Abstract
Leishmaniasis is a serious problem that affects mostly poor countries. Various species of Leishmania are the agents of the disease, which take different clinical manifestations. The parasite is transmitted by sandflies, predominantly from the Phlebotomus genus in the Old World and Lutzomyia in the New World. During development in the gut, Leishmania must survive various challenges, which include avoiding being expelled with blood remnants after digestion. It is believed that attachment to the gut epithelium is a necessary step for vector infection, and molecules from parasites and sand flies have been implicated in this attachment. In previous work, monoclonal antibodies were produced against Leishmania. Among these an antibody was obtained against Leishmania braziliensis flagella, which blocked the attachment of Leishmania panamensis flagella to Phlebotomus papatasi guts. The protein recognized by this antibody was identified and named FLAG1, and the complete FLAG1 gene sequence was obtained. This protein was later independently identified as a small, myristoylated protein and called SMP1, so from now on it will be denominated FLAG1/SMP1. The FLAG1/SMP1 gene is expressed in all developmental stages of the parasite, but has higher expression in promastigotes. The anti-FLAG1/SMP1 antibody recognized the flagellum of all Leishmania species tested and generated the expected band by western blots. This antibody was used in attachment and infection blocking experiments. Using the New World vector Lutzomyia longipalpis and Leishmania infantum chagasi, no inhibition of attachment ex vivo or infection in vivo was seen. On the other hand, when the Old World vectors P. papatasi and Leishmania major were used, a significant decrease of both attachment and infection were seen in the presence of the antibody. We propose that FLAG1/SMP1 is involved in the attachment/infection of Leishmania in the strict vector P. papatasi and not the permissive vector L. longipalpis.
Key Words: : Leishmania, Lutzomyia longipalpis, Phlebotomus papatasi, Flagellum, FLAG1/SMP1, Leishmaniasis, Sand fly, Vector–parasite interaction
Introduction
Phlebotomine (Diptera: Psychodidae) sand flies are the main vectors of leishmaniasis, a disease ranging from self-healing skin lesions to fatal, if left untreated, visceral disease. Different Leishmania species are associated with distinct disease outcome, and from the hundreds of sand flies identified to date only a limited number have been proven to be bona fide vectors of Leishmania (Killick-Kendrick 1990). Some sand fly species are considered permissive (e.g., Lutzomyia longipalpis) because they can be infected by several Leishmania, whereas other vectors can be infected only with the Leishmania species they carry in nature and are thus considered restrictive (e.g., Phlebotomus papatasi) (Sacks and Kamhawi 2001, Volf and Myskova 2007). Several parasite and vector molecules allow the parasite's infection, survival, and multiplication within the midgut of the sand fly and eventual transmission to a vertebrate host during a blood meal. Leishmania can manipulate the sand fly, potentially threatening digestive proteases activity (Borovsky and Schlein 1983, Schlein et al. 1983, Dillon and Lane 1993, Schlein and Jacobson 1998, Telleria et al. 2010) and also can cause damage to the stomodeal valve of the fly (Schlein et al. 1992, Rogers et al. 2008), potentiating transmission. On the other hand, sand flies can mount an immune response to Leishmania infection (Boulanger et al. 2004, Ramalho-Ortigão et al. 2007, Jochim et al. 2008, Pitaluga et al. 2009, Diaz-Albiter et al. 2012, Telleria et al. 2012).
Although Leishmania do not invade the midgut cells, adhesion to epithelial cells is well documented (Killick-Kendrick and Rioux, 1991). In the case of the P. papatasi attachment of Leishmania major can be promoted by the lipophosphoglycan (LPG) that covers the parasite (Sacks and Kamhawi 2001), for which the midgut galactose-binding protein PpGalec was shown to be a receptor (Kamhawi et al. 2004). The function of LPG in attachment was confirmed in strict vector–parasite pairs by the use of LPG-deficient Leishmania that failed to adhere to midguts ex vivo and in vivo after blood digestion (Sacks et al. 1995, Sacks et al. 2000). However, in permissive vectors, LPG-deficient Leishmania infected the insects normally, indicating an alternative attachment mechanism (Svárovská et al. 2010, Jecna et al. 2013). LPG-independent midgut binding has been suggested in association with the degree of glycosylation of proteins expressed by midgut epithelial cells (Myskova et al. 2007).
It is possible that other unknown molecules also have a role in midgut attachment. We have previously shown that a monoclonal antibody developed against Leishmania braziliensis flagella (Ismach et al. 1989) was capable of inhibiting attachment of Leishmania panamensis or L. major to dissected guts of P. papatasi (Warburg et al. 1989). We identified the 15-kD flagellar protein FLAG1/SMP1 recognized by this antibody (Córdova-Rojas 1998). Later, this protein was identified as a small myristoylated protein (SMP1) by another group (Tull et al. 2004). Here we show that the Leishmania flagellar protein FLAG1/SMP1 has a role in parasite interaction with the vector, in the case of the strict vector P. papatasi.
Materials and Methods
Leishmania
Leishmania infantum chagasi (MHOM/BR/1974/PP75), L. infantum (MHOM/ES/00/UCM-1), L. pifanoi (MHOM/VE/1975/LL1), L. amazonensis (MHOM/BR/1967/PH8), L. major (MHOM/SU/1973-ASKH), L. donovani (MHOM/ET/1967/HU3), and L. mexicana (MHOM/BZ/1982/BEL21) were obtained from the Instituto Oswaldo Cruz Leishmania Culture Collection. Promastigote-stage parasites were maintained by weekly transfers using M199 medium (pH 7.0) supplemented with 10% fetal bovine serum (FBS). L. pifanoi amastigotes were maintained at 30°C in F29 medium supplemented with FBS (Pan 1984). To obtain a metacyclic enriched population of parasites, a Ficoll (PM400, Sigma) gradient was used as described (Späth and Beverley 2001, Yao et al. 2008).
Sand flies and infection with Leishmania
L. longipalpis originating from Lapinha Cave, Minas Gerais, Brazil, F1 adults or insects from a laboratory colony maintained at the Department of Entomology, Kansas State University for several years (LLLP strain), were used in the experiments. Insects were sugar fed on 30% sucrose solution ad libitum and blood fed directly on an anesthetized male hamster when needed. P. papatasi from colonies originating from Israel were maintained in a similar way. Three- to five-day-old sand flies were fed on mouse blood alone of blood containing 5×106 L. major promastigotes/mL or 1×107 L. i. chagasi promastigotes/mL using a feeding apparatus. All procedures involving live animals were approved by the FIOCRUZ bioethics committee (CEUA, protocol number P0-116-02) and by the Committee on Institutional Animal Care and Use of the Kansas State University (KSU-IACUC) (protocol numbers 3080 and 3081). All sand fly feedings on animals and all bleeds were performed on animals under anesthesia, and all efforts were made to minimize suffering.
Isolation and characterization of FLAG1
The FLAG1 protein was purified from Leishmania flagella preparations using previously described methods (Pereira et al. 1977, Ismach et al. 1989). The flagellar-enriched fraction was extracted using Brij-97, and nonsoluble material was eliminated by centrifugation at 39,000×g for 1 h at 4°C. The supernatant was then diluted in cold 20 mM sodium phosphate buffer (pH 7.0) containing protease inhibitors (1 mM of iodoacetamide, 1 μg/mL of 1,10-phenanthroline, and 5 μg/mL phenylmethylsulfonyl fluoride) purified on a F-2 monoclonal antibody immunoaffinity column. The FLAG-1 protein was eluted with 0.1 M glycine-HCl (pH 2.7), which was immediately neutralized with 1 M Tris-HCl (pH 7.2). Purity was assessed using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The sample was subjected to isoelectric focusing (first dimension) followed by SDS-PAGE, as described by O'Farrell (1975).
The two-dimensional SDS-PAGE fractionated protein was electrotransferred onto Immobilon-P membranes (Millipore, Bedford, MA) as described (Towbin et al. 1979) and sent for amino acid composition and peptide sequencing at the Yale University School of Medicine Protein and Nucleic Acid Chemistry Facility. Peptides were generated by chemical cleavage with cyanogen bromide (CNBr) (Jirikowski, 1985) or digested enzymatically with trypsin (Lee and Forstner 1985), and isolated by high-performance liquid chromatography (HPLC) using a reverse-phase Vydac column C18 (Nest Group Inc., Southborough, MA). Selected isolated peptide peaks were sequenced using gas-phase Applied Biosystems models 470 and 477, as described (Matsudaira 1987).
FLAG1/SMP1 sequencing
L. i. chagasi FLAG1/SMP1 was amplified from parasites DNA using primers Flag F (5′-GGA TCC GGC TGC GGT GCT TCT TCT-3′) and Flag R (5′-AAG CTT CTT TTC CTT CTC CGC CTG-3′) and sequenced in the Instituto Oswaldo Cruz (FIOCRUZ) PDTIS Facility.
Bioinformatics analyses
The FLAG1/SMP1 sequences aligned were collected at the National Center for Biotechnology Information (NCBI) genes database with the exception of L. i. chagasi, which was obtained from Sanger sequencing in our laboratory. The protein sequences were predicted with the ExPASy translation tool. For the alignments, we used the program Muscle WS with the score matrix Blossum62 through the platform Jalview. Phylogenetic analysis was performed using the neighbor-joining method, with a bootstrap of 1000 and the model p-distance, and using the MEGA 5.1 program.
Real-time PCR
Total RNA from L. i. chagasi and L. major (promastigotes) and L. pifanoi (amastigotes and promastigotes) was extracted using TRIzol reagent (Invitrogen), according to manufacturer's instructions, followed by DNase I (Promega) treatment.
First-strand cDNA was synthesized from 5 μg of total RNA using SuperScript III First-Strand Synthesis (Invitrogen). Real-time polymerase chain reaction (qPCR) was performed using SYBR Green PCR Master Mix (Applied Biosystems) and the following primers FLAG_Rtime_Fwd (5′-AGT GGG TAG CCT CCG TGG TGG TGT A-3′), FLAG_Rtime_Rev (5′-CTC CGA CAG CGG CAA GGC GTC CAT C-3′), LeishActin_Fwd (5′-GTG GTC GAT AAA GCC GAA GGT GGT T-3′), LeishActin_Rev (5′-TTG GGC CAG ACT CGT CGT ACT CGC T-3′). Expression levels of FLAG1/SMP1 were determined through ΔΔCt, normalized using actin gene expression, yielding the relative expression value (Pfaffl 2001).
Immunofluorescence
Leishmania were concentrated to 108 parasites in 20 μL of phosphate-buffered saline (PBS), applied to poly-l-lysine Poly-Prep™ slides from Sigma Diagnostics, fixed in methanol at −20°C, and blocked in PBS+3% bovine serum albumin (BSA) for 45–60 min. The slides were incubated with the anti-FLAG1/SMP1 antibody (Ismach et al. 1989) in a 1:100 dilution and then with a 1:1000 dilution of an anti-mouse rhodamine-coupled antibody (Jackson ImmunoResearch) in blocking solution for 45–60 min. Images were obtained in the confocal microscope LSM 510 META from the FIOCRUZ, PDTIS Facility.
Western blot
Samples corresponding to 25 μg of Leishmania extracts were separated on 15% SDS-PAGE gels at 120V for 2 h. Proteins were transferred to nitrocellulose membranes (BioRad) at 100V for 1.5 h at 4°C. The membranes were blocked with 5% nonfat milk Tris-buffered saline (TBS)+0.1% Tween 20 (TBST) for at least 1 h. The membranes were then incubated with anti-FLAG1/SMP1 monoclonal antibody (Ismach et al. 1989) (1:3000) for 2 h in the same solution. After three washes of 10 min in TBST, the membranes were incubated with anti-mouse secondary antibody (Jackson ImmunoResearch) at a 1:10,000 dilution for 1 h. Three more washes were performed before the incubation of the membrane with the detection system Pierce SuperSignal West Pico Chemiluminescent Substrate (ThermoScientific).
Ex vivo inhibition of parasite binding by anti-FLAG1/SMP1 antibody
Leishmania culture promastigotes (5×106) were incubated at room temperature for 1 h with anti-FLAG1/SMP1 antibody (1:500) or PBS. Midguts from 3- to 5-day-old non–blood-fed sand flies were dissected and washed in ice-cold PBS. Each midgut was opened longitudinally to expose the midgut epithelium and then incubated for 1 h at room temperature with the FLAG1/SMP1-treated parasites. Individual midguts were then washed three times with cold PBS, and homogenized in 30 μL of PBS followed by parasite counting using a hemocytometer.
Inhibition of infection by anti-FLAG1/SMP1 antibody
L. longipalpis and P. papatasi females were artificially fed with blood containing 107 L. i. chagasi or 5×106 L. major. The same volumes of anti-FLAG1/SMP1 antibody or PBS were added to the blood-containing parasites. After 72 h, groups of approximately seven females were examined and dissected in cold PBS. Individual midguts were homogenized in 30 μL of cold PBS, and parasites were counted using a hemocytometer. Total number of insects used in each experiment is shown in the figures (see below), where each dot represents one insect.
Statistical analysis
The Wilcoxon signed-rank test was used to analyze in vitro binding and in vivo infection. Values were considered significant at the 95% confidence interval.
Results
Sequencing of FLAG1/SMP1
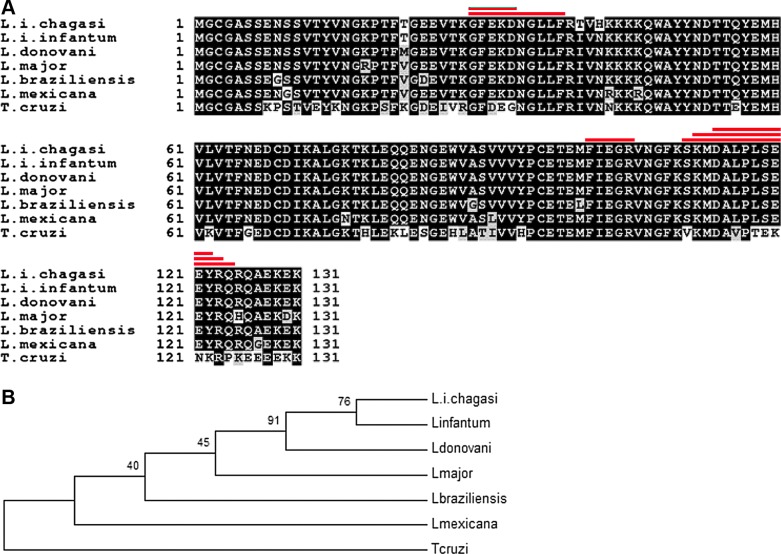
FLAG1/SMP1 protein was isolated and sequence determined (Córdova-Rojas 1998). Briefly, the FLAG1/SMP1 protein was isolated from detergent-solubilized flagellar preparations from L. amazonensis promastigotes using immunoaffinity chromatography and the F-2 monoclonal antibody (Ismach et al. 1989). Peptides from CNBr-trypsin–digested FLAG1/SMP1 protein were isolated by HPLC and sequenced. Peptide sequences are shown in Table 1, and their location on the FLAG1/SMP1 complete sequence is shown in Figure 1.
Table 1.
Peptide Sequence Data from Cyanogen Bromide-Trypsin-Digested FLAG1/SMP1
| Elution peak number | Peptide sequence |
|---|---|
| 33 | FIEGR |
| 52 | a/s/gGxMDALPLSEEYxQ |
| 55 | (d)KMDALPLSEEYR |
| 57 | DALPLSEEY |
| 70 | xFEKDNGLLF |
| 71 | GFEKD |
FIG. 1.
Alignment of various Leishmania species and T. cruzi FLAG1/SMP1 sequences. (A) Alignment of L. i. chagasi, L. infantum (XP_001465265.1), L. donovani (XP_003860476.1), L. major (AAV59017.1), L. braziliensis (XP_003723102.1), L. mexicana (CBZ26701.1), and T. cruzi (XP_806365.1) FLAG1/SMP1-deduced protein sequences. The red lines above the aligned sequences show the position of peptides from Table 1. (B) Phylogenetic tree of the different trypanosomatid FLAG1/SMP1 proteins.
Sequencing of the L. i. chagasi FLAG1/SMP1 gene
Degenerate PCR primers were designed and synthesized based on these internal peptide sequences (Table 1), and partial nucleotide sequences were obtained (Córdoba-Rojas 1998). A complete sequence was obtained as described above and this was aligned with other Leishmania and one Trypanosoma cruzi sequence (Fig. 1). FLAG1/SMP1 is highly conserved among all investigated trypanosomatids, with a higher degree of conservation among the Leishmania, as expected.
Expression of FLAG1/SMP1
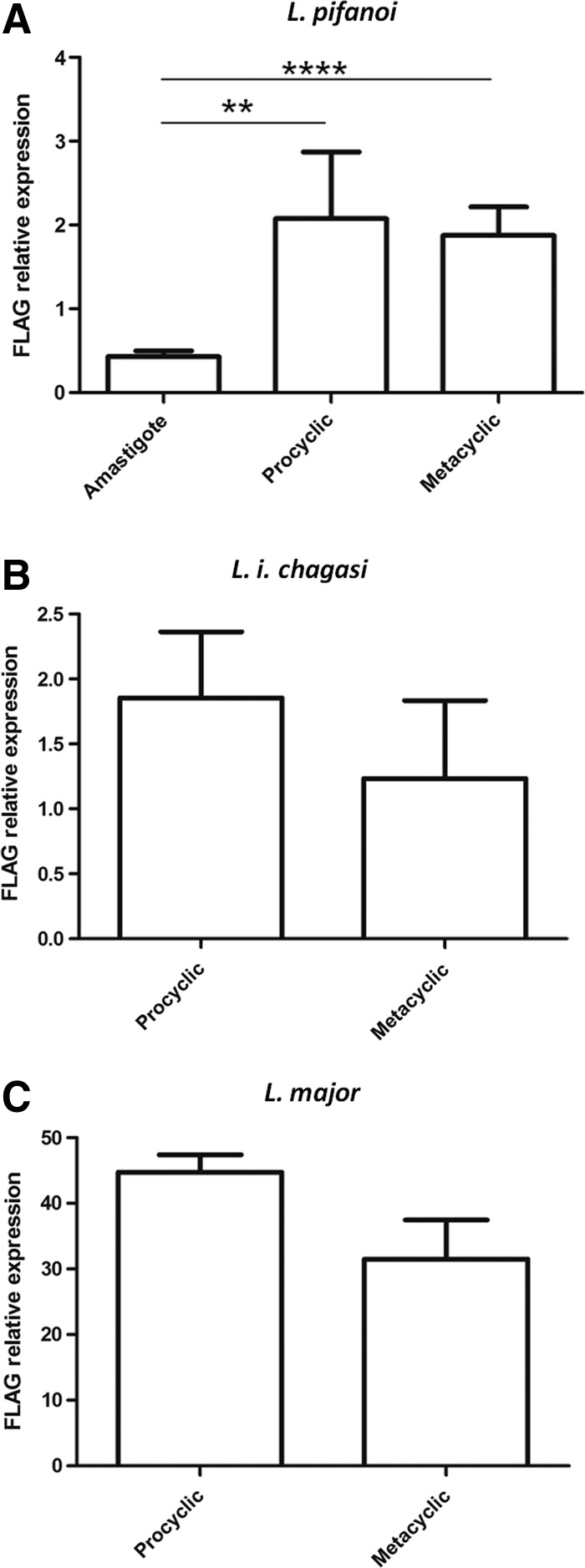
Expression of the FLAG1/SMP1 gene was assessed by qPCR in different developmental stages of L. major, L. i. chagasi, and L. pifanoi, the only Leishmania species for which axenic amastigotes were available. The axenic amastigote forms of L. pifanoi presented a significantly lower expression when compared to promastigote forms (Fig. 2A).
FIG. 2.
FLAG1/SMP1 relative expression in developmental forms of Leishmania. The expression of FLAG1/SMP1 in procyclic, metacyclic and amastigotes forms of L. pifanoi (A), L. i. chagasi (B), and L. major (C) was determined by qPCR. (****) p less than sign 0.0001; (**) p<0.001.
There was no significant difference in FLAG1/SMP1 mRNA expression between promastigote and metacyclic forms of the parasites, although L. i. chagasi and L. major metacyclics presented a tendency toward lower expression (Fig. 2B, C).
Detection of FLAG1/SMP1 in different Leishmania species by western blot and immunofluorescence
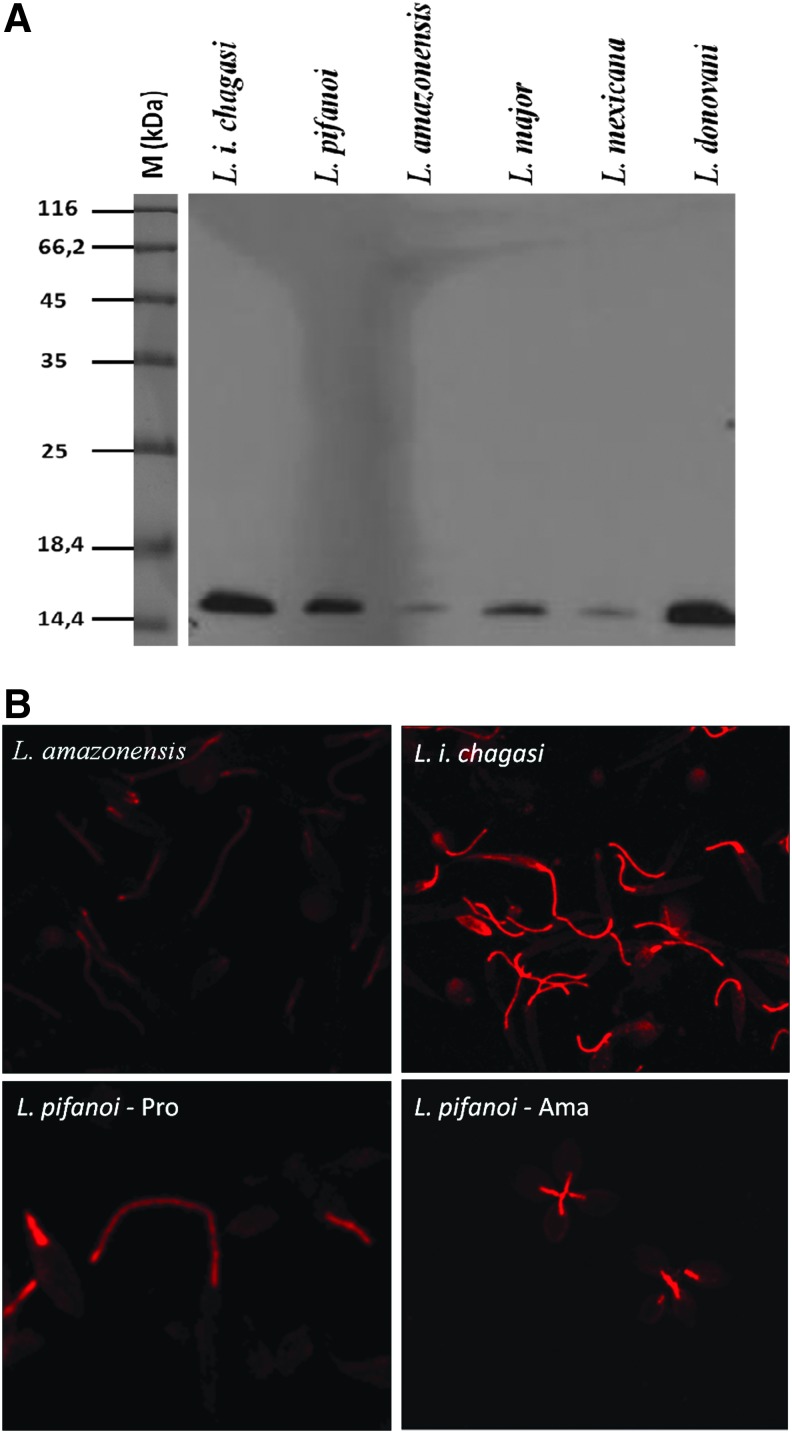
The presence and localization of FLAG1/SMP1 was investigated by western blot and indirect immunofluorescence assays, using the anti-FLAG1/SMP1 monoclonal antibody. FLAG1/SMP1 is expressed in L. i. chagasi, L.pifanoi, L. amazonensis, L. major, L. mexicana, and L. donovani (Fig. 3A) and is localized along the entire length of the flagellum in all species investigated, namely L. amazonensis, L. i. chagasi, and L. pifanoi (Fig. 3B).
FIG. 3.
Presence of FLAG1/SMP1 in different Leishmania species. (A) Western blot of parasite total protein extracts. (B) Indirect immunofluorescence using anti-FLAG1/SMP1 antibody.
Attachment/infection inhibition experiments
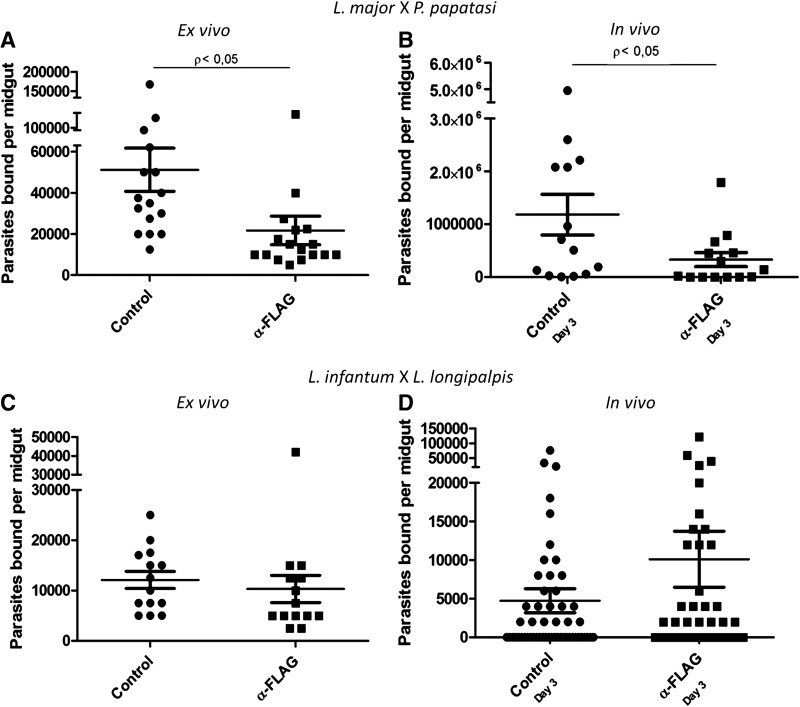
In both ex vivo and in vivo attachment/infection inhibition assays using the pair L. longipalpis and L. i. chagasi, no effect of the antibody was seen on the adhesion/infection process (Fig. 4C, D). Similar results were obtained using F1 L. longipalpis (from the Lapinha Cave) as well as its counterpart maintained in laboratory colonies for many generations (LLLP).
FIG. 4.
Effect of anti-FLAG1/SMP1 antibody on parasite binding to the midgut and sand fly infection. Attachment of L. major to P. papatasi midgut epithelia ex vivo (A) and infection in vivo (B) in the presence or absence of anti-FLAG1/SMP1 antibody were investigated. Attachment of L. infantum to L. longipalpis midgut epithelia ex vivo (C) and sand fly Leishmania infection in vivo (D) in the presence or absence of anti-FLAG1/SMP1 antibody were investigated in the presence or absence of anti-FLAG1/SMP1 antibody. Each dot in the graphs represents one insect; the horizontal line indicates the mean, and the whisker plots represent the standard deviation.
However, ex vivo and in vivo attachment/infection inhibition experiments performed with P. papatasi and L. major showed a different outline. There was a significant decrease in the number of parasites attached or infected insects when they were pretreated with anti-FLAG1/SMP1 monoclonal antibody (Fig. 4A, B).
Discussion
It is widely accepted that a close interaction of the parasite Leishmania to the gut of the sand fly vector, as first seen by Killick-Kendrick et al. (1974a, b), is fundamental for a successful infection. It is also believed that the flagellum is involved in such interactions with a role in the adhesion, both through hemidesmosomes formation with the ectodermic cuticle of the gut (Walters et al. 1989) and through the flagellum insertion between microvilli of the midgut (Molyneux 1977, Walters et al. 1989). Interestingly, promastigotes move anteriorly using the flagellum; thus, as parasites migrate from the midgut toward the anterior gut parts of the sand fly during metacyclogenesis, the salient hypothesis that the flagellum touches, senses, and transduces signals is very appealing. These assumptions were reinforced when Cuvillier et al. (2003) demonstrated that L. amazonensis overexpressing the ADP-ribosylation factor-like protein 3A (LdARL-3A) lacked a flagellum and was incapable of developing in L. longipalpis. In contrast, more recent work indicated that a flagellum-less L. braziliensis isolated from a patient was capable of infecting this same vector (Zauli et al. 2012). These apparently contradicting results suggest a possibly complex interplay of different components during the Leishmania–sand fly interaction and need to be investigated further.
A potential alternative candidate for such interaction was identified several years ago by some of us following the development of an anti-flagellum monoclonal antibody (Ismach et al 1989) shown to block adhesion of L. panamensis flagella to P. papatasi midguts (Warburg et al. 1989). This antibody recognizes a protein called FLAG1 (Rojas 1997) which was independently characterized by another group and named SMP-1. Tull et al. (2004) demonstrated that SMP-1 is myristoylated and palmitoylated, and that these fatty acids are responsible for the flagellar location of the protein. SMP-1 was also shown to stabilize the flagellar membrane, and it is required for flagellar elongation and function. Deletion of the genes encoding SMP-1 led to the production of short flagella and defects in motility (Tull et al. 2009).
We were especially interested in the preliminary results indicating a function of this protein in attachment of parasites to the sand fly gut (Warburg et al. 1989). LPG has been clearly demonstrated to be critical in Leishmania binding to the so-called restrictive vectors (Sacks and Kamhawi 2001). However, in permissive vectors, molecules responsible for such binding remain elusive, in spite of the indication that the level of glycosylation of midgut molecules potentially is related to this binding (Myskova et al. 2007). Hence, we initially investigated the role of FLAG1/SMP1 in the attachment of Leishmania to L. longipalpis. Our results, however, did not support a role for this flagellar protein in attachment in this Leishmania–sand fly pair. On the other hand, our results suggest that FLAG1/SMP1 is involved in the binding of L. major to the restrictive vector P. papatasi. The fact that L. major LPG mutants failed to infect P. papatasi indicates that FLAG1/SMP1 per se is not sufficient to allow survival of the parasite in this vector. LPG covers the entire surface of the parasite whereas FLAG1/SMP1 is localized to the flagellum. The initial interaction of the parasite with the gut occurs by the flagellum, and classical depictions of this interaction show this organelle is deeply embedded into the gut microvilli (Killick-Kendrick et al. 1974a, b). We hypothesize that this initial interaction through the flagellum, via FLAG1/SMP1, may be essential for further and possibly stronger interaction through the LPG molecules.
FLAG1/SMP1 was found to be a well-conserved protein present in all Leishmania investigated so far. By immunofluorescence, the anti- FLAG1/SMP1 monoclonal antibody reacted with the flagellum of L. braziliensis, L. i. chagasi, L. donovani, L. guyanensis, L. major, L. mexicana, L. panamensis, and L. pifanoi (Córdova-Rojas 1998, Tull et al. 2010; our results). Western blots of L. amazonensis, L. i. chagasi, L. donovani, L. major, L. mexicana, and L. pifanoi using the anti-FLAG1/SMP1 antibody produced the expected band of approximately 15 kD. Differences in intensity observed both in the immunofluorescence and western blots may be reflective of the abundance of protein present in different parasites or due to differences in epitope recognition of the different FLAG1/SMP1 proteins by the antibody. Expression of the FLAG1/SMP1 gene was compared among different developmental forms of some parasite species. As expected, there was lower mRNA expression in axenic amastigotes of L. pifanoi and a slight but nonsignificant decreased expression in metacyclic versus procyclic forms of L. pifanoi, L. i. chagasi, and L. major. When the FLAG1/SMP1 sequences available in the database were compared, a high degree of homology between the various sequences was evident, suggesting a strong functional conservation. It is interesting to speculate how such a conserved protein might have a strikingly different function in vector–parasite interactions. This might be due to the small sequence differences observed among Leishmania species, although these do not seem to cause big structural changes in the molecule, as can be seen in Figure 1.
Conclusions
We have shown that FLAG1/SMP1 is involved in the attachment of Leishmania to the strict vector P. papatasi and not to the permissive vector L. longipalpis. We are currently attempting to identify molecule(s) in the restrictive P. papatasi sand fly that are able to interact with FLAG1/SMP1. We predict that such a molecule will be present in significantly greater concentrations in this fly in comparison to L. longipalpis, or altogether absent in the latter.
Acknowledgment
We are grateful to Drs. Gregg N. Milligan and Peter W. Mason from the University of Texas Medical Branch for the production of Anti-FLAG1 monoclonal antibody. This work was supported by the National Council for Research (CNPq), Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), PAPES-Fiocruz.
Author Disclosure Statement
No competing financial interests exist.
References
- Borovsky DP, Schlein Y. Trypsin and chymotrypsin-like enzymes of the sandfly Phlebotomus papatasi infected with Leishmania and their possible role in vector competence. Med Vet Entomol 1987; 1:235–242 [DOI] [PubMed] [Google Scholar]
- Boulanger N, Lowenberger C, Volf P, Ursic R, et al. Characterization of a defensin from the sand fly Phlebotomus duboscqi induced by challenge with bacteria or the protozoan parasite Leishmania major. Infect Immun 2004; 72:7140–7146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Córdova-Rojas JL. Biological, biochemical and molecular characterization of Flag-1,a Leishmania membrane-associated flagellar protein. PhD Thesis, Yale University, 1998 [Google Scholar]
- Cuvillier A, Miranda JC, Ambit A, Barral A, et al. Abortive infection of Lutzomyia longipalpis insect vectors by aflagellated LdARL-3A-Q70L overexpressing Leishmania amazonensis parasites. Cell Microbiol 2003; 5:717–728 [DOI] [PubMed] [Google Scholar]
- Diaz-Albiter H, Sant'Anna MR, Genta FA, Dillon RJ. Reactive oxygen species-mediated immunity against Leishmania mexicana and Serratia marcescens in the phlebotomine sand fly Lutzomyia longipalpis. J Biol Chem 2012; 287:23995–24003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon RJ, Lane RP. Influence of Leishmania infection on blood-meal digestion in the sandflies Phlebotomus papatasi and P. langeroni. Parasitol Res 1993; 79:492–496 [DOI] [PubMed] [Google Scholar]
- Ismach R, Cianci CML, Caulfield JP, Lange PJ, et al. Flagellar membrane and paraxial rod proteins of Leishmania: Characterization employing monoclonal antibodies. J Protozool 1989; 36:617–624 [DOI] [PubMed] [Google Scholar]
- Jecna L, Dostalova A, Wilson R, Seblova V. The role of surface glycoconjugates in Leishmania midgut attachment examined by competitive binding assays and experimental development in sand flies. Parasitology 2013; 140:1026–1032 [DOI] [PubMed] [Google Scholar]
- Jirikowski G. Cyanogen bromide cleavage of methionine residues as a control method for enkephalin immunocytochemistry. Histochemistry 1985; 83:93–95 [DOI] [PubMed] [Google Scholar]
- Jochim RC, Teixeira CR, Laughinghouse A, Mu J, et al. The midgut transcriptome of Lutzomyia longipalpis: Comparative analysis of cDNA libraries from sugar-fed, blood-fed, post-digested and L. i. chagasi-infected sand flies. BMC Genomics 2008; 9:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamahawi S, Ramalho-Ortigao M, Pham VM, Kumar S, et al. A role for insect galectins in parasite survival. Cell 2004;119:329–341 [DOI] [PubMed] [Google Scholar]
- Killick-Kendrick R. Phlebotomine vectors of the leishmaniases: A review. Med Vet Entomol 1990; 4:1–24 [DOI] [PubMed] [Google Scholar]
- Killick-Kendrick R, Rioux JA. Intravectorial cycle of Leishmania in sandflies. Ann Parasitol Hum Comp 1991; 66:71–74 [PubMed] [Google Scholar]
- Killick-Kendrick R, Molyneux DH, Ashford RW. Ultrastructural observations on the attachment of Leishmania in the sandfly. Trans R Soc Trop Med Hyg 1974a; 68:269. [PubMed] [Google Scholar]
- Killick-Kendrick R, Molyneux DH, Ashford RW. Leishmania in phlebotomid sandflies. I. Modifications of the flagellum associated with attachment to the mid-gut and oesophageal valve of the sandfly. Proc R Soc Lond B Biol Sci 1974b; 187:409–419 [DOI] [PubMed] [Google Scholar]
- Lee L, Forstner G. Mapping of proteolytic and cyanogen bromide peptides from subunits of intestinal maltase-glucoamylase: Evidence for significant homology. Can J Biochem Cell Biol 1985; 63:257–262 [DOI] [PubMed] [Google Scholar]
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem 1987; 262:10035–10038 [PubMed] [Google Scholar]
- Molyneux DH. Vector relationships in the Trypanosomatidae. Adv Parasitol 1977; 15:1–82 [DOI] [PubMed] [Google Scholar]
- Myskova J, Svobodova M, Beverley S, Volf P. A lipophosphoglycan-independent development of Leishmania in permissive sand flies. Microbes Infect 2007; 9:317–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem 1975; 250:4007–4021 [PMC free article] [PubMed] [Google Scholar]
- Pan AA. Leishmania mexicana: Serial cultivation of intracellular stages in a cell-free medium. Exp Parasitol 1984; 58:72–80 [DOI] [PubMed] [Google Scholar]
- Pereira NM, de Souza W, Machado RD, de Castro FT. Isolation and properties of flagella of trypanosomatids. J Protozool 1977; 24:511–514 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time qPCR. Nucleic Acids Res 2001; 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitaluga AN, Beteille V, Lobo AR, Ortigao-Farias JR, et al. EST sequencing of blood-fed and Leishmania-infected midgut of Lutzomyia longipalpis, the principal visceral leishmaniasis vector in the Americas. Mol Genet Genomics 2009; 282:307–317 [DOI] [PubMed] [Google Scholar]
- Ramalho-Ortigao M, Jochim RC, Anderson JM, Lawyer PG, et al. Exploring the midgut transcriptome of Phlebotomus papatasi: Comparative analysis of expression profiles of sugar-fed, blood-fed and Leishmania major-infected sandflies. BMC Genomics 2007; 8:300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalho-Ortigao M, Saraiva EM, Traub-Csekö YM. Sand fly-Leishmania interactions: Long relationships are not necessarily easy. Open Parasitol J 2010; 4:195–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers ME, Hajmová M, Joshi MB, Sadlova J, et al. Leishmania chitinase facilitates colonization of sand fly vectors and enhances transmission to mice. Cell Microbiol 2008; 10:1363–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacks D, Kamhawi S. Molecular aspects of parasite-vector and vector-host interactions in leishmaniasis. Annu Rev Microbiol 2001; 55:453–483 [DOI] [PubMed] [Google Scholar]
- Sacks DL, Pimenta PF, McConville MJ, Schneider P, et al. Stage-specific binding of Leishmania donovani to the sand fly vector midgut is regulated by conformational changes in the abundant surface lipophosphoglycan. J Exp Med 1995; 181:685–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacks DL, Modi G, Rowton E, Spath G, et al. The role of phosphoglycans in Leishmania-sand fly interactions. Proc Natl Acad Sci USA 2000; 97:406–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlein Y, Jacobson RL. Resistance of Phlebotomus papatasi to infection with Leishmania donovani is modulated by components of the infective bloodmeal. Parasitology 1998; 117:467–473 [DOI] [PubMed] [Google Scholar]
- Schlein Y, Warburg A, Schnur LF, Shlomai J. Vector compatibility of Phlebotomus papatasi dependent on differentially induced digestion. Acta Trop 1983; 40:65–70 [PubMed] [Google Scholar]
- Schlein Y, Jacobson R, Messer G. Leishmania infections damage the feeding mechanism of the sandfly vector and implement parasite transmission by bite. Proc Natl Acad Sci USA 1992; 89:9944–9948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Späth GF, Beverley SM. A lipophosphoglycan-independent method for isolation of infective Leishmania metacyclic promastigotes by density gradient centrifugation. Exp Parasitol 2001; 99:97–103 [DOI] [PubMed] [Google Scholar]
- Svárovská A, Ant TH, Seblová V, Jecná L, et al. Leishmania major Svárovská: A Glycosylation mutants require phosphoglycans (lpg2-) but not lipophosphoglycan (lpg1-) for survival in permissive sand fly vectors. PLoS Negl Trop Dis 2010; 4:e580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telleria EL, de Araújo AP, Secundino NF, d'Avila-Levy CM, et al. Trypsin-like serine proteases in Lutzomyia longipalpis—expression, activity and possible modulation by Leishmania infantum chagasi. PLoS One 2010; 5:e10697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telleria EL, Sant'Anna MR, Ortigão-Farias JR, Pitaluga AN, et al. Caspar-like gene depletion reduces Leishmania infection in sand fly host Lutzomyia longipalpis. J Biol Chem 2012; 287:12985–12993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordo J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc Natl Acad Sci USA 1979; 76:4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tull D, Vince JE, Callaghan JM, Naderer T, et al. SMP-1, a member of a new family of small myristoylated proteins in kinetoplastid parasites, is targeted to the flagellum membrane in Leishmania. Mol Biol Cell 2004; 11:4775–4786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tull D, Naderer T, Spurck T, Mertens HD, et al. Membrane protein SMP-1 is required for normal flagellum function in Leishmania. J Cell Sci 2010; 123:544–554 [DOI] [PubMed] [Google Scholar]
- Volf P, Myskova J. Sand flies and Leishmania: Specific versus permissive vectors. Trends Parasitol 2007; 23:91–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters LL, Modi GH, Chaplin GL, Tesh RB. Ultrastructural development of Leishmania chagasi in its vector, Lutzomyia longipalpis (Diptera: Psychodidae). Am J Trop Med Hyg 1989; 41:295–317 [PubMed] [Google Scholar]
- Warburg A, Tesh RB, McMahon-Pratt D. Studies on the attachment of Leishmania flagella to sandfly midgut epithelium. J Protozool 1989; 36:613–617 [DOI] [PubMed] [Google Scholar]
- Yao C, Chen Y, Sudan B, Donelson JE, et al. Leishmania chagasi: Homogenous metacyclic promastigotes isolated by buoyant density are highly virulent in a mouse model. Exp Parasitol 2008; 118:129–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zauli RC, Yokoyama-Yasunaka JK, Miguel DC, Moura AS, et al. A dysflagellar mutant of Leishmania (Viannia) braziliensis isolated from a cutaneous leishmaniasis patient. Parasit Vectors 2012; 5:11. [DOI] [PMC free article] [PubMed] [Google Scholar]