Abstract
Campylobacter species are commonly transmitted to humans through consumption of contaminated foods such as milk and meat. The aim of this study was to investigate the prevalence, antimicrobial resistance, and genetic determinants of resistance of Campylobacter isolated from raw milk and beef carcasses in Tanzania. The antimicrobial resistance genes tested included blaOXA-61 (ampicillin), aph-3-1 (aminoglycoside), tet(O) (tetracycline), and cmeB (multi-drug efflux pump). The prevalence of Campylobacter was 9.5% in beef carcasses and 13.4% in raw milk, respectively. Using multiplex-polymerase chain reaction (PCR), we identified 58.1% of the isolates as Campylobacter jejuni, 30.7% as Campylobacter coli, and 9.7% as other Campylobacter spp. One isolate (1.6%) was positive for both C. jejuni and C. coli specific PCR. Antimicrobial susceptibility testing using the disk diffusion assay and the broth microdilution method showed resistance to: ampicillin (63% and 94.1%), ciprofloxacin (9.3% and 11.8%), erythromycin (53.7% and 70.6%), gentamicin (0% and 15.7%), streptomycin (35.2% and 84.3%), and tetracycline (18.5% and 17.7%), respectively. Resistance to azithromycin (42.6%), nalidixic acid (64.8%), and chloramphenicol (13%) was determined using the disk diffusion assay only, while resistance to tylosin (90.2%) was quantified using the broth microdilution method. The blaOXA-61 (52.6% and 28.1%), cmeB (26.3% and 31.3%), tet(O) (26.3% and 31.3%), and aph-3-1 (5.3% and 3.0%) were detected in C. coli and C. jejuni. These findings highlight the extent of antimicrobial resistance in Campylobacter occurring in important foods in Tanzania. The potential risks to consumers emphasize the need for adequate control approaches, including the prudent use of antimicrobials to minimize the spread of antimicrobial-resistant Campylobacter.
Introduction
Many high-risk pathogens that cause diseases in humans are transmitted through various foods. Subsequently, the microbiological safety of food is an important issue for consumers, industry, and regulatory agencies.7 Of the 25 species within the genus Campylobacter, C. jejuni and C. coli are responsible for the majority of human campylobacteriosis.3,22,58 These species are considered important food-borne bacterial pathogens that can cause gastroenteritis worldwide.93 The most significant sources of Campylobacter-associated infections include the consumption and/or handling of raw or undercooked poultry meat or other sources of meat, raw milk, contaminated water, seafood, and vegetables. Furthermore, cross-contamination of ready-to-eat foods during food preparation as well as direct contact with feces from infected humans and domestic pets have been recognized as risk factors.3,40
In the developing world, water and milk remain predominant means of transmission of campylobacteriosis.20 Therefore, it is clear that reducing Campylobacter in foods and water is a desirable target for maintaining the safety of essential resources.
The control of Campylobacter is complicated by several factors, including the distribution of these pathogens in different food-animals/products and the environment. For example, studies have revealed a range of Campylobacter prevalence (up to 70%) in cattle.31,74–76 Furthermore, although the prevalence of Campylobacter in cattle carcasses and milk samples can be relatively low in comparison to other sources, studies have shown that approximately 15% of beef carcasses/meat can be contaminated with these pathogens.38,92 In addition, a Campylobacter prevalence of 41.7%, 10.2%, and 4.6% had been previously reported in raw bulk tank milk in Northern Italy, Pakistan, and Poland, respectively, while 6.25% of raw cow milk samples from retail stores were contaminated with Campylobacter in Iran.9,41,69,94
Milk can be considered a re-emerging risk factor, because the consumption of unpasteurized raw milk and/or products made from raw milk is becoming more popular in most countries.5,77 Taken together, these observations pose a particular concern for countries where milk is consumed raw on a regular basis or where no effort exists for prevention and control of carcass contamination.
While Campylobacter infections in humans are sporadic and often self-limiting, antimicrobial treatment is indicated in severe and prolonged cases of enteritis, in immunosuppressed individuals, young children, elderly, or pregnant women.3 In these circumstances, macrolides (e.g., erythromycin [ERY]) and fluoroquinolones (e.g., ciprofloxacin [CIP]) are often the drugs of choice.4 Other antimicrobials such as gentamicin (GEN), tetracycline (TET), clindamycin, and ampicillin (AMP) may be used as alternative drugs for the treatment of systemic Campylobacter infections.10 However, over the years, various studies have reported an increase in the resistance of Campylobacter to these drugs.8,17,19,59,71,82,85
The rise of antimicrobial resistance among Campylobacter spp. has been linked to the use of antimicrobials in veterinary medicine and in farming practices, mainly as prophylactic agents and growth promoters88,97 This is important, because some of the resistant isolates have been suspected to spread from food animals to humans.71,79 While antimicrobial-resistant Campylobacter spp. has been reported worldwide, the situation might be more severe in developing countries, where there is widespread and largely uncontrolled use of antimicrobials.13,46
The prevalence of Campylobacter species in poultry and humans have been documented in Tanzania.24,43,56,57,61 However, limited data are available regarding the prevalence of Campylobacter in other foods, particularly in meat and milk, and even scantier information exists regarding the antimicrobial resistance properties of these pathogens. Prompted by the lack of data, we investigated the prevalence, and phenotypic and genotypic antimicrobial resistance profiles of Campylobacter spp. recovered from raw milk and meat widely available in Tanzania. Our results will constitute the basis for much-needed surveillance programs to monitor the trends of antimicrobial resistance in these food-borne pathogens.
Materials and Methods
Study area and sample collection
To investigate the prevalence of Campylobacter spp., a cross-sectional study was conducted from April 2013 through March 2014. A total of 537 samples, including unpasteurized raw milk (n=284) and cattle dressed carcass swab samples (n=253), were collected from three geographically different municipalities, namely Arusha (n=205; 105 carcasses and 100 milk samples) in Northern Tanzania, Iringa (n=120; 48 carcasses and 72 milk samples) in Southwestern Tanzania, and Morogoro (n=212; 100 carcasses and 112 milk samples) in Eastern Tanzania.
Milk samples were collected from two sources: vendors selling milk (n=174; 67 from Morogoro, 70 Arusha, and 37 Iringa) or from bulk tanks at milk collecting centers (n=110; 45 from Morogoro, 30 Arusha, and 35 Iringa) from each of the three municipalities. In each sampling, 40 ml of milk were collected aseptically and stored in a cool box with ice packs before transportation to the laboratory for analyses.
For sampling beef carcasses, surface swabbing was performed using a previously described technique.15 Briefly, a sterile gauze pad (7.6×7.6 cm; Johnson & Johnson Medical, Inc.) premoistened with sterile maximum recovery diluents (MRD; Oxoid) was used to swab four parts of the carcass (neck, brisket, flank, and rump) in a downward motion. Each gauze pad was then placed in a sterile plastic bag (Ziploc®; SC Johnson). The air was squeezed out of the bag, which was then closed tightly, labeled, and stored in a cool box with ice packs before transportation to the laboratory for further analyses within a maximum of 48 hr. On arrival at the Faculty of Veterinary Medicine, Sokoine University of Agriculture, each sample was processed using standard procedures for isolation of thermophilic Campylobacter spp. as described next.
Bacterial isolation and growth conditions
For isolation of Campylobacter species, milk was homogenized and 5 ml was added to 10 ml of Preston enrichment broth containing Campylobacter growth supplements (CM067, SR048, SR117, and SR232; Oxoid). The enrichment tubes were incubated at 42°C for 48 hr under microaerophilic condition (5% O2, 10% CO2, and 85% N2), which was generated using airtight jars containing the Campy Pouch System (Becton Dickinson and Co.).
For isolation of Campylobacter species from carcass samples, 50 ml of MRD was added to each swab and homogenized by squeezing for 3 min. A 5 ml aliquot of the resulting suspension was removed and added to 10 ml of Preston enrichment broth as described earlier.
After enrichment of the milk and carcass samples, 100 μl from each culture was spread onto modified charcoal cefaperazone deoxycolate agar (mCCDA) (CM0739; Oxoid) containing the Campylobacter CCDA selective supplement, SR155E (Oxoid). The agar plates were then incubated at 42°C for 48 hr under microaerophilic conditions. Approximately three presumptive Campylobacter colonies from each mCCDA plate were then subcultured onto Mueller-Hinton (MH; Difco) agar containing the Campylobacter Selective Supplement, SR117 (Oxoid) and incubated at 42°C for 48 hr under microaerophilic conditions. Putative Campylobacter colonies were frozen at −80°C in MH broth supplemented with 30% glycerol (v/v) until species differentiation using polymerase chain reaction (PCR) was performed.
DNA extraction and PCR analysis for Campylobacter identification and speciation
Bacterial DNA lysates were prepared from fresh Campylobacter cultures using the boiling method as previously described.25 Briefly, half of a loopful of bacterial growth from the plates were suspended in 100 μl of sterile RNase/DNase-free water, heated at 95°C for 10 min, cooled, and centrifuged at 13,000 g for 10 min. The supernatants containing the nucleic acids were transferred to new tubes and stored at −20°C. In cases where no PCR products were detected, template DNA was prepared again using QIAamp DNA Mini Kit (Qiagen) according to the manufacturer's instructions.
Confirmation of Campylobacter and speciation were performed by multiplex-PCR (mPCR) as previously described.95 The mPCR conditions and the genus- and species-specific primers were previously described.23,47,54 Isolates that were positive for the genus-specific PCR fragment but negative for the C. coli and C. jejuni-specific PCR fragments were designated as other thermophilic Campylobacter. C. jejuni 81–176 (wild-type strain) and C. coli (ATCC 33559) were used as positive controls, while standard-grade laboratory water was used as a template negative control.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was carried out using the Kirby–Bauer disk-diffusion and the broth microdilution methods.18,32,52 Although the disk diffusion method is more convenient, flexible, cheap, and widely used for testing pathogens in resource-limited conditions, it does not allow the determination of minimum inhibitory concentrations (MIC) for each of the antimicrobials.52,55,84 Therefore, we adopted both methods to generate robust antimicrobial resistance analysis and reduce methodological bias. Both tests were performed in accordance to the recommendations of the Clinical Laboratory Standards Institute (CLSI)18 and using the CLSI breakpoint interpretive criteria. In the cases when CLSI recommendations were not available, the ROSCO MIC for veterinary isolates was used to determine the breakpoints70 (Table 1).
Table 1.
The Guidelines Used to Determine the Antimicrobial Resistance Breakpoints Using the Disk-Diffusion and Broth Microdilution Methods
| Result for method | ||||||||
|---|---|---|---|---|---|---|---|---|
| Broth microdilution | Disk diffusion | |||||||
| MIC breakpoints (μg/ml) | Zone diameter breakpoint (mm) | |||||||
| Antimicrobial agent | Test range (μg/ml) | S | I | R | Disk conc. (μg) | S | I | R |
| Ampicillin | 0.03–64.0 | ≤8 | 16 | ≥32 | 10 | ≥17 | 14–16 | ≤13 |
| Ciprofloxacin | 0.03–64.0 | ≤1 | 2 | ≥4 | 5 | ≥21 | 16–20 | ≤15 |
| Erythromycin | 0.03–64.0 | ≤8 | 16 | ≥32 | 15 | ≥23 | 14–22 | ≤13 |
| Gentamicin | 0.03–64.0 | ≤2 | 4 | ≥8 | 10 | ≥15 | 13–14 | ≤12 |
| Streptomycin | 0.03–64.0 | ≤2 | 4 | ≥8 | 10 | ≥15 | 12–14 | ≤11 |
| Tetracycline | 0.03–64.0 | ≤4 | 8 | ≥16 | 30 | ≥15 | 12–14 | ≤11 |
| Tylosin | 0.03–64.0 | ≤4 | 8 | ≥16 | NT | — | — | — |
| Azithromycin | NT | — | — | — | 15 | ≥18 | 14–17 | ≤13 |
| Chloramphenicol | NT | — | — | — | 30 | ≥18 | 13–17 | ≤12 |
| Nalidixic acid | NT | — | — | — | 30 | ≥19 | 14–18 | ≤13 |
I, intermediate; MIC, minimum inhibitory concentrations; NT, not tested; R, resistance; S, susceptible.
The Kirby–Bauer disk-diffusion method was performed on MH agar plates. In this assay, nine antimicrobials (Oxoid) were tested at the following concentrations: 10 μg AMP, 5 μg CIP, 15 μg ERY, 30 μg nalidixic acid (NAL), 10 μg streptomycin (STR), 30 μg TET, 15 μg azithromycin (AZM), 10 μg GEN, and 30 μg chloramphenicol (CHL). These antimicrobials are representatives of the drugs used for humans and in the animal industry in Tanzania. Briefly, Campylobacter cultures were suspended in sterile normal saline and the turbidity was adjusted to the 0.5 McFarland standard using the Vitek colorimeter (Lenexa). A sterile cotton swab was then dipped into a suspension, which was spread evenly on the entire surface of an MH agar plate. The plate was allowed to dry for 5 min. Antimicrobial discs were placed on the surface of plate and after 24 hr of microaerobic incubation at 42°C, the diameter of the zone of inhibition was measured. Staphylococcus aureus (ATCC 25922) and Escherichia coli (ATCC 29213) were used as reference strains. Zones of growth inhibition were interpreted according to the CLSI guidelines.18
Using the broth microdilution method, seven antimicrobial agents (Sigma-Aldrich Co.), ampicillin, ciprofloxacin, erythromycin, gentamicin, tylosin (TYL), streptomycin, and tetracycline, were tested. Briefly, Campylobacter isolates were suspended in MH broth to achieve an OD600 of 0.05. One hundred microliters of a suspension were added to each well of the 96-well plate containing twofold serial dilutions of the antimicrobial agents. After incubation under microaerobic conditions at 42°C for 24 hr, the plates were scored by visual examination of growth inhibition. MIC values were defined as the lowest concentration of an antimicrobial agent that produced no visible growth. In this experiment, C. jejuni (81–176) and C. coli (ATCC 33559) were used as control strains. Multi-drug resistance (MDR) was defined as resistance to three or more antimicrobial agents as previously described.37
Detection of antimicrobial resistance genes
Campylobacter isolates were screened for the presence of four antimicrobial resistance genes, tet(O), aph-3-1, cmeB, and blaOXA-61, as previously described by Obeng et al.63 The mPCR was performed with 5 min initial denaturation at 94°C, followed by 39 cycles of denaturation at 94°C for 30 sec, annealing at 54°C for 45 sec, extension at 72°C for 1 min, and final extension at 72°C for 10 min.63 Purified water was used as a negative control.
Sequence analysis of blaOXA-61 and tet(O)
To investigate the potential heterogeneity in blaOXA-61, gene sequencing was performed on five Campylobacter isolates (two C. jejuni and three C. coli) that exhibited different MIC levels against ampicillin (MIC at 8, 32, 64, and >64 μg/μl, respectively). These strains were subjected to PCR that targeted the whole blaOXA-61 sequence (∼896 bp) using the following primers: F-5′-CGATGGATCCCTTTAATGGTTAC-3′ and R-5′-TACGGGATCCTCACTAG CCATC-3′.2 The PCR products were purified using the QIAquick PCR Purification Kit (Qiagen) and commercially sequenced (Eurofins Genomics) in both directions. The sequences were then analyzed, edited using Chromas (www.technelysium.com.au/chromas2.html), and aligned using Clustal Omega (www.ebi.ac.uk/Tools/msa/clustalo/) to generate the full-length sequence for each of the genes.
To determine whether there was sequence heterogeneity, the full-length blaOXA-61 genes were compared using Clustal Omega. C. jejuni class D β-lactamase (cam1) gene (accession numbers AY587956.1) was used as a reference during sequence analysis. Blast N was used to identify active and conserved sites that are indicative of class D β-lactamase genes.
Furthermore, tet(O) in C. jejuni (n=2) and C. coli (n=1) that exhibited different resistance to tetracycline (MIC at 8, 32, and 64 μg/μl, respectively) were sequenced as described earlier. However, since tet(O) is ∼1998 bp in length, we used two sets of overlapping primers: F-5′-TGCGGCAAGGTATTCTTAAAT-3′ with R-5′-ATGGACAACCCGACAGAAG-3′and F-5′-GCGTTTTGTTTATGTGCG-3′with R-5′-ATTTTATATGACTTTTGCAAGCTG-3′ to cover the whole sequence.63,90 The fragments were edited, aligned, and analyzed as described earlier. The tet(O) gene from C. jejuni strain F8-H1-S1 (accession numbers AM884250) was used as a reference during sequence analysis. The tet(O) sequences were further compared by performing restriction analysis using Restriction Mapper (www.restrictionmapper.org/) and the restriction sites that were identified elsewhere.51
Statistical analysis
The prevalence and antimicrobial resistance of Campylobacter spp. from milk and beef carcasses obtained in all three municipalities were compared statistically using χ2 analysis. A value of p<0.05 was considered statistically significant. Agreement between the antimicrobial resistance tests was determined using the Kappa statistic. A Kappa value of 1 (100%) indicates total agreement between the classifiers.
Results
Occurrence and distribution of Campylobacter spp.
Campylobacter spp. were detected in 13.4% (38 of 284 isolates) of the milk samples and 9.5% (24 of 253) of the beef carcass samples (Table 2). PCR analysis revealed that 58.1%; (36 of 62) of the Campylobacter isolates were C. jejuni, whereas 30.7% (19 of 62) were C. coli. In addition, 9.7% (6 of 62) of isolates were Campylobacter spp. other than C. jejuni or C. coli, while 1 isolate (1.6%) was positive for both C. jejuni and C. coli.
Table 2.
Distribution of Campylobacter spp. Recovered from Milk and Carcass Samples from Three Municipalities in Tanzania
| Microorganisms | |||||||
|---|---|---|---|---|---|---|---|
| Source of isolates | Sampling location | No. of samples | Total isolates (%) | Campylobacter jejuni (%) | Campylobacter coli (%) | C. jejuni/C. coli (%) | OTC (%)a |
| Cattle carcass | Morogoro | 100 | 15 (15.0) | 10 (10.0) | 4 (4.0) | 1 (1.0) | 0 |
| Arusha | 105 | 7 (6.6) | 4 (3.8) | 2 (1.9) | 0 | 1 (1.0) | |
| Iringa | 48 | 2 (4.2) | 1 (2.1) | 1 (2.1) | 0 | 0 | |
| Raw milk | Morogoro | 112 | 24 (21.4) | 17 (15.2) | 5 (4.5) | 0 | 2 (1.8) |
| Arusha | 100 | 7 (7.0) | 0 | 5 (5.0) | 0 | 2 (2.0) | |
| Iringa | 72 | 7 (9.7) | 4 (5.6) | 2 (2.8) | 0 | 1 (1.4) | |
| Total | 537 | 62 (11.6) | 36 (96.7) | 19 (3.5) | 1 (0.2) | 6 (1.1) | |
OTC, other Campylobacter spp. different from C. jejuni and C. coli.
Furthermore, 55.3% (21 of 38) of Campylobacter isolates recovered from milk were C. jejuni, 31.6% (12 of 38) were C. coli, and 1.8% (5 of 38) were other Campylobacter spp. Similarly, 62.5% (15 of 24) of isolates from carcass samples were C. jejuni, 29.2% (7 of 24) were C. coli, and 4.2% (1 of 24) were Campylobacter spp. other than C. jejuni or C. coli. Collectively, the overall isolation rate of C. jejuni (6.7%) from all samples was significantly higher (χ2=5.039; 95% confidence interval [CI]=5.039; p=0.0248) in comparison to 3.5% of C. coli.
The occurrence of Campylobacter in animal products varied according to the geographic location of the sampling sites (Table 2). Specifically, contamination of carcasses with Campylobacter spp. comprised 15% (10 C. jejuni, 4 C. coli, and 1 isolate that was positive for both C. jejuni and C. coli), 4.8% (1 C. jejuni and 1 C. coli), and 6.6% (4 C. jejuni, 2 C. coli and 1 other Campylobacter spp.) for the samples collected from Morogoro, Iringa, and Arusha, respectively.
Similarly, milk samples were the most frequently contaminated with Campylobacter spp. with mean isolation rates of 21.4% (17 C. jejuni, 5 C. coli, and 2 other Campylobacter spp.), 9.7% (4 C. jejuni, 2 C. coli, and 1 other Campylobacter spp.), and 7.0% (0 C. jejuni, 5 C. coli, and 2 other Campylobacter spp.) from Morogoro, Iringa, and Arusha, respectively. Campylobacter contamination (in both milk and beef carcass samples) in Morogoro was higher (p<0.05) than that found in Arusha. There was no significant difference (p>0.05) in Campylobacter prevalence (in both milk and carcass samples) between Arusha and Iringa, and between Morogoro and Iringa.
Antimicrobial susceptibility testing
To better assess the potential public health impact of Campylobacter spp., a total of 54 of 55 isolates (35 C. jejuni and 19 C. coli; 1 C. jejuni could not be retrieved from glycerol stocks) were assayed for their potential to resist nine antimicrobials by the Kirby–Bauer disk-diffusion method. A total of 51 of 55 isolates (32 C. jejuni and 19 C. coli) were also tested by the broth microdilution method against a panel of seven antimicrobial agents that are of both clinical and veterinary importance.
Analysis of the Kirby–Bauer disk diffusion assay showed that 53 of the 54 isolates (98.2%) were resistant to one or more antimicrobial agents, whereas one (1.9%) of the isolates was pan-susceptible to all antimicrobials tested (Table 3). Eleven isolates (20.4%; 5 C. jejuni and 6 C. coli) were resistant to a single antimicrobial agent, and 14 isolates (25.9%; 10 C. jejuni and 4 C. coli) showed resistance to two antimicrobial agents. All isolates were susceptible to GEN, and 9.3% of the isolates (three C. jejuni and two C. coli) were found resistant to CIP (Table 4).
Table 3.
Antimicrobial Resistance of C. jejuni and C. coli Isolated from Raw Milk and Carcass Samples
| Source of isolate and number of resistance (%) | |||||||
|---|---|---|---|---|---|---|---|
| Milk | Carcass | ||||||
| Resistance profile | No. of resistant isolates (%) | C. jejuni | C. coli | Total | C. jejuni | C. coli | Total |
| Pan-susceptible | 1 (1.9) | 0 | 0 | 0 | 1 (7.1) | 0 | 1 (4.8) |
| ERY | 2 (3.7) | 0 | 0 | 0 | 2 (14.3) | 0 | 2 (9.5) |
| TET | 1 (1.9) | 0 | 1 (8.3) | 1 (3.0) | 0 | 0 | 0 |
| AMP | 3 (5.6) | 0 | 2 (16.7) | 2 (6.1) | 1 (7.1) | 0 | 1 (4.8) |
| AZM | 1 (1.9) | 0 | 1 (8.3) | 1 (3.0) | 0 | 0 | 0 |
| NAL | 4 (7.4) | 1 (4.0) | 3 (25.0) | 4 (12.1) | 0 | 0 | 0 |
| CIP/NAL | 1 (1.9) | 0 | 0 | 0 | 1 (7.1) | 0 | 1 (4.8) |
| ERY/AZM | 1 (1.9) | 0 | 0 | 0 | 0 | 1 (14.3) | 1 (4.8) |
| NAL/STR | 2 (3.7) | 0 | 0 | 0 | 1 (7.1) | 1 (14.3) | 2 (9.5) |
| AMP/NAL | 5 (9.3) | 2 (9.5) | 1 (8.3) | 3 (9.1) | 2 (14.3) | 0 | 2 (9.5) |
| AZM/STR | 1 (1.9) | 0 | 1 (8.3) | 1 (3.0) | 0 | 0 | 0 |
| AMP/CHL | 1 (1.9) | 1 (4.0) | 0 | 1 (3.0) | 0 | 0 | 0 |
| AMP/AZM | 2 (3.7) | 2 (9.5) | 0 | 2 (6.1) | 0 | 0 | 0 |
| AMP/STR | 1 (1.9) | 0 | 0 | 0 | 1 (7.1) | 0 | 1 (4.8) |
| CIP/AMP/NAL | 1 (1.9) | 0 | 0 | 0 | 0 | 1 (14.3) | 1 (4.8) |
| CIP/NAL/STR | 1 (1.9) | 0 | 0 | 0 | 0 | 1 (14.3) | 1 (4.8) |
| ERY/AZM/STR | 1 (1.9) | 0 | 0 | 0 | 0 | 1 (14.3) | 1 (4.8) |
| ERY/NAL/STR | 3 (5.6) | 2 (9.5) | 0 | 2 (6.1) | 1 (7.1) | 0 | 1 (4.8) |
| ERY/AZM/NAL | 1 (1.9) | 1 (4.0) | 0 | 1 (3.0) | 0 | 0 | 0 |
| ERY/TET/AMP | 1 (1.9) | 0 | 1 (8.3) | 1 (3.0) | 0 | 0 | 0 |
| ERY/AMP/AZM | 1 (1.9) | 1 (4.0) | 0 | 1 (3.0) | 0 | 0 | 0 |
| ERY/AMP/AZM/NAL | 7 (13.0) | 5 (23.8) | 1 (8.3) | 6 (18.2) | 0 | 1 (14.3) | 1 (4.8) |
| ERY/AMP/NAL/STR | 1 (1.9) | 0 | 0 | 0 | 0 | 1 (14.3) | 1 (4.8) |
| ERY/TET/AMP/AZM | 2 (3.7) | 2 (9.5) | 0 | 2 (6.1) | 0 | 0 | 0 |
| ERY/AZM/AMP/NAL/STR | 2 (3.7) | 1 (4.0) | 1 (8.3) | 2 (6.1) | 0 | 0 | 0 |
| ERY/AMP/AZM/CHL/NAL | 1 (1.9) | 0 | 1 (8.3) | 1 (3.0) | 0 | 0 | 0 |
| ERY/TET/AMP/AZM/NAL/STR | 4 (7.4) | 3 (14.3) | 0 | 3 (9.1) | 1 (7.1) | 0 | 1 (4.8) |
| CIP/ERY/AMP/AZM/CHL/NAL/STR | 2 (3.7) | 0 | 0 | 0 | 2 (14.3) | 0 | 2 (9.5) |
Antimicrobial resistance was determined using the disk-diffusion method. Results are shown as numbers of isolates with percentage given in parentheses.
AMP, ampicillin; AZM, azithromycin; CHL, chloramphenicol; CIP, ciprofloxacin; ERY, erythromycin; GEN, gentamicin; NAL, nalidixic acid; STR, streptomycin; TET, tetracycline.
Table 4.
Comparison of Antimicrobial Resistance of Campylobacter spp. Identified by Disk-Diffusion and Broth Microdilution Methods
| Disk diffusion | Broth microdilution | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| No. of isolates | No. of isolates | Agreement between methods | |||||||
| Antimicrobial agent | S | I | R | No. of resistant isolates (%) | S | I | R | No. of resistant isolates (%) | Kappa value |
| Aminoglycosides | |||||||||
| Gentamicin | 44 | 10 | 0 | 0 | 18 | 27 | 6 | 11.8 | 0.1560 |
| Streptomycin | 25 | 10 | 19 | 35.2 | 6 | 2 | 43 | 84.3 | 0.2380 |
| β-Lactam | |||||||||
| Ampicillin | 18 | 2 | 34 | 63.0 | 3 | 0 | 48 | 94.1 | 0.53467 |
| Macrolides | |||||||||
| Azithromycin | 20 | 11 | 23 | 42.6 | — | — | — | — | |
| Erythromycin | 20 | 5 | 29 | 53.7 | 10 | 5 | 36 | 70.6 | 0.6230 |
| Tylosin | — | — | — | — | 1 | 4 | 46 | 90.2 | |
| Quinolones | |||||||||
| Ciprofloxacin | 40 | 9 | 5 | 9.3 | 28 | 17 | 6 | 11.8 | 0.6831 |
| Nalidixic acid | 16 | 3 | 35 | 64.8 | — | — | — | — | |
| Phenicol | |||||||||
| Chloramphenicol | 41 | 6 | 7 | 13.0 | — | — | — | — | |
| Tetracycline | |||||||||
| Tetracycline | 41 | 3 | 10 | 18.5 | 18 | 24 | 9 | 17.7 | 0.9132 |
A Kappa value of 1 (100%) indicates total agreement between the assays.
About 13% of Campylobacter isolates (six C. jejuni and one C. coli) were resistant to CHL, and 18.5% (eight C. jejuni and two C. coli) were resistant to TET. Furthermore, 19 (35.2%) isolates were shown to be resistant to STR, 23 (42.6%) to AZM, and 29 (53.7%) to ERY. In addition, 63% (25 C. jejuni and 9 C. coli) of the isolates were resistant to AMP, while 64.8% of the isolates were resistant to NAL (23 C. jejuni and 12 C. coli) (Table 4). Resistance to CIP was significantly higher (p<0.05) in isolates recovered from milk in comparison to carcass isolates, while resistance to AMP was significantly higher in isolates from carcass samples (p<0.05). Significant differences between milk and carcass isolates were not observed for the remaining antimicrobial agents.
Twenty-eight of 54 (46.3%) isolates (19 C. jejuni and 9 C. coli) showed MDR. Of the MDR isolates, 10 (47.6%) were from beef carcass and 18 (54.6%) were recovered from raw milk. The most common MDR patterns observed among isolates were resistance to ERY/AMP/AZM/NAL (13%), ERY/NAL/STR (5.6%), and ERY/TET/AMP/AZM/NAL/STR (5.6%) (Table 3).
Analyses using the broth microdilution method showed that of 51 isolates, 2 C. jejuni were pan-susceptible to all tested antimicrobials (Table 5). Forty-eight (94.1%) isolates showed resistance to AMP, 46 (90%) to TYL, 45 (88.2%) to STR, 36 (70.6%) to ERY, 9 (17.7%) to TET, 6 (11.8%) to GEN, and 6 (11.8%) to CIP (Table 4). The most prevalent resistance pattern (n=33; 64.7%) among isolates was for macrolides (ERY and TYL), aminoglycoside (STR), and β-lactam (AMP), respectively (Table 5). Only 17.7% and 11.8% of the isolates were resistant to TET and CIP, respectively (Table 4). The highest MDR, irrespective of the antimicrobials, was found among milk isolates, where more than 90% (n=28) of the isolates were resistant to three or more antimicrobials (Table 5).
Table 5.
Antimicrobial Resistance of C. jejuni and C. coli Isolated from Raw Milk and Carcass Samples
| Source of isolate and number of resistance (%) | |||||||
|---|---|---|---|---|---|---|---|
| Milk | Carcass | ||||||
| Resistance profile | No. of resistant isolates (%) | C. jejuni | C. coli | Total | C. jejuni | C. coli | Total |
| Pan-susceptible | 2 (3.9) | 1 (5.6) | 0 | 1 (3.3) | 1 (7.1) | 0 | 1 (4.8) |
| TYL | 1 (2.0) | 0 | 0 | 0 | 1 (7.1) | 0 | 1 (4.8) |
| AMP | 1 (2.0) | 0 | 0 | 0 | 1 (7.1) | 0 | 1 (4.8) |
| STR/AMP | 2 (3.9) | 1 (5.6) | 0 | 1 (3.3) | 0 | 1 (14.3) | 1 (4.8) |
| GEN/TYL/AMP | 1 (2.0) | 0 | 0 | 0 | 1 (7.1) | 0 | 1 (4.8) |
| ERY/TYL/AMP | 3 (5.9) | 1 (5.6) | 0 | 1 (3.3) | 1 (7.1) | 1 (14.3) | 2 (9.5) |
| TYL/STR/AMP | 5 (9.8) | 1 (5.6) | 1 (8.3) | 2 (6.7) | 3 (21.4) | 0 | 3 (14.3) |
| ERY/TYL/STR/AMP | 22 (43.1) | 10 (55.6) | 3 (25.0) | 13 (43.3) | 4 (28.6) | 5 (71.4) | 9 (42.9) |
| GEN/TYL/STR/AMP | 2 (3.9) | 0 | 1 (8.3) | 1 (3.3) | 0 | 1 (14.3) | 1 (4.8) |
| CIP/TYL/STR/AMP | 1 (2.0) | 0 | 0 | 0 | 1 (7.1) | 0 | 1 (4.8) |
| GEN/ERY/TYL/STR/AMP | 2 (3.9) | 1 (5.6) | 1 (8.3) | 2 (6.7) | 0 | 0 | 0 |
| ERY/TYL/STR/AMP/TET | 4 (7.8) | 3 (16.7) | 1 (8.3) | 4 (13.3) | 0 | 0 | 0 |
| CIP/GEN/ERY/TYL/STR/AMP | 1 (2.0) | 0 | 1 (8.3) | 1 (3.3) | 0 | 0 | 0 |
| CIP/ERY/TYL/STR/AMP/TET | 4 (7.8) | 2 (11.1) | 2 (16.7) | 4 (13.3) | 0 | 0 | 0 |
Antimicrobial resistance was determined using the broth microdilution method. Results are shown as numbers of isolates with percentage given in parentheses.
TYL, tylosin.
A comparison between the disk-diffusion and microdilution methods showed no significant differences (p>0.05) in the number of isolates that were resistant to CIP, ERY, and TET, respectively (Table 4). Significant differences (p<0.05) were found for GEN, STR, and AMP (Table 4). Furthermore, using the Kappa statistics, the lowest agreement between disk diffusion and broth microdilution was noted for GEN and STR (Kappa value=0.1560 and 0.2380, respectively), while the highest agreement was noted for TET (Kappa value=0.9132) (Table 4).
Antimicrobial resistance genes
The antimicrobial resistance genes tet(O), aph-3-1, blaOXA-61, and cmeB were identified in 10.5%, 5.3%, 52.6%, and 26.3% of the resistant C. coli isolates, respectively (Table 6). tet(O), aph-3-1, blaOXA-61, and cmeB were detected in 12.5%, 0%, 25%, and 18.8% of the resistant C. jejuni isolates, respectively (Table 6).
Table 6.
Distribution of Antimicrobial Resistance Genes in C. jejuni and C. coli Isolates from Raw Milk and Carcass Samples
| No. of isolates (%) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| tet(O) | aph-3-1 | blaOXA-61 | cmeB | ||||||||||
| Species | Resistant phenotype and the occurrence of cognate gene | Total | Milk | Carcass | Total | Milk | Carcass | Total | Milk | Carcass | Total | Milk | Carcass |
| C. jejuni (n=32) | Resistant with genes | 4 (12.5) | 4 (22.2) | 0 | 0 | 0 | 0 | 8 (25.0) | 6 (33.3) | 2 (14.3) | 6 (18.8) | 4 (22.2) | 2 (14.3) |
| Resistant without genes | 2 (6.3) | 2 (11.1) | 0 | 3 (9.4) | 3 (16.7) | 0 | 21 (65.6) | 15 (83.3) | 6 (42.9) | 16 (50.0) | 12 (66.7) | 4 (28.6) | |
| Susceptible with genes | 6 (18.8) | 4 (22.2) | 2 (14.3) | 1 (3.1) | 0 | 0 | 1 (3.1) | 0 | 1 (5.6) | 4 (12.5) | 6 (33.3) | 3 (21.4) | |
| C. coli (n=19) | Resistant with genes | 2 (10.5) | 2 (16.7) | 0 | 1 (5.3) | 0 | 1 (14.3) | 10 (52.6) | 6 (50.0) | 4 (57.1) | 5 (26.3) | 3 (25.0) | 2 (28.6) |
| Resistant without genes | 1 (5.3) | 1 (8.3) | 0 | 4 (21.1) | 3 (25.0) | 1 (14.3) | 9 (47.4) | 8 (66.7) | 1 (14.3) | 9 (47.4) | 6 (50.0) | 3 (42.9) | |
| Susceptible with genes | 3 (15.8) | 1 (8.3) | 2 (28.6) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
Twelve isolates (seven C. jejuni and five C. coli) harbored more than one resistance gene. In addition, 15 (10 C. jejuni and 5 C. coli) isolates with an MDR phenotype were identified to carry the multidrug efflux gene cmeB. However, in many instances, an antimicrobial resistance gene was not detected in the isolates exhibiting resistance to the cognate antimicrobial agent. For example, blaOXA-61 was not detected in 21 C. jejuni and 9 C. coli that were phenotypically resistant to ampicillin. Similarly, tet(O), aph-3-1, and cmeB were not detected in phenotypically resistant Campylobacter isolates (Table 6). Only tet(O) was identified in 15.8% of the susceptible C. coli isolates, while tet(O), aph-3-1, blaOXA-61, and cmeB occurred in 18.8%, 3.1%, 3.1%, and 12.5% of the susceptible C. jejuni isolates, respectively (Table 6).
Sequence analysis of blaOXA-61 and tet(O)
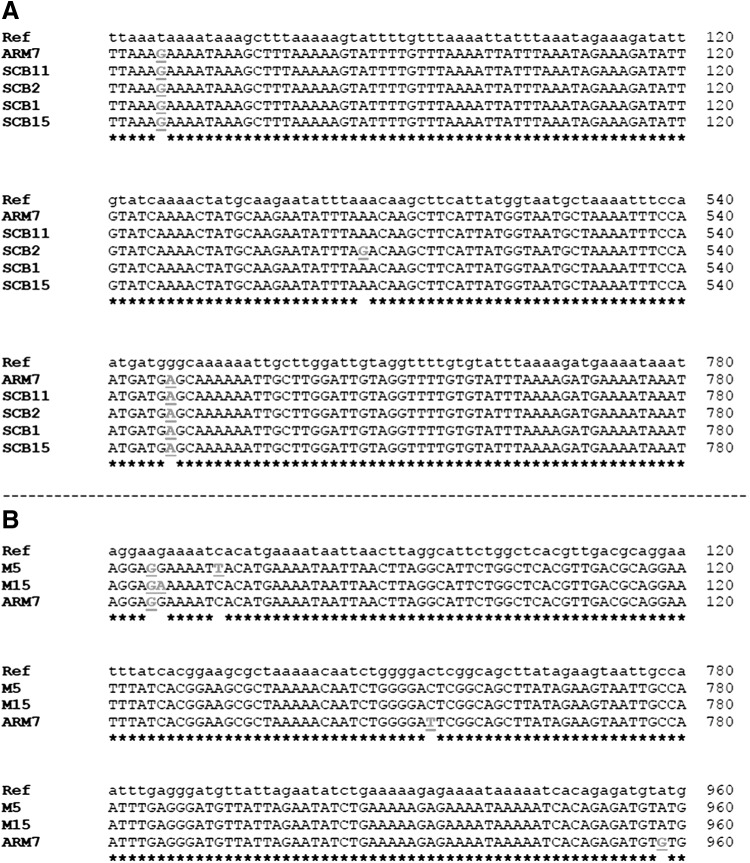
Sequence analysis showed that all the blaOXA-61 genes had the sequences encoding the serine β-lactamase active-site STFK (positions 58–61) and the conserved motifs, YGN (positions 133–135) and KTG (positions 196–198), that are characteristic of class D β-lactamase. Therefore, we confirmed that the blaOXA sequences in our tested samples belonged to class D β-lactamase genes. Further analysis revealed that the five genes could be divided into two main alleles based on base pair substitutions (Fig. 1). One allele was found in SCB11 (ampicillin sensitive C. jejuni isolated from carcass samples; MIC=8 μg/μl), ARM7 (ampicillin resistant C. jejuni isolated from milk samples; MIC>64 μg/μl), SCB1 (ampicillin resistant C. coli isolated from carcass samples; MIC=64 μg/μl), and SCB15 (ampicillin-resistant C. coli isolated from carcass samples; MIC=32 μg/μl).
FIG. 1.
Analysis of sequences of antimicrobial resistance genes. (A) Alignment of the sequence segments of the blaOXA-61 along with a reference gene (Ref) that shows differences in bases. (B) Alignment of the sequence segments of the tet(O) along with a reference gene (Ref) that shows differences in bases. Differing bases are bolded and underlined. Stars indicate identical bases.
The second allele was found in CB15 (ampicillin-resistant C. coli isolated from carcass samples; MIC>64 μg/μl). Collectively, our data show that identical blaOXA-61 alleles can occur in both C. jejuni and C. coli. Furthermore, there was no correlation between the occurrence of a certain allele and consistently increased resistance to ampicillin. Although the allele in CB15 was associated with high ampicillin resistance, the limited number of sequenced samples in this particular case precluded definitive correlations.
All three tet(O) genes analyzed and the reference gene showed identical enzyme restriction profiles that are characteristic of tet(O). The restriction profile was as follows: HincII (restriction frequency: 2; cut positions: 110 and 284 bp); DdeI (restriction frequency: 3; cut positions: 89, 1347 and 1777 bp); HindIII (restriction frequency: 1; cut positions: 513 bp); and NdeI (restriction frequency: 2; cut positions: 307 and 1972 bp). Further analysis revealed that the tet(O) genes were divided into three alleles based on base pair substitutions (Fig. 1). Specifically, the alleles were distributed in M5 (C. jejuni from milk samples with intermediate resistance to tetracycline; MIC=8 μg/μl), ARM7 (tetracycline resistant C. jejuni; MIC=64 μg/μl), and M15 (tetracycline-resistant C. jejuni from milk samples; MIC=32 μg/μl). Although two of the alleles were associated with high tetracycline resistance, the limited number of sequences precluded definitive correlations.
Discussion
Cattle are typically carriers of C. jejuni; therefore, milk and carcass cross-contamination may occur from the feces of the animal or different animals during milking or slaughtering.38 While only one previous study addressed the prevalence of Campylobacter in slaughtered cattle in Morogoro,61 here, we report the prevalence and antimicrobial resistance profiles of Campylobacter spp. recovered from raw milk and beef carcasses in three municipalities in Tanzania. The prevalence of Campylobacter spp. of 9.5% on beef carcasses is in close agreement with a previous study that reported Campylobacter on 9.3% of dressed beef carcasses at the Morogoro municipal abattoir inTanzania.61
In some countries, the prevalence of Campylobacter in beef carcasses was different and usually lower in comparison to this study. For example, the prevalence of Campylobacter in beef carcasses in Poland ranged between 2.7% and 14.9%,19,91 while 3.5%, 3.3%, and 1.5% of the carcasses were contaminated in Finland,38 Belgium,33 and Canada,11 respectively. Furthermore, the overall recovery rate of Campylobacter isolates from beef carcasses in this study was significantly different among the three sampling location/regions. These differences may be due to several variables, including geographical location, type of feeds, husbandry, and/or slaughter practices, which have been previously suggested to account for the variation in the prevalence of Campylobacter spp.38,80
Transmission of Campylobacter infections to humans via the consumption of raw milk has been reported in numerous outbreaks.39,40,60,89 In this study, 38 out of 284 (13.4%) of all milk samples were positive for Campylobacter spp., which corroborates findings reported in some countries, including Pakistan and Italy.9,35,41 However, the prevalence of Campylobacter in milk in this study was higher in comparison with samples from Nigeria, Iran, Egypt, and Poland27,69,73,94 and lower than that reported in China.96 Therefore, the prevalence of Campylobacter in milk appears to vary between different countries.
While the precise cause of the latter is not clear, handling and distribution of milk along with sample collection and processing strategies and Campylobacter isolation might account for the variation in prevalence between countries. In Tanzania, the consumption of raw/unpasteurized milk in rural and peri-urban areas is common.49 Hence, the occurrence of Campylobacter in these samples constitutes a tangible risk to public health.
Although C. jejuni was the most prevalent species isolated in this study (55.3% and 62.5% of raw milk and carcass samples, respectively), C. coli (29.2% and 31.6%) was also frequently isolated from these samples. This is not surprising, because it has been reported that C. jejuni can be more prevalent in some food animals (e.g., poultry and cattle) and in dairy products in comparison to C. coli, which is usually predominant in swine.33,38,41 Furthermore, the prevalence of C. coli in this study is similar to that reported in other countries.21,62,69,92 The distribution of C. jejuni and C. coli can be influenced by many factors, including seasonality, age of the animal, and geographical locations. For example, Sanad et al.75 reported a higher prevalence of C. coli in cattle fecal samples collected from the Southern United States in comparison to those from Northern, Midwestern, and Eastern regions.
Most of the isolates in this study (>98%) showed resistance to one or more of the tested antimicrobial agents. The relatively high percentages of resistance to most antimicrobial agents tested may be due to relatively unrestricted use of antimicrobial agents in animal treatment that is practiced in most of the developing countries.16
Fluoroquinolones such as ciprofloxacin are the drugs of choice for the treatment of campylobacteriosis in humans. In this study, 9.3% (n=5) and 11.8% (n=6) of all Campylobacter isolates were resistant to ciprofloxacin by using the disk-diffusion and broth microdilution methods, respectively. Ciprofloxacin resistance observed in this study is significantly lower than that reported elsewhere.81,92 However, considering the fact that fluoroquinolones are not approved for use in dairy cattle in the Tanzania, this finding is of great interest and concern. This could also imply that the Campylobacter identified from animals and their products could have originated from humans where ciprofloxacin is commonly used. However, identifying the source of these isolates will require further analysis.
In this study, resistance to nalidixic acid was found in both C. jejuni (65.7%) and C. coli (63.2%) isolates. A high number of nalidixic acid-resistant Campylobacter (up to 75% of tested isolates) was previously observed in samples from a variety of sources, including chicken and beef meat and carcasses.12,21,81
The resistance of Campylobacter to fluoroquinolones can be associated with discrete point mutations in gyrA, the gene that encodes the DNA gyrase subunit A.26,65 For example, a gyrA mutation leading to a Thr86-Ile substitution confers resistance to both ciprofloxacin and nalidixic acid.87 However, resistance to nalidixic acid alone was associated with a single gyrA mutation leading to a Thr86-Ala substitution.6,44 Therefore, resistance to nalidixic acid alone can develop independently from resistance to ciprofloxacin.6,44 It follows that gyrA mutations might explain disparate resistance to nalidixic acid and ciprofloxacin in this study. However, further experimental evidence is needed to support this conclusion.
The resistance to macrolides ranged from 33% to 88% for erythromycin and 90% for tylosin. This was expected, because resistance to erythromycin72,85 and tylosin34 had been previously observed in Campylobacter isolated from food animals from different countries. In addition, one study reported that more than 80% of 136 Campylobacter isolates associated with humans in Tanzania were resistant to erythromycin.50 Resistance to macrolides (erythromycin and tylosin) could have been induced by extensive use of tylosin in Tanzania as a therapeutic agent for treatment of cattle respiratory conditions such as Mycoplasma infection. The usage of tylosin in animals for the purpose of either treatment or growth promotion contributes to the selection of resistant Campylobacter strains to erythromycin as well as to resistance to ciprofloxacin and nalidixic acid.45
Chloramphenicol and gentamicin resistance in Campylobacter spp. have been reported to be low.30,48 This is in agreement with the results of this study, which showed that 13% and 11.8% of the isolates were resistant to chloramphenicol and gentamicin, respectively. Chloramphenicol resistance in Campylobacter is associated with a plasmid-borne acetyltransferase, which modifies chloramphenicol, impeding its binding to ribosomes.78,86 However, whether this plasmid occurs in some of the isolates in this study or whether these strains may carry chromosomally encoded resistance gene is not currently known and requires experimental verification. Furthermore, none of the isolates were resistant to gentamicin when the disk diffusion assay was used; hence, these results should be interpreted with caution and methodological biases should be considered.
Campylobacter spp. are inherently resistant to β-lactams (including ampicillin) due to their ability to produce β-lactamases, low affinity binding of β-lactams to the target (penicillin-binding proteins [PBPs]), or failure of the drugs to penetrate the outer membrane porins.29,53 The high resistance to ampicillin observed in this study might be due to the frequent use of β-lactams including penicillin or a combination of penicillin with streptomycin in treatment of pneumonia and mastitis cases in cattle.
In this study, six out of nine (67%) tetracycline-resistant isolates were found to possess tet(O), 35% of isolates with MDR profiles had cmeB, and about 18% of ampicillin-resistant isolates had blaOXA-61 (Table 6). The frequent detection of tet(O) in tetracycline-resistant isolates is expected and matches with studies on resistant Campylobacter isolates recovered from humans and poultry.66 However, tet(O) was also detected in nine tetracycline-susceptible Campylobacter (six C. jejuni and three C. coli), and the gene was not detected in three tetracycline-resistant isolates (two C. jejuni and one C. coli). This is rare but not surprising, because (1) it has been previously reported that the tet(O) might be present in the resistant strains but might be undetected by the primers used as described by Guévremont et al.,36 and (2) it was recently reported that tet(A) can occur in some Campylobacter isolates.1
The relatively low prevalence of cmeB in Campylobacter isolates with MDR profiles might be due to the high sequence variation in cmeB. It is also probable that the primers failed to detect the regions subjected to modification as previously reported.14 Furthermore, MDR is associated with the overexpression of cmeB67 and mutations in the cmeR that regulates expression of CmeABC; hence, the occurrence of the cmeB, its sequence and expression should be noted for a complete interpretation of the antimicrobial resistance. Similarly, Campylobacter resistance to β-lactam such as ampicillin is not only associated with enzymatic inactivation by blaOXA-61, but also other mechanisms such as reduced uptake due to alterations in outer membrane porins and efflux pump system might play a role.42
Since 1990, there have been dramatic increases in the occurrence of MDR strains of zoonotic pathogens causing infections in humans in many countries. MDR poses a threat to humans by limiting therapeutic choice of antimicrobials. The MDR Campylobacter spp. (51.4% C. jejuni and 36.8% C. coli) in this study may suggest a considerable public health hazard. The results are comparable to those reported by other investigators.12,62,64,68,92 Macrolides (erythromycin and tylosin) resistance was primarily associated with most (91.7%) of the MDR isolates. A similar association has been reported in previous studies, although no genetic basis has been described.28,83
In vitro antimicrobial susceptibility testing involves measuring the antimicrobial's activity against the test microorganism by determining the MIC or inhibition zone diameter.32 Although the disk-diffusion method is more convenient, flexible, cheap, and widely used for testing pathogens, several researchers have reported differing results when the method was compared with the broth microdilution method.52,55,84
In this study, 50% of the Campylobacter isolates that were classified as susceptible to ampicillin by the disk diffusion method were found to be resistant using the broth microdilution methods. It is important to note that these isolates harbored the blaOXA-61 gene. Similarly, 54.5% of the Campylobacter isolates that were classified as susceptible to tetracycline by the disk-diffusion method were found to exhibit either intermediate or complete resistance using the broth microdilution method. Again, these isolates were positive for tet(O) using PCR. In addition, resistance was detected against gentamicin only when the broth microdilution method was used. Since accurate determination of Campylobacter susceptibility is of vital importance to ensure an adequate therapy and effectively monitor the antimicrobial resistance trends worldwide,52 it is important to use multiple approaches to limit methodological biases and to interpret the data adequately.
In conclusion, this study revealed that animal products, raw milk and beef meat, are often contaminated with thermophilic Campylobacter spp., suggesting a possible risk of infection to consumers of raw milk and undercooked meat. This study revealed that resistance was detected against most of the antimicrobial agents tested, with many of the Campylobacter isolates showing resistance to three or more antimicrobial agents. The severity of resistance observed in this study may be the result of indiscriminate usage of antimicrobial agents in treatment of food animals in Tanzania. Alternatively, indirect transmission of already resistant organisms from humans and the environment to carcasses or milk products might also be a contributing factor.
The occurrence of MDR isolates in raw milk and meat poses a threat to humans by further limiting therapeutic options. Therefore, coordinated actions are recommended to reduce or eliminate the risk posed by Campylobacter spp. at a number of stages in the food chain. These include implementing Good Agricultural Practice (GAP) and Hazard Analysis of Critical Control Points (HACCP) at every stage of the meat and milk chain production, from the farm, through the abattoir and milk bulk centers, to the retailers/vendors, and those involved with the handling and processing of such animal products in the house environment.
Acknowledgments
The authors are grateful to the Veterinary Officers of the three Municipalities and all Laboratory technicians of the Faculty of Veterinary Medicine (SUA) and Food Animal hearth Research Program (OARDC, OSU) for their keen support during sample collection and laboratory work. This work was supported by the WHO Advisory Group on Integrated Surveillance of Antimicrobial Resistance (AGISAR), the NIH Fogarty fellowship program on Molecular Epidemiology and Key Issues in Food-borne Pathogens in Eastern Africa, and the United States Agency for International Development (USAID) through the Feed the Future Tanzania-based Innovative Agricultural Research Initiative (iAGRI).
Disclosure Statement
No competing financial interests exist.
References
- 1.Abdi-Hachesoo B., Khoshbakht R., Sharifiyazdi H., Tabatabaei M., Hosseinzadeh S., and Asasi K. 2014. Tetracycline resistance genes in Campylobacter jejuni and C. coli isolated from poultry Carcasses. Jundishapur J. Microbiol. 7:e12129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alfredson D. 2005. Characterisation of the B-Lactamase Gene From Campylobacter jejuni. PhD Thesis. Available at www120.secure.griffith.edu.au/rch/file/29902e76-dd9e-74fd-45ef-d19eb9a4b86b/1/01Front.pdf, accessed on February13, 2015 Australia: Institute for Glycomics, Griffith University. (Online.) [Google Scholar]
- 3.Allos B.M. 2001. Campylobacter jejuni infections: update on emerging issues and trends. Clin. Infect. Dis. 32:1201–1206 [DOI] [PubMed] [Google Scholar]
- 4.Allos B.M., and Blaser M.J. (eds.). 2009. Mandell, Douglas and Bennett's Principles and Practice of Infectious Diseases, 7th ed. Elsevier Publishing Co., Philidelphia, PA, Chapter 216 [Google Scholar]
- 5.Angulo F.J., Lejeune J.T., and Rajala-Schultz P.J. 2009. Unpasteurized milk: a continued public health threat. Clin. Infect. Dis. 48:93–100 [DOI] [PubMed] [Google Scholar]
- 6.Bachoual R., Ouabdesselam S., Mory F., Lascols C., Soussy C.J., and Tankovic J. 2001. Single or double mutational alterations of gyrA associated with fluoroquinolone resistance in Campylobacter jejuni and Campylobacter coli. Microb. Drug Resist. 7:257–261 [DOI] [PubMed] [Google Scholar]
- 7.Bai S., Zhao J., Zhang Y., Huang W., Xu S., Chen H., Fan L.M., Chen Y., and Deng X.W. 2010. Rapid and reliable detection of 11 food-borne pathogens using thin-film biosensor chips. Appl. Microbiol. Biotechnol. 86:983–990 [DOI] [PubMed] [Google Scholar]
- 8.Bester L.A., and Essack S.Y. 2008. Prevalence of antibiotic resistance in Campylobacter isolates from commercial poultry suppliers in KwaZulu-Natal, South Africa. J. Antimicrob. Chemother. 62:1298–1300 [DOI] [PubMed] [Google Scholar]
- 9.Bianchini V., Borella L., Benedetti V., Parisi A., Miccolupo A., Santoro E., Recordati C., and Luini M. 2014. Prevalence in bulk tank milk and epidemiology of Campylobacter jejuni in dairy herds in Northern Italy. Appl. Environ. Microbiol. 80:1832–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blaser M.J. 2000. Campylobacter jejuni and related species. In Mandell G.L., Bennett J.E., and Dolin R. (eds.), Mandell, Douglas, and Bennett's Principles and Practice of Infectious Diseases, vol. 2, 5th ed. Churchill Livingstone, Philadelphia, PA, pp. 2276–2285 [Google Scholar]
- 11.Bohaychuk V.M., Gensler G.E., and Barrios P.R. 2011. Microbiological baseline study of beef and pork carcasses from provincially inspected abattoirs in Alberta, Canada. Can. Vet. J. 52:1095–1100 [PMC free article] [PubMed] [Google Scholar]
- 12.Bostan K., Aydin A., and Ang M.K. 2009. Prevalence and antibiotic susceptibility of thermophilic Campylobacter species on beef, mutton, and chicken carcasses in Istanbul, Turkey. Microb. Drug Resist. 15:143–149 [DOI] [PubMed] [Google Scholar]
- 13.Byarugaba D.K. 2004. A view on antmicrobial resistance in developing countries and responsible risk factors. Int. J. Antimicrob. Agents 24:105–110 [DOI] [PubMed] [Google Scholar]
- 14.Cagliero C., Cloix L., Cloeckaert A., and Payot S. 2006. High genetic variation in the multidrug transporter cmeB gene in Campylobacter jejuni and Campylobacter coli. J. Antimicrob. Chemother. 58:168–172 [DOI] [PubMed] [Google Scholar]
- 15.Capita R., Prieto M., and Alonso-Calleja C. 2004. Sampling methods for microbiological analysis of red meat and poultry carcasses. J. Food Prot. 67:1303–1308 [DOI] [PubMed] [Google Scholar]
- 16.Cavaco L.M., and Aarestrup F.M. 2013. Resistance in bacteria of the food chain: epidemiology and control strategies. In Krcméry V. (ed.), Microbial Drug Resistance, Future Medicine Ltd, Brastislava, Slovakia, pp. 136–158 [Google Scholar]
- 17.Chen X., Naren G.W., Wu C.M., Wang Y., Dai L., Xia L.N., Luo P.J., and Zhang Q. 2010. Prevalence and antimicrobial resistance of Campylobacter isolates in broilers from China. Vet. Microbiol. 144:133–139 [DOI] [PubMed] [Google Scholar]
- 18.Clinical and Laboratory Standards Institute. 2012. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Second Informational Supplement M11-S22. CLSI; Wayne, PA [Google Scholar]
- 19.Cody A.J., Clarke L., Bowler I.C., and Dingle K.E. 2010. Ciprofloxacin-resistant Campylobacteriosis in the UK. Lancet 376:1987. [DOI] [PubMed] [Google Scholar]
- 20.Coker A.O., Isokpehi R.D., Thomas B.N., Amisu K.O., and Obi C.L. 2002. Human Campylobacteriosis in developing countries. Emerg. Infect. Dis. 8:237–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dabiri H., Aghamohammad S., Goudarzi H., Noori M., Hedayati M.A., and Ghoreyshiamiri S.M. 2014. Prevalence and antibiotic susceptibility of Campylobacter species isolated from chicken and beef meat. Int. J. Enteric Pathog. 2:e17087 [Google Scholar]
- 22.Debruyne L., Gevers D., and Vandamme P. 2008. Taxonomy of the family Campylobacteraceae. In Nachamkin I., Szymanski C.M., and Blaser M.J. (eds.), Campylobacter, 3rd ed., American Society for Microbiology, Washington, DC, pp. 3–26 [Google Scholar]
- 23.Denis M., Soumet C., Rivoal K., Ermel G., Blivet D., Salvat G., and Colin P. 1999. Development of a m-PCR for simultaneous identification of Campylobacter jejuni and C. coli. Lett. Appl. Microbiol. 29:406–410 [DOI] [PubMed] [Google Scholar]
- 24.Deogratias A., Mushi M.F., Paterno L., Tappe D., Seni J., Kabymera R., Kidenya B.R., and Mshana S.E. 2014. Prevalence and determinants of Campylobacter infection among under five children with acute watery diarrhea in Mwanza, North Tanzania. Arch. Public Health 72:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dingle K.T., Colles F.M., Falush D., and Maiden M.C.J. 2005. Sequence typing and comparison of population biology of Campylobacter coli and Campylobacter jejuni. J. Clin. Microbiol. 43:340–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drlica K., and Zhao X. 1997. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev. 61:377–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.El-Sharoud W.M. 2009. Prevalence and survival of Campylobacter in Egyptian dairy products. Food Res. Int. 42:622–626 [Google Scholar]
- 28.Engberg J., Aarestrup F.M., Taylor D.E., Gerner-Smidt P., and Nachamkin I. 2001. Quinolone and macrolide resistance in Campylobacter jejuni and C. coli: resistance mechanisms and trends in human isolates. Emerg. Infect. Dis. 7:24–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Engberg J., Keelan M., Gerner-Smidt P., and Taylor D.E. 2006. Antimicrobial resistance in Campylobacter. In Aarestrup F.M. (ed.), Antimicrobial Resistance in Bacteria of Animal Origin, ASM Press, Washington DC, pp. 274–277 [Google Scholar]
- 30.Fallon R., O'Sullivan N., Maher M., and Carroll C. 2003. Antimicrobial resistance of Campylobacter jejuni and Campylobacter coli isolates from broiler chickens isolated at an Irish poultry processing plant. Lett. Appl. Microbiol. 36:277–281 [DOI] [PubMed] [Google Scholar]
- 31.Fernandez H., and Hitschfeld M. 2009. Occurrence of Campylobacter jejuni and Campylobacter coli and their biotypes in beef and dairy cattle from the south of Chile. Braz. J. Microbiol. 40:450–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ge B., Wang F., Sjölund-Karlsson M., and McDermott P.F. 2013. Antimicrobial resistance in Campylobacter: susceptibility testing methods and resistance trends. J. Microbiol. Methods 95:57–67 [DOI] [PubMed] [Google Scholar]
- 33.Ghafir Y., China B., Dierick K., De Zutter L., and Daube G.A. 2007. Seven-year survey of Campylobacter contamination in meat at different production stages in Belgium. Int. J. Food Microbiol. 116:111–120 [DOI] [PubMed] [Google Scholar]
- 34.Giacomelli M., Salata C., Martini M., Montesissa C., and Piccirillo A. 2014. Antimicrobial resistance of Campylobacter jejuni and Campylobacter coli from poultry in Italy. Microb. Drug Resist. 20:181–188 [DOI] [PubMed] [Google Scholar]
- 35.Giacometti F., Serraino A., Bonilauri P., Ostanello F., Daminelli P., Finazzi G., Losio M.N., Marchetti G., Liuzzo G., Zanoni R.G., and Rosmini R. 2012. Quantitative risk assessment of verocytotoxin-producing Escherichia coli O157 and Campylobacter jejuni related to consumption of raw milk in a province in Northern Italy. J. Food Prot. 75:2031–2038 [DOI] [PubMed] [Google Scholar]
- 36.Guévremont E., Nadeau E., Sirois M., and Quessy S. 2006. Antimicrobial susceptibilities of thermophilic Campylobacter from humans, swine, and chicken broilers. Can. J. Vet. Res. 70:81–86 [PMC free article] [PubMed] [Google Scholar]
- 37.Hakanen A.J., Lehtopolku M., Siitonen A., Huovinen P., and Kotilainen P. 2003. Multidrug resistance in Campylobacter jejuni strains collected from Finnish patients during 1995–2000. J. Antimicrob. Chemother. 52:1035–1039 [DOI] [PubMed] [Google Scholar]
- 38.Hakkinen M., Heiska H., and Hänninen M. 2007. Prevalence of Campylobacter spp. in cattle in Finland and antimicrobial susceptibilities of bovine Campylobacter jejuni strains. Appl. Environ. Microbiol. 73:3232–3238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harrington P., Archer J., Davis J.P., Croft D.R., and Varma J.K. 2002. Outbreak of Campylobacter jejuni infections associated with drinking unpasteurized milk produced through a cow-leasing program–Wisconsin, 2001. MMWR Morb. Mortal. Wkly. Rep. 51:548–549 [PubMed] [Google Scholar]
- 40.Heuvelink A.E., Van Heerwaarden C., Zwartkruis-Nahuis A., Tilburg J.J., Bos M.H., Heilmann F.G., Hofhuis A., Hoekstra T., and de Boer E. 2009. Two outbreaks of campylobacteriosis associated with the consumption of raw cow's milk. Int. J. Food Microbiol. 134:70–74 [DOI] [PubMed] [Google Scholar]
- 41.Hussain I., Mahmooda M.H., Akhtarb M., and Khanc A. 2007. Prevalence of Campylobacter species in meat, milk and other food commodities in Pakistan. Food Microbiol. 24:219–222 [DOI] [PubMed] [Google Scholar]
- 42.Iovine N.M. 2013. Resistance mechanisms in Campylobacter jejuni. Virulence 4:230–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jacob P., Mdegela R.H., and Nonga H.E. 2011. Comparison of Cape Town and Skirrow's Campylobacter isolation protocols in humans and broilers in Morogoro, Tanzania. Trop. Anim. Health Prod. 43:1007–1013 [DOI] [PubMed] [Google Scholar]
- 44.Jesse T.W., Englen M.D., Pittenger-Alley L.G., and Fedorka-Cray P.J. 2006. Two distinct mutations in gyrA lead to ciprofloxacin and nalidixic acid resistance in Campylobacter coli and Campylobacter jejuni isolated from chickens and beef cattle. J. Appl. Microbiol. 100:682–688 [DOI] [PubMed] [Google Scholar]
- 45.Juntunen P., Heiska H., Olkkola S., Myllyniemi A.L., and Hänninen M.L. 2010. Antimicrobial resistance in Campylobacter coli selected by tylosin treatment at a pig farm. Vet. Microbiol. 146:90–97 [DOI] [PubMed] [Google Scholar]
- 46.Kariuki S. 2010. Antimicrobial resistance in enteric pathogens in developing countries. In Sosa A.J., Byarugaba D.K., Amábile-Cuevas C.F., Hsueh P.-R., Kariuki S., and Okeke I.N. (eds). Antimicrobial Resistance in Developing Countries, Springer Publishing Co., New York, pp. 177–197 [Google Scholar]
- 47.Kashoma I.P.B., Kumar A., Sanad Y.M., Gebreyes W., Kazwala R.R., Garabed R., and Rajashekara G. 2014. Phenotypic and genotypic diversity of thermophilic Campylobacter spp. in commercial Turkey flocks: a longitudinal study. Foodborne Pathog. Dis. 11:850–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kassa T., Gebre-Selassie S., and Asrat D. 2007. Antimicrobial susceptibility patterns of thermotolerant Campylobacter strains isolated from food animals in Ethiopia. Vet. Microbiol. 119:82–87 [DOI] [PubMed] [Google Scholar]
- 49.Kivaria F.M., Noordhuizen J.P., and Kapaga A.M. 2006. Evaluation of the hygienic quality and associated public health hazards of raw milk marketed by smallholder dairy producers in the Dar es Salaam region, Tanzania. Trop. Anim. Health Prod. 38:185–194 [DOI] [PubMed] [Google Scholar]
- 50.Komba E.V., Mdegela R.H., Msoffe P.L., Nielsen L.N., and Ingmer H. 2015. Prevalence, antimicrobial resistance and risk factors for thermophilic Campylobacter infections in symptomatic and asymptomatic humans in Tanzania. Zoonoses Public Health [Epub ahead of print]; DOI: 10.1111/zph.12185 [DOI] [PubMed] [Google Scholar]
- 51.LeBlanc D.J., Lee L.N., Titmas B.M., Smith C.J., and Tenover F.C. 1988. Nucleotide sequence analysis of tetracycline resistance gene tet(O) from Streptococcus mutans DL5. J. Bacteriol. 170:3618–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lehtopolku M., Kotilainen P., Puukka P., Nakari U., Siitonen A., Eerola E., Huovinen P., and Hakanena A.J. 2012. Inaccuracy of the disk diffusion method compared with the agar dilution method for susceptibility testing of Campylobacter spp. J. Clin. Microbiol. 50:52–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li X-Z., Mehrotra M., Ghimire S., and Adewoye L. 2007. β-Lactam resistance and β-lactamases in bacteria of animal origin. Vet. Microbiol. 121:197–214 [DOI] [PubMed] [Google Scholar]
- 54.Linton D., Lawson A.J., Owen R.J., and Stanley J. 1997. PCR detection, identification to species level, and fingerprinting of Campylobacter jejuni and Campylobacter coli direct from diarrheic samples. J. Clin. Microbiol. 35:2568–2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luangtongkum T., Morishita T.Y., El-Tayeb A.B., Ison A.J., and Zhang Q. 2007. Comparison of antimicrobial susceptibility testing of Campylobacter spp. by the agar dilution and the agar disk diffusion methods. J. Clin. Microbiol. 45:590–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mdegela R.H., Nonga H.E., Ngowi H.A., and Kazwala R.R. 2006. Prevalence of thermophilic Campylobacter infections in humans, chickens and crows in Morogoro, Tanzania. J. Vet. Med. 53:116–121 [DOI] [PubMed] [Google Scholar]
- 57.Mdegela R.H., Laurence K., Jacob P., and Nonga H.E. 2011. Occurrences of thermophilic Campylobacter in pigs slaughtered at Morogoro slaughter slabs, Tanzania. Trop. Anim. Health Prod. 43:83–87 [DOI] [PubMed] [Google Scholar]
- 58.Moore J.E., Corcoran D., Dooley J.S., Fanning S., Lucey B., Matsuda M., McDowell D.A., Mégraud F., Millar B.C., O'Mahony R., O'Riordan L., O'Rourke M., Rao J.R., Rooney P.J., Sails A., and Whyte P. 2005. Campylobacter. Vet. Res. 36:351–382 [DOI] [PubMed] [Google Scholar]
- 59.Moore J.E., Barton M.D., Blair I.S., Corcoran D., Dooley J.S.G., Fanning S., Kempff I., and Lastovica A.J. 2006. The epidemiology of antibiotic resistance in Campylobacter. Microb. Infect. 8:1955–1966 [DOI] [PubMed] [Google Scholar]
- 60.Newkirk R., Hedberg C., and Bender J. 2011. Establishing a milk-borne disease outbreak profile: potential food defense implications. Foodborne Pathog. Dis. 8:433–437 [DOI] [PubMed] [Google Scholar]
- 61.Nonga E.H., Sells P., and Karimuribo E.D. 2009. Occurrences of thermophilic Campylobacter in cattle slaughtered at Morogoro municipal abattoir, Tanzania. Trop. Anim. Health Prod. 42:73–78 [DOI] [PubMed] [Google Scholar]
- 62.Noormohamed A., and Fakhr M.K. 2013. A higher prevalence rate of Campylobacter in retail beef livers compared to other beef and pork meat cuts. Int. J. Environ. Res. Public Health 10:2058–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Obeng A.S., Rickard H., Sexton M., Pang Y., Peng H., and Barton M. 2012. Antimicrobial susceptibilities and resistance genes in Campylobacter strains isolated from poultry and pigs in Australia. J. Appl. Microbiol. 113:294–307 [DOI] [PubMed] [Google Scholar]
- 64.Pezzotti G., Serafin A., Luzzi I., Mioni R., Milan M., and Perin R. 2003. Occurrence and resistance to antibiotics of Campylobacter jejuni in animals and meat in northeastern Italy. Int. J. Food Microbiol. 82:281–287 [DOI] [PubMed] [Google Scholar]
- 65.Piddock L.J.V., Ricci V., Pumbwe L., Everett M.J., and Griggs D. 2003. Fluoroquinolone resistance in Campylobacter species from man and animals: detection of mutations in topoisomerase genes. J. Antimicrob. Chemother. 51:19–26 [DOI] [PubMed] [Google Scholar]
- 66.Pratt A., and Korolik V. 2005. Tetracycline resistance of Australian Campylobacter jejuni and Campylobacter coli isolates. J. Antimicrob. Chemother. 55:452–460 [DOI] [PubMed] [Google Scholar]
- 67.Pumbwe L., Randall L.P., Woodward M.J., and Piddock L.J. 2004. Expression of the efflux pump genes cmeB, cmeF and the porin gene porA in multiple-antibiotic-resistant Campylobacter jejuni. J. Antimicrob. Chemother. 54:341–347 [DOI] [PubMed] [Google Scholar]
- 68.Rahimi E., Ameri M., and Kazemeini H.R. 2010. Prevalence and Antimicrobial resistance of Campylobacter species isolated from raw camel, beef, lamb, and goat meat in Iran. Foodborne Pathog. Dis.7:443–447 [DOI] [PubMed] [Google Scholar]
- 69.Rahimi E., Sepehri S., and Momtaz H. 2013. Prevalence of Campylobacter species in milk and dairy products in Iran. Rev. Méd. Vét. 164:283–288 [Google Scholar]
- 70.ROSCO. 2007. Veterinary Practice According to CLSI Breakpoints. Available at https://rosco.docontrol, accessed November2014. (Online.)
- 71.Rozynek E., Antos-Bielska M., Dzierzanowska-Fangrat K., Szczepanska B., and Trafny E.A. 2010. Genetic similarity of Campylobacter isolates in humans, food, and water sources in Central Poland. Foodborne Pathog. Dis. 7:597–600 [DOI] [PubMed] [Google Scholar]
- 72.Saenz Y., Zarazaga M., Lantero M., Gastanares M.J., Baquero F., and Torres C. 2000. Antibiotic resistance in Campylobacter strains isolated from animals, foods, and humans in Spain in 1997–1998. Antimicrob. Agents Chemother. 44:267–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Salihu M.D., Junaidu A.U., Magaji A.A., and Rabiu Z.M. 2010. Study of Campylobacter in raw cow milk in Sokoto State, Nigeria. Br. J. Dairy Sci. 1:1–5 [Google Scholar]
- 74.Sanad Y.M., Closs G.J., Kumar A., LeJeune J.T., and Rajashekara G. 2013. Molecular epidemiology and public health relevance of Campylobacter isolated from dairy cattle and European starlings in Ohio, USA. Foodborne Pathog. Dis. 10:229–236 [DOI] [PubMed] [Google Scholar]
- 75.Sanad Y.M., Kassem I.I., Abley M., Gebreyes W., LeJeune J.T., and Rajashekara G. 2011. Genotypic and phenotypic properties of cattle-associated Campylobacter and their implications to public health in the USA. PLoS One. 10:e25778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sasaki Y., Murakami M., Haruna M., Maruyama N., Mori T., Ito K., and Yamada Y. 2013. Prevalence and characterization of foodborne pathogens in dairy cattle in the eastern part of Japan. Vet. Med. Sci. 75:543–546 [DOI] [PubMed] [Google Scholar]
- 77.Schildt M., Savolainen S., and Hänninen M.L. 2006. Long-lasting Campylobacter jejuni contamination of milk associated with gastrointestinal illness in a farming family. Epidemiol. Infect. 134:401–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schwarz S., Kehrenberg C., Doublet B., and Cloeckaert A. 2004. Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol. Rev. 28:519–542 [DOI] [PubMed] [Google Scholar]
- 79.Smith K.E., Besser J.M., Hedberg C.W., Leano F.T., Bender J.B., Wicklund J.H., Johnson B.P., and Moore K.A. 1999. Quinolone-resistant Campylobacter jejuni infections in Minnesota, 1992–1998. N. Engl. J. Med. 340:1525–1532 [DOI] [PubMed] [Google Scholar]
- 80.Stanley K. and Jones K. 2003. Cattle and sheep farms as reservoirs of Campylobacter. J. Appl. Microbiol. 94(Suppl):104S–113S [DOI] [PubMed] [Google Scholar]
- 81.Taremi M., Mehdi M., Dallal S., Gachkar L., MoezArdalan S., Zolfagharian K., and Zali M.R. 2006. Prevalence and antimicrobial resistance of Campylobacter isolated from retail raw chicken and beef meat, Tehran, Iran. Int. J. Food Microbiol. 108:401–403 [DOI] [PubMed] [Google Scholar]
- 82.U.S. Food and Drug Administration. 2009. U.S. Food and Drug Administration Executive Report 2009. Available at www.fda.gov/downloads/AnimalVeterinary/SafetyHealth/Antimicrobial Resistance/NationalAntimicrobial Resistance Monitoring System/ucm 269042.pdf, accessed January15. 2015
- 83.Vacher S., Ménard A., Bernard E., and Mégraud F. 2003. PCR-restriction fragment length polymorphism analysis for detection of point mutations associated with macrolide resistance in Campylobacter spp. Antimicrob. Agents Chemother. 47:1125–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Van der Beek M.T., Claas E.C., Mevius D.J., van Pelt W., Wagenaar J.A., Kuijper E.J. 2010. Inaccuracy of routine susceptibility tests for detection of erythromycin resistance of Campylobacter jejuni and Campylobacter coli. Clin. Microbiol. Infect. 16:51–56 [DOI] [PubMed] [Google Scholar]
- 85.Van Looveren M., Daube G., De Zutter L., Dumont J.M., Lammens C., Wijdooghe M., Vandamme P., and Jouret M. 2001. Antimicrobial susceptibilities of Campylobacter strains isolated from food animals in Belgium. J. Antimicrob. Chemother. 48:235–240 [DOI] [PubMed] [Google Scholar]
- 86.Wang Y., and Taylor D. E. 1990. Chloramphenicol resistance in Campylobacter coli: nucleotide sequence, expression, and cloning vector construction. Gene. 94:23–28, 1990. [DOI] [PubMed] [Google Scholar]
- 87.Wang Y., Huang W.M., and Taylor D.E. 1993. Cloning and nucleotide sequence of the Campylobacter jejuni gyrA gene and characterization of quinolone resistance mutations. Antimicrob. Agents Chemother. 37:457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.White D.G., Zhao S., Simjee S., Wagner D.D., and Mcdermott P.F. 2002. Antimicrobial resistance of foodborne pathogens. Microbes Infect. 4:405–412 [DOI] [PubMed] [Google Scholar]
- 89.Whyte P., McGill K., Cowley D., Madden R.H., Moran L., Scates P., Carroll C., O'Leary A., Fanning S., Collins J.D., McNamara E., Moore J.E., and Cormican M. 2004. Occurrence of Campylobacter in retail foods in Ireland. Int. J. Food Microbiol. 95:111–118 [DOI] [PubMed] [Google Scholar]
- 90.Widdowson C.A., Klugman K.P., and Hanslo D. 1996. Identification of the tetracycline resistance gene, tet(O), in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 40:2891–2893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wieczorek K., Denis E., Lynch O., and Osek J. 2013. Molecular characterization and antibiotic resistance profiling of Campylobacter isolated from cattle in Polish slaughterhouses. Food Microbiol. 34:130–136 [DOI] [PubMed] [Google Scholar]
- 92.Wieczorek K., and Osek J. 2013. Characteristics and antimicrobial resistance of Campylobacter isolated from pig and cattle carcasses in Poland. Polish J. Vet. Sci. 16:501–508 [DOI] [PubMed] [Google Scholar]
- 93.World Health Organization. 2013. The global view of Campylobacteriosis. Report of Expert Consultation. Utrecht, Netherlands, 9–11 July 2012 [Google Scholar]
- 94.Wysok B., Wiszniewska-Łaszczych A., Uradziński J., and Szteyn J. 2011. Prevalence and antimicrobial resistance of Campylobacter in raw milk in the selected areas of Poland. Polish J. Vet. Sci. 14:473–477 [DOI] [PubMed] [Google Scholar]
- 95.Yamazaki-Matsune W., Taguchi M., Seto K., Kawahara R., Kawatsu K., Kumeda Y., Kitazato M., Nukina M., Misawa N., and Tsukamoto T. 2007. Development of a multiplex PCR assay for identification of Campylobacter coli, Campylobacter fetus, Campylobacter hyointestinalis subsp. hyointestinalis, Campylobacter jejuni, Campylobacter lari and Campylobacter upsaliensis. J. Med. Microbiol. 56:1467–1473 [DOI] [PubMed] [Google Scholar]
- 96.Yang C., Jiang Y., Huang K., Zhu C., and Yin Y. 2003. Application of real-time PCR for quantitative detection of Campylobacter jejuni in poultry, milk and environmental water. FEMS Immunol. Med. Microbiol. 38:265–271 [DOI] [PubMed] [Google Scholar]
- 97.Zhu J., Zhang Y., Hua X., Hou J., and Jiang Y. 2006. Antibiotic resistance in Campylobacter. Rev. Med. Microbiol. 17:107–112 [Google Scholar]