Abstract
Parishin is a phenolic glucoside isolated from Gastrodia elata, which is an important traditional Chinese medicine; this glucoside significantly extended the replicative lifespan of K6001 yeast at 3, 10, and 30 μM. To clarify its mechanism of action, assessment of oxidative stress resistance, superoxide dismutase (SOD) activity, malondialdehyde (MDA), and reactive oxygen species (ROS) assays, replicative lifespans of sod1, sod2, uth1, and skn7 yeast mutants, and real-time quantitative PCR (RT-PCR) analysis were conducted. The significant increase of cell survival rate in oxidative stress condition was observed in parishin-treated groups. Silent information regulator 2 (Sir2) gene expression and SOD activity were significantly increased after treating parishin in normal condition. Meanwhile, the levels of ROS and MDA in yeast were significantly decreased. The replicative lifespans of sod1, sod2, uth1, and skn7 mutants of K6001 yeast were not affected by parishin. We also found that parishin could decrease the gene expression of TORC1, ribosomal protein S26A (RPS26A), and ribosomal protein L9A (RPL9A) in the target of rapamycin (TOR) signaling pathway. Gene expression levels of RPS26A and RPL9A in uth1, as well as in uth1, sir2 double mutants, were significantly lower than those of the control group. Besides, TORC1 gene expression in uth1 mutant of K6001 yeast was inhibited significantly. These results suggested that parishin exhibited antiaging effects via regulation of Sir2/Uth1/TOR signaling pathway.
1. Introduction
The proportion of the world's population over 60 years old will be 22% in 2050 [1]. Aging-related diseases, such as Alzheimer's and Parkinson's diseases and diabetes, are becoming a severe threat to human health in aging society. Although many commercial available drugs are used to treat these diseases [2], they can only alleviate clinical symptoms and cannot cure the diseases. Therefore, a novel therapeutic strategy, such as antiaging, will be a promising technique to delay and prevent the occurrence of aging-related diseases.
Sir2 proteins are a family of proteins influencing the physiological responses and affecting the treatment of aging-related diseases. Increase of SIR2 gene expression and activity can extend the life span of various model organisms [3–5]. More importantly, it can regulate oxidative stress by binding and deacetylation of FOXO transcription factors, which play a central role in regulating stress response [6]. Recently, sirtuin family has been considered a drug target for aging, metabolism, and aging-related diseases [7].
Most eukaryotes express two intracellular SODs, a Mn containing SOD2 in the mitochondrial matrix and a highly abundant Cu/Zn SOD1 that is largely cytosolic but is also found in the mitochondrial intermembrane space. SOD1 has function of protecting cells, regulating cell viability, and metabolism [8]. SOD2 takes an important role for antioxidative stress and scavenger of free radical.
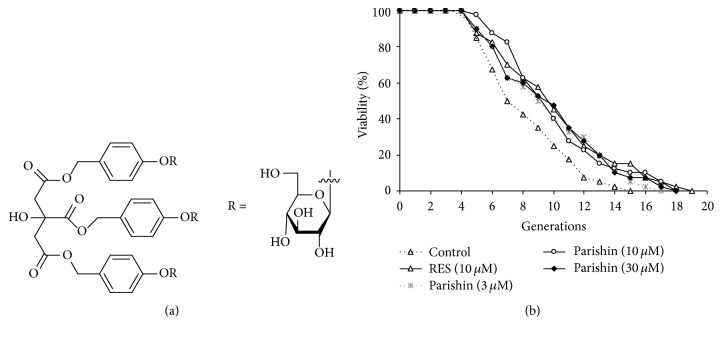
Gastrodia elata (Tian Ma in Chinese) is an important traditional Chinese medicine. This herb has anticonvulsant, analgesia, calmness, hypnosis, nootropic, and anti-brain-aging functions for the central nervous system in traditional therapy of Chinese medicine [9]. In addition, it can promote the energy metabolism of myocardial cells; G. elata also has anti-inflammation effect and increases immunity [10, 11]. Many active ingredients, such as gastrodin, 4-hydroxybenzyl alcohol, 4-hydroxybenzaldehyde, benzyl alcohol, 4-hydroxy-3-methoxybenzaldehyde, 4-hydroxy-3-methoxybenzyl alcohol, parishin, and parishin B and parishin C, have been isolated from G. elata [12–16]. Among these compounds, gastrodin is a major active compound and has been developed to be a commercially available drug that is mainly used to treat neurasthenia-induced headache [17]. Parishin (Figure 1(a)), one of the major compositions of G. elata, can alleviate asthma [15]. However, its antiaging effects and mechanism of action have not been reported yet.
Figure 1.
Chemical structure of parishin (a) and antiaging effects of parishin (b). For replicative lifespan assay, the yeast cells incubated in galactose medium were spread on glucose medium plates containing different concentrations of parishin. The daughter cells of 40 microcolonies in each plate were counted randomly. The assay was repeated at least thrice. The average lifespan of untreated K6001 was 7.38 ± 0.44 generations; resveratrol (RES) at 10 μM, 9.23 ± 0.59∗; parishin at 3 μM, 8.83 ± 0.56∗; parishin at 10 μM, 9.20 ± 0.52∗∗; and parishin at 30 μM, 8.98 ± 0.58∗. ∗ and ∗∗ indicate significant difference relative to the control (p < 0.05, p < 0.01).
Yeast is a well-known bioassay model in antiaging research [18]. Recently, multiple conserved longevity pathways have been discovered in budding yeast [19]. Since it has the characters of short generation time, genetic tractability, and low costs, the budding yeast has become a premier model organism for aging research [20]. Replicative aging and chronological aging are used to assess longevity of yeast. The standard replicative lifespan assay needs to use micromanipulator to remove the daughter cells produced by one mother cell for every two hours. It is time consuming and labor intensive. Thus, it has become a rate-limited step on the progress of aging research. In 2004, Jarolim et al. established the replicative lifespan assay with K6001 yeast strain to improve the lifespan assay [18]. Recently, microfluidic technology for yeast replicative lifespan has also developed to address this problem [20].
In our previous studies, antiaging compounds, such as ganodermasides A–D, phloridzin, and nolinospiroside F, were isolated from Ganoderma lucidum, apple branches, and Ophiopogon japonicus, respectively, under a K6001 yeast bioassay system [21–24]. In the present study, parishin was isolated as a major antiaging composition from G. elata according to the same system. We report the isolation, structure elucidation, biological activity, and mechanism of action of parishin.
2. Materials and Methods
2.1. Isolation and Structure Elucidation of Parishin
The rhizomes of G. elata (dry weight: 200 g) were bought from Chengdu, Sichuan Province, China, and the identification of G. elata was confirmed. A voucher specimen (number 20110521) was kept at College of Pharmaceutical Sciences, Zhejiang University. They were ground and extracted with MeOH. The supernatant was separated via filtration and concentrated to obtain the methanol extract. The extract was partitioned between EtOAc and H2O. The H2O layer was concentrated to give 26 g of dried sample which was chromatographed on ODS (Cosmosil 75 C18-OPN, Nacalai Tesque, Ohtsu, Japan) and eluted with MeOH/H2O (20 : 80, 25 : 75, 30 : 70, 35 : 65, 40 : 60, 60 : 40, and 80 : 20) to afford 43 fractions. The active sample (1.4 g), which was eluted with MeOH/H2O (25 : 75, 30 : 70, and 35 : 65), was separated on silica gel (200–300 mesh, Yantai Chemical Industry Research Institute, Yantai, China) and eluted with CHCl3/MeOH (9 : 1, 8 : 2, 6 : 4, 5 : 5, 4 : 6, 3 : 7, 2 : 8, and 0 : 10) to afford 71 fractions. A portion (150 mg) of the active sample (921.0 mg), eluted with CHCl3/MeOH (6 : 4, 5 : 5, 4 : 6), was subjected to HPLC (Develosil ODS-UG-5 (ϕ 20/250 mm), Nomura Chemical, flow rate: 8 mL/min, MeOH/H2O (28 : 72)) to yield a pure active compound (100.0 mg, t R = 36 min). The chemical structure of the compound was determined to be parishin by comparing 1H NMR, MS, and optical rotation data with those reported [14, 16]. 1H NMR (500 MHz, CD3OD): δ 2.78 (d, 2H, J = 15.0 Hz), 2.94 (d, 2H, J = 15.5 Hz), 3.40–3.52 (m, 12H), 3.70 (dd, 3H, J = 5.0, 12.0 Hz), 3.87 (dd, 3H, J = 1.5, 12.0 Hz), 4.90–5.01 (m, 9H), 7.04 (d, 2H, J = 8.5 Hz), 7.07 (dd, 4H, J = 1.5, 8.5 Hz), 7.16 (d, 2H, J = 8.5 Hz), 7.24 (dd, 4H, J = 2.5, 8.5 Hz); MS m/z 1019 (M + Na)+; [α]D 25 = −73.0 (c 1.0, MeOH).
2.2. Yeast Strains, Media, and Lifespan Assay
The yeast strains used in present study were described in Table 1. The lifespan assay method and medium were similar to those previously reported [18]. To get enough yeast to do experiment, briefly, K6001 yeast strain was resuscitated in 5 mL of galactose medium and incubated in a shaking incubator at 160 rpm for 24–28 h at 28°C. About 1 mL of yeast culture was centrifuged for 3 min at 1,500 rpm. The yeast pellet was washed three times and diluted with phosphate buffer solution (PBS). After counting with a hemocytometer, approximately 4,000 cells were plated on glucose medium agar plates containing resveratrol (positive control, 10 μM) or parishin (0, 3, 10, and 30 μM). The plates were incubated at 28°C for 2 days, and 40 microcolonies formed on the plates were randomly observed under a microscope. The daughter cells produced by the mother cell were counted. The bioassay method of replicative lifespan of sod1, sod2, uth1, and skn7 mutants with a K6001 background was identical to that of the K6001 strain.
Table 1.
Strains used in this study.
| Strains | Genotype | Source or reference |
|---|---|---|
| K6001 | MATa, ade2-1, trp1-1, can1-100, leu2-3, 112, his3-11, 15, GAL, psi+, ho::HO::CDC6 (at HO), cdc6::hisG, ura3::URA3 GAL-ubiR-CDC6 (at URA3) | [18, 35] |
|
| ||
| Δuth1 of K6001 | Replace the UTH1 gene in K6001 with kanamycin gene | Constructed by Professor Akira Matsuura |
|
| ||
| Δskn7 of K6001 | Replace the SKN7 gene in K6001 with kanamycin gene | Constructed by Professor Akira Matsuura |
|
| ||
| Δsod1 of K6001 | Replace the SOD1 gene in K6001 with kanamycin gene | Constructed by Professor Akira Matsuura |
|
| ||
| Δsod2 of K6001 | Replace the SOD2 gene in K6001 with kanamycin gene | Constructed by Professor Akira Matsuura |
|
| ||
| BY4741 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0 | [36] |
|
| ||
| Δuth1 of BY4741 | Replace the UTH1 gene in BY4741 with kanamycin gene | Constructed by Professor Akira Matsuura |
|
| ||
| Δuth1, sir2 of BY4741 | Replace the UTH1 gene and SIR2 gene in BY4741 with kanamycin gene | Constructed by Professor Akira Matsuura |
2.3. Antioxidative Stress Assay
BY4741 yeast was treated with resveratrol (10 μM) as a positive control or parishin (0, 3, 10, and 30 μM) at 28°C for 48 h. Subsequently, about 0.1 OD of yeast cultures in each group was spotted on agar plates containing 9 mM of H2O2. The growth of yeast on the plate was observed and photographed after incubation at 28°C for 3 days.
To validate the accuracy of experiment, we used another method to examine the antioxidative stress of parishin again. BY4741 yeast was incubated for 24 h after it was treated with 0, 3, 10, and 30 μM parishin or 10 μM resveratrol; then it was treated with H2O2 at doses of 0 or 180 mM for 3 h. Approximately 0.1 OD of yeast in each group was washed with cold PBS buffer three times and treated in 15% ethanol for 20 min. The treated yeast cells were incubated with 10 μg/mL propidium iodide at 37°C for 20 min in dark after washing with PBS buffer. Fluorescence microscope (Leica DMI 3000 B, Wetzlar, Germany) was used to observe the change of yeast cells under oxidative stress condition using an excitation wavelength of 535 nm and an emission wavelength of 615 nm. Approximately 100 cells were used to calculate the survival rate.
2.4. Determination of SOD Enzyme Activity, MDA, and ROS Level
BY4741 yeast cells were cultured in glucose medium after adding 0, 3, 10, or 30 μM of parishin for 24 or 48 h. The SOD and MDA assays were performed as in the previous study [24]. After counting the yeast with a hemocytometer, approximately 1 × 109 cells were washed thrice with PBS and resuspended in 1 mL of PBS. The cells were ultrasonicated (1 min for each time) for five times, followed by freeze and thaw (5 min in liquid nitrogen and subsequently 2 min in water bath at 37°C) and repeated sonication for five times. The cell lysates were centrifuged at 12,000 rpm at 4°C for 15 min, and the supernatant was removed to test the SOD activity and MDA level using SOD and MDA assay kits (Nanjing Jiancheng Bioengineering Institute, Nanjing, China), according to the manufacturer's instructions.
ROS assay was carried out using the method in the previous study [23]. BY4741 yeast cells were incubated in glucose medium with parishin (0, 3, 10, and 30 μM) in a shaker incubator at 28°C for 23 h. Subsequently, DCFH-DA was added to 1 mL of the cells to get the final concentration of 40 μM, and the cells were incubated in a shaker at 28°C in dark for 1 h. The cells were then washed thrice with PBS quickly, and the DCF fluorescence magnitude of 1 × 107 cells was detected by a fluorescent plate reader using excitation and emission wavelengths of 488 and 525 nm, respectively.
2.5. RT-PCR Analysis
BY4741, uth1, and uth1, sir2 double mutants with a BY4741 background were treated with control or different concentrations of parishin and cultured in glucose medium in an incubator at 28°C overnight with shaking. Wild type and uth1 mutant of K6001 were cultured in galactose medium in a shaker at 28°C overnight. Cells were collected, and RNA was extracted via the hot-phenol method. RNA was purified with a RNApure tissue kit (Beijing Cowin Biotech Company, Beijing, China), and reverse transcription was performed using a HiFi-MMLV cDNA kit (Beijing Cowin Biotech Company, Beijing, China) with 5 μg of total RNA. RT-PCR was conducted similarly to that of the previous study [23]. CFX96-Touch (Bio-rad, Hercules, USA) and SYBR Premix EX Taq (Takara, Otsu, Japan) were used. The thermal cycling parameters for RPS26A and RPL9A are as follows: 40 cycles, 95°C for 15 s and 60°C for 35 s; for SIR2, the parameters are as follows: 40 cycles, 94°C for 15 s, 60°C for 25 s, and 72°C for 20 s; for TORC1, the parameters are as follows: 40 cycles, 95°C for 15 s, 59°C for 25 s, and 72°C for 20 s. The primers used for RT-PCR are as follows: for RPS26A, sense 5′-TCA GAA ACA TTG TTG AAG CCG C-3′ and antisense 5′-ACA ATT CTG GCG TGA ATA GCA C-3′; for RPL9A, sense 5′-ATG GTG CCA AAT TCA TTG AAG TC-3′ and antisense 5′-AGT TAC CTG ACA AGA CAA TTT CG-3′; for SIR2, sense 5′-CGT TCC CCA AGT CCT GAT TA-3′ and antisense 5′-CCA CAT TTT TGG GCT ACC AT-3′; for TORC1, sense 5′-TTG GTA CAA GGC ATG GCA TA-3′ and antisense 5′-TAC CGT CAA TCC GCA CAT TA-3′; and for TUB1, sense 5′-CCA AGG GCT ATT TAC GTG GA-3′ and antisense 5′-GGT GTA ATG GCC TCT TGC AT-3′. Relative gene expression data were analyzed using 2−ΔΔCt method. The amounts of RPS26A, RPL9A, TORC1, and SIR2 mRNA were normalized to that of TUB1.
2.6. Statistical Analysis
Significant differences among groups in all experiments were determined by analysis of variance, followed by two-tailed multiple t-tests with Bonferroni correction using SPSS biostatistics software. A p value of less than 0.05 was considered statistically significant.
3. Results and Discussion
3.1. Parishin Extends the Replicative Lifespan of Yeast
K6001, a mutant strain of yeast with W303 background expresses CDC6, an essential gene for growth, under control of the mother-specific HO promoter and a galactose-dependent promoter GAL1-10. When K6001 cells are cultured in galactose, GAL1-10::CDC6 is expressed both in mother and in daughter cells; however, when the expression of GAL1-10::CDC6 gene is repressed by glucose, only the mother cell-specific expression of HO::CDC6 remains to support growth [18]. Due to its specificity above, the replicative lifespan assay of K6001 is much more efficient and easier to manipulate than that of the other yeasts. It has been used to evaluate antiaging activity of compounds. By employing this bioassay system, several antiaging substances such as ganodermasides A–D, phloridzin, and nolinospiroside F were isolated. In the present study, this bioassay system was used to guide the isolation of an antiaging substance from G. elata. The changes on the replicative lifespan of K6001 yeast after parishin treatment at various doses are displayed in Figure 1(b). Parishin significantly extended the replicative lifespan of K6001 at 3, 10, and 30 μM (p < 0.05, p < 0.01, and p < 0.05, resp.). These results suggested that parishin had antiaging effects. In addition, we also performed kinetics of growth assay of yeast under the influence of parishin. Significant changes were not observed in resveratrol treatment group and parishin treatment group (see Supplementary Figure 1 in Supplementary Material available online at http://dx.doi.org/10.1155/2016/4074690). At this point, it is possible that the kinetics of growth assay is not suitable to assess the replicative lifespan of yeast.
3.2. Parishin Enhances Gene Expression of SIR2
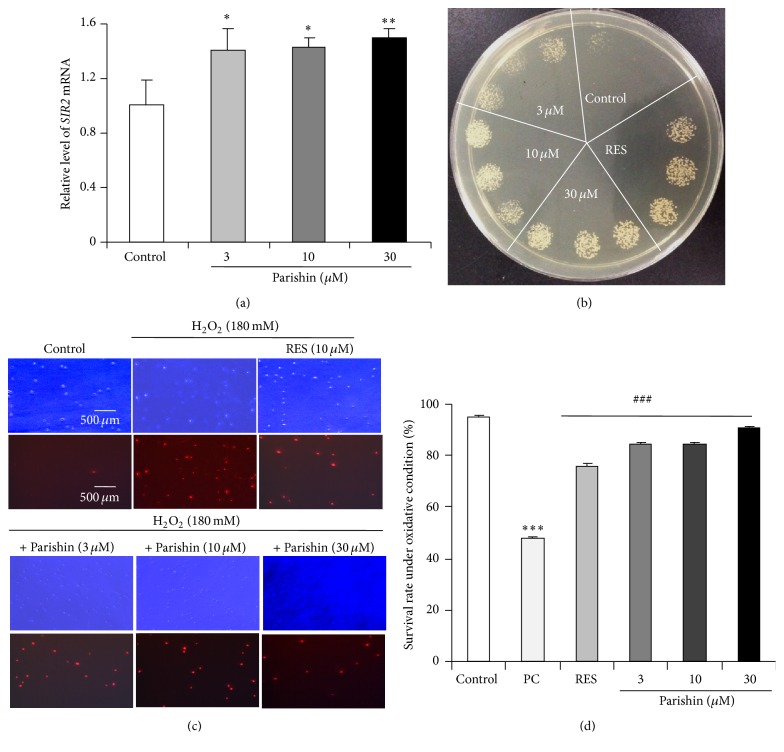
SIR2 gene is one of the most important longevity genes. The increase of SIR2 gene expression or enzyme activity could extend the yeast lifespan [3]. Therefore, we examined the gene expression of SIR2 in yeast treated with parishin. As expected, the gene expression levels of SIR2 in parishin treatment groups were significantly increased (Figure 2(a); p < 0.05, p < 0.05, and p < 0.01). This result suggested that SIR2 gene was involved in the antiaging effects of parishin.
Figure 2.
Effects of parishin on the gene expression of SIR2 (a) in normal condition and growth of BY4741 yeast under oxidative stress conditions (b, c, and d). The change on SIR2 gene expression in yeast after treatment with parishin at doses of 0, 3, 10, and 30 μM (a). Amount of SIR2 mRNA was normalized to that of TUB1. The effects of parishin on yeast growth under oxidative stress induced by H2O2 (b). BY4741 yeast was incubated for 48 h after it was treated with 0, 3, 10, and 30 μM parishin or 10 μM resveratrol; then about 5 μL of the same concentration of yeast was dropped onto glucose medium agar plates containing 9 mM H2O2. The yeast was incubated for 3 d at 28°C and photographed. The micrographs (c) and survival rate (d) of yeast under oxidative stress condition. BY4741 yeast was incubated for 24 h after it was treated with 0, 3, 10, and 30 μM parishin or 10 μM resveratrol; then it was treated by H2O2 at doses of 0 or 180 mM for 3 h. Approximately 0.1 OD of yeast in each group was washed with cold PBS for three times and treated in 15% ethanol for 20 min. The treated yeast cells were incubated with propidium iodide at 10 μg/mL for 20 min after washing with PBS. Fluorescence microscope was used to observe the change of yeast cells under oxidative stress condition using an excitation wavelength of 535 nm and an emission wavelength of 615 nm. Approximately 100 cells were used to calculate the survival rate. Each experiment was performed at least three times. PC represents positive control treated with 180 mM H2O2 for 3 h. ∗, ∗∗, and ∗∗∗ indicate significant difference relative to the corresponding control (p < 0.05, p < 0.01, and p < 0.001, resp.). ### represents significant difference relative to the positive control (p < 0.001).
3.3. Parishin Improves the Survival Rate of Yeast under Oxidative Stress Conditions
Oxidative stress is one of the most important factors for aging, and oxidative free radicals do harm to cellular constituents, such as DNA, proteins, carbohydrates, and lipids [25]. Therefore, we focused on this point to measure the parameters related to antioxidation in yeast. As shown in Figure 2(b), parishin significantly increased the number of colonies of yeast. Moreover, the viability of yeast after treatment with parishin at doses of 3, 10, and 30 μM was notably increased compared with positive control group under oxidative stress condition (Figures 2(c) and 2(d); p < 0.001, p < 0.001 and p < 0.001, resp.). We also used agar plates to examine the survival rate of yeast under oxidative stress, and the same results were obtained (Supplementary Figure 2). These results suggested that antioxidation played an important role in the antiaging effect of parishin.
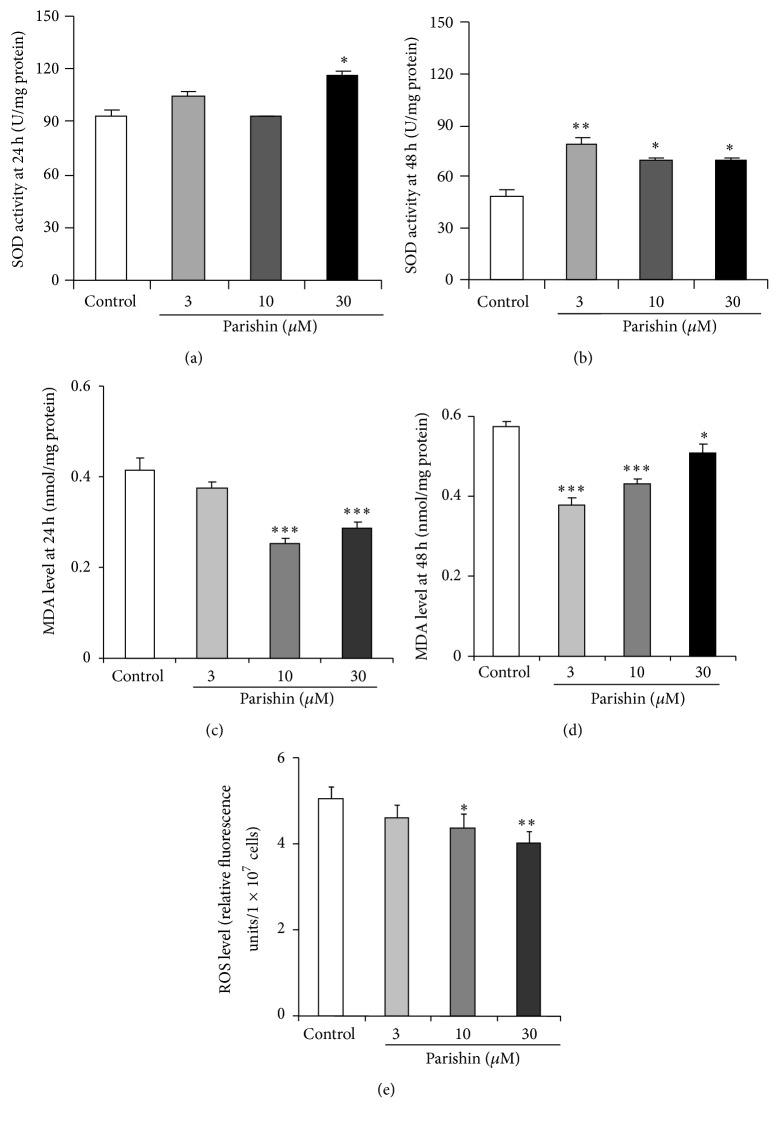
3.4. Parishin Increases SOD Enzyme Activity of Yeast and Decreases ROS and MDA Levels
SOD is an important enzyme that participates in free radical scavenging. Thus, we measured the SOD activity in yeast after parishin treatment. The SOD enzyme activity of yeast was only significantly increased in yeast after treatment with 30 μM of parishin for 24 h (Figure 3(a); p < 0.05). However, the significant increases of SOD activity in yeast were observed after administrating parishin at 3, 10, and 30 μM for 48 h (Figure 3(b); p < 0.01, p < 0.05, and p < 0.05, resp.).
Figure 3.
Effects of parishin on SOD enzyme activity (a, b), MDA (c, d), and ROS levels (e). The change on SOD enzyme activity after treating parishin at various doses for 24 h (a) or 48 h (b), respectively. BY4741 yeast cells were disintegrated by ultrasonication and freeze-thawing for five times, followed by repeated ultrasonication for five times. Cell lysate was centrifuged and the supernatant was removed for measurement of SOD activity using a SOD assay kit. Effect of parishin on MDA in yeast after parishin treatment at various doses for 24 h (c) or 48 h (d). BY4741 yeast cells were cultured for 24 h or 48 h and disintegrated as described in SOD assay, and changes in MDA level were measured with an MDA assay kit. Effect of parishin on ROS level of yeast (e). BY4741 yeast cells were incubated with parishin for 23 h. Subsequently, DCFH-DA was added into culture medium to a final concentration of 40 μM and incubated for 1 h. The intensity of DCF of yeast was detected with a fluorescence plate reader. ∗, ∗∗, and ∗∗∗ indicate significant difference from corresponding control (p < 0.05, p < 0.01, and p < 0.001).
ROS are byproducts of oxidative metabolism and important cause of aging [25]. Hence, we tested the change on ROS accumulation in yeast after parishin treatment. The ROS levels in yeast were decreased significantly at 10 and 30 μM of parishin treatment (Figure 3(e); p < 0.05 and p < 0.01, resp.).
MDA, as the main degradation product of polyunsaturated lipids [26], causes considerable harm to organisms. It can damage membrane, add fluidity to cells, and also influence the DNA [26–28]. Therefore, we investigated the change on MDA level in yeast after treatment with parishin at 24 and 48 h. The MDA levels of yeast at 24 h in Figure 3(c) were significantly decreased after treatment with parishin at doses of 10 and 30 μM (p < 0.001, p < 0.001), compared with control group. The significant reduction of MDA levels in 3, 10, and 30 μM parishin-treated groups were observed at 48 h (Figure 3(d); p < 0.001, p < 0.001, and p < 0.05). These results again suggested that antioxidative stress had an important role in the antiaging activity of parishin.
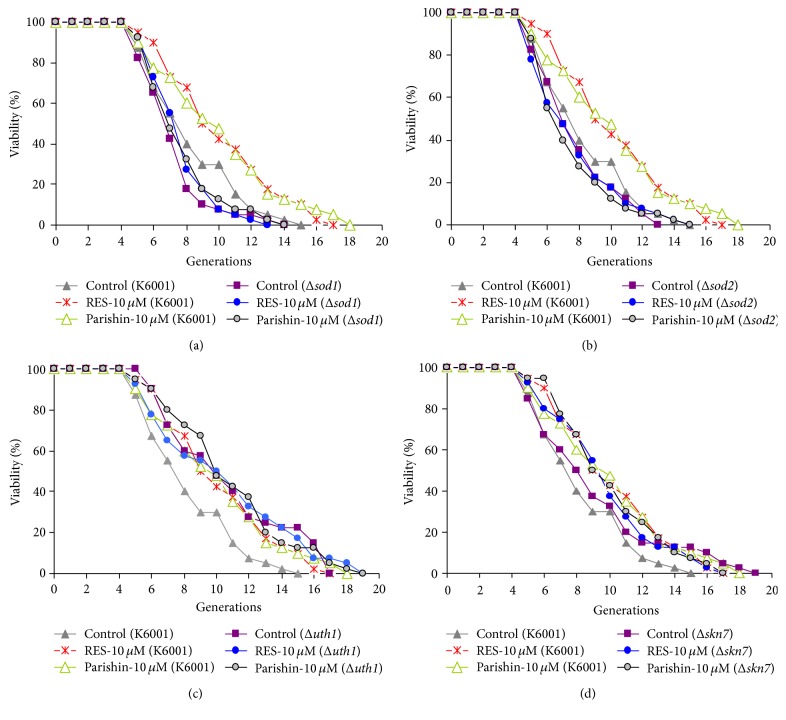
3.5. Parishin Does Not Affect the Lifespans of Sod1, Sod2, Uth1, and Skn7 Mutants with a K6001 Background
SOD gene is an important antioxidative stress gene that participates in free radical scavenging. To confirm whether SOD gene participated in the antiaging effect of parishin, we used sod1 and sod2 mutants with a K6001 background to examine the effects of parishin on replicative lifespan. The lifespan of sod1 and sod2 mutants was shorter than that of K6001 yeast strains, and parishin did not affect the replicative lifespan of these mutants (Figures 4(a) and 4(b)). These results indicated that SOD gene played an important role in the antiaging effect of parishin.
Figure 4.
Effects of parishin on the replicative lifespan of sod1 (a), sod2 (b), uth1 (c), and skn7 (d) mutant yeast strains with a K6001 background. The daughter cells of 40 microcolonies of each experiment were counted. The assay was repeated at least thrice. The results were displayed as mean ± SEM. The average lifespan of untreated K6001 was 7.25 ± 0.26 generations; RES at 10 μM, 9.25 ± 0.29∗∗; and parishin at 10 μM, 9.13 ± 0.30∗∗. (a) Δsod1 was 6.35 ± 0.22; RES at 10 μM, 6.80 ± 0.22 and; parishin at 10 μM, 6.83 ± 0.23. (b) Δsod2 was 6.90 ± 0.24; RES at 10 μM, 6.80 ± 0.26; and parishin at 10 μM, 6.60 ± 0.24. (c) Δuth1 was 9.73 ± 0.31; RES at 10 μM, 9.58 ± 0.32; and parishin at 10 μM, 9.93 ± 0.30. (d) Δskn7 was 8.20 ± 0.31; RES at 10 μM, 8.83 ± 0.28; and parishin at 10 μM, 9.05 ± 0.29. Δsod1, Δsod2, Δuth1, and Δskn7 represent sod1, sod2, uth1, and skn7 mutant yeast strain with a K6001 background, respectively. ∗∗ indicates significant difference compared with untreated K6001 (p < 0.01).
UTH1 was an aging gene related to oxidative stress, and UTH1 inactivation increased resistance to oxidants [29]. In addition, SKN7 is the transcriptional activator of UTH1. Thus, we used uth1 and skn7 mutants with a K6001 background to investigate whether these two genes were involved in the lifespan extension of parishin. The longer replicative lifespan of uth1 mutant of K6001 yeast was observed in our present study as another report [30]. After administrating parishin, the changes on replicative lifespan of these mutants were not observed (Figures 4(c) and 4(d)). These results revealed that UTH1 and SKN7 genes were involved in the antiaging effect of parishin.
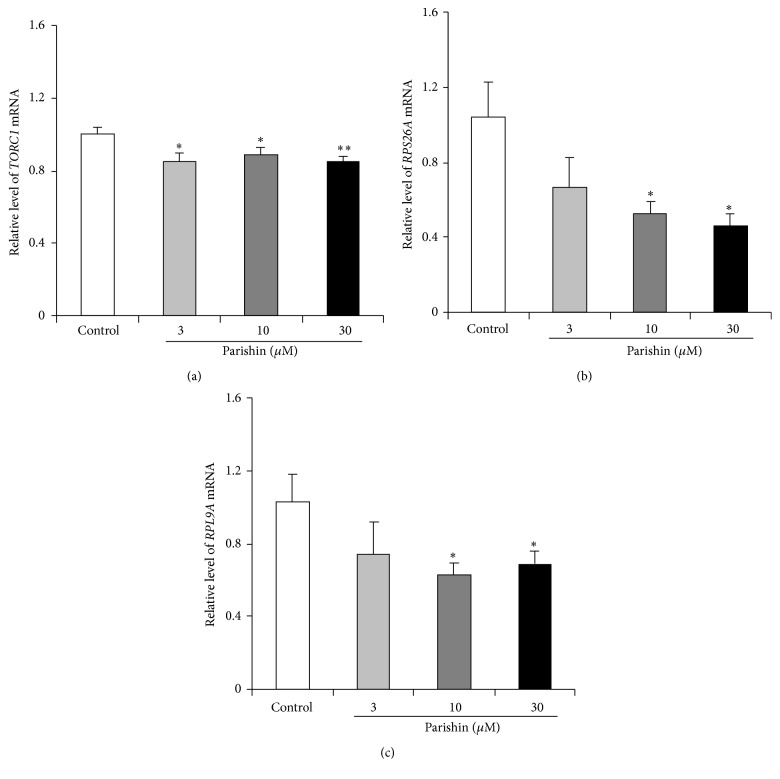
3.6. Parishin Inhibits the TORC1, RPS26A, and RPL9A Gene Expressions, Downstream of the TOR Signaling Pathway
TOR signaling pathway has prominent importance in regulating the process of aging. This signaling pathway controls growth-related processes, including regulation of translation, ribosome biogenesis, amino acid permease stability, and induction of autophagy [31]. TOR inhibition leads to the decreasing expression of some ribosomal protein genes, such as RPS26A and RPL9A, and increasing expression of some genes coding permeases for nitrogenous compounds, such as GAP1 and MEP2 [32]. TOR also regulates ribosome maturation via the nuclear GTP-binding protein NOG1 [33]. Therefore, we detected the effects of parishin on TOR signaling pathway using RT-PCR analysis. The changes on TORC1, RPS26A, and RPL9A gene expressions were presented in Figures 5(a)–5(c). TORC1 gene expression was significantly decreased after parishin treatment at doses of 3, 10, and 30 μM (Figure 5(a); p < 0.05, p < 0.05, and p < 0.01, resp.), and the gene expressions of RPS26A and RPL9A were also inhibited by parishin at doses of 10 and 30 μM (Figures 5(b) and 5(c); p < 0.05 and p < 0.05). These results revealed that TOR signaling pathway may be involved in the antiaging effects of parishin. However, significant difference was not observed in NOG1, GAP1, and MEP2 gene expressions in yeast after parishin treatment (data not shown). Possibly, parishin did not block the TOR signaling pathway completely.
Figure 5.
Effects of parishin on target of rapamycin (TOR) signaling pathway. Parishin significantly reduced the gene expressions of TORC1 (a), RPS26A (b), and RPL9A (c). Amounts of the mRNA above were normalized to that of TUB1. The results were displayed as mean ± SEM for three independent experiments. ∗ and ∗∗ indicate significant difference between the control and treatment groups (p < 0.05, p < 0.01).
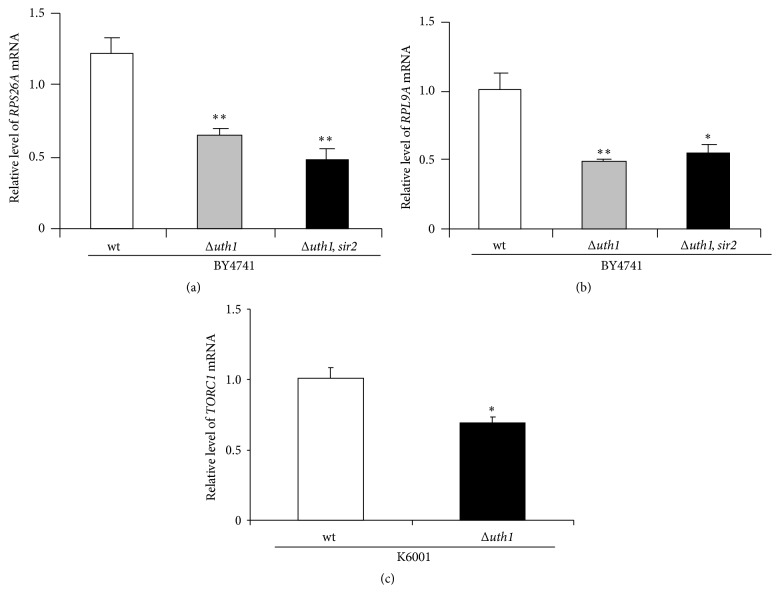
3.7. UTH1 Gene Regulates TOR Signaling Pathway
In the present study, both Uth1 and TOR signaling pathways participated in the antiaging effects of parishin. To investigate whether they had interactions, we investigated the gene expression of RPS26A and RPL9A in uth1 as well as in uth1, sir2 double mutant with BY4741 background. As expected, their gene expression in these mutants was significantly decreased comparing with control group, respectively (Figures 6(a) and 6(b); p < 0.01, p < 0.01, and p < 0.01, p < 0.05, resp.). Furthermore, we detected the gene expression of TORC1 in uth1 mutants with K6001 background. Significant reduction of TORC1 gene expression was observed in the uth1 mutant with K6001 background (Figure 6(c); p < 0.05). In addition, another research also indicated that UTH1 could affect TOR signaling pathway [34]. These results clarified that Uth1 could regulate TOR signaling pathway. Similarly, we did not find significant changes on NOG1, GAP1, and MEP2 gene expression in uth1 mutant as well as uth1, sir2 double mutants of BY4741 (data not shown).
Figure 6.
Interaction between UTH1 gene and TOR signaling pathway. The change on RPS26A (a) and RPL9A (b) gene expressions in wild type, uth1 mutant, and uth1, sir2 double mutant with BY4741 background. The change on TORC1 gene expressions in wild type and uth1 mutants with K6001 background (c). Amounts of the mRNA above were normalized to that of TUB1. Each experiment was carried out in triplicate independently. ∗ and ∗∗ indicate the significant difference between the control and treatment groups (p < 0.05, p < 0.01).
4. Conclusion
In summary, we isolated parishin, an antiaging compound from G. elata, using the replicative lifespan assay of K6001. Parishin could significantly extend the lifespan of yeast via antioxidative stress, increase the SIR2 gene expression, and inhibit the Uth1/TOR signaling pathway. Furthermore, Uth1 can upregulate TOR signaling pathway. Thus, parishin might be a valuable lead compound for drug discovery against age-related diseases.
Supplementary Material
In the supplementary material, the methods and the results of kinetics of K6001 yeast growth and another antioxidative stress assay were presented.
Acknowledgments
The authors thank Professor Michael Breitenbach (Salzburg University, Austria) for gifts of K6001 yeast strains. This work was supported by International Science and Technology Cooperation Program of China (Grant no. 2014DFG32690), National Natural Science Foundation of China (Grants nos. 81072536 and 81273385), and Doctoral Fund of Ministry of Education (Grant no. 20130101110093).
Competing Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Lutz W., Sanderson W., Scherbov S. The coming acceleration of global population ageing. Nature. 2008;451(7):716–719. doi: 10.1038/nature06516. [DOI] [PubMed] [Google Scholar]
- 2.Lleó A., Greenberg S. M., Growdon J. H. Current pharmacotherapy for Alzheimer's disease. Annual Review of Medicine. 2006;57:513–533. doi: 10.1146/annurev.med.57.121304.131442. [DOI] [PubMed] [Google Scholar]
- 3.Howitz K. T., Bitterman K. J., Cohen H. Y., et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425(6954):191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 4.Tissenbaum H. A., Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans . Nature. 2001;410(6825):227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 5.Wood J. G., Rogina B., Lavu S., et al. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430(7000):686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- 6.Daitoku H., Hatta M., Matsuzaki H., et al. Silent information regulator 2 potentiates Foxo 1-mediated transcription through its deacetylase activity. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(27):10042–10047. doi: 10.1073/pnas.0400593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lancelot J., Caby S., Dubois-Abdesselem F., et al. Schistosoma mansoni sirtuins: characterization and potential as chemotherapeutic targets. PLoS Neglected Tropical Diseases. 2013;7(9) doi: 10.1371/journal.pntd.0002428.e2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reddi A. R., Culotta V. C. SOD1 integrates signals from oxygen and glucose to repress respiration. Cell. 2013;152(1-2):224–235. doi: 10.1016/j.cell.2012.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.National Pharmacopoeia Committee. Pharmacopoeia of People's Republic of China. part 1. Beijing, China: Chemical Industry Press; 2010. [Google Scholar]
- 10.Zhou Y., Cao D. B., Han Y. Y., Sun J. H., Yang S. R. Protective effects of Gastrodia elata on myocardial cells of mice with viral myocarditis. Journal of Clinical Pediatrics. 2011;29(8):766–768. [Google Scholar]
- 11.Ahn E.-K., Jeon H.-J., Lim E.-J., Jung H.-J., Park E.-H. Anti-inflammatory and anti-angiogenic activities of Gastrodia elata Blume. Journal of Ethnopharmacology. 2007;110(3):476–482. doi: 10.1016/j.jep.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 12.Zhou J., Pu X. Y., Yang Y. B., Yang T. R. The chemistry of Gastrodia elata BL. IV. the phenolic compounds of some Chinese species of Gastrodia . Acta Botanica Yunnanica. 1983;5(4):443–444. [Google Scholar]
- 13.Taguchi H., Yosioka I., Yamasaki K., Kim I. H. Studies on the constituents of Gastrodia elata Blume. Chemical & Pharmaceutical Bulletin. 1981;29(1):55–62. doi: 10.1248/cpb.29.55. [DOI] [Google Scholar]
- 14.Lin J.-H., Liu Y.-C., Hau J.-P., Wen K.-C. Parishins B and C from rhizomes of Gastrodia elata . Phytochemistry. 1996;42(2):549–551. doi: 10.1016/0031-9422(95)00955-8. [DOI] [Google Scholar]
- 15.Jang Y. W., Lee J. Y., Kim C. J. Anti-asthmatic activity of phenolic compounds from the roots of Gastrodia elata Bl. International Immunopharmacology. 2010;10(2):147–154. doi: 10.1016/j.intimp.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 16.Baek N.-I., Choi S. Y., Park J. K., et al. Isolation and identification of succinic semialdehyde dehydrogenase inhibitory compound from the rhizome of Gastrodia elata Blume. Archives of Pharmacal Research. 1999;22(2):219–224. doi: 10.1007/bf02976550. [DOI] [PubMed] [Google Scholar]
- 17.Qiu F., Liu T.-T., Qu Z.-W., Qiu C.-Y., Yang Z. F., Hu W.-P. Gastrodin inhibits the activity of acid-sensing ion channels in rat primary sensory neurons. European Journal of Pharmacology. 2014;731:50–57. doi: 10.1016/j.ejphar.2014.02.044. [DOI] [PubMed] [Google Scholar]
- 18.Jarolim S., Millen J., Heeren G., Laun P., Goldfarb D. S., Breitenbach M. A novel assay for replicative lifespan in Saccharomyces cerevisiae . FEMS Yeast Research. 2004;5(2):169–177. doi: 10.1016/j.femsyr.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 19.Wasko B. M., Kaeberlein M. Yeast replicative aging: a paradigm for defining conserved longevity interventions. FEMS Yeast Research. 2014;14(1):148–159. doi: 10.1111/1567-1364.12104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen K. L., Crane M. M., Kaeberlein M. Microfluidic technologies for yeast replicative lifespan studies. Mechanisms of Ageing and Development. 2016 doi: 10.1016/j.mad.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weng Y. F., Xiang L., Matsuura A., Zhang Y., Huang Q. M., Qi J. H. Ganodermasides A and B, two novel anti-aging ergosterols from spores of a medicinal mushroom Ganoderma lucidum on yeast via UTH1 gene. Bioorganic & Medicinal Chemistry. 2010;18(3):999–1002. doi: 10.1016/j.bmc.2009.12.070. [DOI] [PubMed] [Google Scholar]
- 22.Weng Y. F., Lu J., Xiang L., et al. Ganodermasides C and D, two new anti-aging ergosterols from spores of the medicinal mushroom Ganoderma lucidum . Bioscience, Biotechnology and Biochemistry. 2011;75(4):800–803. doi: 10.1271/bbb.100918. [DOI] [PubMed] [Google Scholar]
- 23.Xiang L., Sun K. Y., Lu J., et al. Anti-aging effects of phloridzin, an apple polyphenol, on yeast via the SOD and Sir2 genes. Bioscience, Biotechnology and Biochemistry. 2011;75(5):854–858. doi: 10.1271/bbb.100774. [DOI] [PubMed] [Google Scholar]
- 24.Sun K. Y., Cao S. N., Pei L., Matsuura A., Xiang L., Qi J. H. A steroidal saponin from Ophiopogon japonicus extends the lifespan of yeast via the pathway involved in SOD and UTH1. International Journal of Molecular Sciences. 2013;14(3):4461–4475. doi: 10.3390/ijms14034461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu B. P. Aging and oxidative stress: modulation by dietary restriction. Free Radical Biology and Medicine. 1996;21(5):651–668. doi: 10.1016/0891-5849(96)00162-1. [DOI] [PubMed] [Google Scholar]
- 26.Marnett L. J. Lipid peroxidation—DNA damage by malondialdehyde. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 1999;424(1-2):83–95. doi: 10.1016/s0027-5107(99)00010-x. [DOI] [PubMed] [Google Scholar]
- 27.Debouzy J.-C., Fauvelle F., Vezin H., Brasme B., Chancerelle Y. Interaction of the malonyldialdehyde molecule with membranes: a differential scanning calorimetry, 1H-, 31P-NMR and ESR study. Biochemical Pharmacology. 1992;44(9):1787–1793. doi: 10.1016/0006-2952(92)90073-r. [DOI] [PubMed] [Google Scholar]
- 28.Ohyashiki T., Sakata N., Matsui K. A decrease of lipid fluidity of the porcine intestinal brush-border membranes by treatment with malondialdehyde. Journal of Biochemistry. 1992;111(3):419–423. doi: 10.1093/oxfordjournals.jbchem.a123772. [DOI] [PubMed] [Google Scholar]
- 29.Bandara P. D. S., Flattery-O'Brien J. A., Grant C. M., Dawes I. W. Involvement of the Saccharomyces cerevisiae UTH1 gene in the oxidative-stress response. Current Genetics. 1998;34(4):259–268. doi: 10.1007/s002940050395. [DOI] [PubMed] [Google Scholar]
- 30.Camougrand N., Kiššová I., Velours G., Manon S. Uth1p: a yeast mitochondrial protein at the crossroads of stress, degradation and cell death. FEMS Yeast Research. 2004;5(2):133–140. doi: 10.1016/j.femsyr.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 31.Wullschleger S., Loewith R., Hall M. N. TOR signaling in growth and metabolism. Cell. 2006;124(3):471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 32.Matsui A., Kamada Y., Matsuura A. The role of autophagy in genome stability through suppression of abnormal mitosis under starvation. PLoS Genetics. 2013;9(1) doi: 10.1371/journal.pgen.1003245.e1003245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Honma Y., Kitamura A., Shioda R., et al. TOR regulates late steps of ribosome maturation in the nucleoplasm via Nog1 in response to nutrients. EMBO Journal. 2006;25(16):3832–3842. doi: 10.1038/sj.emboj.7601262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Camougrand N., Grelaud-Coq A., Marza E., Priault M., Bessoule J.-J., Manon S. The product of the UTH1 gene, required for Bax-induced cell death in yeast, is involved in the response to rapamycin. Molecular Microbiology. 2003;47(2):495–506. doi: 10.1046/j.1365-2958.2003.03311.x. [DOI] [PubMed] [Google Scholar]
- 35.Bobola N., Jansen R., Shin T. H., Nasmyth K. Asymmetric accumulation of Ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating-type switching to mother cells. Cell. 1996;84(5):699–709. doi: 10.1016/S0092-8674(00)81048-X. [DOI] [PubMed] [Google Scholar]
- 36.Paciello L., de Alteriis E., Mazzoni C., Palermo V., Zueco J., Parascandola P. Performance of the auxotrophic Saccharomyces cerevisiae BY4741 as host for the production of IL-1β in aerated fed-batch reactor: role of ACA supplementation, strain viability, and maintenance energy. Microbial Cell Factories. 2009;8, article 70:13. doi: 10.1186/1475-2859-8-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In the supplementary material, the methods and the results of kinetics of K6001 yeast growth and another antioxidative stress assay were presented.