Abstract
Post-translational modifications (PTMs) of histones regulate chromatin structure and function. Because nucleosomes contain two copies each of the four core histones, the establishment of different PTMs on individual ‘sister’ histones in the same nucleosomal context, i.e. asymmetric histone PTMs, are difficult to analyze. Here, we generated differentially isotope-labeled nucleosomes to study asymmetric histone modification crosstalk by time-resolved NMR spectroscopy. Specifically, we delineate mechanistic insights into nucleosomal histone H3 modification reactions in cis and in trans, hence within individual H3 copies, or between them. We validated our approach using the H3S10phK14ac crosstalk mechanism, mediated by Gcn5 acetyltransferase. Moreover, phosphorylation assays on methylated substrates identified Haspin kinase able under certain conditions to produce nucleosomes decorated asymmetrically with two distinct types of PTMs.
Keywords: NMR spectroscopy, protein modifications, chromatin biology, epigenetics, asymmetrically modified nucleosomes
PTMs of histones work in concert through the formation of combinatorial patterns or histone codes to regulate a plethora of fundamental biological processes, including transcription, replication, recombination and DNA repair. [1,2] Most of these PTMs occur in the unstructured N- or C-terminal tails of core histones and at closely spaced modification sites, giving rise to functional networks of reciprocal PTM crosstalk. [3] Although a few PTMs are deposited on free histone substrates, the vast majority are placed on nucleosome-incorporated histones. The nucleosome is a symmetric structure consisting of ~147bp of DNA wrapped around a histone octamer made up of two copies each of the four-core histones. [4] Historically, and because of the inherent symmetry of the nucleosomal architecture, histone PTMs were thought to exclusively occur in a symmetrical fashion, with both copies of each of the core histones modified in exactly the same manner. This notion was recently challenged by two studies demonstrating the mechanistic basis for establishing asymmetric histone H3 phosphorylations in vitro by certain kinases, [5] and the existence of asymmetrically methylated nucleosomes in embryonic stem cells, fibroblasts and cancer cells. [6] As such, asymmetrically modified nucleosomes represent a novel concept in chromatin biology and their existence raises important biological questions with regard to their establishment and crosstalk in cis, i.e. within individual histone tails, and in trans, i.e. between the two copies of sister histones that this asymmetry might impose on the propagation of histone marks.
Most analytical methods to investigate histone PTMs on nucleosomal substrates fall short in verifying the copy-specific origin of the detected modifications. By proteolytically processing modified nucleosomes for mass spectrometry analysis, for example, individual histone tails are cleaved off from core particles and detected in mixtures of modified peptides, which effectively ‘erases’ all information about whether they originated from the same, or from different nucleosomes. To preserve this information, histone PTMs ought to be studied in a native nucleosomal context and with analytical tools that provide means to distinguish between individual copies of assembled core histones, a veritable experimental challenge altogether.
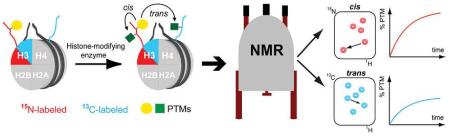
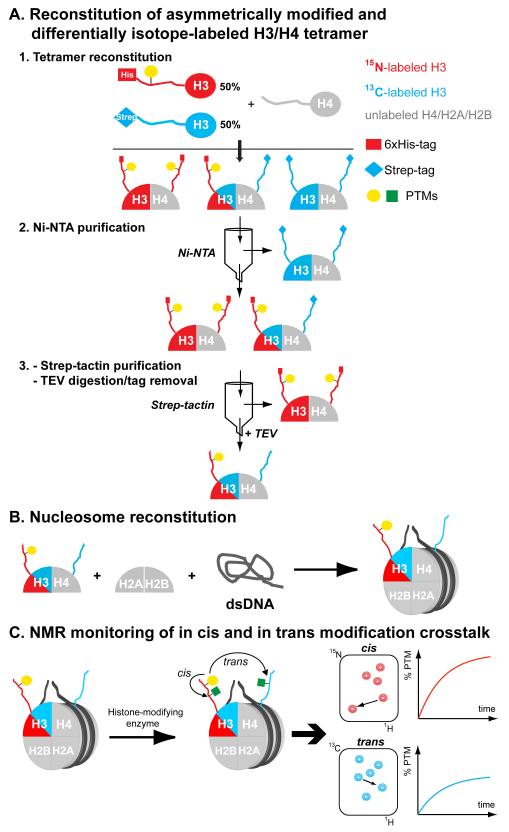
Here, we present the generation of both differentially isotope-labeled and asymmetrically modified nucleosomes (from hereon ‘asNucs’) to study modification crosstalk in cis and in trans. Practically, we reconstituted nucleosomes containing the individual copies of histone H3 incorporated in a 15N- or 13C-enriched form. Additionally, one of the two H3 copies was incorporated in a pre-modified mode. Thus, 15N-, or 13C-edited NMR experiments allow for the selective detection of either nucleosomal copy of histone H3 in a completely unbiased and direct manner. Accordingly, by employing NMR routines to detect different types of PTMs, developed over the past years in our laboratory, [7,8] we illustrate the unique ability to monitor individual histone modifications and modification crosstalk in cis and in trans by time-resolved NMR readouts. To prepare asNucs, we took advantage of a tandem affinity purification scheme of asymmetric histone sub-complexes previously introduced by Voigt et al.[6] Similarly, we prepared two versions of full-length histone H3, carrying either a His- or a Strep-affinity tag at their N-termini and being asymmetrically modified after reacting one of the two pools with a specific enzyme. Additionally, we recombinantly expressed and purified the two H3 pools from either 15N or 13C isotope-enriched growth media. After following standard protocols, [9] we reconstituted H3/H4 tetramers and subsequently asNucs using unlabeled histones H2A, H2B, H4 and DNA (Scheme 1a,b). Next, we reacted those asNucs with H3-modifying enzymes, followed by time-resolved NMR monitoring of the 15N or 13C ‘channel’ to report on cis or trans modification crosstalk, respectively (Scheme 1c).
Scheme 1.
Reconstitution of asymmetrically modified and isotope-labeled nucleosomes for NMR-monitoring of asymmetric modification patterns. A) Tandem affinity purification with Ni-NTA and Strep-tactin chromatography of asymmetrically modified and differentially isotope-labeled H3/H4 tetramers (asH3/H4). B) Nucleosome reconstitution by combining asH3/H4 with unlabeled H2A/H2B dimers and a DNA nucleosome-positioning sequence. C) Mixing of asNucs with histone-modifying enzymes enables NMR-monitoring of modification crosstalk in cis and in trans.
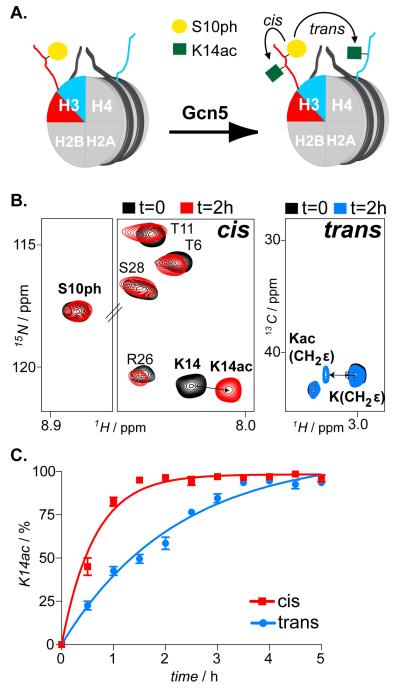
Figure 1.
In cis and in trans modification crosstalk imposed by H3S10ph on H3K14 acetylation activity of Gcn5. A) Schematic illustration of asymmetrically phosphorylated on H3S10 nucleosomes reacted with Gcn5 acetyltransferase enables H3K14ac mapping, in cis and in trans. B) 1H/15N SOFAST-HMQC and 1H/13C-HSQC spectra (selected regions) of asNucs before and after reaction with Gcn5. C) Time-resolved NMR readouts of H3K14ac by Gcn5 on H3S10ph asNucs.
To exemplify the power of this approach in delineating subtle differences on histone copy-specific modification rates, we studied acetylation of H3K14 by the acetyltransferase Gcn5 in the absence, or presence of H3 S10 phosphorylation, which has previously been shown to enhance Gcn5 activity both in vitro [8] and in vivo. [10] To this end, we reconstituted asNucs phosphorylated on S10 of histone H3. Additionally, we incorporated different isotopes (the H3S10ph species was 15N-and the unmodified species 13C-labeled). To prepare fully phosphorylated H3 on S10, we reacted the 15N-labeled H3 with Aurora B kinase and assessed complete phosphorylation by following the characteristic shift of the S10 amide NMR signal. (Figure S1 and Supporting methods). We used this H3 pool to initially reconstitute asymmetric H3/H4 tetramers. To verify that the tandem purification scheme resulted on highly pure asymmetric complexes, we took advantage of the only tryptophan that exists in the system, which comes from the Strep-tag, since histones are lacking this amino acid. We followed the characteristic NMR resonance of the tryptophan side chain NHε throughout the purification protocol, assessing the presence of the Strep-tag, hence the 13C-labeled H3 copy. Indeed, pure asymmetric H3/H4 tetramers were isolated, as concluded after comparing the proton (1H) NMR spectra of H3/H4 tetramer pools from different purification phases (Figure S2). The latter were mixed with H2A/H2B dimers and a 165bp-long DNA fragment containing a strong nucleosome-positioning sequence and using the gradient-dialysis method [9] we reconstituted asNucs, phosphorylated on H3S10 (Figure S3). Subsequently, we reacted this preparation with Gcn5 and we monitored H3K14ac on both H3 tails by recording interleaved 1H/15N and 1H/13C correlation spectra (Figure 1a,b and Supporting methods). We followed H3K14ac of Ser10-phosporylated, 15N isotope-enriched H3 by monitoring characteristic chemical shift changes of K14 amide signal, whereas CH2ε resonance displacements reported on K14ac of the unmodified, 13C-enriched copy of nucleosomal H3. The latter was deduced from a test acetylation reaction with a free H3 tail peptide (Figure S4). By comparing the K14ac levels over time on individual copies of H3, we confirmed that H3S10ph exerted a clear stimulatory effect on the Gcn5 reaction at K14, in cis (Figure 1c).
To further demonstrate the flexibility of the employed labeling approach, we performed the same experiment, but having this time pre-phosphorylated the 13C-enriched H3, using the Cβ group of S10 to assess efficient phosphorylation (Figure S5 and Supporting note). Accordingly, we reconstituted nucleosomes and re-determined K14ac levels by Gcn5. As before, kinetic analyses with these nucleosomes showed faster acetylation of the S10 pre-phosphorylated H3 copy (Figure S6), thus confirming the stimulatory role of S10ph on Gcn5 activity in cis compared to trans. This effect is nucleosome-specific since Gcn5 processes both unmodified and H3S10ph tail peptides similarly [5] and is directly linked to the influence that charge-modulating PTMs have on electrostatic H3 tail/DNA interactions. Particularly, H3S10ph disrupts the aforementioned transient contacts, hence promotes K14ac by rendering H3 tail more accessible to Gcn5. [8]
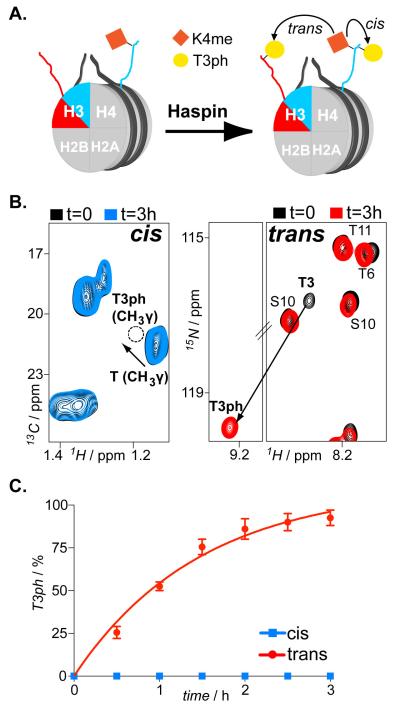
Having established the functionality of our approach, we set out to test its applicability for another important histone modification, lysine methylation. We focused on the methylphospho crosstalk between the adjacent T3 and K4 on histone H3. H3T3ph is a characteristic PTM of mitotic chromatin. [11] H3K4me has been correlated with promoters of active genes, [12] even though certain methylated states of H3K4 have been mapped also in centromeric chromatin, [13] with their levels persisting during mitosis. [14] We reacted 13C-labeled histone H3 with SET7/9 methyltransferase that methylates specifically and solely K4. [15] Following the corresponding CH2ε peak of lysines we monitored efficient H3K4 monomethylation (Figure S7 and Supporting methods). Subsequently, using an unmodified/15N-labeled His-H3 pool, we reconstituted asNucs on H3K4me1 (Figure S8). We reacted the latter with the Haspin kinase that specifically phosphorylates H3T3. [11] We monitored efficient phosphorylation of the unmodified copy of H3, whereas we detected no modification of the K4-methylated sister histone (Figure 2 and Supporting note). In the latter case, H3K4me1 completely abolished Haspin activity. This behavior was similarly displayed with isolated H3 tail peptides (Figure S9). Thus, the observed negative regulation resulted from an effect of H3K4me1 on the catalytic activity or substrate recognition by Haspin and was not imposed by the nucleosomal environment, as was the case with Gcn5 and the H3S10phK14ac crosstalk. Even though we could not test the effect of higher H3K4 methylation states due to inefficient activity of SET7/9 to produce relevant substrates, it is fair to expect a similar inhibitory effect also in those cases. Interestingly, asymmetrically modified H3K4me3 nucleosomes have been found in embryonic stem cells and their levels were partially retained upon differentiation.[6] Based on our data, if these nucleosomes encounter Haspin during mitosis, they will become asymmetrically modified also regarding H3T3ph. Do such nucleosomes containing two different types of PTMs −K methylation and T phosphorylation-, albeit on different sister histones, exist in native chromatin? Immunoaffinity purification coupled with quantitative LC/MS analysis [6] of mitotic extracts could initially identify such species before setting out to address their biological role.
Figure 2.
In cis and in trans modification crosstalk imposed by H3K4me on H3T3 phosphorylation activity of Haspin. A) Schematic illustration of asymmetrically methylated on H3K4 nucleosomes reacted with Haspin kinase enables H3T3ph mapping, in cis and in trans. B) 1H/15N SOFAST-HMQC and 1H/13C HSQC spectra (selected regions) of asNucs before and after reaction with Haspin kinase. C) Time-resolved NMR readouts of H3T3ph by Haspin on H3K4me asNucs
In summary, we introduce an approach to selectively detect and monitor asymmetric modification states of individual histone copies in fully assembled nucleosomes. By employing differential isotope-labeling and site-selective incorporation of histone PTMs, asymmetrically modified nucleosomes can be generated for time-resolved studies, addressing PTM crosstalk in cis and in trans. We used a well-characterized crosstalk example to test the functionality of our scheme. Additionally, we showed that Haspin kinase under certain conditions could generate asymmetrically phosphorylated and methylated nucleosomes. While we have outlined the applicability of our method for monitoring PTM reactions on individual sites of histone H3, this can be extended to other histones, or combinations of different histones. We analyzed crosstalks that involved the most common PTMs (S/T phosphorylation, K acetylation/methylation), but we have already delineated the NMR characteristics of equally or less abundant PTMs, such as arginine methylation, lysine crotonylation/propionylation, tyrosine phosphorylation. [16] Of outmost importance for the utility of our approach is the ability to prepare specifically and quantitatively modified histones that can be used for asNucs reconstitution. In cases where histone modifications cannot be established enzymatically, alternative schemes for the specific incorporation of modified amino acids can be employed, including non-natural amino acid technologies [17] or the use of cysteine mutant-derived PTM mimetics (histones are devoid of cysteines). [18] In addition, heteromeric DNA ligation of two mononucleosomes can be exploited as to construct dinucleosome templates that can be used to analyze internucleosomal effects. [19] Currently, limited data are available and only few PTMs have been found to occur asymmetrically in vivo, but many more are expected to come in the near future. At the same time, new chemical tools are emerging for reconstitution of asymmetrically modified nucleosomes to be used for functional studies. [20] Into this highly growing field, our approach aims to uncover mechanistic facets on the establishment and propagation of asymmetrically modified nucleosomes.
Supplementary Material
Acknowledgements
We thank Dr. Philipp Selenko (FMP-Berlin) for providing his lab to perform the study and for critically reading the manuscript and Dr. Peter Schmieder and Monica Beerbaum for excellent NMR infrastructure maintenance. This work was supported by a research grant from the Deutsche Forschungsgemeinschaft to S.L. (LI 2402/2-1) and from the National Institutes of Health to S.T. (GM088236 and GM11165).
Contributor Information
Dr. Stamatios Liokatis, Department of Structural Biology, Leibniz-Institut für Molekulare Pharmakologie Robert-Rössle-Strasse 10, 13125 Berlin (Germany).
Dr. Rebecca Klingberg, Department of Chemical Biology, Leibniz-Institut für Molekulare Pharmakologie Robert-Rössle-Strasse 10, 13125 Berlin (Germany)
Prof. Dr. Song Tan, Department of Biochemistry and Molecular Biology, The Pennsylvania State University University Park, PA 16802-1014 (USA)
Prof. Dr. Dirk Schwarzer, Department of Chemical Biology, Leibniz-Institut für Molekulare Pharmakologie Robert-Rössle-Strasse 10, 13125 Berlin (Germany)
References
- [1] a).Workman JL, Suganuma T. Annu. Rev. Biochem. 2011;80:473–99. doi: 10.1146/annurev-biochem-061809-175347. [DOI] [PubMed] [Google Scholar]; b) Kouzarides T. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- [2] a).Strahl BD, Allis CD. Nature. 2000;403:41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]; b) Su Z, Denu JM. ACS Chem. Biol. 2016;11:564–74. doi: 10.1021/acschembio.5b00864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lee JS, Smith E, Shilatifard A. Cell. 2010;142:682–685. doi: 10.1016/j.cell.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Luger K, Maeder AW, Richmond RK, Sargent DF, Richmond TJ. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- [5].Liokatis S, Stuetzer A, Elsaesser SJ, Theiillet FX, Klingberg R, van Rossum B, Schwarzer D, Allis CD, Fischle W, Selenko P. Nat. Struct. Mol. Biol. 2012;19:819–23. doi: 10.1038/nsmb.2310. [DOI] [PubMed] [Google Scholar]
- [6].Voigt P, LeRoy G, Drury WJ, III., Zee BM, Son J, Beck DB, Young NL, Garcia BA, Reinberg D. Cell. 2012;151:181–93. doi: 10.1016/j.cell.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7] a).Liokatis S, Dose A, Schwarzer D, Selenko P. J. Am. Chem. Soc. 2010;132:14704–5. doi: 10.1021/ja106764y. [DOI] [PubMed] [Google Scholar]; b) Theillet FX, Liokatis S, Jost JO, Bekei B, Rose HM, Binolfi A, Schwarzer D, Selenko P. J. Am. Chem. Soc. 2012;134:7616–9. doi: 10.1021/ja301895f. [DOI] [PubMed] [Google Scholar]
- [8].Stuetzer A, Liokatis S, Kiesel A, Schwarzer D, Sprangers R, Soeding J, Selenko P, Fischle W. Mol. Cell. 2016;61:247–59. doi: 10.1016/j.molcel.2015.12.015. [DOI] [PubMed] [Google Scholar]
- [9].Luger K, Rechsteiner TJ, Richmond TJ. Methods Mol. Biol. 1999;119:1–16. doi: 10.1385/1-59259-681-9:1. [DOI] [PubMed] [Google Scholar]
- [10] a).Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, Allis CD. Mol. Cell. 2000;5:905–915. doi: 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]; b) Lo WS, Trievel RC, Rojas JR, Duggan L, Hsu JY, Allis CD, Marmorstein R, Berger SL. Mol. Cell. 2000;5:917–26. doi: 10.1016/s1097-2765(00)80257-9. [DOI] [PubMed] [Google Scholar]
- [11].Dai J, Sultan S, Taylor SS, Higgins MG. Genes Dev. 2005;19:472–88. doi: 10.1101/gad.1267105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Barski A, Cuddapah S, Cui K, Roth TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- [13].Sullivan BA, Karpen GH. Nat. Struct. Mol. Biol. 2004;11:1076–83. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kouskouti A, Talianidis I. EMBO J. 2005;24:347–57. doi: 10.1038/sj.emboj.7600516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15] a).Nishioka K, Chuikov S, Sarma K, Erdjument-Bromage H, Allis CD, Tempst P, Reinberg D. Genes Dev. 2002;16:479–89. doi: 10.1101/gad.967202. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wang H, Cao R, Xia L, Erdjument-Bromage H, Borchers C, Tempst P, Zhang Y. Mol. Cell. 2001;8:1207–17. doi: 10.1016/s1097-2765(01)00405-1. [DOI] [PubMed] [Google Scholar]
- [16].Theillet FX, Smet-Nocca C, Liokatis S, Thongwichian R, Kosten J, Yoon MK, Kriwacki RW, Landrieu I, Lippens G, Selenko P. J. Biomol. NMR. 2012;54:217–36. doi: 10.1007/s10858-012-9674-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17] a).Neumann H, Peak-Chew SY, Chin JW. Nat. Chem. Biol. 2008;4:232–4. doi: 10.1038/nchembio.73. [DOI] [PubMed] [Google Scholar]; b) Lang K, Chin JW. Chem. Rev. 2014;114:4764–806. doi: 10.1021/cr400355w. [DOI] [PubMed] [Google Scholar]
- [18] a).Simon MD, Chu F, Racki LR, de la Cruz CC, Burlingame AL, Panning B, Narlikar GJ, Shokat KM. Cell. 2007;128:1003–12. doi: 10.1016/j.cell.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Le DD, Cortesi AT, Myers SA, Burlingame AL, Fujimori DG. J. Am. Chem. Soc. 2013;135:2879–82. doi: 10.1021/ja3108214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19] a).Zheng C, Hayes JJ. Methods Enzymol. 2004;375:179–93. doi: 10.1016/s0076-6879(03)75012-5. [DOI] [PubMed] [Google Scholar]; b) McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Nature. 2008;453:812–6. doi: 10.1038/nature06906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lechner CC, Agashe ND, Fierz B. Angew. Chem. Int. Ed. 2016;55:2903–6. doi: 10.1002/anie.201510996. [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2016;128:2954–58. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.