Abstract
Polyacrylamide hydrogels have been used extensively to study cell responses to the mechanical and biochemical properties of their substrates. A key step in fabricating these substrates is the conjugation of cell adhesion proteins to the polyacrylamide surfaces, which typically involves nonspecifically anchoring these proteins via side-chain functional groups. This can result in a loss of presentation control and altered bioactivity. Here, we describe a new functionalization strategy in which we anchor full-length extracellular matrix proteins to polyacrylamide substrates using 2-pyridinecarboxaldehyde, which can be co-polymerized into polyacrylamide gels and used to immobilize proteins by their N-termini. This one-step reaction proceeds under mild aqueous conditions and does not require additional reagents. We demonstrate that these substrates can readily conjugate to various extracellular matrix proteins, as well as promote cell adhesion and spreading. Notably, this chemistry supports the assembly and cellular remodeling of large collagen fibers, which is not observed using conventional side-chain amine-conjugation chemistry.
Graphical abstract
1. Introduction
Cells are sensitive to the mechanical properties of their environment. In particular, the stiffness of the extracellular matrix (ECM) substrate to which cells adhere can affect cell morphology, adhesion, migration and stem cell differentiation.1–6 To study this behavior in culture, polyacrylamide (PAAm) hydrogels are most commonly used because the compliance of this material can easily be tuned to mimic stiffness values of soft tissues.2–4, 7–12 Additionally, PAAm does not promote protein adsorption or cell adhesion, enabling improved control over substrate functionalization with cell-adhesive ligands.8, 10, 11
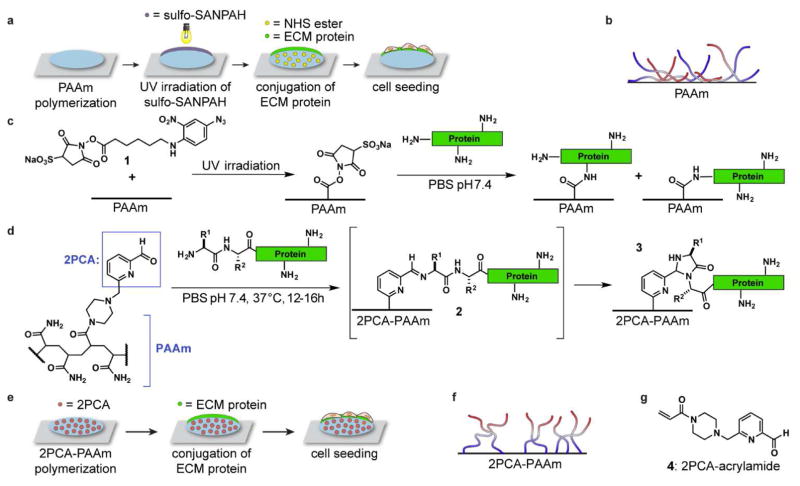
The functionalization of PAAm with the ECM proteins required for cell adhesion has been achieved using pendant N-hydroxysuccinimide (NHS) esters that acylate lysine residues,13 hydrazides that couple to periodate-oxidized glycans on proteins,14 non-covalent adsorption,15 and biotin–streptavidin interactions.16 Perhaps the most common strategy uses a heterobifunctional crosslinker, sulfo-SANPAH (1, Fig. 1a–c), which inserts into the PAAm backbone following photolysis of the aryl azide moiety.7, 8 The resulting polymer-linked NHS ester is then used to acylate amine groups on proteins. Although widely used, this process can create significant batch-to-batch variability, and the immobilized NHS esters are subject to competitive hydrolysis under protein attachment conditions. Moreover, NHS-esters can react with any amines on the proteins, resulting in nonspecific tethering at multiple sites with uncontrollable and unpredictable adhesive ligand presentation (Fig. 1b). The orientation of immobilized ligand has been shown to influence the accessibility of epitopes and affect cell behavior.17 Furthermore, protein structure and activity may be affected if functionally and structurally important lysine residues are engaged with the surface.
Fig. 1.
ECM protein conjugation on polyacrylamide (PAAm) substrates. (a–c) A scheme is shown for ECM protein conjugation using sulfo-SANPAH (1). This reaction targets all available amines, resulting in heterogeneous protein attachment. (d–f) In this work, protein N-termini are coupled to 2-pyridinecarboxaldehyde (2PCA)-modified PAAm gels, resulting in a more uniform presentation. (g) A 2PCA derivative for acrylamide copolymerization is shown. Protein N-termini are shown in blue in (b) and (f).
This conjugation approach has also contributed to a major controversy in the field, questioning whether the density of these lateral tethers directly affects cell behavior and function.18–20 It has been hypothesized that due to smaller pore size, stiffer substrates have shorter distances between surface anchorage points. This reduces the local deformability of these proteins relative to softer substrates, which have comparatively greater surface porosity and longer tethering distances, implying that local tether density controls cell behavior.18 In contrast, a subsequent study showed that systematically varying PAAm porosity without altering stiffness does not significantly influence protein tethering, substrate deformation, or stem cell differentiation, implying that cells respond to bulk substrate stiffness rather than the degree of protein tethering.19 Nevertheless, anchoring density was still observed to increase with increasing sulfo-SANPAH crosslinker concentration, complicating data interpretation.19 Such studies have stimulated strong interest to develop ECM-hydrogel conjugation strategies with predictable and well-controlled attachment chemistry and ligand presentation.
This problem can be addressed by developing immobilization strategies that only anchor the ECM proteins to the hydrogel surface once, thereby ensuring that each protein is tethered predictably and with minimal perturbation of protein structure and function. Site-selective protein immobilization strategies using native cysteine residues, small molecule or peptide fusion tags have been developed for substrates other than PAAm, but they are generally incompatible with commercially-available tissue-purified ECM proteins.21–23 As a result, there remains a significant need for site-specific strategies to immobilize ECM proteins to hydrogel surfaces for cell culture applications.
We report herein a well-defined immobilization strategy in which native proteins are conjugated to PAAm hydrogels specifically through their N-termini using recently published 2-pyridinecarboxyaldehyde (2PCA) conjugation chemistry (Fig. 1d–f).24 We demonstrate that this immobilization strategy is applicable to multiple widely used ECM proteins. Cells readily adhere and spread on these substrates, and despite the presence of only a single protein-hydrogel tethering point, cells recapitulate stiffness-dependent behavior. Moreover, 2PCA-conjugated surfaces remarkably and uniquely support the assembly of attached collagen chains into large fibers, which can be remodeled and bundled by attached cells in a manner reminiscent of collagen remodeling in tissue.
2. Materials and methods
2.1. Reagents and instruments
Unless stated otherwise, all reagents and solvents used were of analytical grade and were used as received from commercial sources. Type I bovine collagen (PureCol, Advanced BioMatrix), human plasma fibronectin (Millipore) and mouse laminin (Gibco) were also used as received. PBS, pH 7.4, was purchased from Fisher Scientific.
NMR spectra were recorded on a Bruker AVQ-400 spectrometer. 1H NMR chemical shifts are reported as δ in units of parts per million (ppm) relative to CDCl3 (δ 7.26). Multiplicities are reported as: s (singlet), br.s (broad singlet), d (doublet), t (triplet), q (quartet), dd (doublet of doublets), dt (doublet of triplets), br (broad) or m (multiplet). Coupling constants are reported as a J value in Hertz (Hz). 13C NMR chemical shifts are reported as δ in units of parts per million (ppm) relative to CDCl3 (δ77.2). High-resolution electrospray ionization mass spectrometry (HR-ESI-MS) data were obtained at the UC Berkeley QB3/Chemistry Mass Spectrometry Facility.
Epifluorescence images were acquired using an inverted Nikon Eclipse Ti microscope equipped with a 10x air objective. DIC images were acquired using an inverted Nikon TE2000-E2 microscope equipped with a 60x oil objective. Time-lapse images were acquired every 15 minutes using an inverted Nikon TE2000-E2 microscope equipped with a 20x air objective. Both microscopes are equipped with a programmable stage and an incubator chamber to maintain constant temperature, humidity and CO2 levels. High magnification images were acquired using a Prairie Technologies upright swept-field confocal microscope equipped with a 60x lens.
2.2. ECM protein conjugation with 2PCA-fluorescein in solution
2PCA-fluorescein was synthesized as previously reported.24 Stock solutions of 50 mM fluorescein (free acid, Sigma) and 2PCA-fluorescein were prepared using DMSO (Sigma). A 250 mM stock solution of benzylalkoxyamine (Sigma, CDS001502) was prepared by the addition of an appropriate volume of water and adjustment to pH 7.5 using 5 M NaOH. In a 50 μL reaction, 0.5 mg/mL ECM protein was incubated with 2PCA-fluorescein or fluorescein (final concentration 5 mM) in 40 mM phosphate buffer, pH 7.5, at 37 °C without agitation. 2PCA-fluorescein was pre-incubated with benzylalkoxyamine (final concentration, 50 mM) for 4 h at 37 °C before the addition of an ECM protein. After 12 h, all samples were centrifuged at 12,000 rpm to remove any precipitate. Collagen and fibronectin samples were purified using a 10 kDa MWCO Amicon Ultra-4 centrifugal filter spin concentrator (Millipore). Laminin samples were purified by acetone precipitation. Samples were analyzed using SDS-PAGE (7.5% Mini-PROTEAN TGX gel, Bio-Rad, or Bolt 8% Bis-Tris Plus gel, Life Technologies), Typhoon 9410 variable mode imager (Amersham Biosciences) before staining with Coomassie Brilliant Blue R-250 (Bio-Rad) and Gel Doc EZ System (Bio-Rad) after staining.
2.3. Synthesis of 2PCA-acrylamide
6-(piperazin-1-ylmethyl)-2-pyridinecarboxaldehyde (S5) was synthesized as previously reported.24 In an oven-dried round bottom flask, 5.5 mmol (1.0 equiv.) of 6-(piperazin-1-ylmethyl)-2-pyridinecarboxaldehyde (S5) was stirred in 30 mL dichloromethane. The solution was cooled in an ice-water bath and turned from cloudy to clear upon the addition of triethylamine (Sigma, 1.82 g, 2.5 mL, 18 mmol, 3.3 equiv.). Acryloyl chloride (Sigma, 0.54 g, 487 μL, 6.0 mmol, 1.1 equiv.) was then added dropwise at 0 °C. The reaction mixture was warmed to room temperature and stirred overnight. After evaporation of solvent and triethylamine under reduced pressure, the resulting material was purified by flash chromatography using 1.5% to 3% methanol in dichloromethane to afford product as a colorless viscous oil (0.86 g, 60% yield). 1H NMR (400 MHz, CDCl3): δ, 9.93 (s, 1H), 7.81–7.70 (m, 2H), 7.60 (d, J = 7.2 Hz, 1H), 6.46 (dd, J = 16.8, 10.6 Hz, 1H), 6.14 (d, J = 16.8 Hz, 1H), 5.57 (d, J = 10.6 Hz, 1H), 3.68 (s, 2H), 3.61 (s, 2H), 3.50 (s, 2H), 2.48–2.42 (m, 4H). 13C NMR (100 MHz, CDCl3): δ, 193.4, 165.22, 159.0, 152.2, 137.5, 127.8, 127.4, 127.3, 120.4, 63.7, 53.3, 52.7, 45.6, 41.7. HRMS (ESI) calculated for C14H18N3O2 ([M+H]+) 260.1394, found 260.1388.
2.4. Preparation of 2PCA-PAAm substrates
12 mm #1 circular glass coverslips were plasma cleaned (Harrick Plasma, PDC-32G), then silanized using a solution of 5% v/v acetic acid (Sigma) and 0.3% v/v PlusOne Bind-Silane (GE Healthcare) in ethanol for 3 min at room temperature. The silanized coverslips were then rinsed with 70% ethanol in water before drying with a Kimwipe. To ease the detachment of polymerized PAAm gels, a flat piece of glass was made hydrophobic by spraying with Rain-X (original glass water repellent) and drying with a Kimwipe. 40% acrylamide and 2% bisacrylamide (Bio-Rad) were combined in different percentages and diluted to the appropriate volume with ultrapure water. A stock solution of 1.12 M 2PCA-acrylamide (molar equiv. of 40% acrylamide) was prepared in acetone and diluted in ultrapure water (Invitrogen) to appropriate concentrations before adding to acrylamide/bisacrylamide solutions. Ammonium persulfate (Bio-Rad, 10% stock solution in water, final concentration 0.1%) and tetramethylethylenediamine (Bio-Rad, 1:1000 v/v) were added to the solutions immediately before sandwiching 30 μL of the polymerization solution between Rain-X treated glass and silanized coverslips. After the solution was allowed to polymerize for 15 to 30 min at room temperature, the gels were removed from the Rain-X treated glass, placed in 24-well plates (Falcon, cat# 353047) and rinsed in PBS for 10 min × 3 times. Substrates containing 2PCA-acrylamide were directly incubated in 400 μL of 50 μg/mL collagen or 25 μg/mL fibronectin or 25 μg/mL laminin in PBS at 37 °C for 12 to 16 h before rinsing in PBS for 10 min × 3 times and subsequent cell seeding. Substrates without 2PCA-acrylamide were submerged in 0.5 mL of 0.5 mg/mL sulfo-SANPAH (Pierce) in PBS. They were then activated with 8 min UV exposure (350 – 550 nm, Sunray 400 SM) and rinsed in PBS 5 min × 3 times before incubating in ECM protein solutions as described above. The pH of ECM protein solutions was always checked and adjusted to pH 7.4, if necessary, prior to incubation with PAAm substrates.
2.5. Mechanical characterization of PAAm hydrogels
Substrates were prepared as described above, with the exception that for each substrate, a 480 μL solution was polymerized in between and then detached from two Rain-X treated 25 mm glass coverslips. Anton Paar Physica MCR 301 rheometer with 25-mm parallel plate was used to determine substrate stiffness at 37 °C. The linear regime was determined based on amplitude sweeps over the range c = 0.1 to 10%. Frequency sweeps at 1% strain over 0.1 to 20 Hz were recorded to extract storage, loss and complex moduli. Storage moduli at 0.1 Hz of 0.1% 2PCA-PAAm substrates containing final acrylamide/bisacrylamide (A/B) percentages of 3% A/0.1% B, 4% A/0.075% B, 4% A/0.2% B, 8% A/0.3% B and 15% A/1.2% B were measured to be 0.08, 0.46, 1.2, 5.8 and 12 kPa respectively. At least three independent samples were measured per condition. Note that the stiffness reported here is the storage modulus instead of the Young’s modulus.
2.6. Cell line and reagents
U2OS human osteosarcoma cells (ATCC HTB-96) were transduced with RFP-LifeAct to enable live microscopy studies and cell area measurements. Briefly, the immediate early promoter of cytomegalovirus (Pcmv IE) and LifeAct-TagRFP were amplified by polymerase chain reaction from pCMV-LifeAct-TagRFP plasmid (Ibidi) and PacI/EcoRI restriction sites were incorporated at each end. The PCR product was subcloned into pFUG-IP (kindly provided by D.V. Schaffer, University of California, Berkeley, CA) after removal of the hUbC promoter and EGFP by digesting with PacI/EcoRI. Viral particles were packaged in 293T cells and used to infect U2OS cells at a multiplicity of infection of 1.5 IU/cell. Cells expressing the vectors were sorted on a DAKO-Cytomation MoFlo High Speed Sorter based on RFP fluorescence. Cells were cultured in DMEM (Gibco, cat# 11965) with 10% fetal bovine serum (JR Scientific), 1% penicillin/streptomycin (Thermo Fisher Scientific) and 1% MEM non-essential amino acids solution (Life Technologies Corporation) in a 37 °C incubator in the presence of 5% CO2. U2OS cells stably transduced with RFP-LifeAct were used in all cell experiments.
2.7. Cell spreading experiments
Trypsinized U2OS RFP-LifeAct cells were seeded on substrates at a density of 9,000 cells/cm2 in 0.5 mL of serum-containing culture medium describe above. Cells were allowed to adhere and spread for 2 h before removal of non-adhered cells by aspiration. Fresh culture media was added and live cells were imaged 3–5 h after seeding. At least 5 fields of view were acquired per substrate. Using ImageJ, projected cell area was determined based on Life-act-RFP signal.
2.8. Immunofluorescence staining
For immunostaining, all reagents were diluted in PBS. After fixing with 4% paraformaldehyde, cells were washed twice with PBS and permeabilized with 0.5% Triton-X 100 (EMD Biosciences) in 5% goat serum (Gibco) for 15 min at room temperature. Cells were rinsed twice and blocked in 5% goat serum for 45 min at room temperature. Cells were then stained for focal adhesions using mouse monoclonal anti-vinculin IgG (Sigma, V9131, 1:200 dilution) in 1% goat serum overnight at 4 °C. After rinsing in 1% goat serum twice, cells were incubated with Alexa Fluor 633 labeled goat anti-mouse IgG (ThermoFisher, A21052, 1:400 dilution) in 1% goat serum for 1 h at room temperature. Finally, F-actin and the nucleus were stained with Alexa Fluor 546 conjugated phalloidin (Thermo Fisher, A22283, 1:200 dilution) and DAPI (ThermoFisher, 2.5 μg/mL) for 20 min at room temperature.
2.9. Characterization of surface available ligands
To label collagen with a fluorophore, 1 mL of 3 mg/mL type I collagen was mixed with 9 mL of 0.1 M carbonate buffer, pH 9.2, and 13 μL of 26 mM Oregon Green 488 carboxylic acid, succinimidyl ester (Molecular Probes, stock solution prepared in DMF). The collagen solution was kept at 4 °C at all times to prevent gelation. The reaction was placed on a rotator for 14 h at 4 °C before using a 10 kDa MWCO Amicon Ultra-4 centrifugal filter spin concentrator (Millipore) to remove unconjugated fluorophore and exchange into 0.01 N HCl. Oregon Green-labeled collagen (OG-Col) was further purified using a size exclusion column (NAP-10, GE Healthcare). Based on a BCA assay and absorbance at 488 nm, the extent of labeling was estimated to be 4 fluorophores per collagen monomer on average. 2PCA-acrylamide containing substrates were incubated with 50 μg/mL total collagen (1:4, Oregon Green labeled: unlabeled) as described above. As control experiments, 2PCA-PAAm substrates were treated with 10 mM piperazine-2PCA (S5) during OG-Col incubation or preincubated with 40 mM benzylalkoxyamine in 50 mM acetate buffer, pH 5.0, for 20–24 h before OG-Col immobilization. After rinsing, U2OS RFP-LifeAct cells were seeded onto the substrates and allowed to adhere for 3 h before acquiring fluorescence images. To ensure unbiased determination of focal planes, focusing was done on adhered cells using bright field. Fluorescence images were processed using rolling ball background subtraction and overall fluorescence was quantified with ImageJ software (NIH).
2.10. Scanning electron microscopy
PAAm substrates were prepared as described above, with the exception that for each substrate, a 10 μL solution was polymerized in between a 5 mm Rain-X treated coverslip and silicon wafers pre-treated with hydrophobic solution (OMS OptoChemicals). After overnight incubation at 37 °C in 50 μg/mL collagen, the hydrogel substrates were fixed in 2% glutaraldehyde for 1 h, rinsed in buffer (10 min × 3 times), then incubated in 1% osmium tetroxide, for 1 h at room temperature in 0.1 M sodium cacodylate buffer at pH 7.2. After rinsing again in buffer (5 min × 3 times), the samples were dehydrated in ethanol, dried using the critical-point technique (AutoSamdri 815, Tousimis), and sputter-coated with approximately 20 nm of gold and palladium (Tousimis) before acquiring images using a Hitachi S-5000 scanning electron microscope.
2.11. Statistical analysis
All images and data are representative of the results of at least three or more independent biological experiments. Data are reported as mean ± SEM unless stated otherwise. Statistical significance was determined using one-way ANOVA followed by the Tukey multiple comparisons test. A Student’s unpaired t test was used if statistical comparisons were made between two sets of data. The significance level was set at p < 0.05.
3. Results and discussion
3.1. Conjugation of 2PCA to ECM proteins
Our approach for site-specifically immobilizing ECM proteins to PAAm surfaces uses a recently reported onestep N-terminal specific modification of native proteins with 2-pyridinecarboxyaldehyde (2PCA) derivatives (Fig. 1d–f).24 This reaction involves the formation of cyclic imidazolidinone product 3 through the addition of an immediately adjacent amide N-H group to imine intermediate 2 (Fig. 1d). Importantly, this cyclization cannot occur when 2PCA imines are formed with lysine side chain amines, confining the reaction to the N-terminal position. In previous work, proteolytic digest and MS/MS analyses have confirmed the N-terminal specificity of the reaction.24 This site-selective reaction is expected to minimize disruption of biologically active motifs and to improve presentation (Fig. 1f). This is in contrast to conventional conjugation chemistry using sulfo-SANPAH crosslinker, which conjugates matrix proteins to PAAm surfaces via side-chain lysines.
Prior to using 2PCA for ECM protein-PAAm conjugation, we first assessed the ability of 2PCA to conjugate to the N termini of commonly used ECM proteins by incubating fluorescein labeled 2PCA (2PCA-fluorescein) with type I collagen, fibronectin and laminin. We verified by SDS-PAGE and fluorescent imaging that 2PCA-fluorescein was indeed covalently conjugated to all three ECM proteins (Fig. 2). To test whether the conjugation occurs through the 2PCA moiety, benzylalkoxyamine was used to quench the aldehyde of 2PCA through oxime formation. ECM proteins showed greatly reduced fluorescence when excess benzylalkoxyamine was added. Incubation with non-functionalized fluorescein also showed minimal fluorescence, suggesting no non-specific binding of fluorescein (Fig. 2b,d,f, indicated by arrows). Together with our earlier work,24 these results support the use of 2PCA to functionalize these ECM proteins specifically at their N-terminal positions.
Fig. 2.
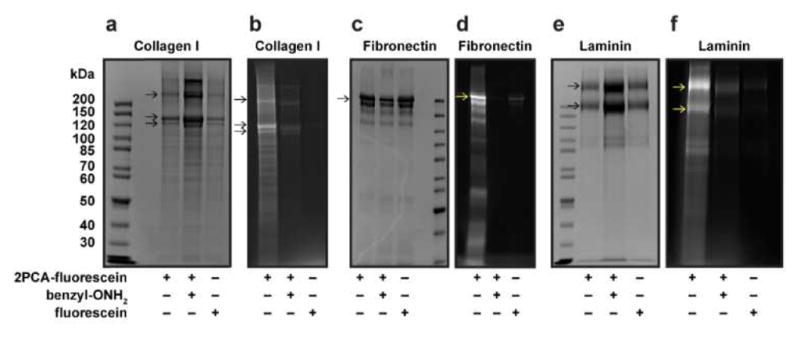
2PCA conjugation of ECM proteins in solution. The N-terminal modification of type I collagen (a, b), fibronectin (c, d) and laminin (e, f) with 2PCA-fluorescein was monitored by SDS-PAGE. The SDS-PAGE gels were fluorescently imaged (b, d, f) to visualize conjugated products and Coomassie stained (a, c, e) to show total protein content. Benzylalkoxyamine (benzyl-ONH2) and fluorescein were used as controls, which show minimal fluorescence. Arrows indicate corresponding ECM protein subunits.
3.2. Mechanical properties of 2PCA-PAAm hydrogels
Having demonstrated that 2PCA can modify the N termini of ECM proteins, we next asked whether we could use this technique for ECM-PAAm immobilization. We decided to directly incorporate 2PCA into the growing PAAm chain during hydrogel polymerization and crosslinking (Fig. 1a) to provide precise control over total 2PCA density, with even surface coverage. We therefore synthesized a 2PCA-acrylamide (4, Fig. 1g) by coupling piperazine-2PCA derivative S5 and acryloyl chloride (Fig. S1). We then added a small amount of the 2PCA-acrylamide monomer into the usual acrylamide/bisacrylamide mixture before polymerization (at 0.1% mole fraction of the acrylamide monomer content). From a solvent screen, we identified that a minimal amount (< 0.1% v/v) of acetone could solubilize 2PCA-acrylamide while keeping the PAAm substrate clear and uniform. Rheological measurements revealed that PAAm hydrogels polymerized with and without 0.1% 2PCA-acrylamide have statistically indistinguishable storage moduli, demonstrating that the presence of 0.1% 2PCA does not alter the bulk stiffness. Additionally, the storage moduli of PAAm hydrogels increased with acrylamide and/or bisacrylamide content whether or not 2PCA was incorporated (Fig. 3 and S2).
Fig. 3.
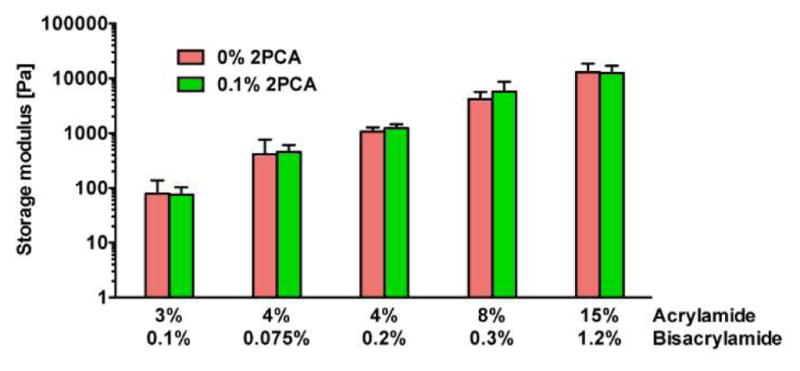
Storage modulus of PAAm substrates with varying acrylamide and bisacrylamide content. For 0.1% 2PCA substrates, 0.1% mole fraction of 2PCA-acrylamide was added relative to acrylamide monomer content. Bars represent mean ± 95% confidence intervals for N ≥ 3 gels per condition. Student’s unpaired t tests of gels with the same acrylamide and bisacrylamide content show no significant difference in storage moduli between PAAm substrates containing 0% and 0.1% 2PCA-acrylamide.
3.3. Cell response to ECM protein density on 2PCA-PAAm substrates
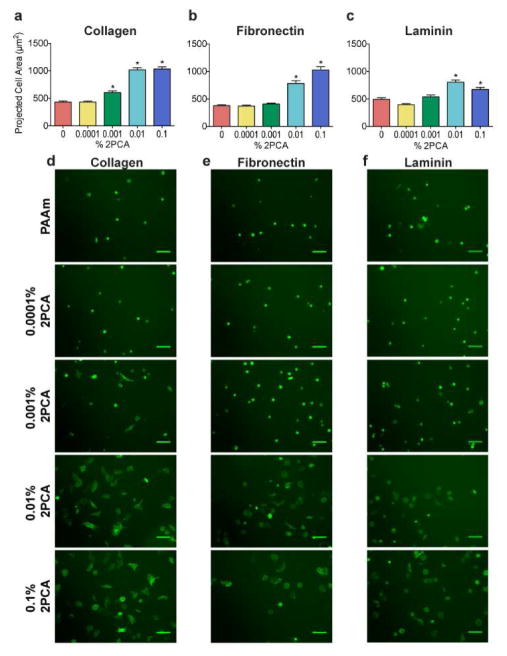
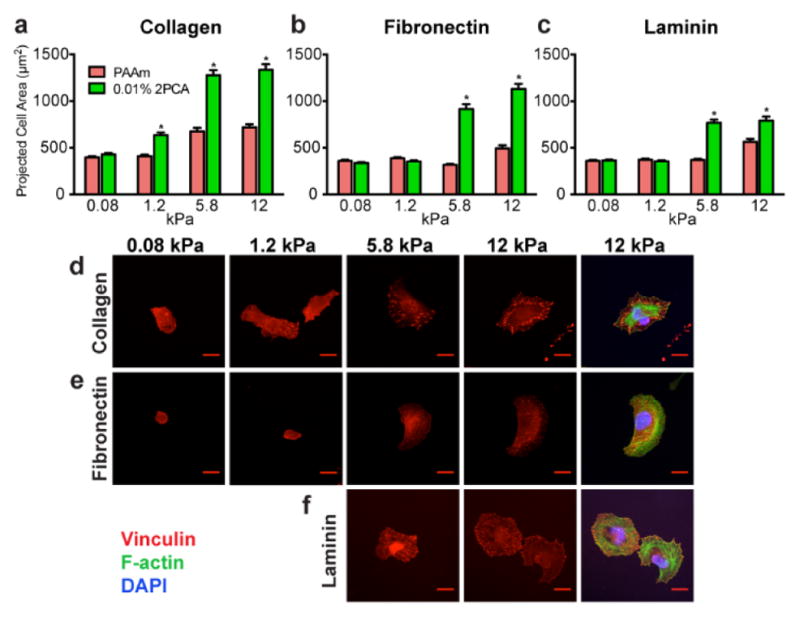
We used U2OS RFP-LifeAct cells as a model system to characterize our substrates, as their adhesion and motility are highly sensitive to substrate stiffness and adhesivity.25 Since the extent of U2OS RFP-LifeAct cell spreading correlates with adhesion protein density, we asked whether we could recapitulate this relationship with 2PCA-based immobilization (Fig. 4). We studied changes in U2OS morphology on collagen-, fibronectin- and laminin-conjugated PAAm substrates as a function of 2PCA-acrylamide content (which is expected to control the amount of immobilized protein) at a constant stiffness of 5.8 kPa. In the absence of 2PCA, cells were poorly adherent and rounded (Fig. 4, red bars and PAAm images), as PAAm substrates do not support passive adsorption of adhesion proteins.8, 10, 11 We next attempted to conjugate the three ECM proteins to 2PCA-PAAm gel surfaces over a range of 2PCA concentrations. For all three proteins, U2OS RFP-LifeAct spread area increased with 2PCA content, which suggests that higher 2PCA content leads to higher density of immobilized ECM proteins (Fig. 4). We did not observe any gross evidence of cell death or other toxicity for U2OS RFP-LifeAct cells cultured on 2PCA-PAAm substrates for periods of > 48 hours. Because U2OS RFP-LifeAct cells spread well on 0.01% 2PCA-PAAm substrates conjugated with all three ECM proteins, we chose this formulation for use in subsequent studies.
Fig. 4.
Dependence of U2OS RFP-LifeAct cell spreading on PAAm substrates containing variable amounts of 2PCA-acrylamide. Quantification (a–c) and representative fields of view (d–f) are shown for U2OS spreading (green = LifeAct) on 5.8 kPa PAAm substrates. Bars represent mean ± SEM; *, p < 0.05 with respect to PAAm; N ≥ 144, 65 and 106 cells per condition for collagen, fibronectin and laminin, respectively. Scale bars = 100 μm.
3.4. Cell response to substrate stiffness
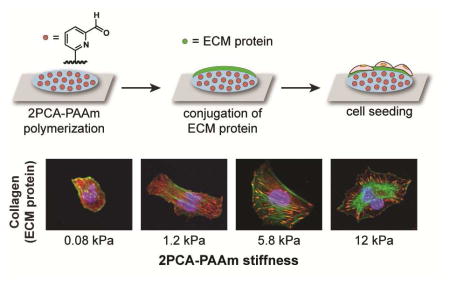
Many cell types, including U2OS cells, increase adhesion and spread area with increases in ECM stiffness.2, 3, 7, 25–28 As an additional proof of principle, we next asked whether cells respond to substrate stiffness when adhesive ligands are end-tethered on the 2PCA-PAAm substrates. We observed greater cell spreading on 2PCA-PAAm substrates relative to non-functionalized PAAm, which further confirms that all three ECM proteins are immobilized on the gel surface through 2PCA. U2OS RFP-LifeAct spreading also dramatically increased with increasing 2PCA-PAAm stiffness, with cells rounded on soft substrates and extensively spread on stiff substrates (Fig. 5a–c). Cells on stiff substrates showed discrete and elongated focal adhesions, which become progressively smaller and more diffuse on softer substrates (Fig. 5d–f), recapitulating the behavior of U2OS cells cultured on alginate substrates.25 Thus, 2PCA-PAAm substrates support the expected rigidity-dependent spreading and adhesion seen on more conventional materials.
Fig. 5.
Influence of substrate rigidity on the spreading of U2OS RFP-LifeAct cells cultured on PAAm gels with 0.01% 2PCA-acrylamide. (a–c) Cell spreading was quantified after U2OS cells were seeded on PAAm substrates conjugated with collagen (a), fibronectin (b) and laminin (c). Bars represent mean ± SEM; *, p < 0.05 with respect to PAAm of corresponding stiffness; N ≥ 122, 90 and 65 cells per condition for collagen, fibronectin and laminin, respectively. (d–f) High-magnification images of representative U2OS cells cultured on 0.01% 2PCA-PAAm substrates conjugated with collagen (d), fibronectin (e) and laminin (f), and subsequently stained for the focal adhesion protein vinculin (red), F-actin (green), and nuclear DNA (blue). Cell adhesion on 0.08 kPa and 1.2 kPa substrates conjugated with laminin were too weak for staining. Scale bars = 20 μm.
3.5. Collagen fiber formation on 2PCA-PAAm substrates
Of the three matrix proteins considered, collagen is of particular interest because of its high abundance in tissue and ability to self-assemble into a remarkable hierarchy of bioactive higher-order structures, including sheets and bundles.29–31 Whereas previous studies have shown that monomeric collagen directly deposited or adsorbed onto rigid surfaces can nucleate the assembly of fibrils and small fibers,32, 33 such structures are in general not observed when collagen is covalently conjugated to PAAm hydrogels using sulfo-SANPAH.34 A possible explanation is that lysine residues required for triple helix formation (e.g., through interchain salt bridges) are blocked in conjugation.35, 36 Moreover, one might anticipate that laterally immobilizing collagen on the hydrogel surface could sterically preclude interchain assembly. In contrast, 2PCA conjugation chemistry, which targets only the N-terminal amine rather than internal lysines and enables the rest of the chain to protrude freely into solution, would circumvent both of these issues. This raises the intriguing possibility that 2PCA-mediated conjugation could support or facilitate collagen fiber assembly.
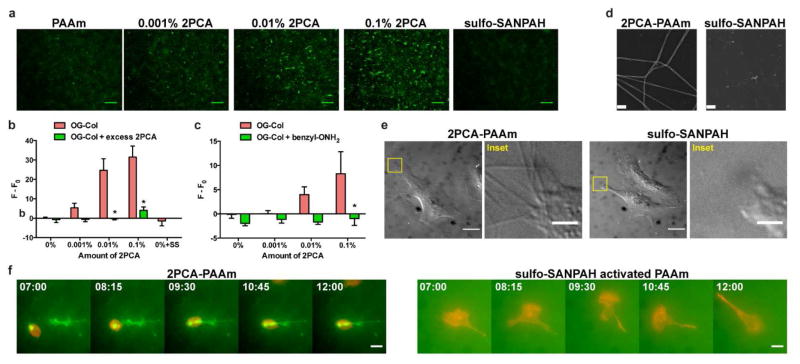
To investigate, we fluorescently labeled type I collagen with Oregon Green NHS ester at low stoichiometry (OG-Col), leaving most amines free for subsequent conjugation (N-terminal amine for 2PCA; N-terminal amine plus internal lysines for sulfo-SANPAH). We then incubated 2PCA-PAAm substrates with OG-Col for covalent conjugation. Fluorescence images revealed the formation of fiber-like structures on the hydrogel surface, with the number and intensity of these structures increasing as the 2PCA-acrylamide content of the substrates increased, suggesting controlled 2PCA-dependent immobilization of collagen (Fig. 6a–c). The presence of more fiber bundles on PAAm substrates with higher 2PCA content is indirect evidence of immobilization of structurally unperturbed collagen on the substrate.
Fig. 6.
Collagen fiber assembly on PAAm substrates. (a) Representative fluorescence images are shown for OG-Col immobilized on 5.8 kPa PAAm gels with various 2PCA-acrylamide content. Scale bars = 100 μm. The reaction was analyzed by fluorescence intensity of OG-Col immobilized on substrates with or without 10 mM piperazine-2PCA (S5) (b) and 40 mM benzylalkoxyamine (benzyl-ONH2) treatment (c). The fluorescence intensity of 0% 2PCA-PAAm without excess 2PCA or benzylalkoxyamine treatment (F0) was subtracted from the fluorescence intensity (F) of the individual samples. Bars represent mean ± SEM; *, p < 0.05 with respect to PAAm of corresponding 2PCA content; N ≥ 6 gels per condition. (d) Representative SEM (d) and DIC (e) images are shown for 50 μg/mL unlabeled collagen immobilized on 5.8 kPa 0.01% 2PCA-PAAm gels or sulfo-SANPAH activated PAAm gels. SEM scale bars = 0.5 μm. DIC scale bars = 20 μm. DIC inset scale bars = 4 μm. (f) Movie stills showing U2OS RFP-LifeAct behavior on OG-Col immobilized PAAm substrates. 50 μg/mL collagen was conjugated to 5.8 kPa 0.0001% 2PCA-PAAm or sulfo-SANPAH activated PAAm before U2OS cell seeding (t = 0). PAAm with low 2PCA content is shown to highlight interactions between the cells and collagen fibers. Fluorescence time-lapse imaging of LifeAct-tag RFP (red) and OG-Col (green) started 7 h after cell seeding. Fluorescence contrast of sulfo-SANPAH activated PAAm substrate is enhanced to show the lack of fiber formation. Time stamp is in hours:minutes. Scale bars = 20 μm.
To verify that collagen was conjugated to the PAAm substrate via the 2PCA moiety rather than noncovalently adsorbed, we conducted two control experiments using (1) excess benzylalkoxyamine to quench the aldehyde group of 2PCA prior to incubation in the collagen solution; and (2) excess 2PCA to block the N-terminus of collagen competitively during incubation in the collagen solution (Fig. 6b,c). Both control surfaces showed reduced fluorescence relative to samples without excess benzylalkoxyamine or 2PCA treatment. The overall fluorescence of control substrates was comparable to that of PAAm substrates without 2PCA. Together, these experiments confirmed that collagen was covalently tethered to 2PCA-PAAm gels through the 2PCA moiety.
Collagen fiber bundles were absent when 2PCA was omitted from the preparation (Fig. 6a, far left) or when OG-Col was conjugated via sulfo-SANPAH to the PAAm surface (Fig. 6a, far right). Fluorescence was not able to quantify the amount of monomeric collagen on the substrates, which is in agreement with previous reports of fluorescent quantification of collagen density on sulfo-SANPAH activated PAAm substrates.19 To verify fiber formation independent of fluorescence, we performed scanning electron microscopy (SEM) and differential interference contrast (DIC) microscopy on unlabeled surfaces, which confirmed the abundance of fibrous structures on the 2PCA-PAAm surfaces. The mean diameter of the collagen fibers is 117 nm (σ = 18 nm) and they span tens of microns (Fig. 6d,e and S3). In contrast, these features were not found on sulfo-SANPAH activated surfaces (Fig. 6d,e).
Notably, despite the lack of observable collagen fibers, U2OS RFP-LifeAct cells were able to adhere, spread and migrate on sulfo-SANPAH activated substrates (Fig. 6f and Supplemental movie 1). This is expected, since several studies have shown that cells can spread and migrate on substrates coated in denatured collagen.32–34 Presumably, however, such surfaces would not easily allow cellular deformation and remodeling of the fibers, which is key to many physiologically critical process including tissue compaction, wound healing and migration.29, 37, 38, 39 Indeed, time-lapse fluorescence imaging of cells cultured on 2PCA-PAAm substrates with fluorescently labeled collagen fibers revealed that cells interact and remodel fibers (Fig. 6f and Supplemental movie 2). Such remodeling events were not observed on surfaces in which collagen was immobilized by sulfo-SANPAH. Thus, 2PCA-PAAm surfaces have the distinct ability to study stiffness-dependent behavior while at the same time presenting the adhesive moiety as a “living” surface that mimics tissue presentation and allows cellular remodeling.
4. Conclusion
We have developed a strategy to end-tether full-length ECM proteins to hydrogel surfaces using 2PCA-mediated N-terminal conjugation. This approach recapitulates widely observed relationships between adhesive ligand density, ECM stiffness, and cell spreading, while uniquely allowing the assembly of collagen fibers that cells can remodel as they migrate. Importantly, this conjugation chemistry eliminates multiple covalent anchoring points on the ECM proteins, thereby fully decoupling the protein lateral tether density from bulk ECM stiffness – a major concern in the preparation of such substrates. In that context, it is notable that we observe strong stiffness-dependent behavior on 2PCA-conjugated PAAm substrates, despite the fact that the protein is anchored at a single point. This is consistent with the notion that ECM stiffness regulates cell adhesion and spreading independently of tether density.19 Irrespective of this interpretation, we expect that this strategy should provide a useful alternative to traditional conjugation chemistries and may be used to affix a broad variety of proteins and peptides to hydrogel surfaces for both basic and translational investigations.
Supplementary Material
Acknowledgments
SK acknowledges the support of the DOD (W911NF-09-1-0507), the NIH (R21EB016359, R21CA174573), and the NSF (CMMI 1105539). MBF acknowledges the NSF (CHE 1413666) for research support. JPL and JIM were supported by the Berkeley Chemical Biology Graduate Program (NIH Training Grant 1 T32 GMO66698). JPL was supported by the Croucher Foundation Scholarship. EK was supported by the Howard Hughes Medical Institute International Student Fellowship. The authors acknowledge D.V. Schaffer for reagent and equipment sharing, as well as the CIRM/QB3 Stem Cell Shared Facility for confocal microscopy access.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 2.Engler A, Bacakova L, Newman C, Hategan A, Griffin M, Discher D. Substrate compliance versus ligand density in cell on gel responses. Biophys J. 2004;86:617–628. doi: 10.1016/S0006-3495(04)74140-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ulrich TA, de Juan Pardo EM, Kumar S. The mechanical rigidity of the extracellular matrix regulates the structure, motility, and proliferation of glioma cells. Cancer Res. 2009;69:4167–4174. doi: 10.1158/0008-5472.CAN-08-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keung AJ, de Juan-Pardo EM, Schaffer DV, Kumar S. Rho GTPases mediate the mechanosensitive lineage commitment of neural stem cells. Stem Cells. 2011;29:1886–1897. doi: 10.1002/stem.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keung AJ, Dong M, Schaffer DV, Kumar S. Panneuronal maturation but not neuronal subtype differentiation of adult neural stem cells is mechanosensitive. Sci Rep. 2013;3:1817. doi: 10.1038/srep01817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahearne M. Introduction to cell-hydrogel mechanosensing. Interface Focus. 2014;4:2. doi: 10.1098/rsfs.2013.0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pelham RJ, Jr, Wang Y. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci U S A. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tse JR, Engler AJ. Preparation of hydrogel substrates with tunable mechanical properties. Curr Protoc Cell Biol. 2010;47:10.16.1–10.16.16. doi: 10.1002/0471143030.cb1016s47. [DOI] [PubMed] [Google Scholar]
- 9.Levental I, Georges PC, Janmey PA. Soft biological materials and their impact on cell function. Soft Matter. 2007;3:299–306. doi: 10.1039/b610522j. [DOI] [PubMed] [Google Scholar]
- 10.Georges PC, Janmey PA. Cell type-specific response to growth on soft materials. J Appl Physiol (1985) 2005;98:1547–1553. doi: 10.1152/japplphysiol.01121.2004. [DOI] [PubMed] [Google Scholar]
- 11.Gribova V, Crouzier T, Picart C. A material’s point of view on recent developments of polymeric biomaterials: control of mechanical and biochemical properties. J Mater Chem. 2011;21:14354–14366. doi: 10.1039/C1JM11372K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kandow CE, Georges PC, Janmey PA, Beningo KA. Polyacrylamide hydrogels for cell mechanics: steps toward optimization and alternative uses. Methods Cell Biol. 2007;83:29–46. doi: 10.1016/S0091-679X(07)83002-0. [DOI] [PubMed] [Google Scholar]
- 13.Pless DD, Lee YC, Roseman S, Schnaar RL. Specific Cell-Adhesion to Immobilized Glycoproteins Demonstrated Using New Reagents for Protein and Glycoprotein Immobilization. J Biol Chem. 1983;258:2340–2349. [PubMed] [Google Scholar]
- 14.Damljanovic V, Lagerholm BC, Jacobson K. Bulk and micropatterned conjugation of extracellular matrix proteins to characterized polyacrylamide substrates for cell mechanotransduction assays. Biotechniques. 2005;39:847–851. doi: 10.2144/000112026. [DOI] [PubMed] [Google Scholar]
- 15.Grevesse T, Versaevel M, Circelli G, Desprez S, Gabriele S. A simple route to functionalize polyacrylamide hydrogels for the independent tuning of mechanotransduction cues. Lab Chip. 2013;13:777–780. doi: 10.1039/c2lc41168g. [DOI] [PubMed] [Google Scholar]
- 16.Hynd MR, Frampton JP, Dowell-Mesfin N, Turner JN, Shain W. Directed cell growth on protein-functionalized hydrogel surfaces. J Neurosci Methods. 2007;162:255–263. doi: 10.1016/j.jneumeth.2007.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia AJ, Vega MD, Boettiger D. Modulation of cell proliferation and differentiation through substrate-dependent changes in fibronectin conformation. Mol Biol Cell. 1999;10:785–798. doi: 10.1091/mbc.10.3.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trappmann B, Gautrot JE, Connelly JT, Strange DG, Li Y, Oyen ML, Cohen Stuart MA, Boehm H, Li B, Vogel V, Spatz JP, Watt FM, Huck WT. Extracellular-matrix tethering regulates stem-cell fate. Nat Mater. 2012;11:642–649. doi: 10.1038/nmat3339. [DOI] [PubMed] [Google Scholar]
- 19.Wen JH, Vincent LG, Fuhrmann A, Choi YS, Hribar KC, Taylor-Weiner H, Chen S, Engler AJ. Interplay of matrix stiffness and protein tethering in stem cell differentiation. Nat Mater. 2014;13:979–987. doi: 10.1038/nmat4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar S. Cellular mechanotransduction: stiffness does matter. Nat Mater. 2014;13:918–920. doi: 10.1038/nmat4094. [DOI] [PubMed] [Google Scholar]
- 21.Chen YX, Triola G, Waldmann H. Bioorthogonal chemistry for site-specific labeling and surface immobilization of proteins. Acc Chem Res. 2011;44:762–773. doi: 10.1021/ar200046h. [DOI] [PubMed] [Google Scholar]
- 22.Wong LS, Khan F, Micklefield J. Selective covalent protein immobilization: strategies and applications. Chem Rev. 2009;109:4025–4053. doi: 10.1021/cr8004668. [DOI] [PubMed] [Google Scholar]
- 23.Bhagawati M, Kumar S. Biofunctionalization of hydrogels for engineering the cellular microenvironment. In: Karp J, Zhao W, editors. Micro- and Nanoengineering of the Cell Surface. William Andrew; Oxford: 2014. pp. 315–348. [Google Scholar]
- 24.MacDonald JI, Munch HK, Moore T, Francis MB. One-step site-specific modification of native proteins with 2-pyridinecarboxyaldehydes. Nat Chem Biol. 2015;11:326–331. doi: 10.1038/nchembio.1792. [DOI] [PubMed] [Google Scholar]
- 25.Chaudhuri O, Gu L, Darnell M, Klumpers D, Bencherif SA, Weaver JC, Huebsch N, Mooney DJ. Substrate stress relaxation regulates cell spreading. Nat Commun. 2015;6:6364. doi: 10.1038/ncomms7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pathak A, Kumar S. Independent regulation of tumor cell migration by matrix stiffness and confinement. Proc Natl Acad Sci U S A. 2012;109:10334–10339. doi: 10.1073/pnas.1118073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim Y, Kumar S. CD44-mediated adhesion to hyaluronic acid contributes to mechanosensing and invasive motility. Mol Cancer Res. 2014;12:1416–1429. doi: 10.1158/1541-7786.MCR-13-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rape AD, Zibinsky M, Murthy N, Kumar S. A synthetic hydrogel for the high-throughput study of cell-ECM interactions. Nat Commun. 2015;6:8129. doi: 10.1038/ncomms9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ulrich TA, Jain A, Tanner K, MacKay JL, Kumar S. Probing cellular mechanobiology in three-dimensional culture with collagen-agarose matrices. Biomaterials. 2010;31:1875–1884. doi: 10.1016/j.biomaterials.2009.10.047. [DOI] [PubMed] [Google Scholar]
- 30.Pathak A, Kumar S. Transforming potential and matrix stiffness co-regulate confinement sensitivity of tumor cell migration. Integr Biol. 2013;5:1067–1075. doi: 10.1039/c3ib40017d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Badylak SF. Regenerative medicine and developmental biology: the role of the extracellular matrix. Anat Rec. 2005;287B:36–41. doi: 10.1002/ar.b.20081. [DOI] [PubMed] [Google Scholar]
- 32.Elliott JT, Tona A, Woodward JT, Jones PL, Plant AL. Thin films of collagen affect smooth muscle cell morphology. Langmuir. 2003;19:1506–1514. [Google Scholar]
- 33.McDaniel DP, Shaw GA, Elliott JT, Bhadriraju K, Meuse C, Chung KH, Plant AL. The stiffness of collagen fibrils influences vascular smooth muscle cell phenotype. Biophys J. 2007;92:1759–1769. doi: 10.1529/biophysj.106.089003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dingal PCDP, Bradshaw AM, Cho S, Raab M, Buxboim A, Swift J, Discher DE. Fractal heterogeneity in minimal matrix models of scars modulates stiff-niche stem-cell responses via nuclear exit of a mechanorepressor. Nat Mater. 2015;14:951–960. doi: 10.1038/nmat4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Persikov AV, Ramshaw JAM, Kirkpatrick A, Brodsky B. Electrostatic interactions involving lysine make major contributions to collagen triple-helix stability. Biochemistry. 2005;44:1414–1422. doi: 10.1021/bi048216r. [DOI] [PubMed] [Google Scholar]
- 36.Gurry T, Nerenberg PS, Stultz CM. The contribution of interchain salt bridges to triple-helical stability in collagen. Biophys J. 2010;98:2634–2643. doi: 10.1016/j.bpj.2010.01.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ulrich TA, Lee TG, Shon HK, Moon DW, Kumar S. Microscale mechanisms of agarose-induced disruption of collagen remodeling. Biomaterials. 2011;32:5633–5642. doi: 10.1016/j.biomaterials.2011.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu P, Takai K, Weaver VM, Werb Z. Extracellular matrix degradation and remodeling in development and disease. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a005058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim DH, Provenzano PP, Smith CL, Levchenko A. Matrix nanotopography as a regulator of cell function. J Cell Biol. 2012;197:351–360. doi: 10.1083/jcb.201108062. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.