Abstract
Induced pluripotent stem cells (iPSCs) have enormous potential in regenerative medicine and disease modeling. It is now felt that clinical trials should be performed with iPSCs derived with nonintegrative constructs. Numerous studies, however, including those describing disease models, are still being published using cells derived from iPSCs generated with integrative constructs. Our experimental work presents the first evidence of spontaneous transgene reactivation in vitro in several cellular types. Our results show that the transgenes were predominantly silent in parent iPSCs, but in mesenchymal and endothelial iPSC derivatives, the transgenes experienced random upregulation of Nanog and c-Myc. Additionally, we provide evidence of spontaneous secondary reprogramming and reversion to pluripotency in mesenchymal stem cells derived from iPSCs. These findings strongly suggest that the studies, which use cellular products derived from iPSCs generated with retro- or lentiviruses, should be evaluated with consideration of the possibility of transgene reactivation. The in vitro model described here provides insight into the earliest events of culture transformation and suggests the hypothesis that reversion to pluripotency may be responsible for the development of tumors in cell replacement experiments. The main goal of this work, however, is to communicate the possibility of transgene reactivation in retro- or lenti-iPSC derivatives and the associated loss of cellular fidelity in vitro, which may impact the outcomes of disease modeling and related experimentation.
Introduction
Human induced pluripotent stem cells (hiPSCs) are considered to be almost identical to human embryonic stem cells (hESCs) [1]. Similar to hESCs [2], hiPSCs are widely used in disease modeling studies [3,4]. While teratoma formation is an essential feature of pluripotent cells, tumor formation, after delivery of differentiated cells, is a concern that is specific to iPSCs developed with integrated constructs because of the possibility of transgene reactivation. Previous work has shown that lenti/retroviral transgenes are largely silenced in pluripotent cells after reprogramming [5]. Constitutive expression of pluripotency genes results in partial reprogramming [6], and the epigenetic silencing of exogenous genes needs to occur to allow iPSC generation and subsequent differentiation [7].
Nevertheless, c-Myc transgene expression was detected in tumors of iPSC-derived chimeric mice [8]. The tumor-forming propensity varied between strains of chimeric mice and may have no correlation with tumorigenic risks of iPSC-based cell therapy in humans due to different species-specific mechanisms underlying tumor formation [9]. In contrast, neither c-Myc reactivation nor tumor formation was observed in chimeric pigs produced from iPSCs [10]. With lower efficiency outcomes, it appeared to be possible to avoid c-Myc in a reprogramming cocktail during generation of iPSCs [11]. Still, a recent study demonstrated tumor development after transplantation of neurospheres derived from iPSCs produced without c-Myc. Oct4 transgene reactivation was detected in this case [12]. The observation of transgene reactivation in iPSC-derived tumors is quite surprising since the opposite event, transgene silencing, typically occurs during embryonic stem cell differentiation [13–15]. The particular pattern of transgene reactivation is unclear, and the earliest events of tumor initiation eluded investigation due to the lack of in vitro models.
We discovered the phenomenon of appearance of pluripotent colonies in one mesenchymal stem cell line (iMSC-DS), which was differentiated from Down syndrome iPSCs (iPSC-DS), and have shown that pluripotent transgene reactivation preceded the reversion to pluripotency. To further study this phenomenon, we analyzed two iPSC lines: iPSC-DS and iPSCs from a healthy adult (iPSC-WT). Both cell lines were established and handled in similar conditions. These iPSC lines were differentiated into iMSCs and endothelial (vascular) cells (iEC). We discovered that the reversion to pluripotency was associated with profound upregulation of Nanog (257-fold) and c-Myc (112-fold) in the MSC derivatives of the iPSC-DS line. The observed reactivation of Nanog (66-fold) in MSC derivatives of the iPSC-WT line, however, did not initiate a reversion to pluripotency.
Our work presents the first evidence of transgene reactivation in vitro in a differentiated cell type, which led to changes in cellular phenotype associated with spontaneous secondary reprogramming (reversion to pluripotency). Although a limited number of cell lines and their derivatives were included in the study, there is a strong possibility that reversion to pluripotency and transgene reactivation have a wider occurrence. Therefore, we believe that transgene reactivation should be assayed in publications communicating the results of studies that used differentiated derivatives.
Materials and Methods
iPSC derivation
The iPSC-WT cell line was derived from MRC-5 fibroblasts (ATCC), and the iPSC-DS clones were derived from AG06872 fibroblasts (Coriell). The fibroblasts were transduced with retroviral vectors (pMXs-cMyc, pMXs-Nanog, pMXs-hOct3-4, and pMXs-Sox2; Addgene) to overexpress Oct4, Sox2, Nanog, and c-Myc transgenes. The retroviral vectors were produced by transient transfection of 293T cells. Following this, the fibroblasts were incubated for 4 h in the viral supernatants containing 5 μg/mL polybrene (Sigma). The transduced cells were then incubated for 3 weeks until development of the pluripotent clones. After isolation, the clones were grown in StemPro medium (Invitrogen) on a Matrigel® substrate (BD Biosciences). The cultures were split mechanically using the StemPro EZ Passage tool (Invitrogen).
Mesenchymal cell derivation (iMSC) by differentiation of iPSCs
Protocol 1
Adapted from Boyd et al. [16], the following protocol was used to derive the iMSC1 line that underwent secondary reprogramming. To differentiate the iPSC-DS cell line into MSCs, hESC culture medium containing knockout DMEM (Invitrogen), 20% knockout serum replacement, l-glutamine, and antibiotics was added to the semiconfluent feeder-free culture at passage 20. After 3 days, the medium was replaced with EGM-2 MV growth medium (Lonza), which was changed every 2–3 days over a period of 30 days. MSCs derived from iPSC-DS (iMSC1) were expanded and maintained in EGM-2 MV growth media at 37°C in 5% CO2. iPSCs derived from a healthy individual (iPSC-WT) were used to derive wild-type mesenchymal stem cells (iMSC1-WT). The WT iPSC differentiation steps are similar to the described DS iPSC differentiation.
Protocol 2
The following protocol was used to derive the iMSC2 cell lines from iPSC-DS (iMSC2-DS) and iPSC-WT (iMSC2-WT). Initially proposed by Vodyanik et al. [17,18], protocol 2 is based on a novel method of iPSC mesenchymal differentiation that requires isolation of a multipotential progenitor at the mesenchymoangioblast stage. For mesendodermal induction, iPSCs at passage 30 were cocultured with OP9 stromal cells. Primitive streak/mesendoderm precursors, which express apelin receptor (APLNR+) [19], were isolated via MACS sorting on day 2 of OP9 coculture. APLNR-APC antibodies and Anti-APC MicroBeads were used (Miltenyi Biotec).
The isolated APLNR+ progenitors were plated as single cells in semisolid colony-forming serum-free medium (CF-SFM) containing 40% ES-Cult M3120 methylcellulose, 25% serum-free expansion medium (SFEM; Stem Cell Technology), 10% BIT 9500 supplement, and other additives. After 2 weeks, the mesenchymal colony-forming units (MS-CFU) were manually picked, and the iMSC2 cells were transferred to an adherent culture and maintained in EGM-2 medium (Lonza).
Endothelial differentiation of iPSCs (iEC derivation)
Endothelial iEC-DS and iEC-WT lines were established from iPSC-DS at passage 32 and iPSC-WT at passage 23. We optimized a monolayer iPSC induction protocol for iEC derivation. The cells were initially grown on Matrigel-coated culture dishes in mTeSR1 medium. Induction medium containing CHIR 99021 (Stem Cell Technologies) was added to the culture to induce differentiation. CD31+CD144+ vascular progenitor cells were isolated on day 6 of differentiation via positive selection of CD144+ cells on a magnetic column (Miltenyi Biotec). The endothelial potential of iEC was evaluated using the following assays: immunostaining for von Willebrand factor (vWF), a tube formation assay, and via acetylated low-density lipoprotein (Ac-LDL) uptake. vWF expression was confirmed by iEC staining using a primary anti-hvWF A2 antibody (R&D Systems) and the appropriate secondary antibody (Alexa Fluor).
For the tube formation assay, isolated CD144+ iEC were seeded at a density of 4.5 × 104 cells per well onto a 12-well plate coated with Matrigel (BD Biosciences). The cells were incubated in VascuLife medium overnight at 37°C in 5% CO2. On the following day, tube networks were visualized under a light microscope. Using a commercial kit (Biomedical Technologies), Ac-LDL uptake was performed in accordance with the manufacturer's instructions. iEC were seeded at a density of 2.5 × 105 cells per well onto a 0.1% gelatin (Sigma)-coated 6-well plate. Ac-LDL was diluted to 10 μg/mL in complete VascuLife medium (Lifeline Cell Technologies) and added to the cells for 3 h. The cells were then washed with VascuLife medium and analyzed via fluorescent microscopy.
Microscopy
Confocal images were acquired using a Zeiss LSM 510 META laser scanning microscope system (Zeiss, Oberkochen, Germany). By varying the detectors' pinhole width, the observed fluorescence was localized to an area of known tissue thickness, and the field depth of the transmitted DIC images was adjusted. Scale bars were integrated into the image during acquisition. Epifluorescent images were acquired on a Leica DM IRB inverted microscope system (Leica, Germany) equipped with a digital camera Retiga 4000R (Qlmaging, Surrey, Canada), which was controlled with Openlab software version 5.0.2 (Perkin-Elmer, Waltham, MA). Scale bars were calibrated to each objective magnification and added after acquisition. Light microscopy images were acquired with a Nikon D100 (Nikon, Tokyo, Japan) digital SLR camera on a Leica DM IRB inverted microscope.
Immunocytochemistry
The cells were washed with PBS, fixed with 4% paraformaldehyde (PFA) for 5 min, and permeabilized with 0.1% Triton X-100 for the same amount of time. After a 30-min incubation in blocking solution (Protein Block; Dako) at 37°C, the cells were incubated with primary antibodies (1:100) for 1 h at room temperature. The secondary antibody (1:250) incubation time was also 30 min at 37°C. Mounting medium containing DAPI (Life Technologies) was used for counterstaining the nuclei. Via an Alkaline Phosphatase Substrate Kit (Vector Laboratories, Inc.), alkaline phosphatase activity was detected according to the manufacturer's instructions. Control cells incubated with only a secondary antibody served as negative controls to exclude nonspecific binding of secondary antibodies.
Flow cytometry
iMSC phenotype was monitored by measuring the expression of characteristic markers for several passages. The CD105, CD90, CD73, and CD31 antibodies were purchased from Miltenyi Biotec. The cells were lifted with 0.05% trypsin, stained with the appropriate conjugated antibodies, and incubated for 30 min at 4°C. Cells were washed in a 0.5% BSA/PBS solution and analyzed via FACSCalibur (BD Biosciences).
Expression analysis
Total RNA was harvested from cells using the commercial RNeasy kit (Qiagen). RNA concentration and purity were measured using a spectrophotometer. RNA (900 ng–1 μg) was transcribed into cDNA using a High Capacity RNA-to-cDNA Kit (Applied Biosystems). Semiquantitative polymerase chain reaction (qPCR) was performed, using Platinum Taq DNA Polymerase (Invitrogen), on an MBS Satellite Thermocycler (Thermo Electron Co.). The sequences of all primers are available upon request.
Real-time qPCR
Via the RNeasy Mini Kit (Qiagen), total RNA was isolated from cells according to the manufacturer's instructions. RNA concentration and purity were measured using a spectrophotometer. RNA (250 ng–1 μg) was transcribed into cDNA using a High Capacity RNA-to-cDNA Kit (Applied Biosystems) in an MBS Satellite Thermocycler (Thermo Electron Co.). Quantitative real time polymerase chain reaction (RT-qPCR) was performed, using primer sets from IDT (sequences available upon request) and SYBR Advantage qPCR Premix (Clonetech), on a 7500 Real Time PCR System (Life Technologies). The 20 μL reaction volume contained 2 μL cDNA, 0.4 μL forward and reverse gene expression primers, 0.4 μL ROX reference dye LMP, and 10 μL PCR master mix (2 × ). Amplification was achieved via the following thermocycler protocol: 1 cycle at 95°C for 3 s and 40 cycles at 60°C for 30 s. All data were analyzed with the 7500 System Software. The expression of each target gene was normalized to an endogenous control gene, GAPDH.
FISH analysis
The cells were fixed in a cold solution of methanol and acetic acid (3:1) and placed on slides to air-dry. The fidelity of 21, Y, and X chromosome segregation was tested by a three-color PN (21, X, Y) FISH probe (Kreatech). After the probe was applied, the slides were denatured for 7 min at 75°C and hybridized overnight at 37°C. The slides were then washed according to the manufacturer's instructions and imaged via a Leica DM IRB.
Molecular karyotype
Genomic DNA was purified with the DNeasy Blood & Tissue Kit (Qiagen). The aCGH procedure was performed using a modified Agilent 4 × 180K aCGH protocol (Agilent Technologies). Genomic DNA was extracted and fragmented by high-intensity sound waves. Gender-mismatched reference samples were labeled with cyanine 5-dUTP, while corresponding patient samples were labeled with cyanine 3-dUTP. After the labeling procedure, the unincorporated label was removed via column purification (Amicon Ultra; Millipore) and resuspended in Tris–EDTA buffer (10 mM Tris and EDTA 1 mM at pH 8.0). A Cy5 reference was paired with a Cy3 testing sample and combined in a microcentrifuge tube. The combined samples were hybridized to the array for 24 h. Following this, they were washed, scanned, and analyzed. The MS200 (Roche Diagnostics) was used to acquire 2-μm array images. ImaGene 9.0 and Nexus CGH 7.5 (Biodiscovery) were used for data analysis.
STR genotyping
Genotyping was performed with the AmpFlSTR Profiler Plus PCR Amplification Kit (Applied Biosystems, Inc.). This kit coamplified the repeat regions of the following nine short tandem repeat autosomal loci: D3S1358, vWA, FGA, D8S1179, D21S11, D18S51, D5S818, D13S317, and D7S820. Amelogenin, a segment of an X-Y homologous gene, was also amplified. Genomic DNA (2–3 ng) was amplified in a 25 μL reaction containing 10.5 μL AmpFlSTR reaction mix, 5.5 μL Profiler Plus™ primer mix, and 0.5 μL AmpliTaq Gold DNA Polymerase. The PCR consisted of the following: 11 min at 95°C, followed by 28 cycles at 94°C for 1 min, 59°C for 1 min, 72°C for 1 min, and finished by 60°C for 45 min. The PCR product (1.5 μL) was added to 24 μL of deionized formamide with a size ladder standard [per sample, 1 μL GeneScan-500 size standard (ROX; Applied Biosystems, Inc.)] The product was separated via capillary electrophoresis on an ABI3130xl platform. Data collection and analysis were performed using GeneMapper™ ID version 3.1 (Applied Biosystems, Inc.).
Results
Atypical cellular morphology and spontaneous reversion to pluripotency in hiPSC-derived mesenchymal cell culture
Human iPSCs were generated from skin fibroblasts that had a normal karyotype and a trisomy 21 karyotype (Down syndrome). The fibroblasts were transduced with retroviral vectors expressing Oct4, Sox2, Nanog, and c-Myc. These iPSC lines are capable of differentiating into neural cells [20], endodermal precursors [21], and iMSCs [22]. We have also previously shown that the iMSCs derived in our laboratory (including iMSC-DS, which later reverted to pluripotency) could differentiate into osteoblasts, chondrocytes, and adipocytes. These cells expressed CD105 and CD90, and they were negative for CD34 and CD45. When seeded onto a decellularized matrix, the cells secreted glycosaminoglycans and collagen; the cells also expressed α-SMA. The gene expression profile of iMSCs was highly comparable with that of MSCs derived from bone marrow [22].
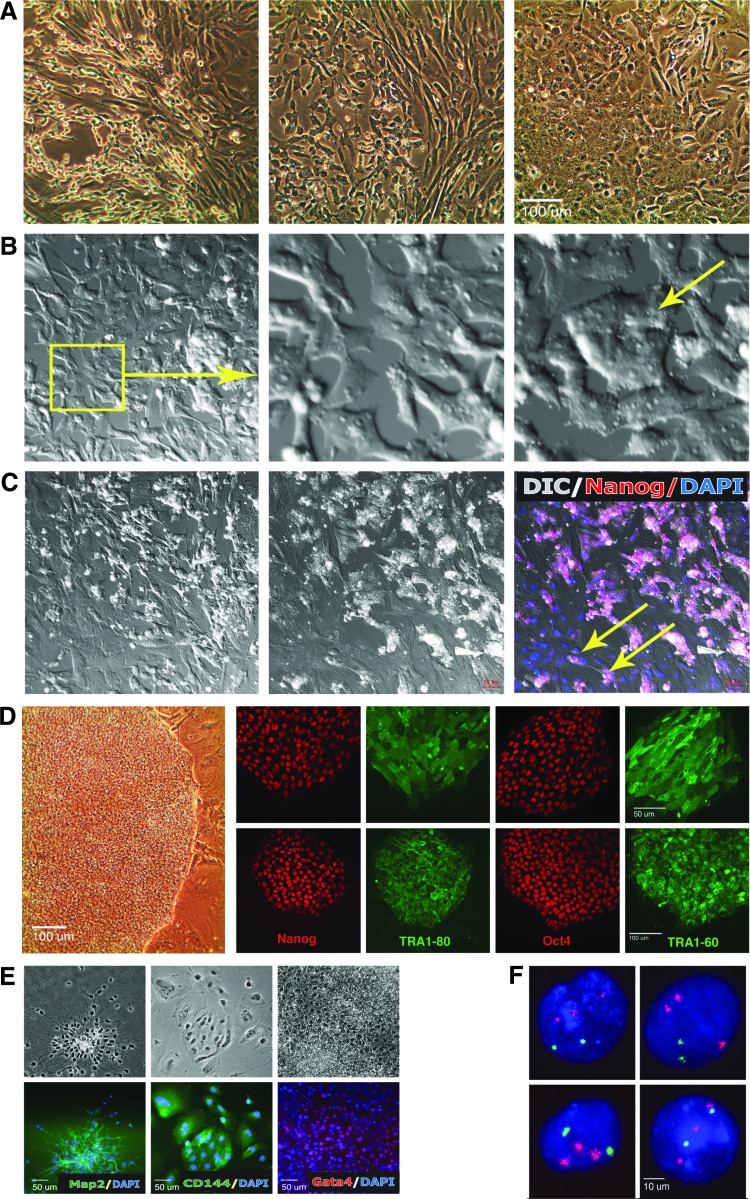
Additionally, iMSC-DS had a typical spindle-shaped mesenchymal morphology (Fig. 1A) and expressed both CD73 and CD146. During subsequent culture, Down syndrome mesenchymal stem cells (iMSC-DS) began to exhibit irregular morphology (Fig. 2A), which was associated with the reappearance of pluripotency markers and the appearance of self-induced pluripotent colonies (discussed below). We referred to these phenomena as spontaneous transgene reactivation and secondary reprogramming (reversion to pluripotency) of iPSC-derived MSCs. In light of such results, we termed these Down syndrome MSCs a “reverted” culture. Additionally, we detected that the transgene upregulation in iMSC-DS preceded reversion to pluripotency.
FIG. 1.
Reversion to pluripotency of mesenchymal stem cells. (A) Morphology of mesenchymal stem cells at a passage before pluripotency reversion; FACS analysis showing absence of pluripotent cells (TRA1-60) and homogeneous expression of mesenchymal marker CD73. Right panel: semiquantitative RT-PCR analysis showing significant upregulation of c-Myc and Oct4 transgene expression in mesenchymal stem cells (iMSC-DS) derived from piPSC-DS. (B) Culture of mesenchymal stem cells during reversal to pluripotency showing elements of the mesenchymal-to-epithelial transition characteristic of reprogramming and activation of pluripotency markers in somatic cells. (I) PC and IF overlay showing Oct4 (green) activation in cells of epithelial morphology; overlay of Oct4 (green)/Nanog (red) showing Oct4 coexpressed with Nanog, although Nanog expression alone is more widespread; overlay of Nanog (red)/DAPI (blue). (II) Enlarged area within the yellow box (I) of the PC/IF overlay showing Oct4 expression (green); DIC/IF overlay showing Nanog expression (red) in cells with epithelial morphology; overlay of Nanog (red) and DAPI (blue). (III) PC and IF TRA1-81 (green) showing .TRA1-81 expression in cells with epithelial morphology (arrow), and overlay of c-Myc (red)/DAPI (blue). (IV) PC and TRA2-39 (green) showing TRA2-39 expression in “reverted” cells; cells with epithelial morphology (arrow), and overlay of Nanog (red)/DAPI (blue). DIC, differential interference contrast; IF, immunofluorescence; iMSC-DS, Down syndrome mesenchymal stem cells; PC, phase contrast; RT-PCR, reverse transcription polymerase chain reaction.
FIG. 2.
Reversion to pluripotency of mesenchymal stem cells and rederivation of secondary (rederived) riPSCs. (A–C) Transformation of mesenchymal stem cell culture during reversion to pluripotency. (A) The culture is showing transformations of cellular shape. (B, C) The start and end frames of time-lapse confocal imaging showing formation of pluripotent clusters via aggregation of “somatic” cells (movies available online). (B) The frames of a 12-h movie (Supplementary Video S1), an enlarged patch showing the beginning and the end of cell aggregation. (C) The frames of a 20-h movie (Supplementary Video S2) showing multiple new cell clusters formed at the end of the documented period; DIC/Nanog/DAPI overlay showing Nanog (red) expression in cell clusters. (D) The PC image of riPSC-DS colony at passage 1 growing on MEF and the IF pluripotency assay. piPSC-DS line (top) and riPSC-DS (bottom) growing on Matrigel express pluripotency markers Nanog, TRA1-80, Oct4, and TRA1-60. (E) Differentiation of riPSC-DS into three embryonic lineages: ectoderm (Map2, green): PC and overlay with DAPI, mesoderm CD144 (VE-cadherin, green): PC and overlay with DAPI, and endoderm (Gata 4, red): PC and overlay with DAPI. (F) FISH analysis of riPSC-DS clone 6 for chromosome 21 (red) and the X chromosome (green) showing trisomy of chromosome 21. DIC, differential interference contrast; IF, immunofluorescence; iPSC, induced pluripotent stem cell; MEF, mouse embryonic fibroblasts; PC, phase contrast.
Validation of riPSC source: reversion to pluripotency in iPSC-derived MSC culture follows the steps described for reprogramming with transcription factors
The surprising phenomena of spontaneous transgene reactivation and reversion to pluripotency have not been previously described. We, therefore, carefully examined the initiation of colony formation to exclude the remote possibility that undifferentiated cells may still persist in MSC culture. First, we analyzed a preceding passage of MSCs via FACS analysis. Immediately before reversion to pluripotency, the cells expressed 100% of the CD73 mesenchymal marker and had no expression of the pluripotent marker TRA1-60. We then used a semiquantitative RT-PCR analysis to compare transgene expression between the reverted mesenchymal cell culture and the “parental” iPSCs (piPSC-DS) from which that culture was derived.
The transgenes were predominantly silent in piPSC-DS. However, there was a notable upregulation of Nanog and c-Myc in MSCs derived from piPSC-DS (Fig. 1A). These findings indicated that transgene reactivation, not contamination with undifferentiated piPSC-DS, was responsible for the appearance of colonies. Thus, we eliminated undifferentiated “pluripotent contamination” as the source of colony formation in iPSC-derived MSCs.
Next, we documented a stepwise transformation of the cellular phenotype characteristic of the iPSC generation, which is believed to undergo the mesenchymal-to-epithelial transition (MET) during the reprogramming process [23,24]. Indeed, before colony formation, we detected an abundance of cells with epithelial and transitional morphology. A nascent stage of reprogramming in somatic cells can be distinguished by the gradual appearance of pluripotency markers. Accordingly, we observed that many somatic epithelial cells in the reverted MSC culture showed activation of pluripotency transcription factors, such as nuclear expression of Nanog and Oct4, and the cells were randomly positive for surface pluripotency markers TRA2-39 (AP) and TRA1-81 (Fig. 1B). Notably, there is some controversy regarding pluripotency marker expression in MSCs, including a report of low-level cytoplasmic but not nuclear Oct4 expression [25,26].
Finally, using time-lapse confocal microscopy, we established that formation of pluripotent clusters was occurring by aggregation of “somatic” cells (Fig. 2B, C and Supplementary Videos S1 and S2; Supplementary Data are available online at www.liebertpub.com/scd). Supplementary Video S1 shows the dynamic of cellular aggregation into a cluster, while Supplementary Video S2 shows the formation of multiple aggregates and documents the acquisition of pluripotency with imunofluorescent costain of DAPI/Nanog at the end of the recording. Markedly similar to the iPSC posttransduction event, many cellular clusters experienced altered morphology and expression of pluripotency TFs but remained partially reprogrammed and did not acquire a typical iPSC morphology (Fig. 2A).
Pluripotent clone isolation and characterization
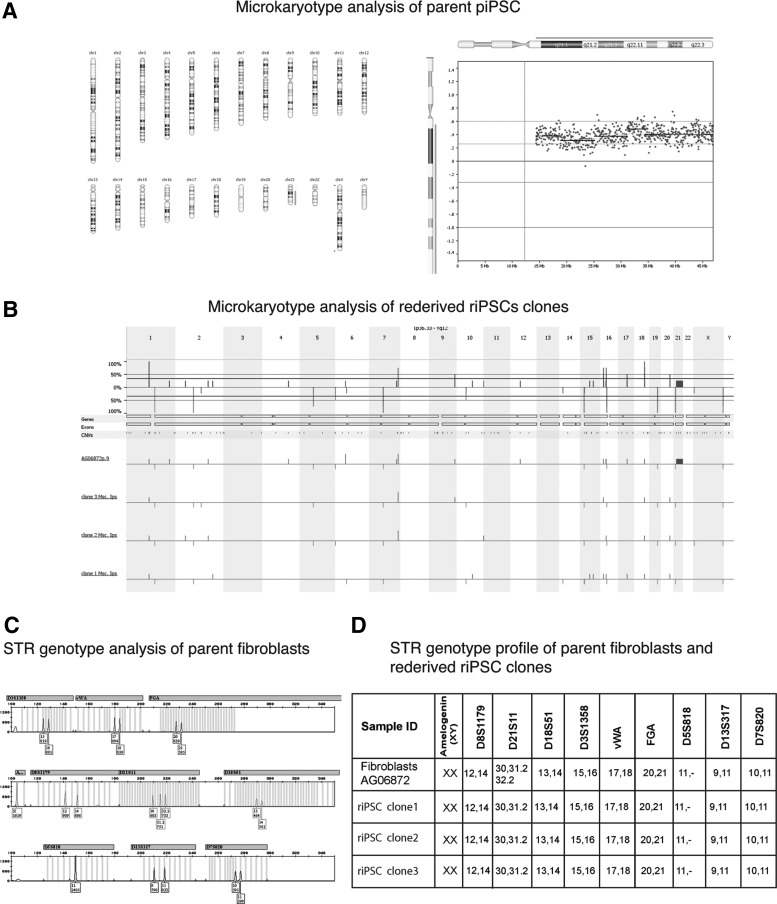
After eliminating the possibility that the source of new pluripotent colonies in reversed culture was due to undifferentiated cells, the self-induced pluripotent clones (n = 8) were collected and expanded for study. They were positive for pluripotency markers and able to differentiate into derivatives of all three embryonic lineages (Fig. 2D, E). In contrast to piPSC clones, which had karyotypes similar to the parental fibroblasts, some riPSC clones retained an extra chromosome 21 (n = 5) (Fig. 2F). Others (n = 3) had a euploid karyotype with the extra chromosome 21 expelled (Fig. 3A, B).
FIG. 3.
Genetic analysis of parent fibroblast, piPSC-DS, and riPSC-DS lines. (A) Molecular karyotype (chromosomal microarray) analysis of parental piPSC-DS showing trisomy of chromosome 21. (B) Chromosomal microarray analysis of parental fibroblasts showing trisomy 21 and 3 reverted riPSC-DS clones showing a euploid karyotype with the extra chromosome 21 expelled. No other clinically significant gains or losses were observed in the three riPSC-DS clones compared with the parental fibroblast line. Benign gains and losses were concordant across all samples, although not all copy number changes reached the software algorithm-calling threshold. (C) Image of fragment analysis showing three peaks (alleles) for chromosome 21 in parental fibroblasts. Testing was performed by PCR amplification of STR (short tandem repeats). (D) Table of STR genotypes of parental fibroblasts and reverted riPSC-DS clones establishing authenticity of the lines and showing the loss of the triploid allele of chromosome 21 in riPSC-DS clones. iPSC, induced pluripotent stem cell.
The cultures with a trisomy of chromosome 21 were previously described as relatively stable, and loss of the extra chromosome 21 in riPSC clones may reflect a physiological challenge during transgene reactivation and culture metamorphosis. For instance, Li et al. [27] achieved a correction of Down syndrome with TKNEO transgene integrated into the extra chromosome when grown in conditions that selected against TKNEO. Genetic identity between the patient's fibroblasts, the piPSC-DS, and the riPSC-DS was established by STR analysis (Fig. 3C, D).
Transgene activation in iPSC derivatives
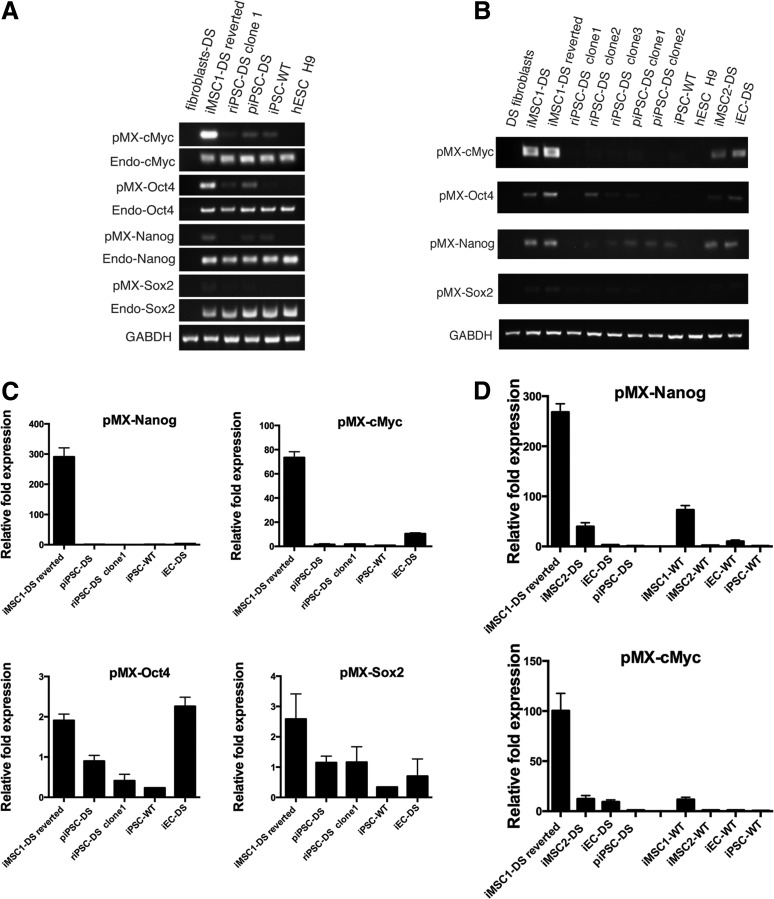
We hypothesized that iMSC-DS reversion to pluripotency was driven by transgene reactivation. Semiquantitative RT-PCR results revealed transgene reactivation in MSCs (Fig. 4A). The transgenes were predominantly silent in piPSC-DS but showed upregulation of Nanog, c-Myc, and Oct4 in MSCs derived from piPSC. The significance of transgene reactivation in differentiated derivatives prompted us to investigate this finding more broadly. We noted that the “secondary” riPSC-DS clone 1, which was collected from MSCs, showed decreased transgene expression compared with piPSC-DS. We then isolated and tested two more riPSC-DS clones to verify the tendency. Apparently, the transgene expression in riPSC-DS clones (1–3) was variable, similar to the clones of the first generation (piPSC-DS 1 and 2). We also observed transgene expression in MSCs of the previous passage, although it was not as high as in the culture that eventually underwent secondary reprogramming (Fig. 4B). We further asked whether MSCs differentiated from iPSC-DS by a different protocol (iMSC2-DS) (Fig. 5A) would show signs of transgene reactivation. For this purpose, we used a novel method of iPSC mesenchymal differentiation based on the isolation of multipotential progenitor at the mesenchymoangioblast stage. We observed some reactivation of the c-Myc transgene in iMSC2-DS, however, to a lesser degree than in iMSC1-DS reverted. To inquire whether transgene expression was cell-type specific, we assayed another type of iPSC-DS derivative—endothelial cells (iEC-DS) (Fig. 5B), and we detected upregulation of c-Myc (Fig. 4B). We then performed a quantitative assessment of transgene upregulation via RT-qPCR. In mesenchymal stem cells (iMSC1-DS) derived from piPSC-DS, we found almost 300-fold upregulation of Nanog and a 77-fold upregulation of c-Myc, and there was no significant upregulation of Oct4 and Sox2. Additionally, we noted a significant (14-fold) upregulation of c-Myc in iMSC2-DS compared with piPSC-DS (Fig. 4C). Finally, to broaden the scope of our findings, we also investigated differentiated iPSC-derived phenotypes from a healthy individual (iPSC-WT). The WT iPSC differentiation steps are similar to the described DS iPSC differentiation. The following differentiated phenotypes were used in the experimental work: MSCs derived by protocol 1 (iMSC1-WT), MSCs derived by protocol 2 (iMSC2-WT), and endothelial cells (iEC-WT).
FIG. 4.
Reactivation of “pluripotency” transcription factors in iPSC derivatives. (A) Semiquantitative RT-PCR assay of transgene expression in iPSC derivatives. Expression of pluripotency transcription factors showing reactivation of transgenes and endogenous expression of c-Myc, Oct4, and Nanog in iMSC-DS. Endogenous transcription factors c-Myc, Oct4, Nanog, and Sox2 are actively expressed in riPSC-DS, piPSC-DS, and iPSC-WTs at a level comparable to hESC (H9), while transgene transcription factors are predominantly silent. Fibroblasts DS—the fibroblasts AG06872 from which piPSC-DS and riPSC-DS were developed. iMSC-DS reverted mesenchymal stem cells that are in the process of forming pluripotent colonies; hESC H9—human embryonic stem cell line H9. (B) Transgene expression of pluripotency transcription factors showing reactivation of transgenes c-Nanog, c-Myc, and Oct4 in iMSC-DS that underwent reversion and formed pluripotent colonies (iMSC1-DS reverted) and in mesenchymal stem cells (iMSC-DS) of a previous passage that did not form colonies. A low level of transgene expression was detected in some of the riPSC-DS and piPSC-DS clones. The transgene reactivation was also detected in other iPSC-DS-differentiated derivatives, such as iMSC2-DS and iEC-DS. Cell lines tested: iMSC2-DS—mesenchymal stem cells differentiated from piPSC-DS cells (single cell/blast colony protocol was used); iEC-DS—endothelial cells developed from piPSC-DS. (C) RT-qPCR showing significant reactivation of transgenes Nanog (nearly 300-fold) and c-Myc (77-fold) in iMSC-DS reverted cells. In contrast, there is a low expression level of Nanog in iEC-DS cells, but the c-Myc expression in these cells increased 10-fold. c-Myc expression is detected in the other samples as well. In addition to this, there is insignificant variable expression of Oct4 and Sox2 transgenes in all samples. (D) RT-qPCR showing Nanog and c-Myc reactivation in differentiated phenotypes derived from both DS and WT iPSCs. In comparison to the iMSC-DS reverted cells, the transgene reactivation in differentiated WT iPS derivatives is of a smaller magnitude. There is a significant 66-fold upregulation of Nanog and a 13-fold upregulation of c-Myc in iMSC1-WT cells. The WT endothelial cells (iEC-WT) have a 12-fold upregulation of Nanog and no significant upregulation of c-Myc. In the iMSC2-WT cells, no significant upregulation of transgenes was detected. Cell lines tested (A–C): piPSC-DS—iPS line developed from AG06872 fibroblasts; iMSC1-DS reverted—iMSC-DS (protocol 1) that underwent reversion and formed pluripotent colonies; riPSC-DS clone 1—the reverted secondary iPSC-DS cells; iMSC2-DS—mesenchymal stem cells differentiated from piPSC-DS cells (protocol 2); iEC-DS—endothelial cells developed from piPSC-DS cells; iPSC-WT—iPSC line developed from MRC5 fibroblasts; iMSC1-WT—mesenchymal stem cells derived from iPSC-WT cells (protocol 1); iMSC2-WT—mesenchymal stem cells derived from iPSC-WT cells (protocol 2); and iEC-WT—endothelial cells developed from iPSC-WT. iEC, endothelial cell; iPSC, induced pluripotent stem cell; iMSC-DS, Down syndrome mesenchymal stem cells; hESC, human embryonic stem cell; RT-PCR, reverse transcription polymerase chain reaction.
FIG. 5.
Summary chart of iPSC derivatives. (A) Mesenchymal differentiation steps (protocol 2). Via this protocol, iMSC2 cells were obtained from both WT and DS iPSCs. From left to right: (1) FACS analysis shows that the differentiated cells are negative for CD34, and more than 99% of the cells express CD73; (2) colony-forming unit assay: iPSC cultures are collected on day 3 of OP9 coculture and grown for 14 days in MethoCult semisolid serum-free media to generate mesoblast progenitors; (3) an enlarged image of a mesoblast colony developed in semisolid media; and (4) morphology of adherent iMSC2 cells, which were expanded from a mesoblast colony. (B) Phenotypic and functional characterization of endothelial cells obtained from both DS and WT iPSCs (iEC). From left to right: (1) FACS analysis shows that more than 99% of the cells coexpress VE-cadherin (CD144) and PECAM1 (CD31); (2) tube formation assay (calcein AM staining); (3) immunostaining: VE-cadherin expression (green), and cells were costained with DAPI (blue); and (4) morphology of adherent iEC at passage 5. (C) Summary chart of the investigated iPSC derivatives, cell lines, and clones. iPSC-WT and iPSC-DS cell lines were subjected to three differentiation processes: MSCs derived by protocol 1 (iMSC1), MSCs derived by protocol 2 (iMSC2), and endothelial cells (iEC). RT-PCR analysis revealed upregulation of Nanog and c-Myc transgenes in the differentiated cell lines. The figure shows, via an arrow representation, which transgene reactivation was detected in the cell lines. The arrow length is proportional to the activation level. The iMSC1-DS cells showed the highest degree of transgene upregulation, which triggered the reversion to pluripotency. Three clones were established from the reverted iPSC cell line. DS, Down syndrome; iPSC, induced pluripotent stem cell; RT-PCR, reverse transcription polymerase chain reaction.
Surprisingly, we detected transgene reactivation in the iMSC1-WT and iEC-WT phenotypes (Fig. 4D). In comparison to the DS reversed MSCs, this reactivation was of a smaller magnitude. There was a significant 66-fold upregulation of Nanog and a 13-fold upregulation of c-Myc in iMSC1-WT cells. The WT endothelial cells (iEC-WT) had a 12-fold upregulation of Nanog and no significant upregulation of c-Myc. Regarding the iMSC2-WT cells, no significant upregulation of transgenes was detected. The upregulation of Nanog (257-fold) and c-Myc (112-fold) in iMSC1-DS only slightly differed from the previous experiment's results. This small difference may have occurred as a result of the use of a different brand of PCR mix. Nonetheless, all these results indicate that transgene reactivation occurs sporadically during various differentiation processes.
Discussion
In this work, we present the first demonstration of profound upregulation of transgene expression in mature differentiated phenotypes, such as endothelial and MSCs. We thoroughly evaluate the reverted culture with regard to undifferentiated cell contamination and firmly eliminate this possibility. By analyzing transgene levels across various cell types, we demonstrate that transgene upregulation leads to morphological changes, and in some cases, it initiates a reversion to pluripotency. The summary chart (Fig. 5C) shows the cell types and clones used in the study as well as the cell lines they were derived from. The figure also shows, via an arrow representation, which transgene reactivation was detected in the cell lines. The arrow length is proportional to the activation level.
We propose that a reversal to pluripotency occurs when some of the transgenes experience an upregulation greater than 100-fold in magnitude (257-fold for Nanog and 112-fold for c-Myc in this study), and the same mechanism may be responsible for the development of in vivo tumors. Furthermore, we propose to introduce the evaluation of the transgene status in iPSC derivatives since its activation may be associated with loss of cellular fidelity and impact the outcomes of disease modeling and related research.
Residual transgene expression in pluripotent cells
We have shown that the transgenes were predominantly silent in our piPSCs. Similarly, Park et al. described that low-level transgene expression (“largely silenced”) is frequently found for disease-specific iPSC lines ADA-iPS2, PD-iPS1, HD-iPS1, DMD-iPS1, SBDS-iPS3, GD-iPS3, and so forth. This residual expression did not interfere with the formation of teratoma and mature cell formation [5,28]. Notably, several groups reported continued transgene expression in domestic animals, such as porcine iPS cells [29–31] and bovine [32], which were nevertheless capable of forming teratoma and contribute to chimeras (reviewed [33]). In contrast, transgene expression may have contributed to the fluctuations of pluripotency observed in spontaneously differentiating cultures [34]. The authors described a cell population (P4 subset, which included a fraction of low and negative cells for CD9/GCTM-2) that expressed pluripotent transcription factors at low levels, which then reverted to medium or high expression. They proposed that residual expression of the viral transgene might drive the secondary reprogramming.
Transgene reactivation in differentiated cells
Numerous studies, including those describing disease models, publish data based on cells derived from iPSCs produced with integrative constructs without assessing transgene reactivation [35–40] because transgene silencing normally takes place during the differentiation of embryonic stem cells. Precisely for the reason of avoiding gene silencing during the differentiation, the human homolog of the ROSA26 locus, used in mouse ESCs, has been identified for constitutive ubiquitous gene expression in human pluripotent cells [13–15]. Our finding demonstrates that transgene activation instead of expected silencing in iPSC derivatives may occur, thus underlying a necessity of assessing transgene reactivation in the cells differentiated from iPSCs.
Factors that could potentially trigger the transgene reactivation
Specifics of differentiation may play a role since mesenchymal differentiation involves activation of epithelial to mesenchymal transition (EMT) genes important in cancer transformation [12,41]. Endothelial cell specification also involves EMT at the progenitor stage of development. The combination of reprogramming factor may play a role [42]. We used a modified (“enhanced”) combination of factors [6,43] with Klf4 replaced by Nanog. The choice of “strong” TFs leads to faster reprogramming, which may be associated with accumulation of aberrations at genetic and epigenetic levels [44]. Activation of endogenous retroelements during reprogramming may change transcriptional control [45–47].
Remarkably, although individuals with DS have a reduced risk of solid tumor development, the reversion to pluripotency occurred in the MSC derivative of the iPSC-DS line with trisomy 21. In contrast, the the euploid iPSC-WT-differentiated derivatives, which were developed and handled in similar conditions, did not show signs of reversion to pluripotency. This result is likely associated with the lower levels of transgene expression detected in iEC-WT and iMSC1-WT cell lines. We are not aware of any indications suggesting possible contribution of trisomy 21 toward transgene reactivation or reversal to the pluripotency.
Reversion to pluripotency: confirmation that riPSC-DS clones developed by reversion to pluripotency of MSCs
Our initial assumption was that undifferentiated cells, which may have remained during differentiation, were the source of pluripotent colony growth in MSC cultures. Careful experimentation and analysis ruled out this possibility. Our data show that secondary reprogramming began after profound upregulation of transgene expression in MSCs. This strongly suggests that transgene reactivation initiated the secondary reprogramming. Similar to piPSC-DS, newly isolated riPSC-DS had no visible transgene expression, which further confirms that pluripotent cells did not contribute to transgene upregulation. Additionally, some of the isolated riPSC-DS clones had a normal karyotype that distinguished them from piPSC-DS. Flow analysis showed the absence of pluripotent cells in the MSC culture passage preceding the reversion. In the reverted culture, we documented the transformation of the cellular phenotype, the appearance of epithelial and transitional morphology, the activation of pluripotency transcription factors, and the events characteristic of iPSC generation, which is believed to undergo the MET reviewed in [23,24]. Time-lapse movies showed somatic cell shape transformation and pluripotent colony aggregation. Based on this evidence, we determined that secondary reprogramming and reversion to pluripotency resulted in the formation of pluripotent colonies in MSCs.
Based on the investigation of several types of iPSC derivatives and the fact that reactivation of transgene expression does not necessarily initiate secondary reprogramming, we hypothesized that higher levels of transgene re-expression (eg, 112-fold is shown for c-Myc and 257-fold for Nanog) are needed to trigger culture transformation and colony formation. This is the first demonstration of an event that led to a spontaneous secondary reprogramming and reversion to pluripotency in characterized MSCs derived from iPSCs in an in vitro cell culture.
Future considerations
Although transgene activation may only be possible in integrative systems, evidence indicates that integration free technology of cellular reprogramming also increases the risk of impaired cellular fidelity associated with the consequences of forced reversal of somatic cells to iPSCs. The comparative studies of pluripotent cells, with hESCs serving as the gold standard, provide important insights. Our group reported earlier evidence of differences between hESCs and iPSCs—specifically the discovery of cancer hallmarks in iPSCs, indicated by perturbations of expression of key members of the Nodal embryonic signaling pathway, which is critical to the maintenance of pluripotency. Furthermore, we uncovered a dramatic difference in expression of cancer-related miRNAs in iPSCs [48].
In another multicenter study, aberrant methylation and numerous deletions of tumor suppressor genes that were absent from the somatic cells of origin were found in iPSCs after induction of pluripotency [49]. Other authors also reported that the same epigenetic memory and copy number variations are exhibited by hiPSC lines produced with nonintegrative approaches [50,51]. One study, which involved integrative and nonintegrative vectors [52], revealed a higher gene expression overlap of the hiPSC derivatives with cancer compared with hESC derivatives. It was recently found that the reprogramming per se triggers endogenous L1 and Alu retrotransposition in hiPSCs, which can change the functional landscape of the hiPSC genome and alter cellular phenotype [45–47]. These preliminary observations indicate that even though errors at the genetic and epigenetic levels have been described in both iPSCs and hESCs [49,53–55], the differentiated derivatives of iPSCs may remain more inclined to cancer transformation than the derivatives of fellow hESCs. As a result, although the use of nonintegrative vectors for reprogramming is well justified, the derivatives of hiPSCs might still bear some neoplastic properties.
It is widely assumed that the development of in vivo tumors, following the introduction of pluripotent derivatives into the tissue of adult animals, originates from residual undifferentiated pluripotent stem cells. Consequently, a number of recommendations have been developed with the aim of eliminating undifferentiated cells from stem cell transplants, for example [56,57] (reviewed [58]). Given the evidence of in vitro reversal of differentiated iPS cells to pluripotency, we offer the hypothesis that a similar process may be responsible for the development of in vivo tumors. In this model, purification of iPSC derivatives from undifferentiated cells alone does not guarantee sufficient safety of a therapeutic product. It also explains the variety of cellular types found in tumors, which cannot be explained by cancer transformation of iPSC-differentiated derivatives.
In this study, we demonstrate the reactivation of Nanog, in addition to the c-Myc transgene reported earlier for in vivo tumors, which emphasizes that reprogrammed iPSCs in the absence of c-Myc might still bear a risk of tumorigenesis. The lines described in this study provide a valuable model for dissecting microenvironmental components that can promote transgene reactivation. We anticipate these lines to be useful for verification of prognostic markers that would distinguish “safe” cellular lineages from those that underwent reprogramming to pluripotency [34,59]. Furthermore, our cells can aid the development of safety evaluations for differentiated derivatives. Most importantly, this new finding strongly suggests that the results of studies that involve iPSCs generated with retro- or lentiviruses should be carefully interpreted and thoroughly examined with respect to transgene reactivation in iPSC derivatives.
Supplementary Material
Acknowledgments
These studies were supported by NIH CA 12205 grant to M.J.C.H., Eisenberg Foundation for Charities (P.M.I.), and Stanley Manne Children's Research Institute's grant to V.G. We thank Kathrin Plath (University of California, Los Angeles) for providing the sequences of PCR primers, Irina Elcheva, Alina Gilgur, McCabe Chris, and George David for technical help, and Vladimir Galat for graphic design.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Blanpain C, Daley GQ, Hochedlinger K, Passegue E, Rossant J. and Yamanaka S. (2012). Stem cells assessed. Nat Rev Mol Cell Biol 13:471–476 [DOI] [PubMed] [Google Scholar]
- 2.Verlinsky Y, Strelchenko N, Kukharenko V, Rechitsky S, Verlinsky O, Galat V. and Kuliev A. (2005). Human embryonic stem cell lines with genetic disorders. Reprod Biomed Online 10:105–110 [DOI] [PubMed] [Google Scholar]
- 3.Sterneckert JL, Reinhardt P. and Scholer HR. (2014). Investigating human disease using stem cell models. Nat Rev Genet 15:625–639 [DOI] [PubMed] [Google Scholar]
- 4.Benam KH, Dauth S, Hassell B, Herland A, Jain A, Jang KJ, Karalis K, Kim HJ, MacQueen L,et al. (2015). Engineered in vitro disease models. Annu Rev Pathol 10:195–262 [DOI] [PubMed] [Google Scholar]
- 5.Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, Lensch MW, Cowan C, Hochedlinger K. and Daley GQ. (2008). Disease-specific induced pluripotent stem cells. Cell 134:877–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takahashi K. and Yamanaka S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126:663–676 [DOI] [PubMed] [Google Scholar]
- 7.Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE. and Jaenisch R. (2007). In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature 448:318–324 [DOI] [PubMed] [Google Scholar]
- 8.Okita K, Ichisaka T. and Yamanaka S. (2007). Generation of germline-competent induced pluripotent stem cells. Nature 448:313–317 [DOI] [PubMed] [Google Scholar]
- 9.Okano H, Nakamura M, Yoshida K, Okada Y, Tsuji O, Nori S, Ikeda E, Yamanaka S. and Miura K. (2013). Steps toward safe cell therapy using induced pluripotent stem cells. Circ Res 112:523–533 [DOI] [PubMed] [Google Scholar]
- 10.West FD, Uhl EW, Liu Y, Stowe H, Lu Y, Yu P, Gallegos-Cardenas A, Pratt SL. and Stice SL. (2011). Brief report: chimeric pigs produced from induced pluripotent stem cells demonstrate germline transmission and no evidence of tumor formation in young pigs. Stem Cells 29:1640–1643 [DOI] [PubMed] [Google Scholar]
- 11.Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N. and Yamanaka S. (2008). Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol 26:101–106 [DOI] [PubMed] [Google Scholar]
- 12.Nori S, Okada Y, Nishimura S, Sasaki T, Itakura G, Kobayashi Y, Renault-Mihara F, Shimizu A, Koya I,et al. (2015). Long-term safety issues of iPSC-based cell therapy in a spinal cord injury model: oncogenic transformation with epithelial-mesenchymal transition. Stem Cell Reports 4:360–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pfaff N, Lachmann N, Ackermann M, Kohlscheen S, Brendel C, Maetzig T, Niemann H, Antoniou MN, Grez M,et al. (2013). A ubiquitous chromatin opening element prevents transgene silencing in pluripotent stem cells and their differentiated progeny. Stem Cells 31:488–499 [DOI] [PubMed] [Google Scholar]
- 14.Ellis J. (2005). Silencing and variegation of gammaretrovirus and lentivirus vectors. Hum Gene Ther 16:1241–1246 [DOI] [PubMed] [Google Scholar]
- 15.Irion S, Luche H, Gadue P, Fehling HJ, Kennedy M. and Keller G. (2007). Identification and targeting of the ROSA26 locus in human embryonic stem cells. Nat Biotechnol 25:1477–1482 [DOI] [PubMed] [Google Scholar]
- 16.Boyd NL, Robbins KR, Dhara SK, West FD. and Stice SL. (2009). Human embryonic stem cell-derived mesoderm-like epithelium transitions to mesenchymal progenitor cells. Tissue Eng Part A 15:1897–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vodyanik MA, Yu J, Zhang X, Tian S, Stewart R, Thomson JA. and Slukvin , II (2010). A mesoderm-derived precursor for mesenchymal stem and endothelial cells. Cell Stem Cell 7:718–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimbrel EA, Kouris NA, Yavanian GJ, Chu J, Qin Y, Chan A, Singh RP, McCurdy D, Gordon L, Levinson RD. and Lanza R. (2014). Mesenchymal stem cell population derived from human pluripotent stem cells displays potent immunomodulatory and therapeutic properties. Stem Cells Dev 23:1611–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slukvin , II and Vodyanik M. (2011). Endothelial origin of mesenchymal stem cells. Cell Cycle 10:1370–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malchenko S, Xie J, de Fatima Bonaldo M, Vanin EF, Bhattacharyya BJ, Belmadani A, Xi G, Galat V, Goossens W,et al. (2014). Onset of rosette formation during spontaneous neural differentiation of hESC and hiPSC colonies. Gene 534:400–407 [DOI] [PubMed] [Google Scholar]
- 21.Galat V, Malchenko S, Galat Y, Ishkin A, Nikolsky Y, Kosak ST, Soares BM, Iannaccone P, Crispino JD. and Hendrix MJ. (2012). A model of early human embryonic stem cell differentiation reveals inter- and intracellular changes on transition to squamous epithelium. Stem Cells Dev 21:1250–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simpson DL, Wehman B, Galat Y, Sharma S, Mishra R, Galat V. and Kaushal S. (2014). Engineering patient-specific valves using stem cells generated from skin biopsy specimens. Ann Thorac Surg 98:947–954 [DOI] [PubMed] [Google Scholar]
- 23.Papp B. and Plath K. (2013). Epigenetics of reprogramming to induced pluripotency. Cell 152:1324–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.David L. and Polo JM. (2014). Phases of reprogramming. Stem Cell Res 12:754–761 [DOI] [PubMed] [Google Scholar]
- 25.Guillot PV, Gotherstrom C, Chan J, Kurata H. and Fisk NM. (2007). Human first-trimester fetal MSC express pluripotency markers and grow faster and have longer telomeres than adult MSC. Stem Cells 25:646–654 [DOI] [PubMed] [Google Scholar]
- 26.Pierantozzi E, Gava B, Manini I, Roviello F, Marotta G, Chiavarelli M. and Sorrentino V. (2011). Pluripotency regulators in human mesenchymal stem cells: expression of NANOG but not of OCT-4 and SOX-2. Stem Cells Dev 20:915–923 [DOI] [PubMed] [Google Scholar]
- 27.Li LB, Chang KH, Wang PR, Hirata RK, Papayannopoulou T. and Russell DW. (2012). Trisomy correction in Down syndrome induced pluripotent stem cells. Cell Stem Cell 11:615–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW. and Daley GQ. (2008). Reprogramming of human somatic cells to pluripotency with defined factors. Nature 451:141–146 [DOI] [PubMed] [Google Scholar]
- 29.West FD, Terlouw SL, Kwon DJ, Mumaw JL, Dhara SK, Hasneen K, Dobrinsky JR. and Stice SL. (2010). Porcine induced pluripotent stem cells produce chimeric offspring. Stem Cells Dev 19:1211–1220 [DOI] [PubMed] [Google Scholar]
- 30.Nakano K, Watanabe M, Matsunari H, Matsuda T, Honda K, Maehara M, Kanai T, Hayashida G, Kobayashi M,et al. (2013). Generating porcine chimeras using inner cell mass cells and parthenogenetic preimplantation embryos. PLoS One 8:e61900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montserrat N, Bahima EG, Batlle L, Hafner S, Rodrigues AM, Gonzalez F. and Izpisua Belmonte JC. (2011). Generation of pig iPS cells: a model for cell therapy. J Cardiovasc Transl Res 4:121–130 [DOI] [PubMed] [Google Scholar]
- 32.Sumer H, Liu J, Malaver-Ortega LF, Lim ML, Khodadadi K. and Verma PJ. (2011). NANOG is a key factor for induction of pluripotency in bovine adult fibroblasts. J Anim Sci 89:2708–2716 [DOI] [PubMed] [Google Scholar]
- 33.Koh S. and Piedrahita JA. (2014). From “ES-like” cells to induced pluripotent stem cells: a historical perspective in domestic animals. Theriogenology 81:103–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Polanco JC, Ho MS, Wang B, Zhou Q, Wolvetang E, Mason E, Wells CA, Kolle G, Grimmond SM,et al. (2013). Identification of unsafe human induced pluripotent stem cell lines using a robust surrogate assay for pluripotency. Stem Cells 31:1498–1510 [DOI] [PubMed] [Google Scholar]
- 35.Singh R, Kuai D, Guziewicz KE, Meyer J, Wilson M, Lu J, Smith M, Clark E, Verhoeven A, Aguirre GD. and Gamm DM. (2015). Pharmacological modulation of photoreceptor outer segment degradation in a human iPS cell model of inherited macular degeneration. Mol Ther 23:1700–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shoji E, Sakurai H, Nishino T, Nakahata T, Heike T, Awaya T, Fujii N, Manabe Y, Matsuo M. and Sehara-Fujisawa A. (2015). Early pathogenesis of Duchenne muscular dystrophy modelled in patient-derived human induced pluripotent stem cells. Sci Rep 5:12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fujiwara N, Shimizu J, Takai K, Arimitsu N, Ueda Y, Wakisaka S, Suzuki T. and Suzuki N. (2015). Cellular and molecular mechanisms of the restoration of human APP transgenic mouse cognitive dysfunction after transplant of human iPS cell-derived neural cells. Exp Neurol 271:423–431 [DOI] [PubMed] [Google Scholar]
- 38.Hossini AM, Megges M, Prigione A, Lichtner B, Toliat MR, Wruck W, Schroter F, Nuernberg P, Kroll H,et al. (2015). Induced pluripotent stem cell-derived neuronal cells from a sporadic Alzheimer's disease donor as a model for investigating AD-associated gene regulatory networks. BMC Genomics 16:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han F, Wang W, Chen B, Chen C, Li S, Lu X, Duan J, Zhang Y, Zhang YA, Guo W. and Li G. (2015). Human induced pluripotent stem cell-derived neurons improve motor asymmetry in a 6-hydroxydopamine-induced rat model of Parkinson's disease. Cytotherapy 17:665–679 [DOI] [PubMed] [Google Scholar]
- 40.Djuric U, Cheung AY, Zhang W, Mok RS, Lai W, Piekna A, Hendry JA, Ross PJ, Pasceri P,et al. , (2015). MECP2e1 isoform mutation affects the form and function of neurons derived from Rett syndrome patient iPS cells. Neurobiol Dis 76:37–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eid JE. and Garcia CB. (2015). Reprogramming of mesenchymal stem cells by oncogenes. Semin Cancer Biol 32:18–31 [DOI] [PubMed] [Google Scholar]
- 42.Buganim Y, Markoulaki S, van Wietmarschen N, Hoke H, Wu T, Ganz K, Akhtar-Zaidi B, He Y, Abraham BJ,et al. , (2014). The developmental potential of iPSCs is greatly influenced by reprogramming factor selection. Cell Stem Cell 15:295–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V,et al. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science 318:1917–1920 [DOI] [PubMed] [Google Scholar]
- 44.Gao Y. and Gao S. (2014). Quality control: H2A.X links to better iPSCs. Cell Stem Cell 15:259–260 [DOI] [PubMed] [Google Scholar]
- 45.Klawitter S, Fuchs NV, Upton KR, Munoz-Lopez M, Shukla R, Wang J, Garcia-Canadas M, Lopez-Ruiz C, Gerhardt DJ,et al. (2016). Reprogramming triggers endogenous L1 and Alu retrotransposition in human induced pluripotent stem cells. Nat Commun 7:10286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wissing S, Munoz-Lopez M, Macia A, Yang Z, Montano M, Collins W, Garcia-Perez JL, Moran JV. and Greene WC. (2012). Reprogramming somatic cells into iPS cells activates LINE-1 retroelement mobility. Hum Mol Genet 21:208–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Friedli M, Turelli P, Kapopoulou A, Rauwel B, Castro-Diaz N, Rowe HM, Ecco G, Unzu C, Planet E,et al. (2014). Loss of transcriptional control over endogenous retroelements during reprogramming to pluripotency. Genome Res 24:1251–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Malchenko S, Galat V, Seftor EA, Vanin EF, Costa FF, Seftor RE, Soares MB. and Hendrix MJ. (2010). Cancer hallmarks in induced pluripotent cells: new insights. J Cell Physiol 225:390–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laurent LC, Ulitsky I, Slavin I, Tran H, Schork A, Morey R, Lynch C, Harness JV, Lee S,et al. (2011). Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell 8:106–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O'Malley R, Castanon R,et al. (2011). Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 471:68–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hussein SM, Batada NN, Vuoristo S, Ching RW, Autio R, Narva E, Ng S, Sourour M, Hamalainen R,et al. (2011). Copy number variation and selection during reprogramming to pluripotency. Nature 471:58–62 [DOI] [PubMed] [Google Scholar]
- 52.Ghosh Z, Huang M, Hu S, Wilson KD, Dey D. and Wu JC. (2011). Dissecting the oncogenic and tumorigenic potential of differentiated human induced pluripotent stem cells and human embryonic stem cells. Cancer Res 71:5030–5039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nazor KL, Altun G, Lynch C, Tran H, Harness JV, Slavin I, Garitaonandia I, Muller FJ, Wang YC,et al. (2012). Recurrent variations in DNA methylation in human pluripotent stem cells and their differentiated derivatives. Cell Stem Cell 10:620–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ma H, Morey R, O'Neil RC, He Y, Daughtry B, Schultz MD, Hariharan M, Nery JR, Castanon R,et al. (2014). Abnormalities in human pluripotent cells due to reprogramming mechanisms. Nature 511:177–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Amps K, Andrews PW, Anyfantis G, Armstrong L, Avery S, Baharvand H, Baker J, Baker D, Munoz MB,et al. (2011). Screening ethnically diverse human embryonic stem cells identifies a chromosome 20 minimal amplicon conferring growth advantage. Nat Biotechnol 29:1132–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tateno H, Onuma Y, Ito Y, Minoshima F, Saito S, Shimizu M, Aiki Y, Asashima M. and Hirabayashi J. (2015). Elimination of tumorigenic human pluripotent stem cells by a recombinant lectin-toxin fusion protein. Stem Cell Reports 4:811–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wyles SP, Yamada S, Oommen S, Maleszewski JJ, Beraldi R, Martinez-Fernandez A, Terzic A. and Nelson TJ. (2014). Inhibition of DNA topoisomerase II selectively reduces the threat of tumorigenicity following induced pluripotent stem cell-based myocardial therapy. Stem Cells Dev 23:2274–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ben-David U. and Benvenisty N. (2014). Chemical ablation of tumor-initiating human pluripotent stem cells. Nat Protoc 9:729–740 [DOI] [PubMed] [Google Scholar]
- 59.Wu T, Liu Y, Wen D, Tseng Z, Tahmasian M, Zhong M, Rafii S, Stadtfeld M, Hochedlinger K. and Xiao A. (2014). Histone variant H2A.X deposition pattern serves as a functional epigenetic mark for distinguishing the developmental potentials of iPSCs. Cell Stem Cell 15:281–294 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.