM, Chokkalingam AP, Chu LW, Clavel-Chapelon F, Colditz GA, Colt JS, Conti D, Cook MB, Cortessis VK, Crawford ED, Cussenot O, Davis FG, De Vivo I, Deng X, Ding T, Dinney CP, Di Stefano AL, Diver WR, Duell EJ, Elena JW, Fan JH, Feigelson HS, Feychting M, Figueroa JD, Flanagan AM, Fraumeni JF, Jr, Freedman ND, Fridley BL, Fuchs CS, Gago-Dominguez M, Gallinger S, Gao YT, Gapstur SM, Garcia-Closas M, et al. Imputation and subset-based association analysis across different cancer types identifies multiple independent risk loci in the TERT-CLPTM1L region on chromosome 5p15.33. Hum Mol Genet. 2014;23:6616–6633. doi: 10.1093/hmg/ddu363. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.