Abstract
Therapeutic targeting of KRAS-mutant lung adenocarcinoma represents a major goal of clinical oncology. KRAS itself has proven difficult to inhibit, and the effectiveness of agents that target key KRAS effectors has been thwarted by activation of compensatory or parallel pathways that limit their efficacy as single agents. Here we take a systematic approach towards identifying combination targets for trametinib, an FDA-approved MEK inhibitor that acts downstream of KRAS to suppress signaling through the mitogen-activated protein kinase (MAPK) cascade. Informed by a short-hairpin RNA (shRNA) screen, we show that trametinib provokes a compensatory response involving the fibroblast growth factor receptor 1 (FGFR1) that leads to signaling rebound and adaptive drug resistance. As a consequence, genetic or pharmacologic inhibition of FGFR1 in combination with trametinib enhances tumor cell death in vitro and in vivo. This compensatory response shows distinct specificities – it is dominated by FGFR1 in KRAS mutant lung and pancreatic cancer cells, but is not activated or involves other mechanisms in KRAS wild-type lung and KRAS-mutant colon cancer cells. Importantly, KRAS-mutant lung cancer cells and patient tumors treated with trametinib show an increase in FRS2 phosphorylation, a biomarker of FGFR activation; this increase is abolished by FGFR1 inhibition and correlates with sensitivity to trametinib and FGFR inhibitor combinations. These results demonstrate that FGFR1 can mediate adaptive resistance to trametinib and validate a combinatorial approach for treating KRAS-mutant lung cancer.
Introduction
KRAS encodes a GTPase that couples growth factor signaling to the MAPK cascade and other effector pathways. Oncogenic KRAS mutations compromise its GTPase activity leading to accumulation of KRAS in the active GTP-bound state, thereby leading to hyperactive signaling that initiates and maintains tumorigenesis1. Owing to the high frequency of KRAS mutations in lung adenocarcinoma and other cancers, strategies to inhibit the KRAS protein or exploit synthetic lethal interactions with a mutant KRAS gene have been widely pursued but have been fraught with technical challenges or produced inconsistent results2–7. Conversely, strategies to target key RAS effectors including MAPK pathway components RAF, MEK, and ERK have been hindered by toxicities associated with their sustained inhibition and/or adaptive resistance mechanisms8–11.
shRNA screen for identifying trametinib sensitizers
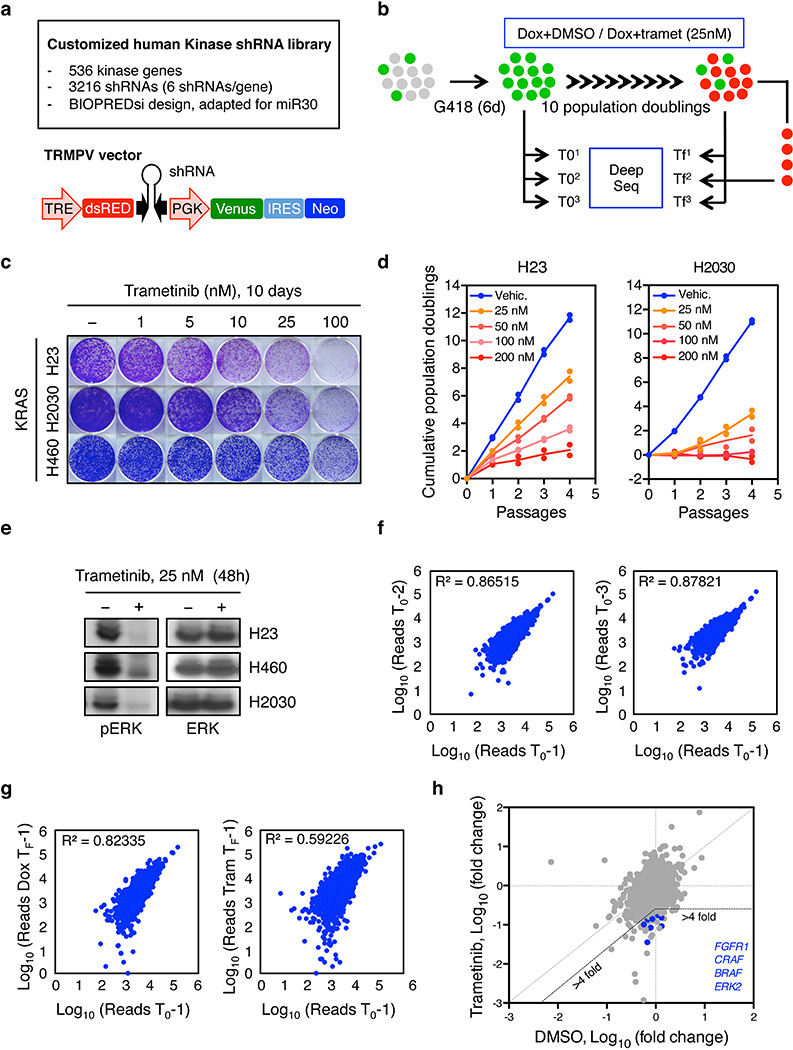
Hypothesizing that sustained MAPK inhibition is necessary, but not sufficient, for targeting KRAS-mutant cancers, we performed a pool-based shRNA screen to identify genes whose inhibition sensitizes KRAS-mutant lung cancer cells to the FDA-approved MEK inhibitor trametinib (Supplementary Table 1). A customized shRNA library targeting the human kinome was introduced into the TRMPVIN vector that we previously optimized for negative selection screening12,13. In this system, cassettes encoding a mir-30 shRNA linked to a dsRed fluorescent reporter are placed downstream of a tetracycline responsive promoter, enabling doxycycline dependent gene silencing and the facile tracking and/or sorting of shRNA expressing cells (Extended Data 1a)12. This library was transduced into H23 KRASG12C mutant lung cancer cells expressing a reverse-tet-transactivator (rtTA3). The transduced populations were then treated with doxycycline in the presence or absence of 25 nM trametinib, a dose that effectively inhibits ERK signaling without substantially affecting proliferation (Extended Data Fig.1b, c, d, e). After ten population doublings, changes in shRNA representation were determined by sequencing of shRNAs amplified from dsRed-sorted cells (Extended Data Fig.1b).
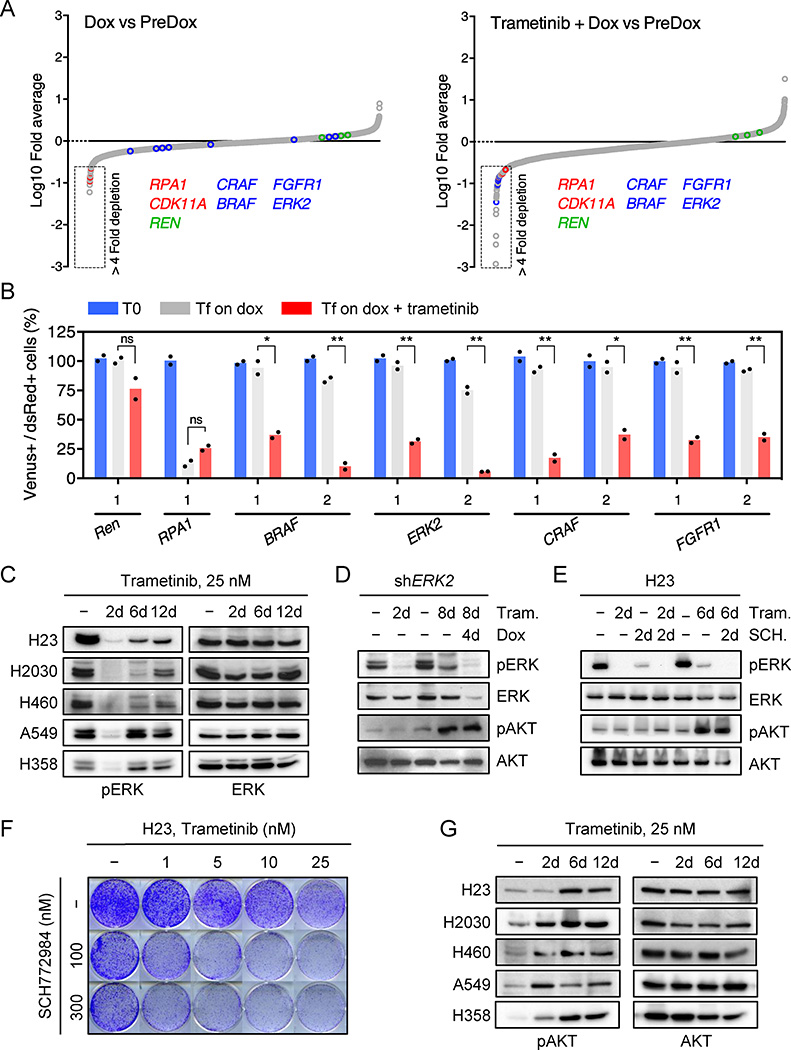
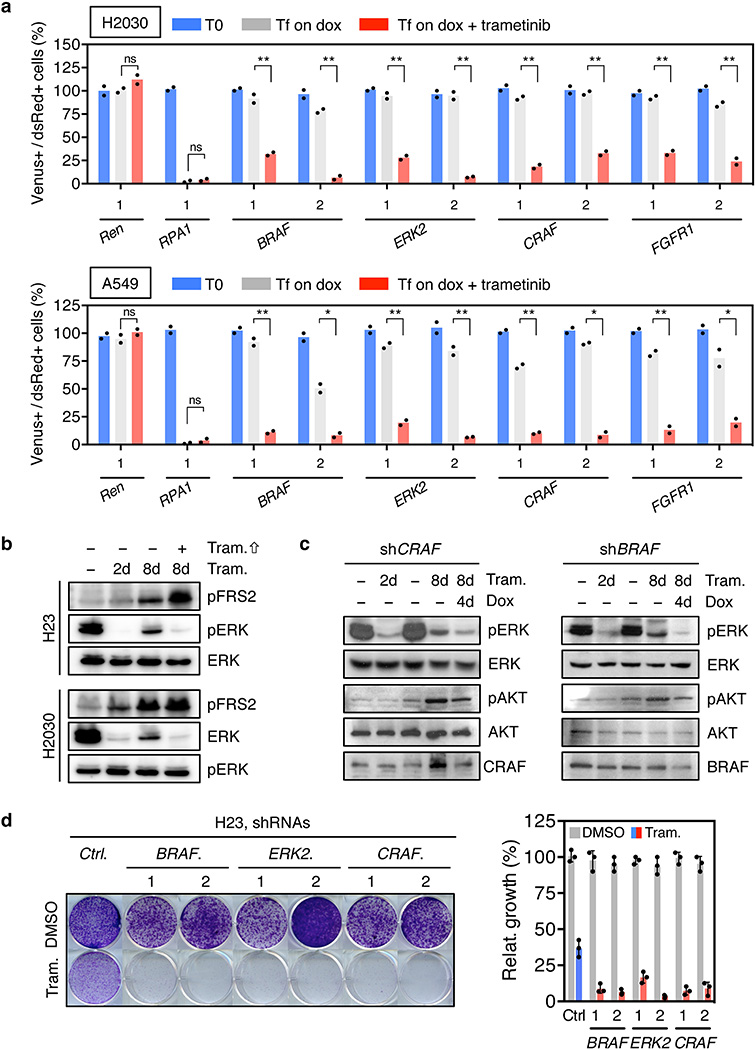
As expected, shRNAs targeting essential genes (RPA1 and CDK11A) were strongly depleted in both vehicle and trametinib-treated cells, whereas the relative representation of neutral non-targeting control shRNAs (Renilla (REN)) remained unchanged (Fig. 1a and Extended Data Fig. 1f, g). Using selection criteria that required an average 4-fold or greater depletion between conditions, we identified 64 shRNAs corresponding to 53 genes that were selectively depleted upon MEK inhibition in trametinib-treated compared to untreated cells (Fig. 1a and Extended Data Fig.1h). Of these, shRNAs targeting the 8 genes for which multiple shRNAs identified as hits were validated using cell competition assays in multiple KRAS-mutant lung lines. These studies identified BRAF, CRAF, ERK2, and FGFR1 as the top candidates in our screen (Fig. 1b and Extended Data Fig. 2a).
Figure 1. Suppression of MAPK signaling effectors and FGFR1 sensitizes KRAS-mutant lung cells to trametinib.
a, Relative abundance of each shRNA in the library in vehicle- or trametinib-treated H23 cells after ten population doublings on doxycycline. The mean of three (vehicle) and two (trametinib) replicates is plotted. Positive and negative controls included shRNAs targeting RPA1 and CDK11A (Red circles), and renilla (REN) luciferase (Green circles). b, Quantification of fluorescent cells in competitive proliferation assays in H23 cells transduced with non-targeting control (Ren) or the indicated shRNAs. Data presented as mean (n = 2). ns: not significant, *P<0.05, **P<0.01 (unpaired two-tailed t-test). c, Immunoblot of KRAS-mutant lung cells treated with 25 nM trametinib for various times. d, Immunoblot of H23 cells transduced with a doxycycline-inducible shRNA targeting ERK2 and treated with trametinib (25 nM) and doxycycline for the times shown. e, Immunoblot of H23 cells treated with trametinib (25 nM), SCH772984 (500 nM), or their combination for the times shown. f, Clonogenic assay of H23 cells treated with trametinib, ERK inhibitor SCH772984, or their combination as indicated. (n = 3). g, Immunoblot of KRAS-mutant lung cancer cells treated with 25 nM trametinib for various times. For gel source data, see supplementary Fig. 1.
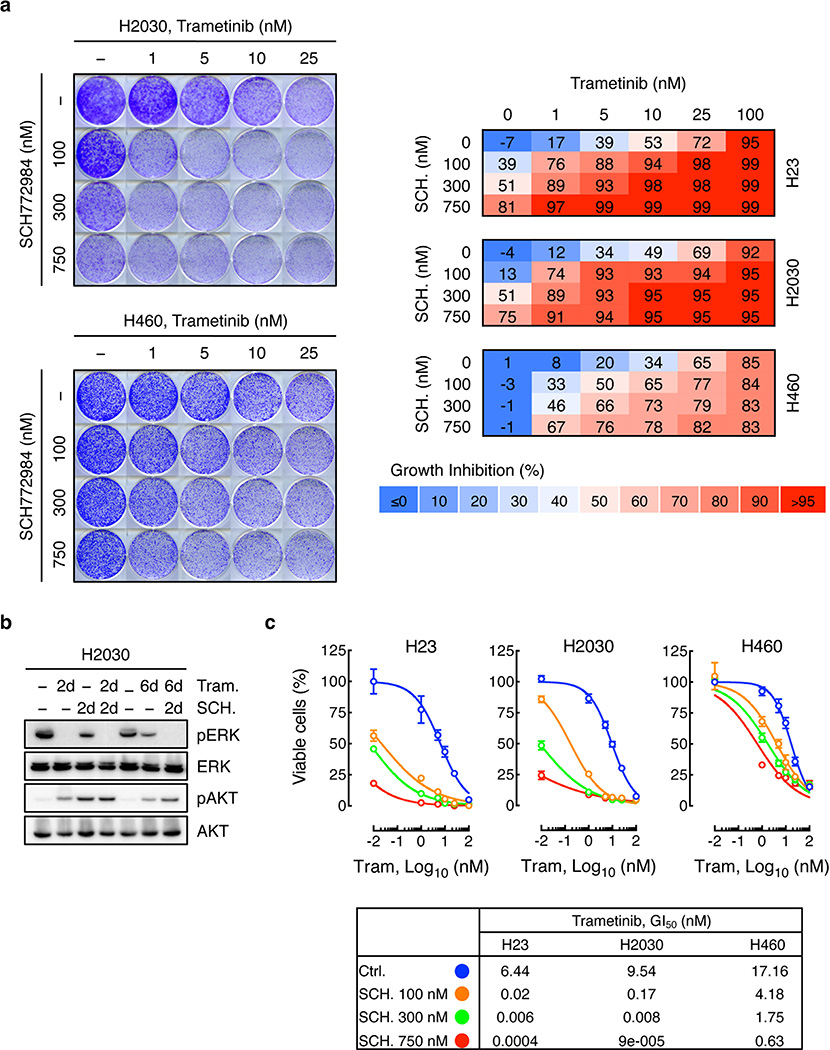
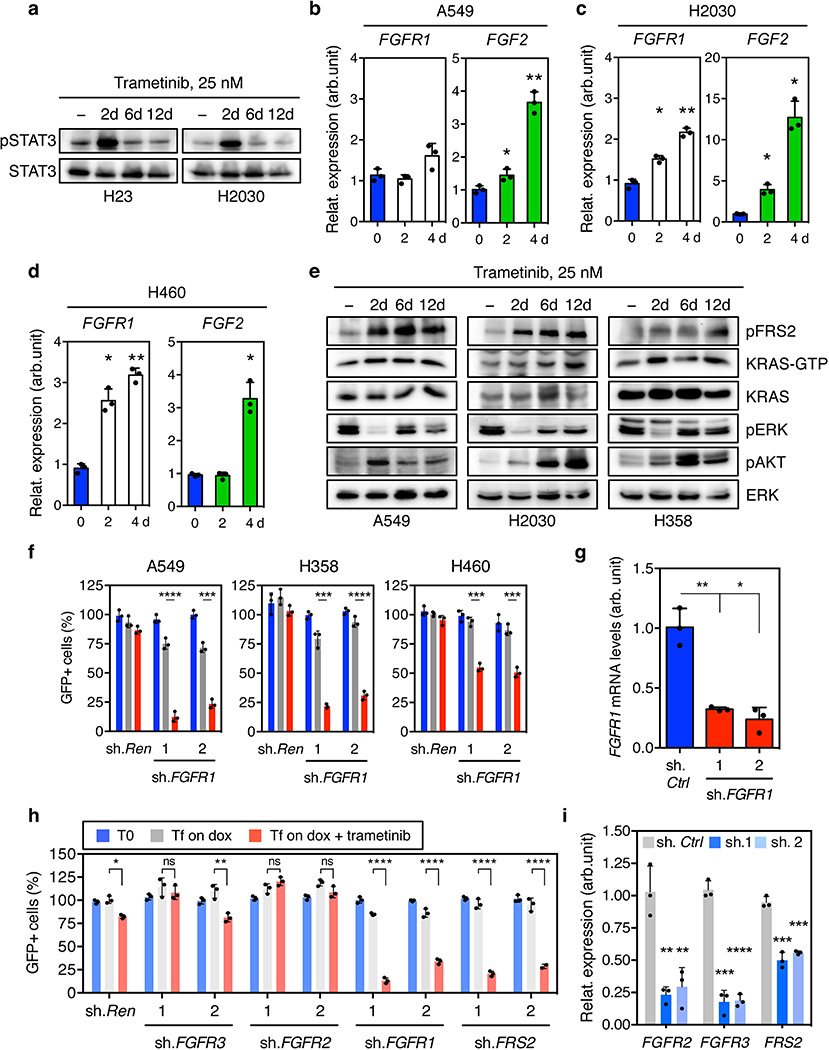
Trametinib has superior pharmacologic properties compared to other MEK inhibitors because it impairs feedback reactivation of ERK10. Still, the fact that MAPK components were identified as hits in our screen implied that pathway reactivation eventually occurs. Indeed, although trametinib stably inhibits ERK signaling at 48-hours – a time where rebound occurs with other agents10 - we observed an increase in phospho-ERK after 6–12 days of drug exposure (Fig. 1c). This rebound was reduced by subsequently increasing the concentration of trametinib, indicating that it is MEK dependent (Extended Data Fig. 2b). Accordingly, inducible knockdown of ERK2, CRAF, and BRAF blocked ERK signaling rebound and reduced clonogenic growth after trametinib treatment (Fig. 1d and Extended Data Fig. 2c, d). Similar effects were observed in KRAS-mutant lung cancer cells treated with trametinib and the ERK inhibitor SCH772984 (Fig. 1e, f, and Extended Data Fig. 3)14. These observations underscore the marked dependency of KRAS-mutant tumors on the MAPK signaling pathway.
In agreement with other studies, KRAS-mutant cells treated with trametinib also displayed compensatory activation of the PI3K and JAK/STAT pathways as assessed by AKT and STAT3 phosphorylation, respectively (Fig. 1d, e, g and Extended Data Fig. 2c, 3b, 4a)11,15. Although the increase in STAT3 phosphorylation was transient (Extended Data Fig. 4a), AKT phosphorylation was sustained (Fig. 1g). In contrast to their effects on ERK signaling rebound, genetic or pharmacologic inhibition of MAPK signaling had little effect on the trametinib-induced increase in pAKT (Fig. 1d, e, and Extended Data Fig. 2c, 3b). The activation of multiple signaling pathways following trametinib-treatment likely reflects a relief in pleiotropic feedback mechanisms produced by hyperactive RAS signaling in KRAS-mutant cells8,9.
FGFR1 mediates adaptive drug resistance
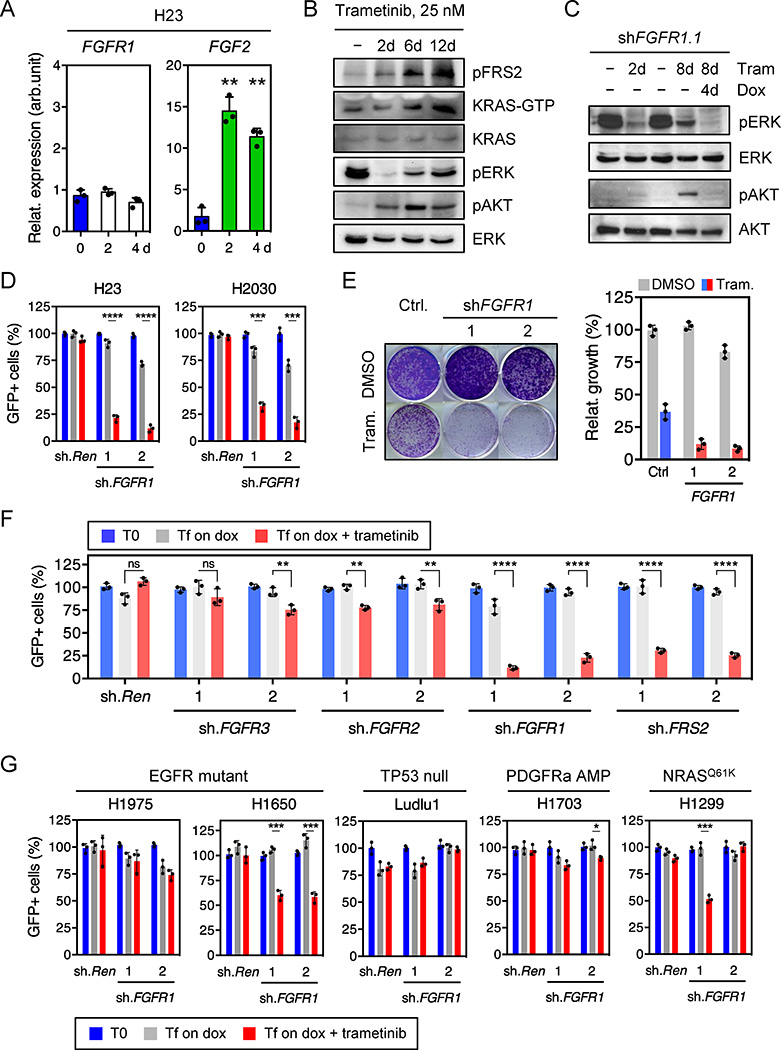
Several RTKs have been implicated in adaptive resistance to RAS pathway antagonist8,9,11,15–20. The identification of FGFR1 shRNAs as trametinib sensitizers raised the possibility that FGFR1 mediates MAPK and PI3K activation in trametinib-treated KRAS-mutant cells. In agreement, treatment of KRAS-mutant lung tumor cell lines with trametinib increased FGFR1 receptor and/or ligand expression together with FGFR pathway activation as assessed by an increase in phosphorylation of the FGFR adaptor protein FRS2 (Fig. 2a, b, and Extended Data Fig. 2b, 4b, c, d, e)21. In turn, FGFR1 activation correlated with an increase in the levels of RAS-GTP, phospho-AKT, and phospho-ERK (Fig. 2b and Extended Data Fig. 4e), which was prevented by FGFR1 knockdown (Fig. 2c). Accordingly, FGFR1 shRNAs did not inhibit the proliferation of KRAS-mutant lung cancer, but displayed synergistic inhibitory effects when combined with trametinib (Fig. 2d, e, and Extended Data Fig. 4f, g).
Figure 2. Feedback activation of FGFR1 mediates adaptive resistance to trametinib in KRAS-mutant lung cells.
a, qRT-PCR for FGFR1 and FGF2 in H23 cells treated with trametinib for the indicated times (n = 3). b, Immunoblot of H23 cells treated with 25 nM of trametinib for various times. c, Immunoblot of H23 cells transduced with a doxycycline-inducible shRNA targeting FGFR1 and treated with trametinib (25 nM) and doxycycline for the times shown. d, Quantification of fluorescent cells in competitive proliferation assay in H23 and H2030 cells transduced with doxycycline-inducible non-targeting control (Ren) or FGFR1 shRNAs (n = 3). e, Clonogenic assay of H23 cells transduced with FGFR1 and non-targeting control shRNAs, and cultured with DMSO or trametinib (25 nM). Relative growth of DMSO- (grey bars) and trametinib-treated cells (blue and red bars) is shown (right) (n = 3). f, g, Quantification of fluorescent cells in competitive proliferation assays in H23 (f) and the indicated lung cancer cells (g) transduced with doxycycline-inducible non-targeting control (Ren (Renilla)) or the indicated shRNAs (n = 3). a, Paired two-tailed t-test. d, f, g, Unpaired two-tailed t-test. Data presented as mean ± s.d. ns: not significant, *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. For gel source data, see supplementary Fig. 1.
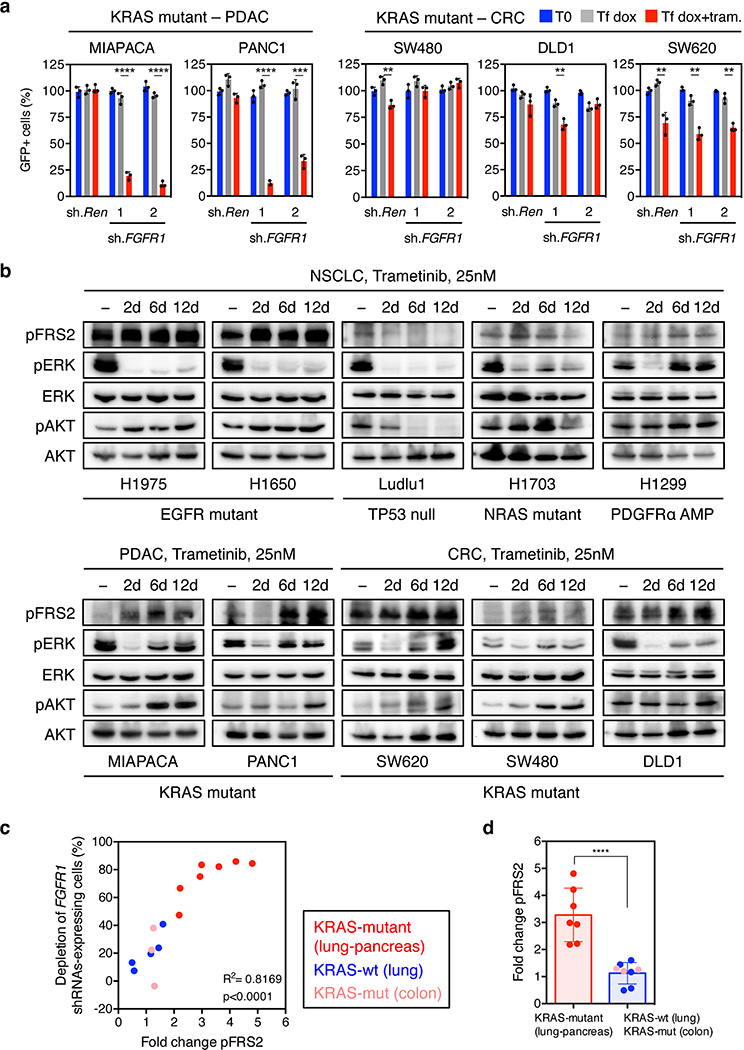
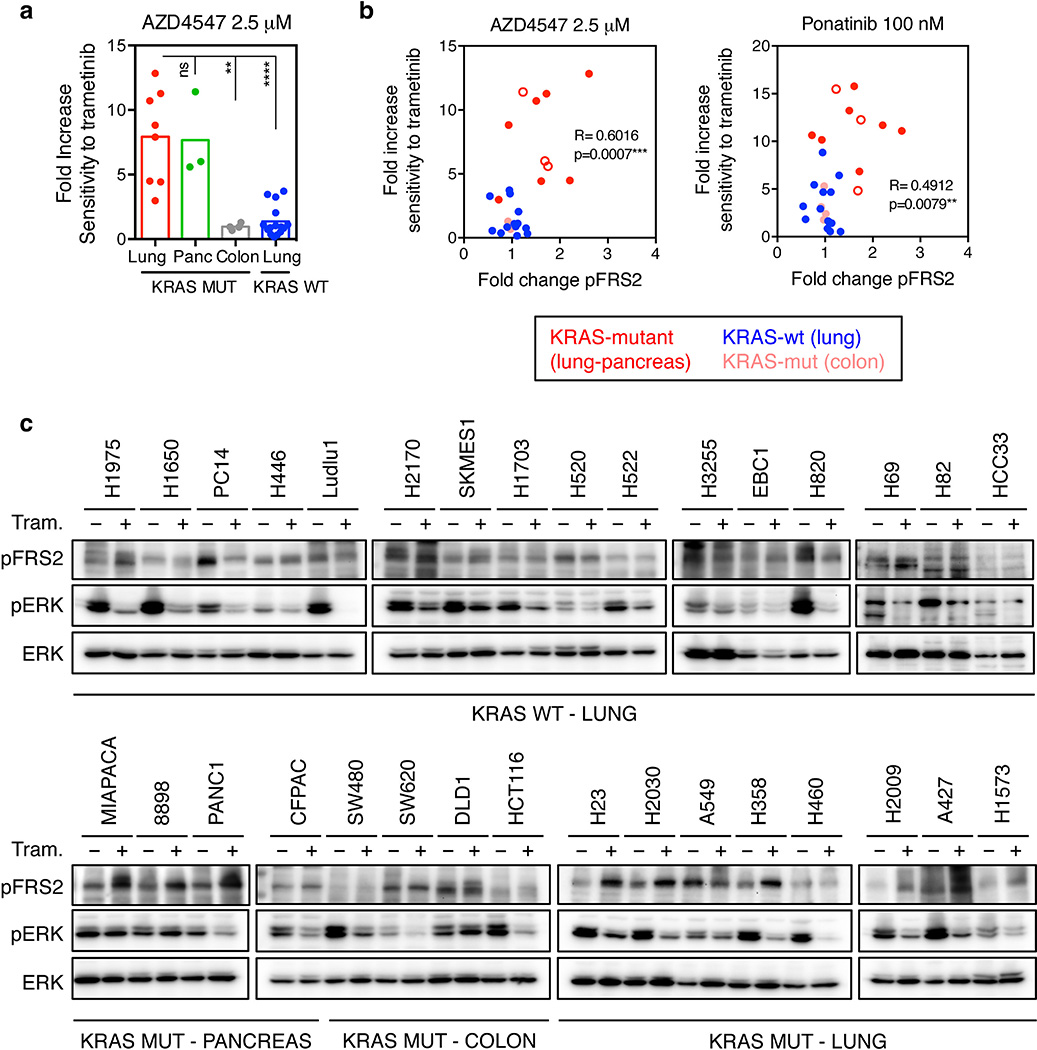
The combinatorial effects of FGFR1 inhibition and trametinib showed distinct specificities: for example, shRNAs targeting FGFR1 or FRS2, but not those targeting FGFR2 and 3, sensitized KRAS-mutant lung cancer cells to trametinib (Fig. 2f and Extended Data Fig. 4h, i). By contrast, FGFR1 knockdown had little impact on trametinib sensitivity in KRAS wild-type lung cancer cells (Fig. 2g). While FGFR1 shRNAs synergized with trametinib in two KRAS-mutant pancreatic cancer cell lines, they showed little activity in trametinib-treated KRAS-mutant colorectal lines (Extended Data Fig. 5a). Importantly, this genotype and tissue specificity correlated with the ability of trametinib to trigger FRS2 phosphorylation when applied as a single agent (Extended Data Fig. 5b, c, d). Therefore, treatment of certain KRAS-mutant tumor types with trametinib induces a dependency on FGFR1 signaling that promotes adaptive drug resistance.
FGFR1 inhibition enhances trametinib effects
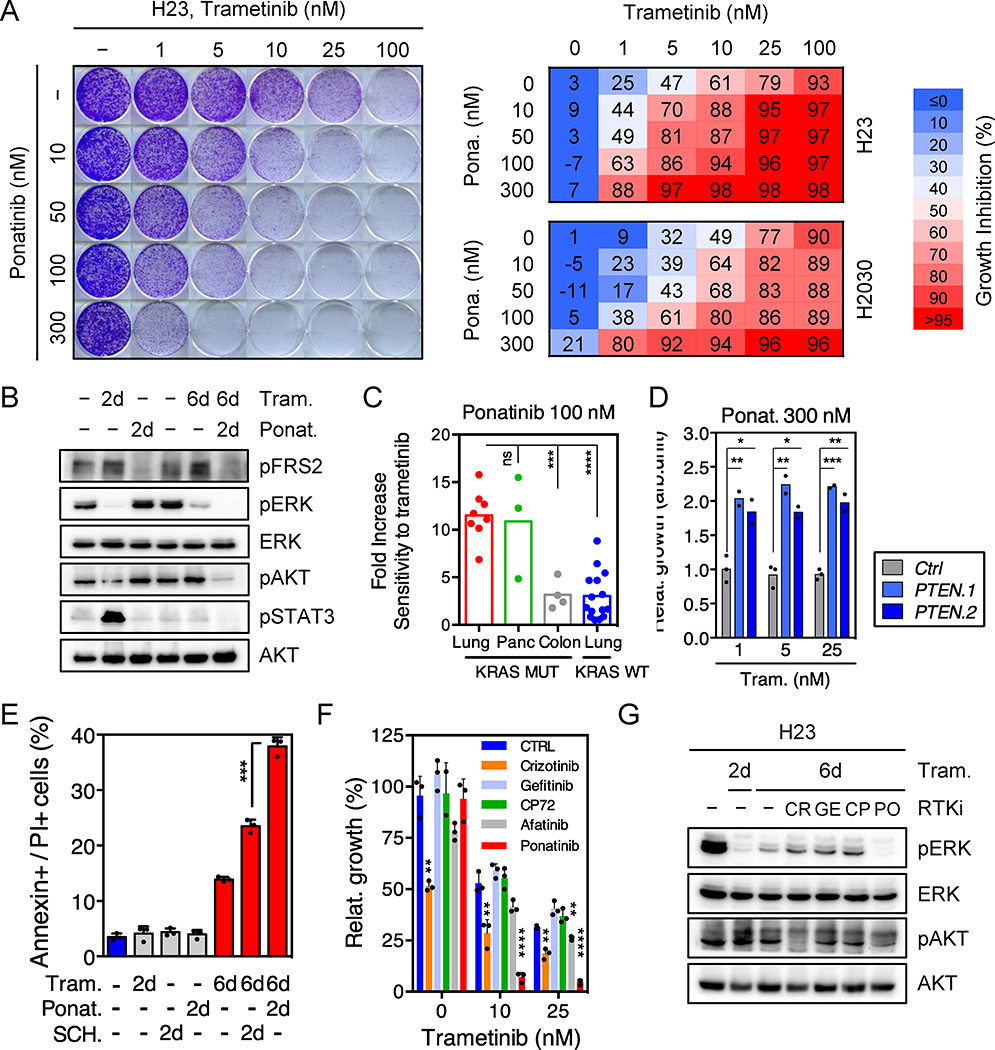
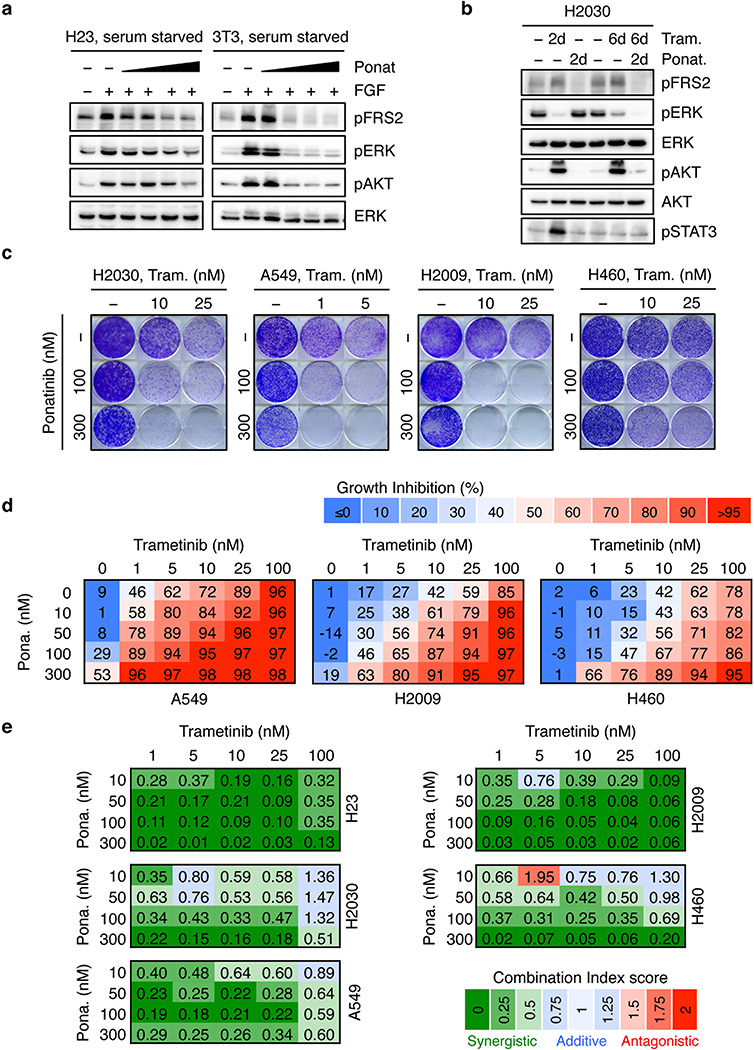
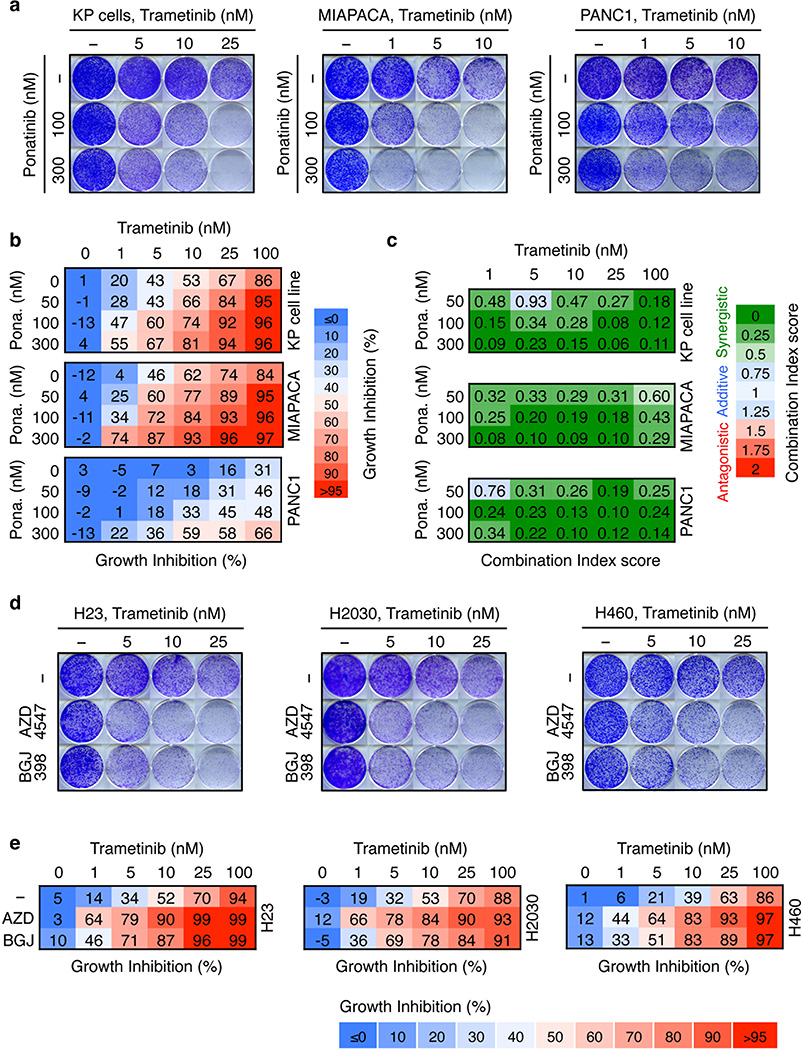
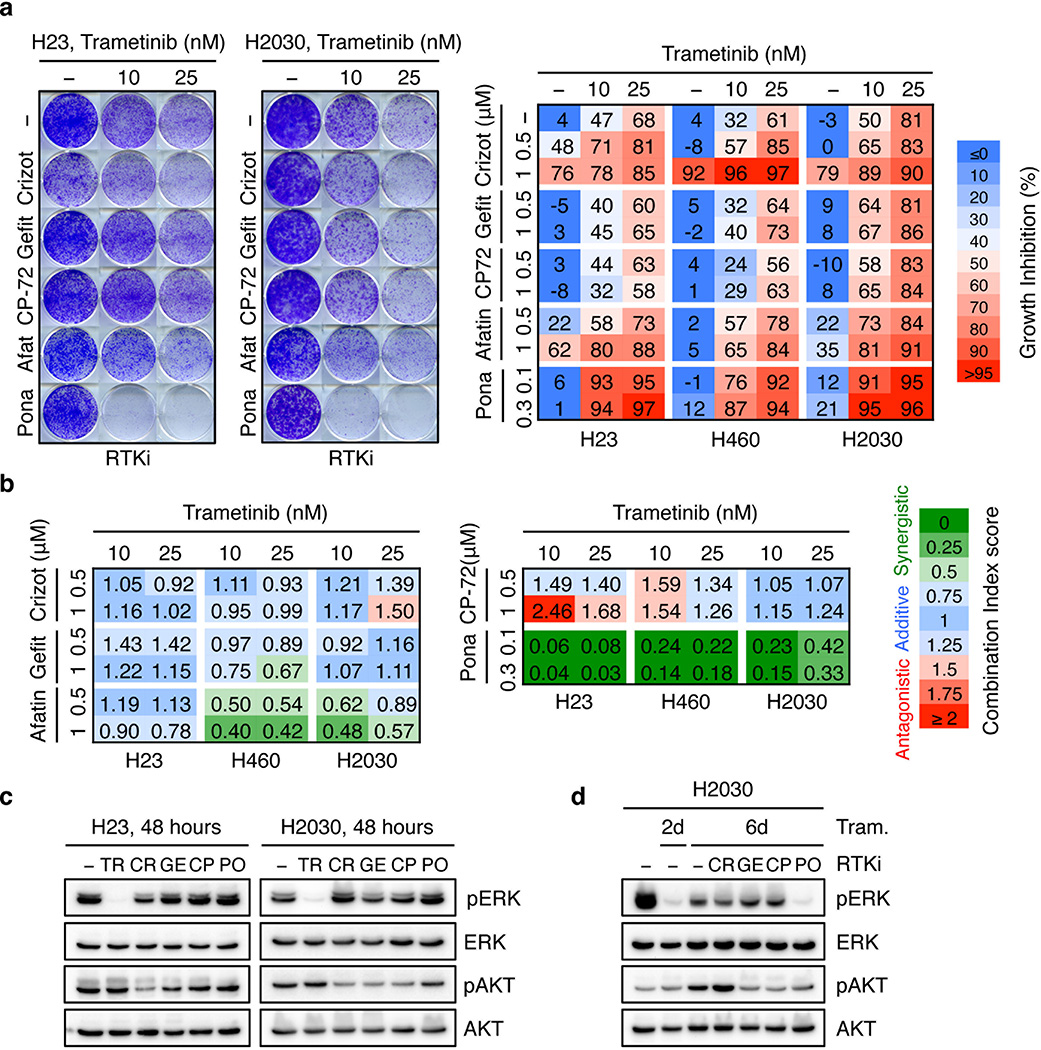
We next tested whether therapeutic strategies combining trametinib with an FGFR1 inhibitor could be effective in treating some KRAS-mutant lung cancers by combining trametinib with ponatinib, an FDA-approved multikinase inhibitor that inhibits FGFR1 and is being tested clinically for activity against FGFR1-amplified lung cancer (Extended Data Fig. 6a)22,23. Ponatinib had little effect on KRAS-mutant cells but countered the trametinib-induced increase in pFRS2, pERK, and pAKT, and synergized with trametinib in inhibiting cell proliferation (Fig. 3a, b, c, and Extended Data Fig. 6b, c, d, e). As observed in our genetic studies, this combination also showed combined activity in human KRAS-mutant pancreatic cancer cells and a Kras-mutant murine lung adenocarcinoma line (Extended Data Fig. 7a, b, c), but to a lesser extent in KRAS wild-type lung cancer cells or KRAS-mutant colon cancer cells (Fig. 3c). Although it remains possible that the synergistic effects of this combination involve ponatinib’s ability to target additional kinases, similar results were observed with two other chemically-distinct FGFR inhibitors (Extended Data Fig. 7d, e, 8a)24,25. Importantly, sensitivity to the combination of trametinib and FGFR inhibition correlated with the degree of pFRS2 induction after trametinib treatment (Extended Data Fig. 8b, c).
Figure 3. Ponatinib synergizes with trametinib at inhibiting cell proliferation of KRAS-mutant lung cells.
a, Clonogenic assay of H23 cells treated with trametinib, ponatinib, or their combination as indicated. Percent inhibition at each concentration of the drugs in H23 and H2030 cells is presented (right). Data presented as mean of three independent experiments. b, Immunoblot of H23 cells treated with trametinib (25 nM), ponatinib (750 nM), or their combination for the times shown. c, Dot plot illustrating the sensitivity increase to trametinib after treatment with ponatinib (100 nM) in a panel of KRAS mutant (n=15) and KRAS wild-type (n=15) cancer cell lines. Data presented as mean of two independent replicates. d, Quantification of the relative growth of H2030 cells transduced with PTEN and non-targeting control shRNAs, and treated with ponatinib (300 nM) in combination with trametinib (1, 5, and 25 nM). Data presented as mean of two independent replicates. e, Quantification of AnnexinV/PI double positive cells in H23 cells treated with trametinib (25 nM), ponatinib (300 nM), SCH772984 (1 µM) or their combination for the times shown (n = 3). f, Quantification of the relative growth of H23 cells treated with trametinib alone or in combination with 500 nM crizotinib, gefitinib, CP-724714, afatinib, or 300 nM ponatinib (n = 3). g, Immunoblot of H23 cells pretreated with trametinib (25 nM) for 4 days, followed by treatment with trametinib (25 nM) alone or in combination with crizotinib (1 µM), gefitinib (1 µM), CP-724714 (1 µM) and ponatinib (750 nM) for 2 days. c–f, Unpaired two-tailed t-test. Error bars represent mean ± s.d. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. For gel source data, see supplementary Fig. 1. Source data for Figure 3.
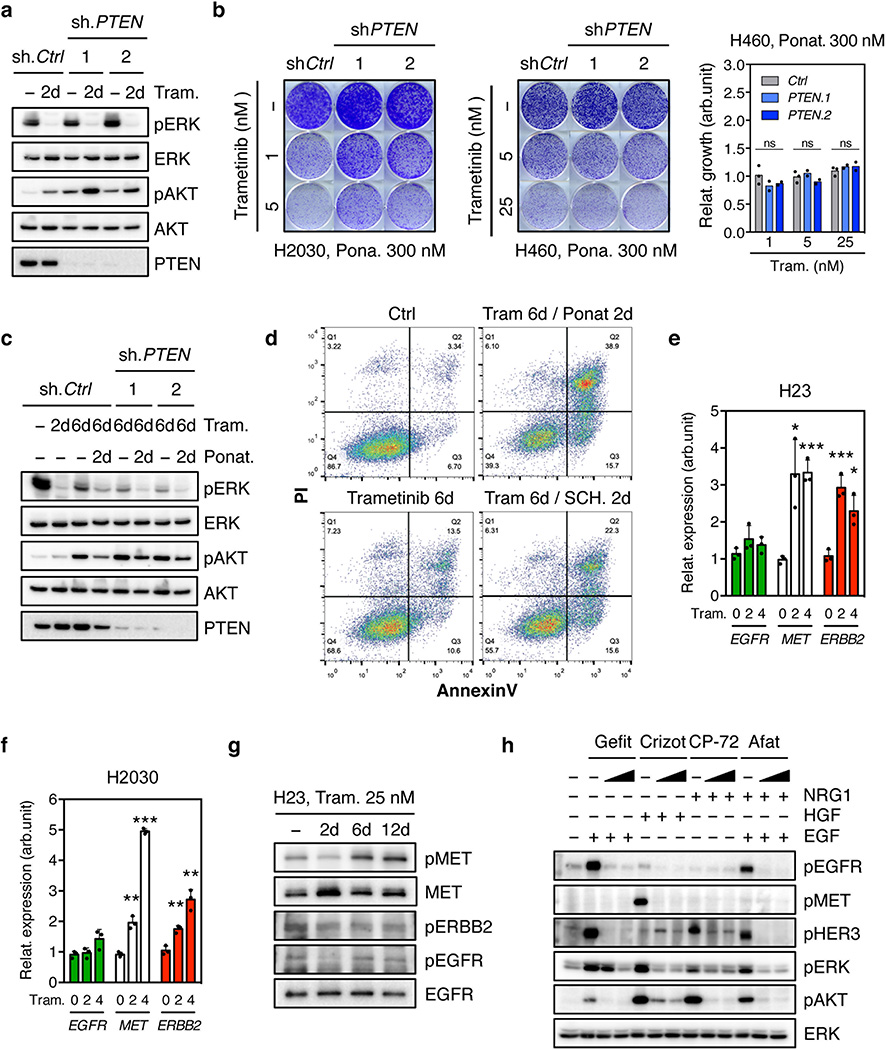
While our genetic and pharmacologic studies establish the importance of the MAPK pathway in adaptive resistance to trametinib, we reasoned that the compensatory increase in PI3K/AKT signaling also plays a role and that its inhibition by ponatinib contributes to the effects of this drug combination. Accordingly, PTEN knockdown, which can increase PI3K signaling independently of RTK activation, promoted partial resistance to the drug combination in KRAS-mutant H2030 cells. This effect was not observed in H460 cells, a KRAS-mutant line that also harbors an activating mutation in the p110α catalytic subunit of PI3K (Fig. 3d and Extended Data Fig. 9a, b, c). Consistent with a role for PI3K signaling in promoting cell survival, co-treatment of H23 cells with trametinib and ponatinib triggered substantial apoptosis in a manner that was not observed following treatment with trametinib alone or in combination with an ERK inhibitor (Fig. 3e and Extended Data Fig. 9d). Thus, the combined ability of ponatinib to impact reactivation of the MAPK and PI3K pathways contributes to its combinatorial activity in KRAS-mutant lung cancer cells.
We also tested whether other RTKs known to be reactivated following MAPK inhibition contribute to adaptive resistance to trametinib in KRAS-mutant lung cancer cells8,9,11,16–20,26,27. While trametinib treatment of H23 and H2030 cells increased MET and ERBB2 (but not EGFR) levels (Extended Data Fig. 9e, f, g), inhibitors targeting these kinases did not synergize with trametinib under the conditions tested (Fig. 3f, Extended Data Fig. 9h, 10a, b). Consistent with previous reports11, the dual EGFR/ERBB2 inhibitor afatinib also showed combinatorial activity with trametinib in some KRAS-mutant lung cancer lines, though in our hands less robustly than the trametinib and ponatinib combination (Fig. 3f and Extended Data Fig. 9h, 10a, b). Accordingly, none of the agents tested prevented the rebound in ERK signaling following trametinib treatment (Fig. 3g and Extended Data Fig. 10c, d). Thus, reactivation of FGFR1 signaling is a prominent mechanism of adaptive resistance to trametinib in KRAS-mutant lung cancer cells.
MEK/FGFR1 inhibition induces KRAS-mutant tumor regression
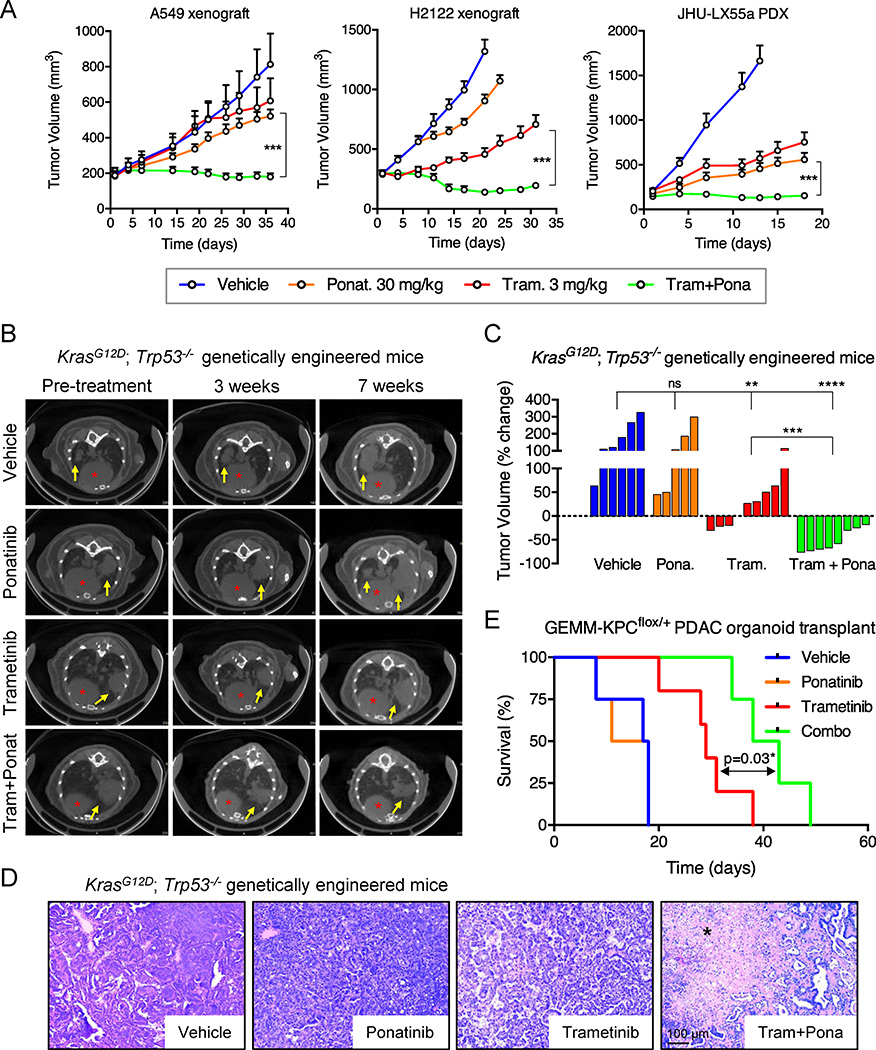
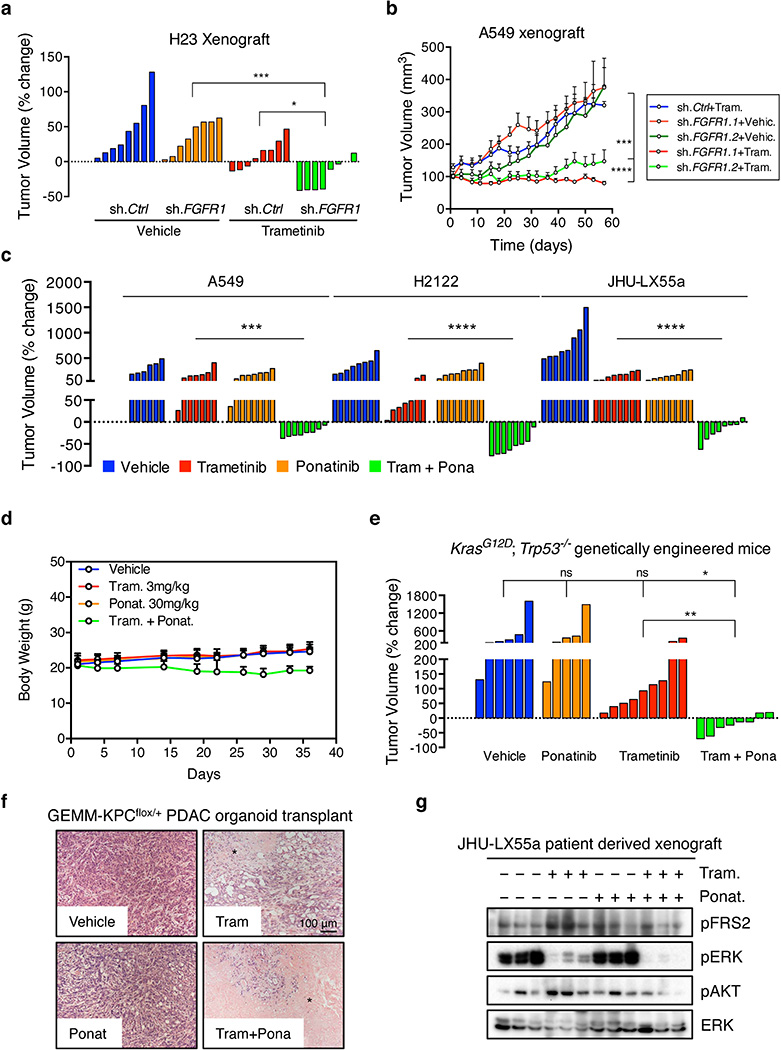
We validated our in vitro results in KRAS-mutant lung cancer xenografts, a KRAS-mutant patient-derived xenograft, and a genetically engineered mouse model (GEMM) of KrasG12D-induced lung adenocarcinoma that accurately resembles the human disease28. A549 and H23 xenografts harboring tet-responsive FGFR1- or control-shRNAs were treated with doxycycline and a daily dose of 3 mg/kg of trametinib when tumors reached ~150 mm3. While knockdown of FGFR1 or treatment with trametinib alone had only minor anti-tumor effects, the combination of FGFR1 knockdown with trametinib potently inhibited tumor growth and typically caused tumor regression (Extended Data Fig. 11a, b). Treatment of the xenografts, PDX and GEMM models with vehicle, trametinib, ponatinib, or the drug combination showed similar results, with only the combination producing marked tumor regressions despite no apparent toxicities (Fig. 4a, b, c and Extended Data Fig. 11c, d, e). Moreover, histological analysis of the residual tumor mass in GEMMs treated with the drug combination showed massive necrosis, an effect not seen with either agent alone (Fig. 4d). Similar results were observed in an organoid based, transplantable model of KrasG12D-driven pancreatic cancer, in which the drug combination produced marked cell death and significantly enhanced survival (Fig. 4e and Extended Data Fig. 11f).
Figure 4. Suppression of FGFR1 in combination with trametinib leads to tumor regression of KRAS-mutant lung tumors.
a, Tumor volumes of mice bearing A549 and H2122 xenografts, and JHU-LX55a patient-derived xenograft tumors and treated with vehicle, trametinib (3 mg/kg), ponatinib (30 mg/kg), or both drugs in combination for the indicated times. Error bars represent mean ± s.e.m. (n ≥ 6 per treatment group). b, c, d, Representative µCT images of the lungs of KrasG12D; Trp53−/− genetically engineered mice treated with vehicle, trametinib (3 mg/kg), ponatinib (30 mg/kg), or both drugs in combination for 3 and 7 weeks. Lung tumors are indicated by yellow arrows and red asterisks mark the hearts (b). A waterfall representation of the response for each tumor after three weeks of treatment is shown. (n ≥ 5 per group). (c). Representative hematoxylin and eosin stains are shown. A black asterisk indicates necrosis (d). e, Kaplan-Meier survival analysis of mice bearing pancreatic tumors resulting from orthotopic transplantation of GEMM-KPCflox/+ PDAC organoids and treated as in b (n ≥ 4 per group) (log-rank test). a, c, Unpaired two-tailed t-test. ns: not significant, *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. Source data for Figure 4.
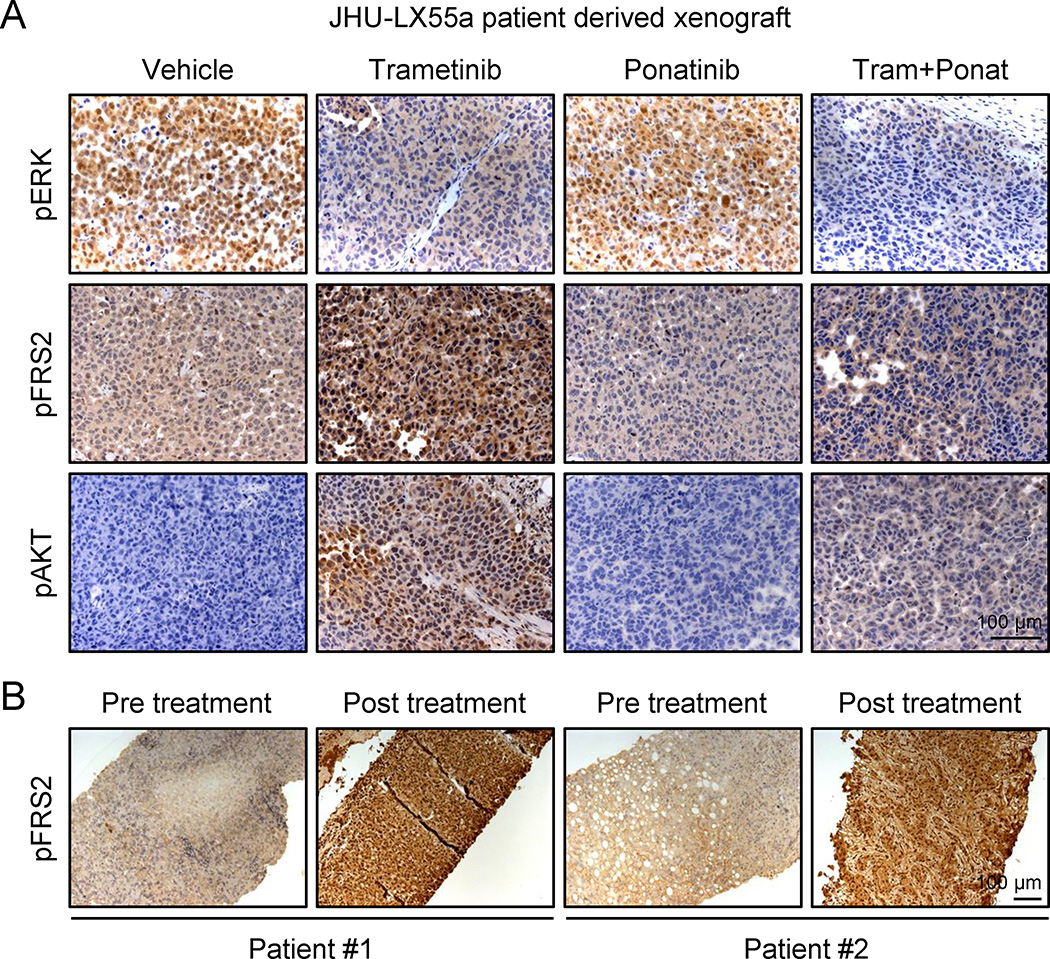
We also examined the ability of trametinib to induce FGFR1 signaling in KRAS-mutant tumors. Consistent with in vitro results, a KRAS-mutant lung PDX model showed a concomitant increase in FRS2, ERK, and AKT phosphorylation following trametinib treatment – an effect that was canceled by ponatinib (Fig. 5a and Extended Data Fig. 11g). Furthermore, FRS2 phosphorylation was dramatically increased following trametinib treatment in two KRAS-mutant lung adenocarcinoma patients (Fig. 5b), indicating that the mechanism of adaptive resistance identified in our preclinical models is clinically relevant.
Figure 5. Trametinib induces FGFR1 signaling in KRAS-mutant lung tumors.
a, Tumor tissue from JHU-LX55a patient derived xenografts treated with vehicle, trametinib (3 mg/kg), ponatinib (30 mg/kg), or both drugs in combination for 18 days was evaluated by IHC for phospho-FRS2, phospho-ERK, and phospho-AKT. b, Paired tumor biopsies from patients having KRAS-mutant lung adenocarcinomas (before and after treatment with the MEK inhibitor trametinib) were evaluated by IHC for phospho-FRS2.
Discussion
In summary, by implementing a stringent approach for negative selection shRNA screening, we identified feedback activation of FGFR1 signaling as a prominent mechanism of adaptive resistance to the MEK inhibitor trametinib in KRAS-mutant lung cancer. The mechanism was specific: only shRNAs targeting FGFR1, but not other FGFR family members or other RTKs tested conferred trametinib sensitivity, and only FGFR1 inhibition blocked compensatory reactivation of both ERK and AKT. In agreement, an unbiased ORF screen identified FGFR1, but not other RTKs, as sufficient to allow proliferation of KRAS-mutant colon cancer cells following KRAS suppression29. In our hands, the synergistic effects of the trametinib/FGFR inhibitor combinations were largely restricted to KRAS-mutant lung and pancreatic cancer cells, but not KRAS wild-type lung or KRAS-mutant colon cancer cells. These results strongly associate sensitivity to the combination with the magnitude of FRS2 phosphorylation following trametinib treatment alone and provide a mechanistic foothold to predict and study cell line and tumor variability.
Our results provide strong mechanistic support for combining trametinib with FGFR1 inhibitors for treating KRAS-mutant lung cancer and pinpoint a biomarker that might eventually be used to identify other patients likely to benefit from this drug combination. Although careful attention to additive or synergistic toxicities will be required for the clinical implementation of these findings, it seems likely that targeting a specific RTK such as FGFR1 will be more tolerable than targeting more pleiotropic factors such as AKT30 and presents a rationale for developing more specific FGFR1 antagonists. Regardless, our study provides further evidence that targeting adaptive resistance mechanisms can improve the efficacy of molecular targeted therapies and provides one path towards developing rational strategies for treating KRAS-mutant lung cancer.
METHODS
Pooled negative-selection RNAi screening
A custom shRNA library targeting 526 human kinases was designed using miR30-adapted DSIR predictions refined with “sensor” rules31 (six shRNAs per gene) and constructed by PCR-cloning a pool of oligonucleotides synthesized on customized arrays (Agilent Technologies and CustomArray) as previously described (Supplementary Table 1)12. The list of genes was obtained from KinBase Database (http://kinase.com/human/kinome/) and was manually curated. After sequence verification, 3156 shRNAs (5–6 per gene) were combined with 20 positive- and negative-control shRNAs at equal concentrations in one pool. This pool was subcloned into the TRMPV-Neo vector and transduced in triplicates into Tet-on H23 KRAS-mutant lung cancer cells using conditions that predominantly lead to a single retroviral integration and represent each shRNA in a calculated number of at least 1,000 cells. Transduced cells were selected for 6 days using 1 mg ml−1 G418 (Invitrogen); at each passage more than 30 million transduced cells were maintained to preserve library representation throughout the experiment. After drug selection, T0 samples were obtained (~30 million cells per replicate (n = 3)) and cells were subsequently cultured in the presence or absence of trametinib (25 nM) and 1 µg ml−1 doxycycline to induce shRNA expression. After ten population doublings (Tf), about fifteen million shRNA-expressing (dsRed+/Venus+) cells were sorted for each replicate using a FACSAriaII (BD Biosciences). Genomic DNA from T0 and Tf samples was isolated by two rounds of phenol extraction using PhaseLock tubes (5prime) followed by isopropanol precipitation. Deep-sequencing template libraries were generated by PCR amplification of shRNA guide strands as previously described12. Libraries were analyzed on an Illumina Genome Analyzer at a final concentration of 8 pM; 50 nucleotides of the guide strand were sequenced using a custom primer (miR30EcoRISeq, TAGCCCCTTGAATTCCGAGGCAGTAGGCA). To provide a sufficient baseline for detecting shRNA depletion in experimental samples, we aimed to acquire >500 reads per shRNA in the T0 sample, which required more than twenty million reads per sample to compensate for disparities in shRNA representation inherent to the pooled plasmid preparation or introduced by PCR biases. With these conditions, we acquired T0 baselines of >500 reads for 3151 (97.9%) shRNAs. Sequence processing was performed using a customized Galaxy platform32.
Using selection criteria that required an shRNA depletion averaging greater than 4-fold after ten population doublings and an effect greater than 4-fold in trametinib-treated cells with respect to untreated ones, 64 shRNAs were identified. The 8 targets for which at least two shRNAs were selectively depleted following trametinib treatment were subject to secondary validation in cell competition assays using multiple KRAS-mutant lung cancer cell lines. 6 targets validated in the cell line in which the primary screen was performed (H23 cells) and 4 (BRAF, CRAF, ERK2, and FGFR1) across a panel of KRAS-mutant lung cancer cells, and as such these represented the top hits of our screen.
Plasmids and recombinant proteins
All vectors were derived from the Murine Stem Cell Virus (MSCV, Clontech) retroviral vector backbone. miR30- and mirE-based shRNAs were designed and cloned as previously described12 and sequences are available in Supplementary Table 1. shRNAs were cloned into the TRMPV-Neo (pSIN-TREdsRed-miR30-PGK-Venus-IRES-NeoR), LT3GEPIR (TRE3G-GFP-miRE-PGK-PuroR-IRES-rtTA3), and MLP (LTR-miR30-PGK-PuroR-IRES-GFP) vectors as previously described12. All constructs were verified by sequencing. Recombinant proteins FGF2 (8910, Cell Signaling), HGF (100-39, Peprotech), EGF (AF-100-15, Peprotech), and NRG1 (100-03, Peprotech) were used at 50 ngml−1 for 10 minutes.
Cell culture, compounds, and competitive proliferation assays
H23, H460, H2030, H358, H2122, H820, H3255, and A549 cells were kindly provided by R. Somwar and H. Varmus (Cornell University, New York). H2009, HCT116, SW480, SW620, DLD-1, PaTu 8988t, 3T3, MIAPACA-2, and PANC-1 cells were purchased from the American Type Culture Collection (ATCC). H69, H82, HCC-33, and H446 were kindly provided by C. Rudin (Memorial Sloan Kettering Cancer Center, New York). H1975, H1650, Ludlu-1, H1703, PC-14, H2170, SK-MES-1, H520, H522, EBC-1, HCC-15, H441, A-427, and H1299 cells were kindly provided by M. Sanchez-Cespedes (IDIBELL, Barcelona). Cell lines were not authenticated. Murine KRASG12D; p53R270H cells were derived from a murine lung adenocarcinoma. Cells were maintained in a humidified incubator at 37 °C with 5% CO2, grown in RPMI 1640 or DMEM supplemented with 10% FBS and 100 IUml−1 penicillin/streptomycin. All cell lines used were negative for mycoplasma.
Trametinib (S2673), SCH772984 (S7101), Gefitinib (S1025), Crizotinib (S1068), CP-724714 (S1167), Afatinib (S1011), BGJ398 (S2183), AZD4547 (S2801), and Ponatinib (S1490) were obtained from Selleckchem. Drugs for in vitro studies were dissolved in dimethyl sulfoxide (DMSO) to yield 5 or 10 mM stock solutions and stored at −80 °C.
For shRNA experiments, when necessary, human cancer cells were modified to express the ecotropic receptor and rtTA3 by retroviral transduction of MSCV-RIEP (MSCV-rtTA3-IRES-EcoR-PGK-Puro) followed by drug selection (1 µgml−1 puromycin for 1 week). Cell lines were transduced with ecotropically packaged TRMPV-Neo-shRNA retroviruses or, alternatively, with amphotropically packaged LT3GEPIR-Puro-shRNA lentiviruses, selected with 1mgml−1 G418 or 1 µgml−1 puromycin for 1 week, and treated with 1 µgml−1 doxycycline to induce shRNA expression.
For competitive proliferation assays shRNA-transduced cells were mixed with non-transduced cells (8:2) and cultured with doxycycline in the presence or absence of trametinib (25 nM). The relative percentage of Venus+/dsRed+ or GFP+ cells was determined before (T0 – blue bars) and after ten population doublings (Tf) (results are relative to T0) (Tf on dox – grey bars / Tf on dox + trametinib – red bars). The quantification of fluorescent cells was monitored on a Guava Easycyte (Millipore). Experiments were performed independently two or three times.
Lentiviral production
Lentiviruses were produced by co-transfection of 293T cells with lentiviral-Cre backbone construct and packaging and envelope vectors (psPAX2 and VSV-G), using the calcium phosphate method. Supernatant was collected 48, 60 and 72 h post-transfection, concentrated by ultracentrifugation at 24,000 r.p.m. for 120 min and resuspended in an appropriate volume of HBSS solution (Gibco).
Clonogenic and apoptosis assay
For clonogenic assays, cells were seeded in triplicate into 6-well plates (5–10 × 103 cells per well) and allowed to adhere overnight in regular growth media. Cells were then cultured in the absence or presence of drug as indicated in complete media for 10–14 days. Growth media with or without drug was replaced every 2 days. Remaining cells were fixed with methanol (1%) and formaldehyde (1%), stained with 0.5% crystal violet, and photographed using a digital scanner. Relative growth was quantified by densitometry after extracting crystal violet from the stained cells using 10% of acetic acid. All experiments were performed at least three times. Representative experiments are shown.
For apoptosis assays, around 1 × 106 cells were seeded into 10-cm plates and cultured in the presence or absence of drugs as indicated. After 6 days, apoptosis and cell death were determined using AnnexinV-APC apoptosis detection kit according to the manufacturer’s instruction (Affymetrix eBioscience). Data were acquired using a FACS Calibur (BD Biosciences). All experiments were performed independently three times.
Quantitative analysis of drug synergy and determination of fold change in sensitivity to trametinib
Drug synergism was analyzed using CompuSyn software (Version 1.0) (http://www.combosyn.com), which is based on the Median-Effect Principle (Chou) and the Combination Index-Isobologram Theorem (Chou-Talalay)33. CompuSyn software generates combination index (CI) values, where CI <1 indicates synergism; CI = 0.75–1.25 indicates additive effects; and CI >1 indicates antagonism. Following the instruction of the software, drug combinations at non-constant ratios were used to calculate Combination Index (CI) in our study.
For calculating the fold change in sensitivity to trametinib, the GI50 for trametinib was determined for a panel of KRAS wild type and mutant cancer cell lines in the absence or presence of ponatinib and AZD4547. Experiments were performed independently two times.
Immunoblotting and RAS-GTP assay
Phospho-lysis buffer (50 mM Tris pH 7.5, 1% Tween-20, 200 mM NaCl, 0.2% NP-40) supplemented with phosphatase inhibitors (5 mM Sodium Fluoride, 1 mM Sodium Orthovanadate, 1 mM Sodium Pyrophosphate, 1 mM β-glycerophosphate), and protease inhibitors (Protease Inhibitor Cocktail Tablets, Roche) was used for cell lysis and protein concentration was determined by Bradford protein Assay kit (Biorad). Proteins were separated by SDS-Page and immunoblotted and transferred to polyvinyl difluoride (PVDF) membranes (Millipore) according to standard protocols. Membranes were immunoblotted with antibodies against pERKT202/Y204 (9101), tERK (9107), pAKTS473 (4060), tAKT (9272), pFRS2Y436 (3861), pSTAT3Y705 (9145), pMEKS217/221 (9154), MEK (4694), pMETY1234/1235 (3077), MET (8198), pERBB2Y1221/1222 (2243), pEGFRY1068 (3777), EGFR (4267), pERBB3Y1289 (4791), and PTEN (9559) from Cell Signaling; CRAF (SC-227), and BRAF (SC-5284) from Santa Cruz Biotechnology; and KRAS (WH0003845M1) from Sigma in 5% BSA in TBS blocking buffer. After primary antibody incubation, membranes were probed with ECL anti-rabbit IgG, anti-mouse IgG or anti-goat IgG secondary antibody (1:10,000) from GE Healthcare Life Science and imaged using the FluorChem M system (protein simple). GTP-bound RAS was measured using the CRAF RAS-binding-domain (RBD) pull down and detection kit (8821, Cell Signaling) as instructed by the manufacturer. All immunoblots were performed independently at least twice.
qRT-PCR
Total RNA was isolated using TRIZOL (Invitrogen), and cDNA was obtained using the TaqMan reverse transcription reagents (Applied Biosystems). Real-time PCR was carried out in triplicate in three independent experiments using SYBR Green PCR Master Mix (Applied Biosystems) on the ViiA™ 7 Real-Time PCR System (Invitrogen). GAPDH or β-actin served as endogenous normalization controls.
Animal studies
All mouse experiments were approved by the Memorial Sloan Kettering Cancer Center (MSKCC) Animal Care and Use Committee (protocol no. 12-04-006). Mice were maintained under specific pathogen-free conditions, and food and water were provided ad libitum. 5–7-week-old, female athymic NCR-NU-NU (Harlan laboratories) mice were used for animal experiments with human cell lines and patient-derived xenografts. For A549, H23, and H2122 xenografts, cells (10 × 106) were harvested on the day of use and injected in growth-factor-reduced Matrigel/PBS (50% final concentration). One flank was injected subcutaneously per mouse. For JHU-LX55a patient-derived xenograft, a poorly differentiated lung adenocarcinoma bearing a KRASG12C mutation, tumors were cut into pieces and inserted into a pocket in the subcutaneous space as previously described34. After inoculation, mice were monitored daily, weighed twice weekly, and caliper measurements begun when tumors became visible. Tumor volume was calculated using the following formula: tumor volume=(D × d2)/2, in which D and d refer to the long and short tumor diameter, respectively. When tumors reached a size of 150–300 mm3, mice were randomized into 5–8 per group and treated with vehicle, trametinib and/or ponatinib per os for 4 consecutive days followed by 3 days off treatment, at 3 mg/kg and 30 mg/kg, respectively. No obvious toxicities were observed in the vehicle- or drug-treated animals as assessed by difference in body weight between vehicle- and drug-treated mice taking tumor size into account. For immunohistochemistry analysis of JHU-LX55a patient-derived xenograft tumors, tumors were harvested 4 hours after dosing on day 18.
For drug efficacy studies using a genetically engineered mouse model of lung cancer, KrasLSL-G12D/+ and Trp53fl/fl mice (8–12 weeks old) were anaesthetized by intraperitoneal injection of ketamine (80 mg per kg) and xylazine (10 mg per kg) and infected intratracheally with 2.5 × 105 infectious particles of Lenti-Cre per mouse, as previously described35. Mice were evaluated by µCT imagingto quantify lung tumor burden before being assigned to various treatment study cohorts. Mice were treated with vehicle, trametinib and/or ponatinib per os for 4 consecutive days followed by 3 days off treatment, at 3 mg/kg and 30 mg/kg, respectively. µCT imaging evaluation was repeated every week during the treatment. Investigators were not blind with respect to treatment.
For drug efficacy studies using an organoid-derived murine model of pancreatic cancer, spherical, duct like organoids were derived and cultured in matrigel and defined media as previously described36 from pancreatic ductal adenocarcinoma (PDAC) occurring in KrasLSL-G12D/+; Trp53fl/+; CHC (Untargeted collagen homing cassette); RIK (Rosa26-LSL-rtTa3-IRES-Kate2); p48Cre mice (GEMM-KPCflox/+) generated via the PDAC-GEMM-ESC approach37. After initially establishing primary organoid cultures, Kate positive cells were sorted and expanded to minimize injection of non-recombined, normal duct cells. For the orthotopic transplantation of PDAC organoids, mice were anesthetized using Isoflurane, and the pancreas was externalized through a small incision made in the left abdominal side near the spleen. Organoids (approximately 250000–500000 cells per mouse) were removed from matrigel and separated into single cells by trypsinization, washed, and finally resuspended in 25 µl of Matrigel (Matrigel, BD) diluted 1:1 with cold PBS. The organoid suspension was injected into the tail region of the pancreas using 28 gauge surgical syringes (Hamilton). Successful injection was verified by the appearance of a fluid bubble without signs of intraperitoneal leakage. The abdominal wall was sutured with absorbable Vicryl suture (Ethicon), and the skin was closed with wound clips (CellPoint Scientific Inc.). Mice were evaluated by ultrasound(Vevo 2100, VisualSonics)to quantify pancreas tumor burden before being randomized to various treatment study cohorts. All the treatment mice had similar initial tumor burden. Mice were treated as described above for drug efficacy studies using a genetically engineered model of lung cancer. Investigators were not blind with respect to treatment.
µCT imaging
µCT Scans were performed on the Mediso Nano SPECT/CT System covering only the lung fields of each mouse. Each scan averaged approximately 6 min using 240 projections with an exposure time of 1,000 ms set at a pitch of 1 degree. The tube energy of the X-ray was 55 kVp and 145 µA. The in-plane voxel sizes chosen were small and thin creating a voxel size of 73 × 73 × 73 µm. The final reconstructed image consisted of 368 × 368 × 1,897 voxels. Scans were analysed with the Osirix software.
Patient samples
Patients with KRAS mutation-positive advanced lung adenocarcinomas were enrolled in the phase I/II clinical study of trametinib and navitoclax (NCT02079740) and the response was assessed per RECIST (response evaluation criteria in solid tumors) criteria. Biopsies were obtained before treatment, and within 2–4 weeks after starting the treatment with trametinib. Specifically, for patient #1, the post-treatment biopsy was obtained after treatment with navitoclax for 7 days, followed by co-treatment with navitoclax and trametinib for 16 days. The post-treatment biopsy from patient #2 was obtained after co-treatment with navitoclax and trametinib for 22 days. All human studies were approved by the Massachusetts General Hospital Institutional Review Board, and informed consent to study was obtained as per protocol from all patients.
Immunohistochemistry
Tissues were fixed overnight in 4% paraformaldehyde, embedded in paraffin, and cut into 5 µm thick sections. Sections were subject to hematoxylin and eosin staining, and immunohistochemical staining following standard protocols. The following primary antibodies were used: pERKT202/Y204 (4370) and pAKTS473 (4060) (Cell signaling), and pFRS2Y436 (ab193363) (Abcam).
Statistical Analysis
Data are expressed as mean ± s.e.m or mean ± s.d. Group size was determined based on the results of preliminary experiments and no statistical method was used to predetermine sample size. The indicated sample size (n) represents biological replicates. Group allocation and outcome assessment were not performed in a blinded manner. All samples that met proper experimental conditions were included in the analysis. Survival was measured using the Kaplan-Meier method. Statistical significance was determined using Student’s t-test, log-rank test, and pearson’s correlation using Prism 6 software (GraphPad Software). Significance was set at P<0.05.
Extended Data
Extended Data Figure 1. A synthetic lethal RNAi screen identifies different MAPK signaling effectors and FGFR1 as sensitizers to MEK inhibition in KRAS-mutant lung cancer cells.
a, Library features and schematic of the TRMPV-Neo vector. b, Schematic outline of the synthetic lethal RNAi screen for identifying sensitizers to trametinib in KRAS-mutant lung cancer cells. c, Clonogenic assay of KRAS-mutant lung cancer cell lines (H23, H460, and H2030) cultured in the presence of increasing concentrations of trametinib. d, Proliferation assay of H23 and H2030 cells in the presence of increasing concentrations of trametinib for 4 passages. Data presented as mean of two independent replicates. e, Immunoblot analysis of KRAS-mutant lung cancer cell lines treated with 25 nM of trametinib for 48 hr. f, g, Scatter plots illustrating the correlation of normalized reads per shRNA between replicates at the beginning of the experiment (d) and replicates at different time points in the absence (left panel) or presence (right panel) of trametinib (25 nM) (e). h, Scatter plot illustrating the fold change in the relative abundance of each shRNA in the library after ten population doublings on doxycycline in the absence or presence of trametinib (25 nM) in H23 cells. Two shRNAs for FGFR1, CRAF, BRAF, and ERK2 were identified as selectively depleted in trametinib-treated cells. For gel source data, see supplementary Fig. 1.
Extended Data Figure 2. Suppression of FGFR1 and different MAPK signaling effectors reduces the proliferation and viability of KRAS-mutant lung cancer cells treated with trametinib.
a, Quantification of fluorescent cells in competitive proliferation assays in H2030 (upper) and A549 (lower) cells transduced with non-targeting control (Ren) or the indicated shRNAs. Data presented as mean (n = 2). Unpaired two-tailed t-test. ns: not significant, *P<0.05, **P<0.01. b, Immunoblot of H23 and H2030 cells pretreated with 25 nM trametinib for various times and subsequently treated with 200 nM trametinib for 2 hours. c, Immunoblot of H23 cells transduced with doxycycline-inducible shRNAs targeting CRAF and BRAF and treated with trametinib (25 nM) and doxycycline for the times shown. H23 cells were pretreated with trametinib for 4 days, followed by treatment with doxycycline and trametinib for 4 days. d, Clonogenic assay of H23 cells transduced with BRAF, CRAF, ERK2, and non-targeting control shRNAs, and cultured with DMSO or trametinib (25 nM) for 10 days. Relative growth of DMSO- (grey bars) and trametinib-treated cells (blue and red bars) is shown (right). Data presented as mean ± s.d. (n = 3). For gel source data, see supplementary Fig. 1.
Extended Data Figure 3. ERK inhibitor SCH772984 enhances the antiproliferative effects of trametinib in KRAS-mutant lung cancer cells.
a, Clonogenic assay of H2030 (upper) and H460 (lower) cells treated with increasing concentrations of trametinib, ERK inhibitor SCH772984, or their combination as indicated. Percent inhibition at each concentration of the drugs in H23, H2030, and H460 cells is presented (right). Data presented as mean of three independent experiments (n = 3). b, Immunoblot analysis of H2030 cells treated with trametinib (25 nM), SCH772984 (500 nM), or their combination for the times shown. H2030 cells were pretreated with trametinib for 4 days, followed by treatment with SCH772984 and trametinib for 2 days. c, Cell viability of H23, H2030, and H460 cells treated with increasing doses of trametinib, ERK inhibitor SCH772984, or their combination for 10 days. Data presented as mean ± s.d. (n = 3). The concentration of trametinib that inhibited cell proliferation by 50% (GI50) was calculated in the absence or the presence of increasing concentrations of SCH772984 (bottom). For gel source data, see supplementary Fig. 1. Source data for Extended Data Figure 3.
Extended Data Figure 4. Feedback activation of FGFR1 signaling leads to adaptive resistance to trametinib in KRAS-mutant lung cancer cells.
a, Immunoblot analysis of KRAS-mutant lung cancer cell lines H23 and H2030 treated with 25 nM trametinib for various times. b, c, d, qRT-PCR for FGFR1 and FGF2 in A549 (b), H2030 (c) and H460 (d) cells treated with trametinib for the indicated times. Data presented as mean normalized for FGFR1 and FGF2 expression ± s.d. (n = 3). e, Immunoblot analysis of A549, H2030, and H358 cells treated with trametinib (25 nM) for various times. f, Quantification of fluorescent cells in competitive proliferation assays in A549, H358, and H460 cells transduced with doxycycline-inducible non-targeting control (Ren) or FGFR1 shRNAs. Data presented as mean ± s.d. (n = 3). g, qRT-PCR for FGFR1 in H23 cells transduced with non-targeting control and FGFR1 shRNAs. Data presented as mean normalized for FGFR1 expression ± s.d. (n = 3). h, Quantification of fluorescent cells in competitive proliferation assays in A549 cells transduced with non-targeting control (Ren) or the indicated shRNAs. Data presented as mean ± s.d. (n = 3). i, qRT-PCR for FGFR2, FGFR3, and FRS2 in A549 cells transduced with non-targeting control, FGFR2, FGFR3 and FRS2 shRNAs. Data presented as mean normalized for FGFR2, FGFR3, and FRS2 expression ± s.d. (n = 3). b–d, paired two-tailed t-test. f–i, unpaired two-tailed t-test. ns: not significant, *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. For gel source data, see supplementary Fig. 1.
Extended Data Figure 5. Trametinib-induced phosphorylation of FRS2 predicts sensitivity to MEK and FGFR1 combined inhibition.
a, Competitive proliferation assays in the indicated KRAS-mutant cancer cell lines transduced with doxycycline-inducible non-targeting control (Ren) or FGFR1 shRNAs. Data presented as mean ± s.d. (n = 3). b, A panel of lung (H1975, H1650, Ludlu-1, H1703, and H1299), pancreas (MIAPACA, PANC1), and colorectal (SW620, SW480, and DLD1) cancer cell lines were treated with 25 nM trametinib for various times. Lysates were subject to immunoblot analysis with the indicated antibodies. c, Scatter plot illustrating the correlation between depletion of FGFR1 shRNAs-expressing cells and fold change in FRS2 phosphorylation following trametinib treatment in human cancer cells lines (n = 15). d, Representation of the fold change in FRS2 phosphorylation following treatment with trametinib for 12 days in human cancer cell lines (n = 15). a, d, unpaired two-tailed t-test. c, two-tailed pearson’s correlation. **P<0.01, ***P<0.001, ****P<0.0001. For gel source data, see supplementary Fig. 1.
Extended Data Figure 6. Trametinib in combination with ponatinib synergizes at inhibiting cell proliferation of KRAS-mutant lung cancer cells.
a, Serum starved H23 (left panel) and 3T3 (right panel) cells were pre-treated with increasing concentration of ponatinib for 24 hr (1, 30, 100, and 300 nM), followed by stimulation with FGF2 (50 ng/ml) for 10 min. Immunoblot analysis for the indicated antibodies is shown. b, Immunoblot analysis of H2030 cells treated with trametinib (25 nM), ponatinib (750 nM), or their combination for the times shown. Cells were pretreated with trametinib for 4 days, followed by co-treatment with ponatinib and trametinib for 2 days. c, Clonogenic assay of H2030, A549, H2009, and H460 cells treated with increasing concentrations of trametinib, ponatinib, or their combination as indicated. d, Percentage of cell growth inhibition at each concentration of trametinib, ponatinib, or their combination in A549, H2009, and H460 cells after is shown. Data presented as mean of three independent experiments (n = 3). e, Combination Index (CI) scores for H23, H2030, A549, H2009, and H460 cells treated with trametinib in combination with ponatinib at the indicated concentrations. Each CI score represents data from at least three independent experiments. For gel source data, see supplementary Fig. 1. Source data for Extended Data Figure 6.
Extended Data Figure 7. Distinct FGFR1 inhibitors sensitize murine and human KRAS-mutant cancer cells to trametinib.
a, Clonogenic assay of a murine lung cancer cell line harboring KrasG12D and Trp53R270H mutations (KP cell line), and human KRAS-mutant pancreatic cancer cell lines (MIAPACA and PANC1). Tumor cells were cultured with increasing concentrations of trametinib, ponatinib, or their combination as indicated. b, Percentage of cell growth inhibition at each concentration of trametinib, ponatinib, or their combination in KP, MIAPACA, and PANC1 cells is shown. Data presented as mean of three independent replicates (n = 3). c, Combination Index (CI) scores for KP, MIAPACA, and PANC1 cells treated with trametinib in combination with ponatinib at the indicated concentrations. Each CI score represents data from at least three independent experiments (n = 3). d, Clonogenic assay of H23, H2030, and H460 cells cultured with increasing concentrations of trametinib alone or in combination with FGFR1 inhibitors BGJ398 (1.5 µM) or AZD4547 (2 µM). e, Percentage of cell growth inhibition at each concentration of trametinib alone or in combination with BGJ398 (1.5 µM) or AZD4547 (2 µM) in H23, H2030, and H460 cells is shown. Data presented as mean of three independent replicates (n = 3). Source data for Extended Data Figure 7.
Extended Data Figure 8. The magnitude of trametinib-induced FRS2 phosphorylation correlates with the sensitivity to trametinib and FGFR1 combined inhibition in human cancer cells.
a, Dot plot illustrating the sensitivity increase to trametinib after the treatment with AZD4547 (2.5 µM) in a panel of KRAS mutant (n=15) and KRAS wild type (n=15) cancer cell lines. Data presented as mean of two independent replicates (n = 2). b, Scatter plot illustrating the correlation between fold increase in sensitivity to trametinib after treatment with AZD4547 (2.5 µM) or ponatinib (100 nM) and fold change in FRS2 phosphorylation following trametinib treatment in a panel of human cancer cells lines. c, Immunoblot analysis of a panel of human cancer cells treated with trametinib (25 nM) for 6 days. a, unpaired two-tailed t-test. b, two-tailed pearson’s correlation. ns: not significant, **P<0.01, ***P<0.001, ****P<0.0001. For gel source data, see supplementary Fig. 1.
Extended Data Figure 9. Ponatinib prevents trametinib-induced reactivation of MAPK and PI3K signaling. Upregulation of distinct RTKs in KRAS-mutant lung cancer cells after trametinib treatment.
a, Immunoblot analysis of H2030 transduced with PTEN and non-targeting control shRNAs, and treated with trametinib (25 nM) for the times shown. b, Clonogenic assay of H2030 (left) and H460 (middle) cells transduced with PTEN and non-targeting control shRNAs. Cells were treated with ponatinib alone (300 nM) or in combination with trametinib at the indicated concentrations. Quantification of the relative cell growth of H460 cells is shown (right). Data presented as mean of two independent experiments. c, Immunoblot analysis of H2030 transduced with PTEN and non-targeting control shRNAs, and treated with trametinib (25 nM) alone or in combination with ponatinib (750 nM) for the times shown. PTEN suppression did not affect ERK signaling or its inhibition following trametinib treatment but instead activated AKT and, more importantly, attenuated the ability of ponatinib to suppress trametinib-induced increase in pAKT. d, AnnexinV/PI double staining assay of H23 cells treated with vehicle, trametinib (25 nM) alone or in combination with ponatinib (300 nM) or SCH772984 (1 µM) for the times shown (n = 3). e, f, qRT-PCR for EGFR, MET, and ERBB2 in H23 (e) and H2030 (f) cells treated with trametinib for 0, 2, and 4 days. Data presented as mean normalized for EGFR, MET, and ERBB2 expression ± s.d. (n = 3). g, Immunoblot analysis of H23 cells treated with 25 nM of trametinib for various times. h, Immunoblot analysis of serum starved H2030 cells pre-treated with 500 nM or 1 µM of gefitinib, crizotinib, CP-724714, or afatinib for 12 hr, followed by stimulation with EGF, HGF, NRG1, or their combination (50 ng/ml) for 10 min. b, e, f, unpaired two-tailed t-test. ns: not significant, *P<0.05, **P<0.01, ***P<0.001. For gel source data, see supplementary Fig. 1.
Extended Data Figure 10. Unresponsiveness of KRAS-mutant lung cancer cells to MEK inhibitor trametinib is predominantly mediated by feedback activation of FGFR1 signaling.
a, Clonogenic assay of H23 and H2030 cells treated with increasing concentration of trametinib alone or in combination with 500 nM crizotinib, gefitinib, CP-724714, and afatinib, or 300 nM ponatinib. Percent inhibition at each concentration of the drugs in H23, H460, and H2030 cells is presented (right). Data presented as mean of at least two independent experiments (n = 2). b, Combination Index (CI) scores for H23, H460, and H23 cells treated with trametinib in combination with crizotinib, gefitinib, CP-724714, afatinib, and ponatinib at the indicated concentrations. Each CI score represents data from at least two independent experiments (n = 2). c, Immunoblot of H23 and H2030 treated with trametinib (25 nM), crizotinib (1 µM), gefitinib (1 µM), CP-724714 (1 µM), and ponatinib (750 nM) for 48 hours. d, Immunoblot analysis of H2030 treated with trametinib (25 nM), crizotinib (1 µM), gefitinib (1 µM), CP-724714 (1 µM), ponatinib (750 nM), or their combination for the times shown. Cells were pretreated with trametinib for 4 days, followed by co-treatment with RTK inhibitors and trametinib for 2 days. For gel source data, see supplementary Fig. 1. Source data for Extended Data Figure 10.
Extended Data Figure 11. Suppression of FGFR1 cooperates with trametinib to inhibit growth of KRAS-mutant lung tumors.
a, b, Mice bearing H23 (a) or H2030 (b) xenografts transduced with FGFR1 or non-targeting control shRNAs were treated with either vehicle or trametinib (3 mg/kg). For H23 xenografts, a waterfall representation of the best response for each tumor is shown (n = 8 per group) (a). For H2030 xenografts, the tumor volumes are shown as a function of time after treatment. Error bars represent mean ± s.e.m. (n ≥ 4 per group) (b). c, Mice bearing A549 and H2122 xenografts, and JHU-LX55a patient-derived xenograft tumors were treated with vehicle, trametinib (3 mg/kg), ponatinib (30 mg/kg), or both drugs in combination. A waterfall representation of the best response for each tumor is shown. (n ≥ 6 per group). d, Body weight of mice bearing A549 xenografts and treated with vehicle, trametinib (3 mg/kg), ponatinib (30 mg/kg), or both drugs in combination for the indicated times (n ≥ 6 per group). e, KrasG12D; Trp53−/− genetically engineered mice harboring lung adenocarcinomas were treated with vehicle, trametinib (3 mg/kg), ponatinib (30 mg/kg), or both drugs in combination for 7 weeks. A waterfall representation of the response for each tumor after 7 weeks of treatment is shown (n ≥ 5). f, Representative hematoxylin and eosin stains of pancreatic tumor tissue resulting from orthotopic transplantation of GEMM-KPCflox/+ PDAC organoids. Mice were treated with vehicle, trametinib (3 mg/kg), ponatinib (30 mg/kg), or both drugs in combination. A black asterisk indicates necrosis. g, Immunoblot analysis of tumor tissue from mice bearing JHU-LX55a patient-derived xenografts treated with vehicle, trametinib (3 mg/kg), ponatinib (30 mg/kg), or both drugs in combination for 18 days. a–c, e, Unpaired two-tailed t-test. ns: not significant, *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. For gel source data, see supplementary Fig. 1. Source data for Extended Data Figure 11.
Supplementary Material
Acknowledgments
We gratefully thank M. Sánchez-Céspedes, R. Somwar, and H. Varmus for generously sharing cell lines; S. Tian, J. Ahn, M. Taylor, A. Shroff, and J. Plevy for technical assistance; C.J. Sherr, L.E. Dow, P. Lito, T. Kastenhuber, and J. Leibold for advice on experimental design and/or for editing the manuscript; and other members of the Lowe laboratory for advice and critical discussions. This work was supported by a program project grant from the National Cancer Institute (S.W.L., N.R.), a grant from the Center of Experimental Therapeutics (ETC) (S.W.L., N.R.), a Stand Up To Cancer grant from the American Association for Cancer Research (N.R., J.A.E., C.R.), and a Cancer Center Support grant to MSKCC. E.M. was supported by The Jane Coffin Childs Memorial Fund for Medical Research and a K99/R00 grant from the NIH/NCI. S.W. was supported by the Annette Kade Fellowship from the Watson School of Biological Sciences. R.W. was supported by a Carl-Duisberg Fellowship from the Bayer Foundation. A.L. was supported by an EMBO Long-Term fellowship. E.d.S. received support through the Geoffrey Beene Cancer Research Center. S.W.L. is the Geoffrey Beene Chair of Cancer Biology and a Howard Hughes Medical Institute investigator.
Footnotes
Supplementary Table 1. Primary screening data. Primary data for Figure 1a and Extended Data Figure 1f, g, h. shRNA screen under doxycycline or doxycycline and trametinib 25 nM.
Supplementary Table 2. Additional shRNA sequences. shRNA sequences for Figure 2f and Extended Data Figure 4h, i.
Supplementary Figure 1. This file contains full scanned blot images with size markers.
AUTHOR CONTRIBUTION
E.M. conceived the project, performed and analyzed experiments and wrote the paper with assistance of all authors. S.W., C.C., and R.W. performed and analyzed in vitro experiments. S.W., J.P.M., and E.d.S. performed and analyzed in vivo experiments. A.L. helped designing and producing the shRNA library. J.T.P and C.R. provided and analyzed patient derived xenografts. J.F.G., R.B.C., and J.A.E. provided human specimens. N.R. conceived the project, supervised experiments and wrote the paper. S.W.L conceived the project, supervised experiments, analyzed data and wrote the paper.
N.R. is a member of the scientific advisory board of Novartis, Astrazeneca, and Chugai Pharmaceutical.
REFERENCES
- 1.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 2.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbie DA, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scholl C, et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell. 2009;137:821–834. doi: 10.1016/j.cell.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 5.Luo J, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137:835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stephen AG, Esposito D, Bagni RK, McCormick F. Dragging ras back in the ring. Cancer cell. 2014;25:272–281. doi: 10.1016/j.ccr.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 7.Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov. 2014;13:828–851. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lito P, Rosen N, Solit DB. Tumor adaptation and resistance to RAF inhibitors. Nature medicine. 2013;19:1401–1409. doi: 10.1038/nm.3392. [DOI] [PubMed] [Google Scholar]
- 9.Samatar AA, Poulikakos PI. Targeting RAS-ERK signalling in cancer: promises and challenges. Nat Rev Drug Discov. 2014;13:928–942. doi: 10.1038/nrd4281. [DOI] [PubMed] [Google Scholar]
- 10.Lito P, et al. Disruption of CRAF-mediated MEK activation is required for effective MEK inhibition in KRAS mutant tumors. Cancer cell. 2014;25:697–710. doi: 10.1016/j.ccr.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun C, et al. Intrinsic resistance to MEK inhibition in KRAS mutant lung and colon cancer through transcriptional induction of ERBB3. Cell reports. 2014;7:86–93. doi: 10.1016/j.celrep.2014.02.045. [DOI] [PubMed] [Google Scholar]
- 12.Zuber J, et al. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nature biotechnology. 2011;29:79–83. doi: 10.1038/nbt.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 14.Morris EJ, et al. Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer discovery. 2013;3:742–750. doi: 10.1158/2159-8290.CD-13-0070. [DOI] [PubMed] [Google Scholar]
- 15.Lee HJ, et al. Drug resistance via feedback activation of Stat3 in oncogene-addicted cancer cells. Cancer cell. 2014;26:207–221. doi: 10.1016/j.ccr.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 16.Nazarian R, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Villanueva J, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-R/PI3K. Cancer cell. 2010;18:683–695. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corcoran RB, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer discovery. 2012;2:227–235. doi: 10.1158/2159-8290.CD-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prahallad A, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 20.Duncan JS, et al. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell. 2012;149:307–321. doi: 10.1016/j.cell.2012.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kouhara H, et al. A lipid-anchored Grb2-binding protein that links FGF-receptor activation to the Ras/MAPK signaling pathway. Cell. 1997;89:693–702. doi: 10.1016/s0092-8674(00)80252-4. [DOI] [PubMed] [Google Scholar]
- 22.Gozgit JM, et al. Ponatinib (AP24534), a multitargeted pan-FGFR inhibitor with activity in multiple FGFR-amplified or mutated cancer models. Mol Cancer Ther. 2012;11:690–699. doi: 10.1158/1535-7163.MCT-11-0450. [DOI] [PubMed] [Google Scholar]
- 23.University of Colorado, Denver. ClinicalTrials.gov [Internet] Bethesda (MD): National Library of Medicine (US); 2000. Study of Ponatinib in Patients With Lung Cancer Preselected Using Different Candidate Predictive Biomarkers. [2015 March 30]. Available from: https://clinicaltrialsgov/show/NCT01935336 NLM Identifier: NCT01935336. [Google Scholar]
- 24.Guagnano V, et al. Discovery of 3-(2,6-dichloro-3,5-dimethoxy-phenyl)-1-{6-[4-(4-ethyl-piperazin-1-yl)-phenylamin o]-pyrimidin-4-yl}-1-methyl-urea (NVP-BGJ398), a potent and selective inhibitor of the fibroblast growth factor receptor family of receptor tyrosine kinase. J Med Chem. 2011;54:7066–7083. doi: 10.1021/jm2006222. [DOI] [PubMed] [Google Scholar]
- 25.Gavine PR, et al. AZD4547: an orally bioavailable, potent, and selective inhibitor of the fibroblast growth factor receptor tyrosine kinase family. Cancer research. 2012;72:2045–2056. doi: 10.1158/0008-5472.CAN-11-3034. [DOI] [PubMed] [Google Scholar]
- 26.Li F, et al. FGFR-Mediated Reactivation of MAPK Signaling Attenuates Antitumor Effects of Imatinib in Gastrointestinal Stromal Tumors. Cancer discovery. 2015;5:438–451. doi: 10.1158/2159-8290.CD-14-0763. [DOI] [PubMed] [Google Scholar]
- 27.Traer E, et al. Ponatinib overcomes FGF2-mediated resistance in CML patients without kinase domain mutations. Blood. 2014;123:1516–1524. doi: 10.1182/blood-2013-07-518381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jackson EL, et al. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer research. 2005;65:10280–10288. doi: 10.1158/0008-5472.CAN-05-2193. [DOI] [PubMed] [Google Scholar]
- 29.Shao DD, et al. KRAS and YAP1 converge to regulate EMT and tumor survival. Cell. 2014;158:171–184. doi: 10.1016/j.cell.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimizu T, et al. The clinical effect of the dual-targeting strategy involving PI3K/AKT/mTOR and RAS/MEK/ERK pathways in patients with advanced cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:2316–2325. doi: 10.1158/1078-0432.CCR-11-2381. [DOI] [PubMed] [Google Scholar]
- 31.Fellmann C, et al. Functional identification of optimized RNAi triggers using a massively parallel sensor assay. Molecular cell. 2011;41:733–746. doi: 10.1016/j.molcel.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taylor J, Schenck I, Blankenberg D, Nekrutenko A. Using galaxy to perform large-scale interactive data analyses. Current protocols in bioinformatics / editoral board, Andreas D. Baxevanis … [et al.] 2007:15. doi: 10.1002/0471250953.bi1005s19. Chapter 10, Unit 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer research. 2010;70:440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 34.Poirier JT, et al. DNA methylation in small cell lung cancer defines distinct disease subtypes and correlates with high expression of EZH2. Oncogene. 2015 doi: 10.1038/onc.2015.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nature protocols. 2009;4:1064–1072. doi: 10.1038/nprot.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huch M, et al. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. The EMBO journal. 2013;32:2708–2721. doi: 10.1038/emboj.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saborowski M, et al. A modular and flexible ESC-based mouse model of pancreatic cancer. Genes & development. 2014;28:85–97. doi: 10.1101/gad.232082.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.