Abstract
New antibacterial compounds, preferentially exploiting novel cellular targets, are urgently needed to fight the increasing resistance of pathogens against conventional antibiotics. Here we demonstrate that Carolacton, a myxobacterial secondary metabolite previously shown to damage Streptococcus mutans biofilms, inhibits planktonic growth of Streptococcus pneumoniae TIGR4 and multidrug-resistant clinical isolates of serotype 19A at nanomolar concentrations. A Carolacton diastereomer is inactive in both streptococci, indicating a highly specific interaction with a conserved cellular target. S. mutans requires the eukaryotic-like serine/threonine protein kinase PknB and the cysteine metabolism regulator CysR for susceptibility to Carolacton, whereas their homologues are not needed in S. pneumoniae, suggesting a specific function for S. mutans biofilms only. A bactericidal effect of Carolacton was observed for S. pneumoniae TIGR4, with a reduction of cell numbers by 3 log units. The clinical pneumonia isolate Sp49 showed immediate growth arrest and cell lysis, suggesting a bacteriolytic effect of Carolacton. Carolacton treatment caused a reduction in membrane potential, but not membrane integrity, and transcriptome analysis revealed compensatory reactions of the cell. Our data show that Carolacton might have potential for treating pneumococcal infections.
The resistance of pathogens against antibiotics is increasing worldwide1, while the number of new antibiotics in the clinical pipeline remains to be low2. As antibiotic treatment often fails against pathogens that have developed resistance, it disturbs the human microbiome at the same time3, which has become a major concern. New antibacterial compounds are urgently needed, preferably compounds that act on novel or underexploited cellular mechanisms and could be used as a starting point for rational drug design4.
Carolacton is a secondary metabolite produced by the myxobacterium Sorangium cellulosum. It is a macrolide ketocarbonic acid5 that can be synthesised de novo6,7. Carolacton was discovered to cause membrane damage of biofilms of the caries pathogen Streptococcus mutans at nanomolar concentrations and is non-toxic to eukaryotic cells8. Dental composite materials incorporating Carolacton maintain a strong in vitro activity against S. mutans biofilms while the mechanical properties of the material are not affected9, suggesting that such composites might be applicable to reduce secondary caries formation. Carolacton has only a weak effect on the growth of S. mutans10, but inhibits growth of Streptococcus oralis11 and also of Escherichia coli TolC with a minimal inhibitory concentration (MIC) as low as 0.06 μg/ml5. Carolacton-treated cultures show increased septum formation, elongated cells, and cell chains, suggesting defects in cell division or cell wall synthesis8,10,11,12. All chemical modifications of Carolacton tested until now rendered the molecule inactive11. Based on these studies, we hypothesised that the primary effect of Carolacton is a disturbance of cell division or cell wall synthesis through a stereospecific interaction with a conserved cellular target. Moreover, we hypothesised that the death of S. mutans biofilm cells, which occurs several hours after Carolacton treatment at low pH only10, is a secondary effect that might be unique to this species due to the strong acidification of mutans streptococci within mature biofilms13.
To investigate these hypotheses and to understand the mode of action of Carolacton in more detail, we studied the response of S. pneumoniae to Carolacton and compared it with that of S. mutans. S. pneumoniae is a ubiquitous colonizer of the nasopharynx14 whose carriers usually remain asymptomatic. Nevertheless, invasive and non-invasive pneumococcal infections account for almost half a million hospitalizations and approximately 22.000 deaths in the United States annually15. A continuous increase in multidrug resistance (MDR, resistant to ≥3 classes of antimicrobials) is being reported16. Particularly the prevalence of S. pneumoniae strains of serotype 19A has increased drastically from 5% in the mid-1990’s to up to 48% in 200817. Serotype 19A isolates have the highest proportion of MDR and extensively drug resistant strains (XDR, resistant to ≥5 classes of antimicrobials)18. Consequently, the search for novel antimicrobial compounds and novel cellular targets remains of crucial importance, especially with regard to pneumococci.
In this study, we focused on S. pneumoniae TIGR4 as an extensively characterized strain19. For the first time, we report complete growth inhibition of planktonically growing cells. We investigated growth inhibition by Carolacton of MDR and XDR clinical isolates of S. pneumoniae classified as serotype 19A. To clarify if S. pneumoniae shares the molecular target of Carolacton with S. mutans, we tested its sensitivity against 9-epi-Carolacton. This Carolacton derivative has the opposite absolute configuration at C-9, the area of macrolactone formation, and is inactive against S. mutans11. Cell death of S. mutans biofilm cells has been shown to be dependent on the eukaryotic-like serine/threonine protein kinase (eSTK) PknB10 and the regulator of cysteine metabolism CysR20. Therefore, their homologues in S. pneumoniae TIGR4 were deleted and their response to Carolacton determined. Finally we investigated the transcriptome, including small regulatory RNAs, of Carolacton-treated cultures of S. pneumoniae TIGR4, constructed knock-out mutants of strongly differentially expressed genes and tested their susceptibility. Our data demonstrate that Carolacton acts via the same molecular target in S. mutans and S. pneumoniae. Death of biofilm cells of S. mutans is a late secondary effect. The fast (within 5 min) and distinct transcriptional response to Carolacton treatment represents a compensatory mechanism, since deletion of those genes does not render the mutants insensitive to Carolacton. Finally, the inhibition of growth of S. pneumoniae by Carolacton shows the characteristics of bactericidal and in some cases even bacteriolytic antibiotics.
Results
Carolacton inhibits growth of S. pneumoniae TIGR4 in planktonic culture
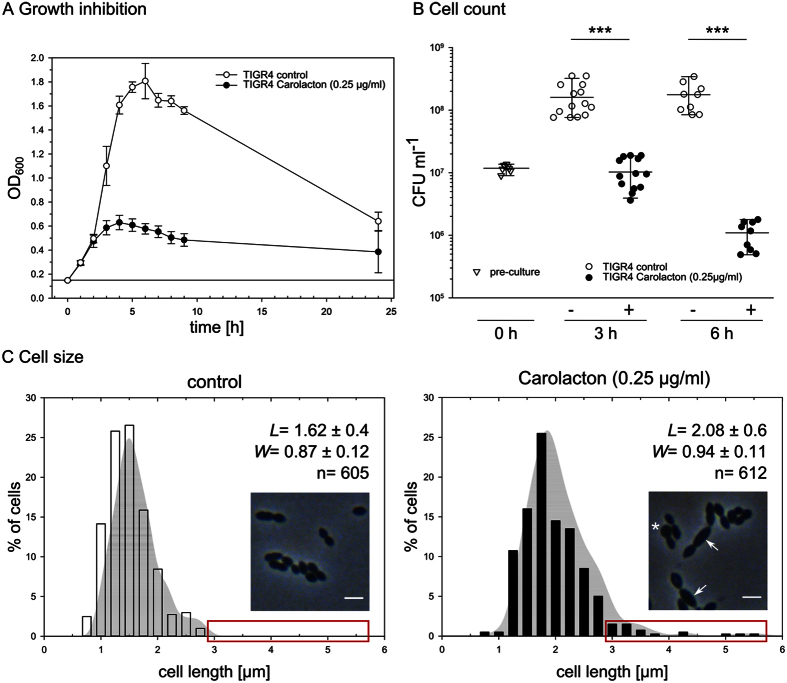
Carolacton (0.25 μg/ml final concentration) was added to cultures of S. pneumoniae TIGR4 prior to the onset of the exponential phase (OD600 = 0.15, termed “t0”). Carolacton-treated cultures grew at the same rate as untreated controls for about 2 h, after which the OD remained constant or decreased (Fig. 1A), suggesting a bactericidal activity21. The Carolacton-treated cultures typically reached a maximal OD600 of 0.55–0.60, while the controls grew up to an OD600 of 1.8. A maximal inhibition of ~67% could be observed after 9 h of growth. Since S. pneumoniae autolyzes after reaching the stationary phase22, final cell densities after 24 h were very low and almost the same in treated and untreated cultures. The number of viable cells was determined by counting colony forming units (CFUs). Figure 1B shows that after 3 h of Carolacton treatment, the number of CFUs was reduced by 1 log unit (92.4%) and decreased further by more than 2 log units (99.4%) after 6 h. Since this decrease in CFUs was not accompanied by a reduction in OD600, the data indicate reduced cell viability in the presence of Carolacton. For S. pneumoniae TIGR4, the minimal inhibitory concentration (MIC) of Carolacton was 0.06 μg/ml (Table 1). Cells grown with Carolacton showed a more voluminous phenotype and a fraction of the cells showed a moderate but significant increase in size (Fig. 1C). On average, cell length increased from 1.62 ± 0.4 μm to 2.08 ± 0.6 μm, while cell width increased from 0.87 ± 0.12 μm to 0.94 ± 0.11 μm. Even a 100-fold increase (0.025–2.5 μg/ml) of the Carolacton concentration did not increase the inhibition of growth considerably (Supplementary Fig. S1). This is in full accordance with Kunze et al. who reported a concentration-independent effect of Carolacton on death of S. mutans biofilms over the same concentration range8.
Figure 1. Treatment with Carolacton inhibits growth of S. pneumoniae TIGR4 and affects cell count and cell size.
(A) The growth curve indicates a growth inhibition of planktonically growing cells of S. pneumoniae TIGR4 by Carolacton at a final concentration of 0.25 μg/ml. The start OD600 is indicated by a solid horizontal line. Depicted is the average inhibition from three independent experiments (±standard deviation). (B) Determination of viable cell numbers by plating for colony counts. The cell number was determined after plating serial dilutions and counting the CFU. (+) 0.25 μg/ml Carolacton and (−) control. Data are representative for at least three biologically independent experiments (±s.d.). P value: *** = < 0.001; Mann-Whitney U. (C) Cell length distribution of cells grown with 0.25 μg/ml Carolacton for 3 h. A length of ≥ 3 μm was only observed for treated cells (red box). In the microscope image, elongated cells are indicated by arrows, a cell showing a bulky and voluminous morphology is marked by an asterisk (*). The average cell length (L) and width (W) with standard deviations are shown for n cells from three independent experiments. Scale bar: 2.5 μm. Changes in L and W were statistically significant for P < 0.05 as determined by non-parametric Mann-Whitney-U test; P(L) < 0.001, P(W) = 0.011.
Table 1. Minimal inhibitory concentration (MIC) of Carolacton against planktonically growing S. pneumoniae strains.
| Strain | Carolacton (μg/ml) |
|---|---|
| TIGR4 | 0.06 |
| Sp49 (NRZ:3198/36486) | ≤ 0.03 |
| Sp61 (NRZ:3364/39533) | 64 |
| Sp64 (NRZ:3066/35051) | 64 |
Carolacton inhibits planktonic growth of S. pneumoniae serotype 19A clinical isolates
To investigate the effectiveness of Carolacton treatment on S. pneumoniae strains found in pneumococcal disease, three different recent clinical isolates of S. pneumoniae, representing drug resistant serotype 19A strains (Table 2, Supplementary Table S1), were subjected to Carolacton susceptibility testing. Isolate Sp49 had been obtained from the blood of a septic pneumonia patient and was resistant against two classes of antibiotics. Isolate Sp61 and Sp64 were XDR (resistant to ≥5 classes of antibiotics); they were isolated from the thorax drainage of a pleuritis patient and by swab from an ear canal of a non-symptomatic patient, respectively. The growth of all three clinical isolates was inhibited by Carolacton, but to a widely varying extent (Fig. 2). The clinical pneumonia isolate (Sp49) was most sensitive and showed a different behaviour from TIGR4: it started to die immediately and after 2 h its OD was lower than at t0, suggesting a bacteriolytic effect of Carolacton (Fig. 2A). A maximum inhibition of 94.9% could be observed after approximately 6–7 h of growth, this was the same time point at which the control culture of Sp49 typically reached its peak (OD600 ~1.2). For the clinical pleuritis isolate (Sp61) and the non-invasive mid-ear serotype 19A isolate (Sp64), the period of uninhibited growth was longer than for the TIGR4 strain, and they reached higher final culture densities than TIGR4. The pleuritis isolate Sp61 showed a maximal inhibition of growth of about 45% after 6 h (Fig. 2B). For the non-invasive mid-ear isolate Sp64, the maximal inhibition was less than that detected for the pleuritic isolate, reaching a maximum inhibition of 23%. Moreover, the inhibition peak was retarded and observed after 8 h (Fig. 2C). Carolacton had a strong lytic effect on clinical isolate Sp49; the MIC value determined for this isolate was ≤0.03 μg/ml and thus lower than for TIGR4. Drug-resistant isolates Sp61 and Sp64, in contrast, were highly tolerant of Carolacton and were only inhibited at 64 μg/ml Carolacton (Table 1). These results indicate that Carolacton inhibits growth of clinical S. pneumoniae isolates with varying efficiency. The degree of inhibition was not affected by the change of serotype from TIGR4 (serotype 4) to the clinical isolates (serotype 19A). Interestingly, growth inhibition was weaker in the pleuritis isolate Sp61 and in the non-invasive isolate Sp64, which elicit multiple resistances to antibiotics compared to the pneumonia isolate.
Table 2. Bacterial strains, clinical isolates and plasmids used in this study.
| Strain/plasmid | Relevant genotype/serotype | Description | Reference |
|---|---|---|---|
| Streptococcus pneumoniae | |||
| TIGR4 (ATCC BAA-334) | encapsulated serotype 4 | wild type | 19 |
| JD01 | TIGR4 sp_1732::ermR | ΔstkP strain | This work |
| JD02 | TIGR4 sp_0927::ermR | ΔcysR strain | This work |
| JD03 | TIGR4 sp_0119::ermR | Δsp_0119 strain | This work |
| JD04 | TIGR4 sp_0120::ermR | ΔgidA strain | This work |
| JD05 | TIGR4 sp_0119-sp_0120::ermR | Δsp_0119/ΔgidA strain | This work |
| Clinical isolates | |||
| Sp49 | serotype 19A | pneumonia/sepsis isolate | M. van der Linden, NRZ Aachen |
| Sp61 | serotype 19A | pleuritis isolate | M. van der Linden, NRZ Aachen |
| Sp64 | serotype 19A | mid-ear isolate | M. van der Linden, NRZ Aachen |
| Plasmid | |||
| pALN122 | ermR | 58 | |
Figure 2. Activity of Carolacton against S. pneumoniae clinical isolates of serotype 19A.
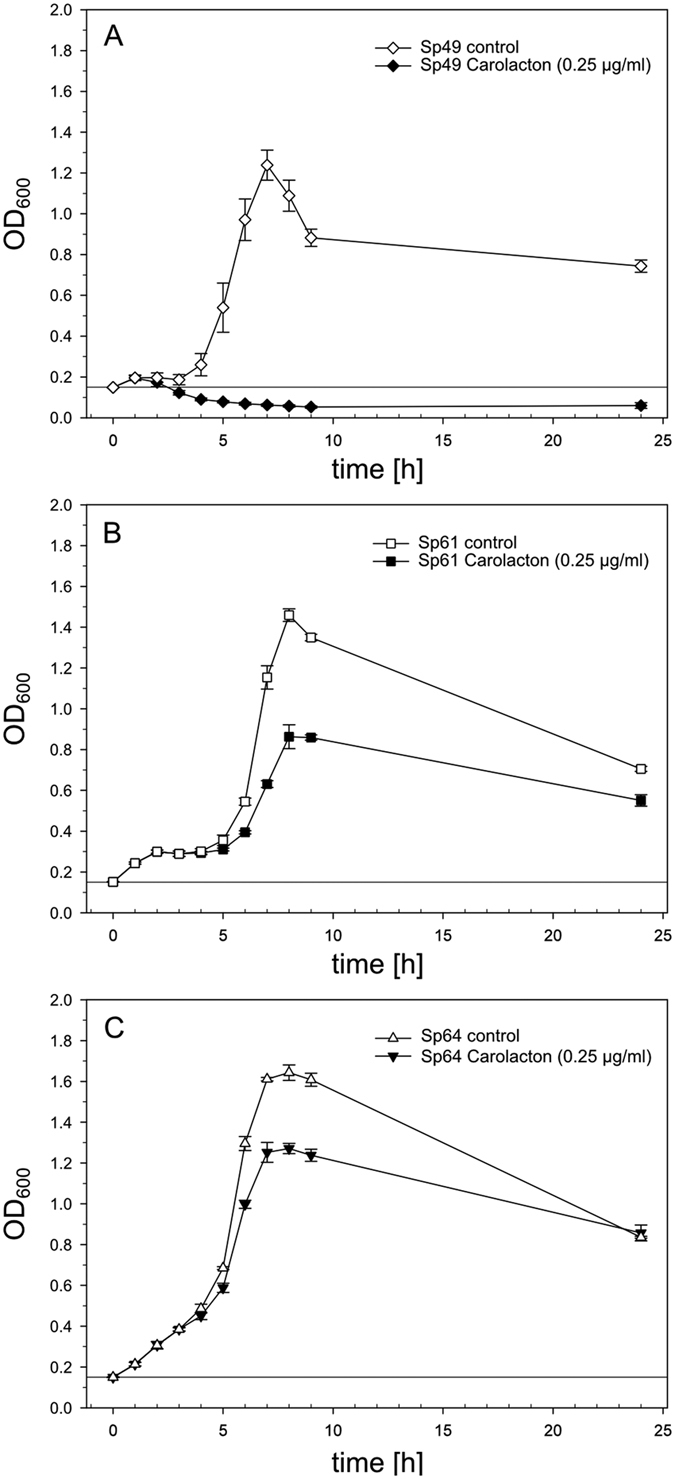
Growth inhibition of pneumococcal clinical isolates Sp49 (pneumonia/sepsis isolate, (A)), Sp61 (pleuritis isolate, (B)) and Sp64 (mid ear isolate, (C)) in THBY. Carolacton was added at a final concentration of 0.25 μg/ml prior to the onset of exponential growth at an OD600 of 0.15. The start OD600 is indicated by a solid horizontal line. The growth curves show the average inhibition calculated from three independent experiments (±s.d.).
The interaction of Carolacton with its cellular target is highly stereo-specific
The Carolacton derivative 9(R)-Carolacton (epi-Carolacton) is a diastereomer of Carolacton exhibiting an inversion at C–9 [(S)→(R)] (Fig. 3A). Although the structural changes are minute, epi-Carolacton is biologically inactive against S. mutans in biofilms11. Likewise, 0.25 μg/ml epi-Carolacton, a concentration which is generally applied in our growth experiments with streptococci, did not inhibit growth of S. pneumoniae TIGR4 (Fig. 3B). Thus the interaction between Carolacton and its molecular target must be highly specific with respect to the stereochemistry at position 9 of the molecule, heavily influencing the topology of the macrocycle. Due to the stereospecificity of this interaction we suggest that the cellular target of Carolacton is likely to be identical between S. mutans and S. pneumoniae, in spite of the differences of their physiological responses.
Figure 3. Effect of epi-Carolacton on growth of S. pneumoniae TIGR4.
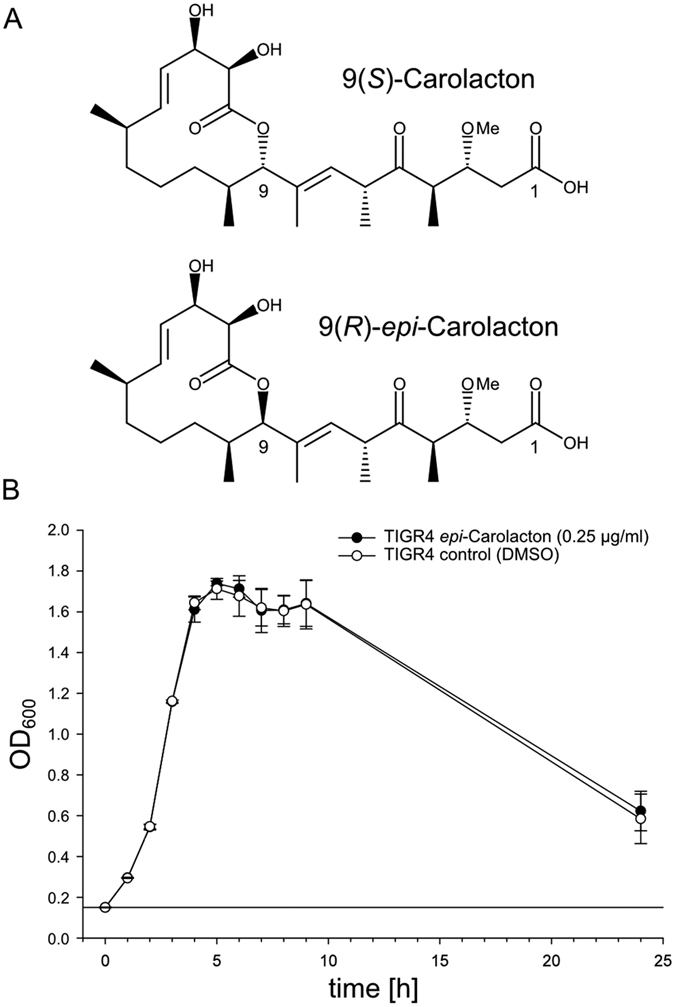
(A) Chemical structures of native 9(S)-Carolacton (top) and the 9(R)-epimer (epi-Carolacton, bottom) obtained by total synthesis. (B) Cells were treated with epi-Carolacton (black dots) at a final concentration of 0.25 μg/ml at an OD600 of 0.15. As control, pneumococci were cultivated with an equivalent volume of solvent (DMSO, white dots). The experiment was repeated independently at least three times.
The serine/threonine protein kinase StkP and the regulator CysR are not required for growth inhibition by Carolacton
Gene deletion of the signalling kinase PknB induces Carolacton resistance of S. mutans biofilms10. The S. pneumoniae genome encodes stkP (sp_1732) as corresponding homologue to S. mutans pknB. Therefore, the role of the homologue of PknB in S. pneumoniae, StkP, in Carolacton susceptibility of S. pneumoniae TIGR4 was tested. Since the molecular target of Carolacton appears to be highly conserved in those two organisms, this analysis will clarify whether a bacterial eukaryotic-like serine/threonine protein kinase (eSTK) is the direct molecular target of Carolacton. Due to the strong indications of eSTKs as the primary target of Carolacton10, in addition to PCR amplification and DNA-sequencing, the absence of the StkP protein in theΔstkP isogenic mutant was confirmed by immunoblotting using polyclonal anti-StkP antibodies23 (Fig. 4A). The StkP deletion strain showed formation of elongated cells and multiple unconstricted septa, as reported previously23. Its growth was highly impaired; nevertheless, Carolacton treatment led to growth inhibition (Fig. 4B). The data show that the ΔstkP TIGR4 strain is susceptible to Carolacton, which is in sharp contrast to the findings described for S. mutans10.
Figure 4. Growth inhibition of S. pneumoniae TIGR4ΔstkP and TIGR4ΔcysR by treatment with Carolacton.
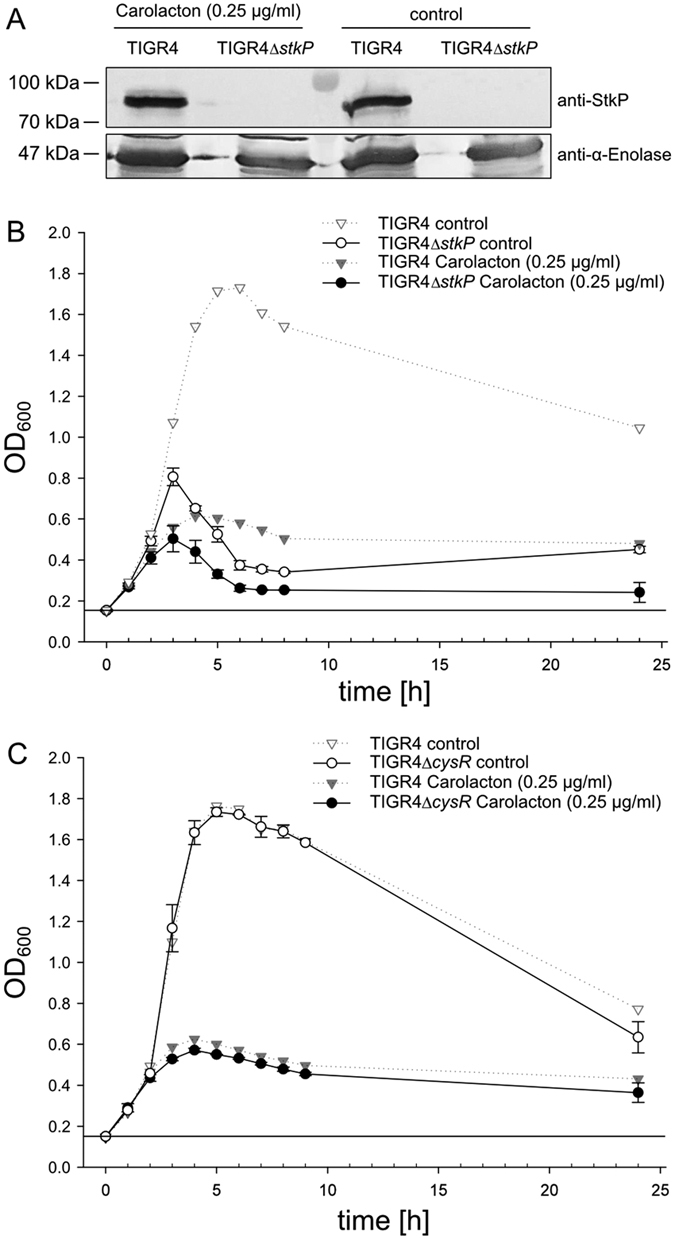
(A) Immunoblot analysis with StkP-specific antibodies of S. pneumoniae TIGR4 and TIGR4ΔstkP confirms absence of StkP-expression in the TIGR4ΔstkP deletion mutant. Pneumococcal α-enolase was detected with specific antibodies as loading control. Presented are two parts of the same blot treated with different primary antibodies. (B) Growth inhibition of a TIGR4ΔstkP mutant (solid lines) is plotted after addition of 0.25 μg/ml Carolacton (black dots) and without Carolacton as control (white dots). (C) Inhibition of a planktonically growing TIGR4ΔcysR mutant with (black dots) and without (white dots) Carolacton. Inhibition of the TIGR4 wild type strain (dotted lines), used as controls during both assays (Carolacton-treated = black triangles, control = white triangles), is displayed as a semi-transparent plot in (A) and (B). All growth curves present the average of three independent biological replicates.
Deletion of the cysteine metabolism regulator CysR also resulted in a Carolacton-insensitive mutant in S. mutans20. The CysR homologue (SP_0927) was deleted in S. pneumoniae and tested for susceptibility to Carolacton. Similar to the ΔstkP strain, growth of the ΔcysR mutant was strongly inhibited by Carolacton (Fig. 4C). Thus, the pneumococcal homologues to both proteins mediating the susceptibility to Carolacton in S. mutans, StkP and CysR, are most likely not directly involved in the Carolacton response in S. pneumoniae, in spite of the presumably conserved molecular target of Carolacton in those two species. We conclude that PknB and CysR are involved in the mechanisms of membrane damage at low pH in S. mutans, which is a late secondary response to Carolacton treatment. In order to confirm this hypothesis, the effect of Carolacton on membrane integrity and membrane potential of S. pneumoniae TIGR4 was analysed.
Effect of Carolacton on cytoplasmic membrane integrity and membrane potential in S. pneumoniae
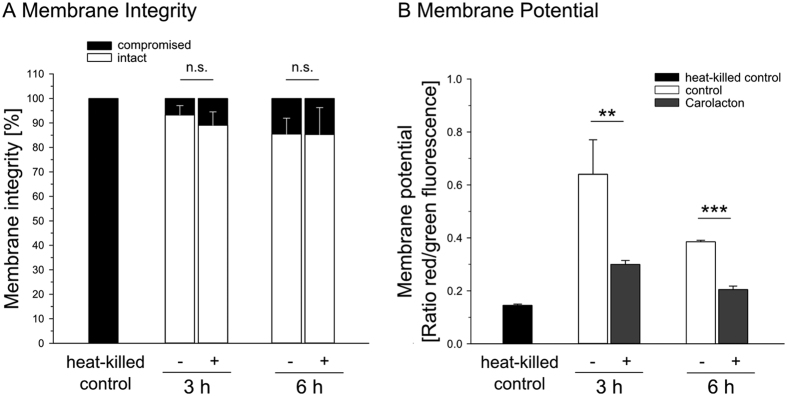
For S. mutans it has been reported that Carolacton leads to death of biofilm cells at low pH by disturbing the integrity of the plasma membrane and thereby causing leakage of cytoplasmic proteins and DNA10. Membrane damage of S. pneumoniae cells was similarly determined by LIVE/DEAD staining during exponential and stationary phase, 3 h and 6 h after addition of Carolacton, respectively. Heat-killed S. pneumoniae cells were included as a control and a calibration curve was recorded using flow cytometry (Supplementary Fig. S2). No indication of S. pneumoniae cell death by Carolacton treatment was detected (Supplementary Fig. S3). Controls and Carolacton-treated cultures both contained a maximum of 14% cells with defective plasma membranes (Fig. 5A).
Figure 5. Effect of Carolacton-treatment on the membrane integrity and membrane potential of S. pneumoniae TIGR4.
(A) Cells grown with 0.25 μg/ml Carolacton (+) and controls (−) were stained with Syto 9/PI and analysed using flow cytometry as exemplified in Fig. S8. Error bars of stained heat-killed controls (red bars) are not visible, as continuously more than 99.9% of all heat-treated cells were killed and emitted red fluorescence. (B) Changes in membrane potential were quantified by flow cytometric analysis of DiOC2(3)-stained planktonic cells. The relative membrane potential was calculated ratiometrically. Analyses were carried out with 30 μM DiOC2(3). Heat-killed cells served as a control for fully depolarized cells. The bar charts present the average of three biological replicates (±s.d.). P value: n.s. = > 0.05; ** = < 0.01; *** = < 0.001; two-tailed Student’s t-test.
We then used staining with the fluorescent dye 3,3-Diethyloxacarbocyanine iodide (DiOC2(3)) to determine changes in the MP upon Carolacton treatment. Since the measured fluorescence intensities are directly influenced by the size of the stained particles, and Carolacton treatment affects the cell size, a size-independent ratiometric technique was applied24. Carolacton treatment caused a time-dependent depolarization of the membrane potential (Fig. 5B, Supplementary Fig. S4). Approximately 48% of the cells showed a membrane depolarization, indicated by a reduction of the red/green fluorescent ratio, after 3 h. Membrane depolarization continued during the next 3 h, though not as fast as during the first time interval. After 6 h, the red/green fluorescent ratio dropped to ~34% of that measured for an exponentially growing control after 3 h, and was reduced to ~56% when compared to the control at the same time point. These results indicate a Carolacton-dependent disturbance of the membrane potential during growth that is not the result of a porous plasma membrane.
Transcriptional response of S. pneumoniae TIGR4 to Carolacton
We analysed the transcriptome of S. pneumoniae TIGR4 cells growing with Carolacton starting 5 min after addition of the drug, until 3 h after Carolacton addition. Total RNA was fractionated into two size classes: >200 nt and ≤200 nt, and differential regulation of mRNAs (30–4776 nt), small RNAs (37–486 nt) and tRNAs (71–90 nt) was assessed individually. We noted a high number of reads for the previously identified housekeeping sRNAs 6S RNA (194 nt)25 and RNase P (391 nt)/tmRNA (348 nt)26,27 in the samples containing small transcripts (≤200 nt) or in the samples containing large transcripts (>200 nt), respectively, confirming the applicability of our sequencing approach for reliable separation of transcripts according to length (Supplementary Dataset 1). Since differential transcription after Carolacton treatment was exclusively limited to the later time points (t120/t180), we chose to only sequence three time points (t0, t5 and t180) for analysis of short small RNAs/tRNAs. The RNA-seq data of biological replicates showed a very high Pearson correlation between 0.93 for small regulatory RNAs/tRNAs and 0.99 for large regulatory RNAs/mRNAs.
None of the 58 tRNAs of S. pneumoniae TIGR4 were significantly differentially expressed after Carolacton treatment (Supplementary Fig. S5, Supplementary Dataset 1).
Differential transcription of small regulatory RNAs
A total number of 181 known small sRNAs (≤200 nt) and 21 known long sRNAs (>200 nt) were analysed. Transcription of the vast majority of sRNAs was confirmed by obtaining high read counts after mapping (Supplementary Dataset 1). Seven small sRNAs were differentially transcribed at t180 only (Supplementary Fig. S5). The highest upregulated small sRNA trn0057 (log2FC~3.9) is located downstream of the sp_0119-sp_0120 operon and its function is unknown. Ten long sRNAs were differentially regulated at t120 and t180 (Supplementary Fig. S5) of which only two (R12 and F20) have been investigated and were shown to be important for virulence in mice28.
Differential transcription of mRNAs
RNA-seq data was analysed for differential expression of 1985 large (>200 nt) and 120 small (≤200 nt) transcripts of S. pneumoniae TIGR4, accounting for 2105 protein-coding genes (Supplementary Dataset 2). Carolacton caused a maximum of differential gene expression after 120 and 180 min. Overall, only 1.9% of the 2105 protein encoding genes were differentially transcribed (log2FC ≥ ± 2, FDR < 0.01) (Supplementary Fig. S6).
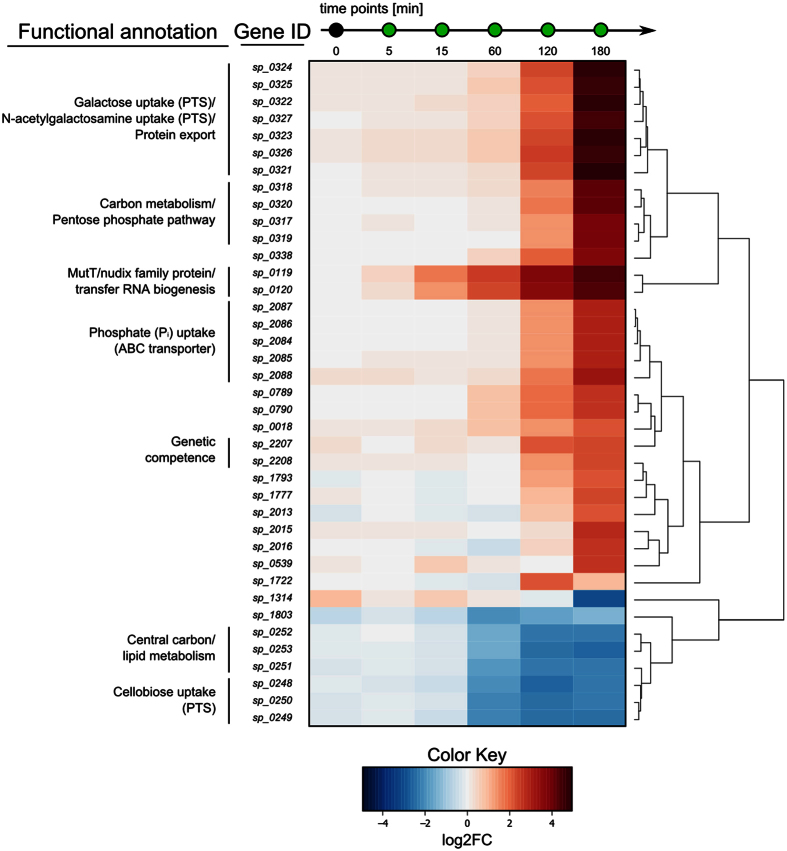
Two transcripts smaller than 200 nt were differentially regulated, a type II DNA modification methyltransferase (sp_0569) and the 50S ribosomal protein L28 (sp_0441) (Supplementary Fig. S5). 39 transcripts larger than 200 nt were strongly differentially expressed (log2FC ≥ ± 2 and FDR < 0.01 for at least one time point) (Fig. 6), 26 of which are encoded within 7 operons on the TIGR4 chromosome (Table 3). Differential expression of sp_0119 and sp_0120 was detected already 5 min after addition of Carolacton and continued to increase during the experiment to a final log2FC of 4.4 and 4.3 after 180 min, respectively. Sp_0119 codes for a MutT/Nudix family protein and sp_0120 encodes the glucose-inhibited division protein A (GidA).
Figure 6. Differentially regulated mRNAs in S. pneumoniae TIGR4 upon treatment with Carolacton.
The heat map illustrates the 39 most differentially expressed genes (mRNA > 200 nt) after addition of Carolacton (0.25 μg/ml). The cut-off for differentially regulated genes was set at log2FC ≥ ± 2 and FDR < 0.01 for at least one sample during the time course. RNA-seq data was analysed using Rockhopper and the edgeR package for R. The heat map was constructed using the R package gplots. Main operon structures are indicated by vertical lines on the left. Information on encoded proteins and their involvement in biological pathways were obtained from KEGG ORTHOLOGY or KEGG BRITE databases.
Table 3. Summary of the most differentially expressed genes upon growth of S. pneumoniae TIGR4 with Carolacton for 180 min.
| Locus tag | Gene product | Gene symbol | log2FCa | FDR |
|---|---|---|---|---|
| Galactose uptake (PTS)/N-acetylgalactosamine uptake (PTS)/Protein export | ||||
| sp_0324 | PTS system transporter subunit IIC | 4.68 | 7.52E-91 | |
| sp_0325 | PTS system transporter subunit IID | 4.56 | 5.10E-123 | |
| sp_0322 | glucuronyl hydrolase | 4.73 | 2.58E-100 | |
| sp_0327 | hypothetical protein | 4.41 | 5.05E-74 | |
| sp_0323 | PTS system transporter subunit IIB | 4.80 | 2.75E-95 | |
| sp_0326 | preprotein translocase subunit | yajC | 4.55 | 5.11E-74 |
| sp_0321 | PTS system transporter subunit IIA | 4.96 | 6.23E-89 | |
| Carbon metabolism/Pentose phosphate pathway | ||||
| sp_0318 | carbohydrate kinase | 4.13 | 1.30E-67 | |
| sp_0320 | gluconate 5-dehydrogenase | 4.08 | 1.82E-78 | |
| sp_0317 | 4-hydroxy-2-oxoglutarate aldolase/2-dehydro-3-deoxyphosphogluconate aldolase | 3.82 | 6.60E-64 | |
| sp_0319 | hypothetical protein | 3.88 | 4.69E-72 | |
| Protein degradation | ||||
| sp_0338 | ATP-dependent Clp protease, ATP-binding subunit | 3.73 | 4.09E-67 | |
| MutT/nudix family protein/transfer RNA biogenesis | ||||
| sp_0119 | MutT/nudix family protein | 4.44 | 2.57E-67 | |
| sp_0120 | tRNA uridine 5-carboxymethylaminomethyl modification protein | gidA | 4.33 | 1.35E-50 |
| Phosphate (Pi) uptake (ABC transporter) | ||||
| sp_2087 | phosphate transporter ATP-binding protein | pstB | 2.96 | 1.16E-45 |
| sp_2086 | phosphate ABC transporter permease | pstA | 2.97 | 1.34E-42 |
| sp_2084 | phosphate ABC transporter permease | pstS | 2.89 | 2.78E-38 |
| sp_2085 | phosphate ABC transporter substrate-binding protein | pstC | 2.96 | 2.78E-33 |
| sp_2088 | phosphate transport system regulatory protein | phoU | 3.19 | 5.26E-42 |
| sp_0789 | conserved hypothetical protein | 2.64 | 2.13E-37 | |
| sp_0790 | conserved hypothetical protein | 2.58 | 2.77E-44 | |
| sp_0018 | hypothetical protein | 2.10 | 1.13E-24 | |
| Genetic competence | ||||
| sp_2207 | competence protein | comFC | 2.21 | 2.18E-05 |
| sp_2208 | helicase | comFA | 2.22 | 3.04E-09 |
| sp_1793 | hypothetical protein | 2.17 | 9.19E-32 | |
| sp_1777 | hypothetical protein | 2.22 | 2.46E-25 | |
| sp_2013 | hypothetical protein | 2.13 | 4.64E-31 | |
| Nicotinate and nicotinamide metabolism | ||||
| sp_2015 | IS630-Spn1, transposase Orf1 | 2.74 | 2.48E-40 | |
| sp_2016 | nicotinate-nucleotide pyrophosphorylase | 2.60 | 1.07E-46 | |
| sp_0539 | bacteriocin | blpM | 2.59 | 1.38E-04 |
| Starch and sucrose metabolism | ||||
| sp_1722 | PTS system IIABC components | 0.87 | 6.04E-04 | |
| sp_1314 | IS66 family element, Orf1 | −3.11 | 7.89E-03 | |
| sp_1803 | hypothetical protein | −1.35 | 3.02E-09 | |
| Central carbon/lipid metabolism | ||||
| sp_0252 | fructose-6-phosphate aldolase | −2.34 | 4.59E-06 | |
| sp_0253 | glycerol dehydrogenase | −2.57 | 2.21E-07 | |
| sp_0251 | formate acetyltransferase | −2.29 | 8.60E-06 | |
| Cellobiose uptake (PTS) | ||||
| sp_0248 | PTS system transporter subunit IIA | −2.30 | 8.11E-05 | |
| sp_0250 | PTS system transporter subunit IIC | −2.28 | 1.85E-05 | |
| sp_0249 | PTS system transporter subunit IIB | −2.42 | 4.79E-05 | |
Horizontal lines indicate separation of transcriptional units.
acut-off: log2FC≥2 or ≤−2 at one point during the time course.
Influence of sp_0119 and sp_0120 on susceptibility of S. pneumoniae TIGR4 to Carolacton
Since the gene products of both sp_0119 and sp_0120 are involved in central cellular processes we wondered whether an uncontrolled transcriptional upregulation of this operon could cause the growth inhibitory effect of Carolacton. Therefore, single deletion mutants of sp_0119 and sp_0120 and a Δsp_0119-sp_0120 double mutant were constructed in S. pneumoniae TIGR4. Growth of all three mutant strains was still significantly inhibited by Carolacton treatment (Supplementary Fig. S7).
Comparison of the transcriptome of S. mutans UA159 and S. pneumoniae TIGR4 in response to Carolacton
Until now, the only other organism for which the influence of Carolacton has been studied in more detail is the human oral pathogen S. mutans. Therefore, the RNA-seq data obtained in this study were compared to microarray data previously obtained for Carolacton-treated S. mutans biofilm cells10. 22.8% of all genes were differentially transcribed at 180 min in S. pneumoniae at a log2FC of ≥ ± 0.8 (Supplementary Fig. S6), which had previously been used for S. mutans10. At this cut-off, the effect of Carolacton was highly pleiotropic. Supplementary Fig. S8 shows a direct comparison between log2FC of the most strongly differentially expressed genes in S. mutans compared to their homologues in S. pneumoniae. Overall, the patterns of transcriptional changes were similar for some, but not all genes, and the timing and extent of the responses shows differences (Supplementary Fig. S8). For example, upregulation of the ComDE two-component system (TCS) for genetic competence is observed in both species, but stronger and earlier in S. pneumoniae, and might represent a general stress response29. The most conspicuous transcriptional response in S. mutans was the instantaneous strong upregulation of genes for pyrimidine biosynthesis. In S. pneumoniae, transcription of these genes showed a different pattern, indicating that their regulation in S. mutans might be related to the membrane damage occurring at later stages of biofilm growth. In S. mutans, downregulation of sortase A was observed. In S. pneumoniae TIGR4, neither sortase A (srtA, sp_1218) nor its three sortase paralogs (srtBCD, sp_0466-sp_0468)30 were differentially transcribed. Also in contrast to S. mutans, the VicKRX TCS was not differentially expressed upon Carolacton treatment in S. pneumoniae. Consequently, the VicKRX regulon also showed a different pattern: only transcripts of the cell shape proteins MreC and MreD were upregulated, although earlier and stronger than in S. mutans. In S. mutans, cell wall metabolism-related genes were upregulated, while the respective homologues in S. pneumoniae TIGR4 were not significantly differentially expressed.
Interestingly, the gidA homologue of S. mutans (smu_2141) was among the most strongly upregulated transcripts like in S. pneumoniae, although its upregulation was weaker and occurred later than in the pneumococcus (Supplementary Fig. S8).
Taken together, the transcriptional response of S. mutans and S. pneumoniae to Carolacton appears to be a pleiotropic stress response, different between species, and not a specific and focused transcriptional response as assumed after studies with S. mutans before10.
Discussion
For the first time we report on the physiological effect of Carolacton on the clinically relevant human pathogen S. pneumoniae. Growth of planktonic cultures of S. pneumoniae is already inhibited by Carolacton at a concentration as low as 0.025 μg/ml (Supplementary Fig. S1). Growth inhibition starts about 2 h after application of the drug; at later time points, the optical density remains constant, but the number of viable cells continues to decrease. Such behaviour is typically observed when treating pneumococci with bactericidal antibiotics21. Growth inhibition by Carolacton is concentration-independent, which could indicate that the primary cellular target of Carolacton is easily saturated and thus of low abundance in cells.
Here we show that growth inhibition by Carolacton occurs for clinical isolates as well as for the virulent laboratory strain TIGR4 and thus appears to be independent of the serotype. Interestingly, the susceptibility to Carolacton was related to the drug resistance of the pathogen. The sepsis isolate Sp49 (which is resistant to two classes of antibiotics only) was instantly killed by Carolacton, while the multidrug-resistant isolates Sp61 and Sp64 were inhibited to a lower degree than the TIGR4 strain. Accordingly, also the MIC values of Carolacton differed notably between S. pneumoniae TIGR4 and among the clinical isolates. Carolacton has a potent inhibitory effect on S. pneumoniae TIGR4 and Sp49 with MIC values in the range of MICs of classical antibiotics used for the therapy of pneumococcal disease, such as e.g. ampicillin, clarithromycin or rifampicin31. The exact MIC of Sp49 remains to be determined and could be even lower than 0.03 μg/ml. Multidrug-resistant isolates Sp61 and Sp64 can tolerate and grow in the presence of high concentrations of Carolacton. Multidrug resistance is often mediated by increased expression or activity of multidrug efflux pumps. Sp61 and Sp64 are resistant to macrolides (clarithromycin, telithromycin)32 and fluoroquinolones (levofloxacin)33, which is mediated by efflux in the pneumococcus. Carolacton is structurally similar to the macrolide class of natural compounds and might be exported in a similar way. Unspecific efflux of Carolacton over time, as a by-product of resistance to macrolides and fluoroquinolones, could explain why the MICs of Sp61 and Sp64 are much higher than that of S. pneumoniae TIGR4. This is in accordance with the observation that E. coli is insensitive to Carolacton, but the growth of E. coli TolC, which lacks a multidrug efflux pump, is completely inhibited5. These findings are important for future development of the compound and potential clinical applications. For example, Carolacton could be used synergistically with structurally unrelated non-macrolide antibiotics.
A Carolacton derivative with an inverted stereocentre at C-9 (epi-Carolacton) was completely inactive against both S. mutans and S. pneumoniae, as well as against several oral streptococci11. All other chemical modifications of the side-chain or the lactone ring tested so far were also inactive11. This indicates a highly stereospecific interaction of Carolacton with its molecular target and led us to hypothesise that the target could be highly conserved and shared between S. pneumoniae and S. mutans. It appears unlikely that a switch of the stereochemistry at C-9 could completely abolish interaction with multiple less conserved targets in both species. If Carolacton would interact with several weakly conserved targets, we would expect a remaining activity of epi-Carolacton in one of the tested species, which is absent as shown here and previously11. A pattern similar to the one described for epi-Carolacton here was found for the antibiotic fosfomycin, which targets the conserved bacterial cell wall hydrolase MurA34, but its stereoisomer lacks antimicrobial activity35. Consequently we investigated the roles of PknB and CysR, which both had been shown to be essential for the damage of biofilm cells of S. mutans at low pH10,20, for Carolacton susceptibility in S. pneumoniae. Our data indicate that neither StkP nor CysR are required for susceptibility of S. pneumoniae to Carolacton and they thus cannot be its molecular target if this is conserved in both streptococci. Another indication that Carolacton acts independently of StkP is the potent activity of Carolacton against E. coli TolC, as E. coli does not possess StkP homologues36. StkP and CysR are apparently exclusively important for S. mutans at low pH in mature biofilms, which is a unique environment fundamentally different from planktonically growing S. pneumoniae cultures.
As a possible mode of action of Carolacton, Reck et al. proposed a disturbance of cell wall regulatory processes, resulting in plasma membrane damage and leakage of cytoplasmic content at a low pH10. Plasma membrane damage is usually paralleled by a loss of membrane potential (MP)37. Treatment with antimicrobial agents such as streptomycin, reutericyclin38 or ampicillin37 can decrease the MP and thereby interfere with the localization of cell division proteins39. By fluorescent staining of Carolacton-treated cells with DiOC2(3), we detected a continuous decrease in MP over time. When assessing the integrity of the plasma membrane, as a possible cause for MP reduction, no damage could be detected. This is in agreement with Kunze et al. who reported no changes in LIVE/DEAD-stained S. mutans cells during planktonic growth with Carolacton8. Thus, Carolacton treatment leaves the plasma membrane intact in both S. mutans and S. pneumoniae during planktonic growth.
To identify components of a possible conserved response in S. mutans and S. pneumoniae, RNA-sequencing was carried out. Among differentially regulated sRNAs, sspn1731.1 and sspn1732.1 are strongly downregulated at late time points. Both contain a T-box motif that is known to stimulate anti-termination of the tyrS gene in Bacillus subtilis by interaction with the acceptor end of tRNA(Tyr)40. T-box leader sequences of non-coding RNAs are part of a negative feedback loop controlling expression of aminoacyl-tRNA synthetase genes41 and components of the amino acid biosynthesis machinery42. Downregulation of sspn1731.1 and sspn1732.1 may present another compensatory mechanism to Carolacton by increasing expression, for example of aminoacyl-tRNA-synthetases. Among differentially regulated mRNAs, an immediate strong upregulation of the sp_0119-sp_0120 operon was identified. Both encoded proteins have a global regulatory role in S. pneumoniae. The sp_0119 gene encodes a MutT/Nudix family-like protein. A homologous protein in E. coli (MutT) functions as an anti-mutator protein, by maintaining replication fidelity through hydrolysis of mutagenic 8-oxo-dGTP43. Other MutT/Nudix family-like proteins in E. coli similarly protect the cell by degradation of toxic nucleotide derivatives44 and have been described as “housecleaning” enzymes45. Thus, it can be assumed that SP_0119 likewise plays a role in protecting the pneumococcus by detoxification of harmful metabolites. GidA (SP_0120) is a highly conserved protein; homologues are present in Proteobacteria and Firmicutes and influence protein biosynthesis by altering mRNA codon recognition through modification of tRNAs at the wobble position46. GidA modulates the virulence of a large number of pathogens47. Consequently, a Salmonella gidA mutant strain is currently being investigated for use in a live-attenuated vaccine48. As an important virulence factor, also in streptococci49, it is conceivable that by altering the transcription of gidA, Carolacton may also interfere with virulence of S. pneumoniae. Expression of virulence factors is tightly controlled in group A streptococci50 and upregulated transcription of virulence factors can have a negative effect on pathogenicity, as shown for Listeria monocytogenes51. The investigation of compounds that are aiming at virulence factors in order to render pathogens harmless, instead of targeting essential cellular components, hold enormous possibilities52. The use of anti-virulence drugs could even counteract resistance development in bacteria altogether53. If S. pneumoniae cells show an altered virulence after treatment with Carolacton needs to be explored further.
Growth of single and double deletion mutants of sp_0119 and sp_0120 was still impaired by Carolacton, thus neither SP_0119 nor SP_0120 qualify as its molecular target. We hypothesise that these proteins mediate a compensatory response to Carolacton treatment. This is further confirmed by comparison of the transcriptional response of S. mutans and S. pneumoniae, which suggests that these responses likely show species-specific stress reactions or compensatory mechanisms and indicate that the effect of Carolacton cannot be compensated by differential regulation of a single specific gene set. For example, competence development, e.g. by upregulating transcription of comDE, is a direct reaction to stress in bacteria, such as streptococci, that lack a classical SOS response54. An upregulation of the VicKRX operon and VicKRX-regulated genes, as seen in S. mutans, possibly presents a direct response to disturbance of cell wall homeostasis55. The proteins MreC and MreD are located at the equators and septa of dividing S. pneumoniae cells and direct peripheral peptidoglycan synthesis56. Their increased transcription after Carolacton treatment could be interpreted as an attempt to compensate defects in peptidoglycan synthesis and cell division.
The data show that Carolacton holds promise as a bactericidal antibiotic. The extent of growth inhibition observed after Carolacton treatment in S. pneumoniae is variable, does not depend on the serotype and can even be bacteriolytic. This calls for further investigation of the primary mode of action of Carolacton and demonstrates the importance to discover the direct molecular target as a pre-requisite for rational drug design.
Methods
Additional methods can be found in the Supplementary Information.
Bacterial strains and growth conditions
Bacterial strains used in this study are listed in Table 2. S. pneumoniae TIGR4 serotype 4 (ATCC BAA-334), used for growth experiments in liquid medium, was routinely grown on Columbia blood agar plates containing 5% sheep blood (BD Biosciences) overnight (o/n) at 37 °C and 5% CO2. Cells were then scraped from plate and used to inoculate Todd-Hewitt-Broth + 1% (w/v) yeast extract (THBY, BD Biosciences) liquid medium, starting from an optical density at 600 nm (OD600) of 0.1. After the initial culture had reached an OD600 of 0.15, it was split into equal volumes and Carolacton or 9(R) epi-Carolacton11 were added to a final concentration of 0.25 μg/ml, if not stated otherwise. Carolacton was solved in methanol and stored 1000-fold concentrated (5.3 mM) in small aliquots in glass vials at −20 °C in the dark. Epi-Carolacton was solved in dimethyl sulfoxide (DMSO) and also stored in concentrated small aliquots (2 mM) at −20 °C in the dark.
Clinical S. pneumoniae isolates and information about clinical background data, serotype and antimicrobial susceptibility were obtained from Dr. Mark van der Linden from the National Reference Laboratory (NRZ) on Streptococcal Disease in Aachen (Germany). All clinical strains were isolated between 5/2008 and 4/2009. For cryo-conservation, S. pneumoniae strains were grown in THBY to early exponential phase (OD600~0.25) and immediately frozen at −80 °C in 10% (v/v) glycerol.
Growth inhibition was calculated as follows:
 |
Determination of MIC values
S. pneumoniae was grown on Columbia blood agar plates o/n as described before. Minimum inhibitory concentration (MIC) values of Carolacton were determined by two-fold serial microdilution and incubation at 37 °C and 5% CO2 for 18–20 h, as described for the pneumococcus previously31. The dilution range of Carolacton tested was 0.03–128 μg/ml. The MIC was determined as the lowest concentration of Carolacton that did not yield visible bacterial growth. The cell number of the initial inoculum was 5 × 105 CFU/ml as determined by serial dilution and plating. Ampicillin, erythromycin, tetracycline and rifampicin were included as controls during MIC determination for S. pneumoniae TIGR4, confirming target MIC values commonly reported for susceptible pneumococci31. MICs were confirmed by at least two independent measurements (with two replicates).
Construction of gene deletion mutants of S. pneumoniae TIGR4
Construction of gene deletion strains in the TIGR4 background was carried out by the Duplicate, cf. end of sentence. PCR ligation mutagenesis method, replacing the gene of interest (GOI) with an erythromycin (ermR) resistance cassette, via double homologous recombination as described previously57. All used enzymes were purchased from New England Biolabs (NEB). First, the up- and downstream regions flanking the GOI were amplified using the primer pairs listed in Supplementary Table S2, introducing an FseI and an AscI restriction site into the PCR products. The ermR cassette was amplified from pALN12258. PCR fragments were digested with the appropriate restriction enzymes and purified using the QIAquick PCR Purification Kit (Qiagen). 100 ng of all DNA fragments was ligated with T4 DNA ligase at 16 °C o/n.
Transformation of S. pneumoniae TIGR4
Transformation of S. pneumoniae was carried out by induction of competence by addition of competence stimulating peptide 2 (CSP-2) as described previously57. Briefly, THBY (pH 6.8) supplemented with 1 mM CaCl2 and 0.2% (w/v) bovine serum albumin (BSA) was inoculated with exponentially growing cells from liquid culture in THBY (OD600~0.5) to an OD600 of 0.005. After reaching the early log phase (OD600~0.15), 1 ml culture was harvested by centrifugation and the cells resuspended in 1 ml THBY (pH 8.0) supplemented with 1 mM CaCl2, 0.2% (w/v) BSA and 400 ng/ml CSP-2. Transformation was achieved by addition of 10 μl of a ligation mix and static incubation at 37 °C for 2 h. Transformants were selected on THBY agar plates containing 0.5 μg/ml erythromycin. Correct integration was checked by PCR and Sanger DNA sequencing.
Verification of the absence of StkP in the TIGR4ΔstkP deletion mutant by immunoblotting
In addition to DNA sequencing, the absence of StkP of JD01 was confirmed by immunoblotting. First, a defined volume (V[μl] = 300/OD600) of S. pneumoniae TIGR4 wild type and TIGR4ΔstkP cells in THBY was boiled in SDS loading dye (95 °C, 15 min) and separated via 12% (v/v) SDS PAGE according to59. Proteins were blotted to a PVDF membrane (Immobilion-P, Milipore) using a semi-dry blotting system (Sigma-Aldrich) and blocked with 5% (w/v) BSA in TBS-T at room temperature (RT) for 1 h. Incubation with the primary antibody, rabbit anti-StkP, was carried out by using a 1:50.000 dilution in 1% (w/v) BSA/TBS-T o/n. Pneumococcal α-enolase (α-eno) served as a loading control60. The α–eno antibody was used in a 1:500 dilution in 1% (w/v) BSA/TBS-T at RT o/n. Alkaline phosphatase-coupled goat anti-rabbit IgG (Sigma-Aldrich) diluted 1:10 000 in TBS-T was applied for 1 h at RT for detection of the primary antibody. Alkaline phosphatase activity was visualized colorimetrically using SIGMA FAST BCIP/NBT tablets according to the manufacturer’s instructions (Sigma-Aldrich).
Isolation and enrichment of mRNA
Processing of RNA-seq data is presented in detail in the Supplementary Information. Total RNA from was isolated using the miRNeasy mini kit (Qiagen) according to the manufacturer’s instructions for separation of small RNA-enriched fractions (≤200 nt) and larger transcripts (>200 nt). DNA library generation was carried out using the ScriptSeq™ v2 RNA-Seq Library Preparation Kit (epicentre) for long transcripts (>200 nt), and the TruSeq Small RNA Library Prep Kit (Illumina) for transcripts ≤200 nt. Libraries were sequenced on the Illumina HiSeq 2500 (Illumina) using 50-bp single-end sequencing.
RNA-seq data analysis
Processing of RNA-seq data is described in Supplementary Information. All analysed small RNAs are documented in Supplementary Dataset S3. False discovery rate (FDR)-adjusted P values were calculated according to61. FDR values of <0.01 were considered significant. For visualization of these results heat maps were generated, taking into account only genes that, in addition to a significant FDR, showed a log2-fold change (log2FC) of transcription of ≥ ± 2 (or ≥ ± 0.8 for small RNAs) at least at one point during a time course. For this, the log2-fold values of transcript abundance obtained in edgeR were used in combination with the heatmap. 2 function of the R package gplots (v2.15.0)62. Raw and processed RNA-seq data have been deposited in NCBI’s Gene Expression Omnibus (GEO) database63 and are accessible through GEO Series accession number GSE76979.
Fluorescent staining of S. pneumoniae
To detect changes in the membrane potential of S. pneumoniae TIGR4 when treated with Carolacton, the bacteria were stained with the fluorescent dye 3,3-Diethyloxacarbocyanine iodide (DiOC2(3)) (BacLight Bacterial Membrane Potential Kit, Life Technologies). For staining with DiOC2(3), cells were diluted from culture to approx. 107 cells/ml and stained with 30 μM DiOC2(3) in the dark for 30 min. In order to assess bacterial membrane integrity, cells were separately co-stained with the fluorescent dyes Syto 9 and propidium iodide (PI) using the LIVE/DEAD BacLight Bacterial Viability Kit (Life Technologies).
For Syto 9/PI staining, cells from a culture grown with 0.25 μg/ml Carolacton and from a control were harvested by centrifugation and washed in 0.85% (w/v) NaCl. Approx. 5 × 106 cells/ml were stained with an equal volume 2× staining solution (10 μM Syto 9, 60 μM PI) at RT in the dark for 30 min. Heat-inactivated cells were used as a control in both staining experiments. Heat-killing of cells was achieved by incubation at 80 °C and shaking (1,400 rpm) for 30 min.
Flow cytometric analysis of fluorescently-labelled S. pneumoniae cells
DiOC2(3) and Syto 9/PI-fluorescently stained cells were analysed in triplicate on a FACSCanto flow cytometer (BD Biosciences) as described in the material section of the Supplementary Information.
Microscopy and quantification of cell size
S. pneumoniae TIGR4 cells growing in THBY were washed with 0.85% (w/v) NaCl solution and mounted to a microscope slide coated with a thin layer of 1% (w/v) agarose. Phase-contrast microscopic analysis of S. pneumoniae was performed using an Olympus BX60 microscope equipped with an Olympus DP50 color camera (5 Mpixel) color view II camera and a 100x magnification oil immersion objective. Pictures and measurements were taken with the Olympus cellSens (v1.6) microscopy software. The length of pneumococcal cells was measured along the length axis. Cell width was determined as the diameter along the equatorial plane, perpendicular to the length axis of the cell.
Quantification of colony forming units
For determination of colony forming units (CFUs), cells were treated with Carolacton (0.25 μg/ml) prior to exponential growth. Ten-fold serial dilutions of cultures were prepared in 0.85% (w/v) NaCl solution (pre-warmed to 37 °C) after 3 h and 6 h of growth. A total volume of 100 μl from each dilution was then plated onto pre-warmed (37 °C) THBY agar plates, which were incubated at 37 °C and 5% CO2 o/n.
Statistical analysis
Comparisons between two groups at a single time were performed with unpaired two-tailed (non-parametric) Mann-Whitney U test64 or with Student’s t-tests for two-tailed distribution (with a critical P value of 0.05). The data were analysed for normality by Shapiro-Wilk test65. Statistical analyses were performed with the statistics options of SigmaPlot (v13.0) software (Systat Software Inc.).
Additional Information
Accession codes: Raw and processed RNA-seq data have been deposited in NCBI’s Gene Expression Omnibus. (GEO) database and are accessible through GEO Series accession number GSE76979. http://www.nature.com/srep
How to cite this article: Donner, J. et al. The biofilm inhibitor Carolacton inhibits planktonic growth of virulent pneumococci via a conserved target. Sci. Rep. 6, 29677; doi: 10.1038/srep29677 (2016).
Supplementary Material
Acknowledgments
We are very grateful to Pavel Branny for sharing his antibody against StkP. We also thank the Genome Analytics Group at the Helmholtz Centre for Infection Research (Braunschweig) for library construction, sequencing and helpful discussions throughout the project and Rolf Jansen from the Microbial Drugs Group at HZI for supplying Carolacton. A.K. thanks “Biofabrication for NIFE”, which is financially supported by the ministry of Lower Saxony and the Volkswagen foundation. This work was supported partly by the Federal Ministry of Education and Research (BMBF) in the program e:Bio (grant number 031 A299) and by the President’s Initiative and Networking Fund of the Helmholtz Association of German Research Centres (HGF) under contract number VH-GS-202.
Footnotes
The authors declare no competing financial interests.
Author Contributions J.D., M.R., S.B. and I.W.-D. designed the experiments. J.D. collected the data. J.D., M.R., S.B., R.M. and I.W.-D. analysed and discussed the data. A.K. provided epi-Carolacton. J.D. and I.W.-D. wrote the main manuscript text. All authors reviewed, discussed and agreed on the final version of the manuscript.
References
- World Health Organization. Antimicrobial resistance: global report on surveillance, World Health Organization, Geneva, Switzerland. (2014) Available online at: http://www.pwho.int/drugresistance/documents/surveillancereport/en/ (Accessed: 3rd March 2016).
- Butler M. S., Blaskovich M. A. & Cooper M. A. Antibiotics in the clinical pipeline in 2013. J Antibiot 66, 571–591, 10.1038/ja.2013.86 (2013). [DOI] [PubMed] [Google Scholar]
- Zaura E. et al. Same Exposure but Two Radically Different Responses to Antibiotics: Resilience of the Salivary Microbiome versus Long-Term Microbial Shifts in Feces. mBio 6, e01693–01615 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver L. L. Challenges of antibacterial discovery. Clin Microbiol Rev 24, 71–109, 10.1128/CMR.00030-10 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R. et al. Carolacton - A Macrolide Ketocarbonic Acid that Reduces Biofilm Formation by the Caries- and Endocarditis-Associated Bacterium Streptococcus mutans. Eur J Org Chem 7, 1284–1289, 10.1002/ejoc.200901126 (2010). [DOI] [Google Scholar]
- Schmidt T. & Kirschning A. Total synthesis of carolacton, a highly potent biofilm inhibitor. Angew Chem Int Ed Engl 51, 1063–1066, 10.1002/anie.201106762 (2012). [DOI] [PubMed] [Google Scholar]
- Hallside M. S., Brzozowski R. S., Wuest W. M. & Phillips A. J. A concise synthesis of carolacton. Org Lett 16, 1148–1151, 10.1021/ol500004k (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze B. et al. Damage of Streptococcus mutans biofilms by carolacton, a secondary metabolite from the myxobacterium Sorangium cellulosum. BMC Microbiol 10, 199, 10.1186/1471-2180-10-199 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel C., Barg A., Rheinberg A., Conrads G. & Wagner-Döbler I. Dental composite materials containing carolacton inhibit biofilm growth of Streptococcus mutans. Dent Mater 29, 1188–1199, 10.1016/j.dental.2013.09.005 (2013). [DOI] [PubMed] [Google Scholar]
- Reck M. et al. The biofilm inhibitor carolacton disturbs membrane integrity and cell division of Streptococcus mutans through the serine/threonine protein kinase PknB. J Bacteriol 193, 5692–5706, 10.1128/JB.05424-11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpp N. et al. Synthesis of new carolacton derivatives and their activity against biofilms of oral bacteria. Org Biomol Chem 13, 5765–5774, 10.1039/C5OB00460H (2015). [DOI] [PubMed] [Google Scholar]
- Reck M. & Wagner-Döbler I. Carolacton treatment causes delocalization of the cell division proteins PknB and DivIVa in Streptococcus mutans in vivo. Front Microbiol 7, 10.3389/fmicb.2016.00684 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N. & Yamada T. Acid-induced acid tolerance and acidogenicity of non-mutans streptococci. Oral Microbiol Immunol 14, 43–48, 10.1034/j.1399-302X.1999.140105.x (1999). [DOI] [PubMed] [Google Scholar]
- Regev-Yochay G. et al. Nasopharyngeal carriage of Streptococcus pneumoniae by adults and children in community and family settings. Clin Infect Dis 38, 632–639, 10.1086/381547 (2004). [DOI] [PubMed] [Google Scholar]
- Huang S. S. et al. Healthcare utilization and cost of pneumococcal disease in the United States. Vaccine 29, 3398–3412, 10.1016/j.vaccine.2011.02.088 (2011). [DOI] [PubMed] [Google Scholar]
- Whitney C. G. et al. Increasing prevalence of multidrug-resistant Streptococcus pneumoniae in the United States. N Engl J Med 343, 1917–1924, 10.1056/NEJM200012283432603 (2000). [DOI] [PubMed] [Google Scholar]
- Sharma D. et al. Pneumococcal carriage and invasive disease in children before introduction of the 13-valent conjugate vaccine: comparison with the era before 7-valent conjugate vaccine. Pediatr Infect Dis J 32, e45–53 (2013). [DOI] [PubMed] [Google Scholar]
- Golden A. R. et al. Characterization of MDR and XDR Streptococcus pneumoniae in Canada, 2007–13. J Antimicrob Chemother 70, 2199–2202, 10.1093/jac/dkv107 (2015). [DOI] [PubMed] [Google Scholar]
- Tettelin H. et al. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293, 498–506 (2001). [DOI] [PubMed] [Google Scholar]
- Sudhakar P. et al. Construction and verification of the transcriptional regulatory response network of Streptococcus mutans upon treatment with the biofilm inhibitor carolacton. BMC Genomics 15, 362, 10.1186/1471-2164-15-362 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorg R. A. & Veening J. W. Microscale insights into pneumococcal antibiotic mutant selection windows. Nat Commun 6, 8773, 10.1038/ncomms9773 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasz A. Biological consequences of the replacement of choline by ethanolamine in the cell wall of Pneumococcus: chanin formation, loss of transformability, and loss of autolysis. Proc Natl Acad Sci USA 59, 86–93, 10.1073/pnas.59.1.86 (1968). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilharz K. et al. Control of cell division in Streptococcus pneumoniae by the conserved Ser/Thr protein kinase StkP. Proc Natl Acad Sci USA 109, E905–913, 10.1073/pnas.1119172109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novo D., Perlmutter N. G., Hunt R. H. & Shapiro H. M. Accurate flow cytometric membrane potential measurement in bacteria using diethyloxacarbocyanine and a ratiometric technique. Cytometry 35, 55–63, (1999). [DOI] [PubMed] [Google Scholar]
- Sakamoto H., Kimura N. & Shimura Y. Processing of transcription products of the gene encoding the RNA component of RNase P. Proc Natl Acad Sci USA 80, 6187–6191, 10.1073/pnas.80.20.6187 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick J. E., Sudarsan N., Weinberg Z., Ruzzo W. L. & Breaker R. R. 6S RNA is a widespread regulator of eubacterial RNA polymerase that resembles an open promoter. RNA 11, 774–784, 10.1261/rna.7286705 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Chao S., Wei C. L. & Lin Y. T. RNase E is required for the maturation of ssrA RNA and normal ssrA RNA peptide-tagging activity. Proc Natl Acad Sci USA 96, 12406–12411, 10.1073/pnas.96.22.12406 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann B. et al. Control of virulence by small RNAs in Streptococcus pneumoniae. PLoS Pathog 8, e1002788, 10.1371/journal.ppat.1002788 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverys J. P., Prudhomme M. & Martin B. Induction of competence regulons as a general response to stress in gram-positive bacteria. Annu Rev Microbiol 60, 451–475, 10.1146/annurev.micro.60.080805.142139 (2006). [DOI] [PubMed] [Google Scholar]
- Hava D. L. & Camilli A. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol Microbiol 45, 1389–1406, 10.1046/j.1365-2958.2002.03106.x (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews J. M. Determination of minimum inhibitory concentrations. J Antimicrob Chemother 48, 5–16, 10.1093/jac/48.suppl_1.5 (2001). [DOI] [PubMed] [Google Scholar]
- Ambrose K. D., Nisbet R. & Stephens D. S. Macrolide Efflux in Streptococcus pneumoniae Is Mediated by a Dual Efflux Pump (mel and mef) and Is Erythromycin Inducible. Antimicrob Agents Chemother 49, 4203–4209, 10.1128/AAC.49.10.4203-4209.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenwald N. P., Appelbaum P., Davies T. & Gill M. J. Evidence for efflux pumps, other than PmrA, associated with fluoroquinolone resistance in Streptococcus pneumoniae. Clin Microbiol Infect 9, 140–143, 10.1046/j.1469-0691.2003.00482.x (2003). [DOI] [PubMed] [Google Scholar]
- Eschenburg S., Priestman M. & Schonbrunn E. Evidence that the fosfomycin target Cys115 in UDP-N-acetylglucosamine enolpyruvyl transferase (MurA) is essential for product release. J Biol Chem 280, 3757–3763, 10.1074/jbc.M411325200 (2005). [DOI] [PubMed] [Google Scholar]
- Hirota K. et al. Fosfomycin Reduces CD15s-Related Antigen Expression of Streptococcus pyogenes. Antimicrob Agents Chemother 42, 1083–1087 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinho M. G., Kjos M. & Veening J.-W. How to get (a)round: mechanisms controlling growth and division of coccoid bacteria. Nat Rev Microbiol 11, 601–614, 10.1038/nrmicro3088 (2013). [DOI] [PubMed] [Google Scholar]
- Novo D. J., Perlmutter N. G., Hunt R. H. & Shapiro H. M. Multiparameter flow cytometric analysis of antibiotic effects on membrane potential, membrane permeability, and bacterial counts of Staphylococcus aureus and Micrococcus luteus. Antimicrob Agents Chemother 44, 827–834, 10.1128/AAC.44.4.827-834 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherian P. T. et al. Chemical Modulation of the Biological Activity of Reutericyclin: a Membrane-Active Antibiotic from Lactobacillus reuteri. Sci Rep 4, 4721, 10.1038/srep04721 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl H. & Hamoen L. W. Membrane potential is important for bacterial cell division. Proc Natl Acad Sci USA 107, 12281–12286, 10.1073/pnas.1005485107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy F. J., Rollins S. M. & Henkin T. M. Interaction between the acceptor end of tRNA and the T box stimulates antitermination in the Bacillus subtilis tyrS gene: a new role for the discriminator base. J. Bacteriol 176, 4518–4526 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green N. J., Grundy F. J. & Henkin T. M. The T box mechanism: tRNA as a regulatory molecule. FEBS Lett 584, 318–324, 10.1016/j.febslet.2009.11.056 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigg J. C. et al. T box RNA decodes both the information content and geometry of tRNA to affect gene expression. Proc Natl Acad Sci USA 110, 7240–7245, 10.1073/pnas.1222214110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler R. G. & Schaaper R. M. The role of the mutT gene of Escherichia coli in maintaining replication fidelity. FEMS Microbiol Rev 21, 43–54, 10.1111/j.1574-6976.1997.tb00344.x (1997). [DOI] [PubMed] [Google Scholar]
- McLennan A. G. The MutT motif family of nucleotide phosphohydrolases in man and human pathogens (review). Int J Mol Med 4, 79–168, 10.3892/ijmm.4.1.79 (1999). [DOI] [PubMed] [Google Scholar]
- Bessman M. J., Frick D. N. & O’Handley S. F. The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J Biol Chem 271, 25059–25062, 10.1074/jbc.271.41.25059 (1996). [DOI] [PubMed] [Google Scholar]
- Moukadiri I. et al. Evolutionarily conserved proteins MnmE and GidA catalyze the formation of two methyluridine derivatives at tRNA wobble positions. Nucleic Acids Res 37, 7177–7193, 10.1093/nar/gkp762 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shippy D. C. & Fadl A. A. tRNA modification enzymes GidA and MnmE: potential role in virulence of bacterial pathogens. Int J Mol Sci 15, 18267–18280, 10.3390/ijms151018267 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shippy D. C. & Fadl A. A. Immunological characterization of a gidA mutant strain of Salmonella for potential use in a live-attenuated vaccine. BMC Microbiol 12, 286, 10.1186/1471-2180-12-286 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho K. H. & Caparon M. G. tRNA modification by GidA/MnmE is necessary for Streptococcus pyogenes virulence: a new strategy to make live attenuated strains. Infect Immun 76, 3176–3186, 10.1128/IAI.01721-07 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patenge N., Fiedler T. F. & Kreikemeyer B. Common regulators of virulence in streptococci. Curr Top Microbiol Immunol 368, 111–153, 10.1007/82_2012_295 (2013). [DOI] [PubMed] [Google Scholar]
- Fang C. et al. Activation of PrfA results in overexpression of virulence factors but does not rescue the pathogenicity of Listeria monocytogenes M7. J Med Microbiol 64, 818–826, 10.1099/jmm.0.000101 (2015). [DOI] [PubMed] [Google Scholar]
- Rasko D. A. & Sperandio V. Anti-virulence strategies to combat bacteria-mediated disease. Nat Rev Drug Discov 9, 117–128, 10.1038/nrd3013 (2010). [DOI] [PubMed] [Google Scholar]
- Allen R. C., Popat R., Diggle S. P. & Brown S. P. Targeting virulence: can we make evolution-proof drugs? Nat Rev Microbiol 12, 300–308, 10.1038/nrmicro3232 (2014). [DOI] [PubMed] [Google Scholar]
- Charpentier X., Polard P. & Claverys J. P. Induction of competence for genetic transformation by antibiotics: convergent evolution of stress responses in distant bacterial species lacking SOS? Curr Opin Microbiol 15, 570–576, 10.1016/j.mib.2012.08.001 (2012). [DOI] [PubMed] [Google Scholar]
- Senadheera D. et al. Inactivation of VicK Affects Acid Production and Acid Survival of Streptococcus mutans. J Bacteriol 191, 6415–6424, 10.1128/JB.00793-09 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land A. D. & Winkler M. E. The Requirement for Pneumococcal MreC and MreD Is Relieved by Inactivation of the Gene Encoding PBP1a. J. Bacteriol 193, 4166–4179, 10.1128/JB.05245-11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau P. C., Sung C. K., Lee J. H., Morrison D. A. & Cvitkovitch D. G. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J Microbiol Methods 49, 193–205, 10.1016/S0167-7012(01)00369-4 (2002). [DOI] [PubMed] [Google Scholar]
- Macrina F. L. et al. Novel shuttle plasmid vehicles for Escherichia-Streptococcus transgeneric cloning. Gene 25, 145–150 (1983). [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 227, 680–685, 10.1038/227680a0 (1970). [DOI] [PubMed] [Google Scholar]
- Bergmann S., Rohde M., Chhatwal G. S. & Hammerschmidt S. alpha-Enolase of Streptococcus pneumoniae is a plasmin(ogen)-binding protein displayed on the bacterial cell surface. Mol Microbiol 40, 1273–1287, 10.1046/j.1365-2958.2001.02448.x (2001). [DOI] [PubMed] [Google Scholar]
- Benjamini Y. & Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 57, 289–300, 10.2307/2346101 (1995). [DOI] [Google Scholar]
- Warnes G. R. et al. gplots: Various R programming tools for plotting data. R package version 2.12. 1, (2013). Available online at: http://CRANR-project.org/package=gplots (Accessed: 24th February 2016).
- Edgar R., Domrachev M. & Lash A. E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30, 207–210, 10.1093/nar/30.1.207 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann H. B. & Whitney D. R. On a Test of Whether one of Two Random Variables is Stochastically Larger than the Other. Ann Math Statist 18 (1947). [Google Scholar]
- Shapiro S. S. & Wilk M. B. An Analysis of Variance Test for Normality (Complete Samples). Biometrika 52, 591–611, 10.1128/JB.02602-14 (1965). [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.