Abstract
Translation is the fundamental biological process converting mRNA information into proteins. Single-molecule imaging in live cells has illuminated the dynamics of RNA transcription; however, it is not yet applicable to translation. Here, we report single-molecule imaging of nascent peptides (SINAPS) to assess translation in live cells. The approach provides direct readout of initiation, elongation, and location of translation. We show that mRNAs coding for endoplasmic reticulum (ER) proteins are translated when they encounter the ER membrane. Single-molecule fluorescence recovery after photobleaching provides direct measurement of elongation speed (5 amino acids per second). In primary neurons, mRNAs are translated in proximal dendrites but repressed in distal dendrites and display “bursting” translation. This technology provides a tool with which to address the spatiotemporal translation mechanism of single mRNAs in living cells.
Genome-wide studies have shown that protein abundance is predominantly controlled at the level of translation (1). Translational regulation allows cells to respond rapidly to environmental cues and synthesize proteins with precise timing and at specific subcellular locations (2, 3). Numerous studies have concentrated on RNA localization and its underlying mechanism (4). However, the translation of localized mRNA in living cells remains poorly understood because unlike transcription, a single-molecule method to directly image the process is lacking (5). Ensemble biochemical measurements such as ribosome profiling can provide a genome-wide measurement of translation (6, 7). Approaches that measure the association of ribosomes with mRNAs are not direct readouts of translation (8). Pulse-labeling in cell culture [stable isotope labeling with amino acids in cell culture (SILAC)] quantifies newly synthesized proteins by means of mass spectrometry (1). Cellular approaches, such as fluorescent noncanonical amino acid tagging (FUNCAT) (9) and puromycylation (10), measure overall protein synthesis but are not gene-specific. Fluorescent protein–based translation assays (11–14) require lengthy maturation of fluorophores. Recently, translating RNA imaging by coat protein knock off (TRICK) (15) distinguished previously translated from untranslated mRNAs. A single-molecule fluorescence imaging approach would complement these cellular and biochemical methods by providing precise spatiotemporal molecular information, including variance among mRNAs. Here, we describe a strategy for directly visualizing the nascent peptide (NAP) on translating mRNA with single-molecule imaging of nascent peptides (SINAPS). It provides precise parameters and reveals unpredicted phenomena of mRNA regulation in the endoplasmic reticulum (ER) and neuronal dendrites.
Development of SINAPS, a single-molecule translation assay in live cells
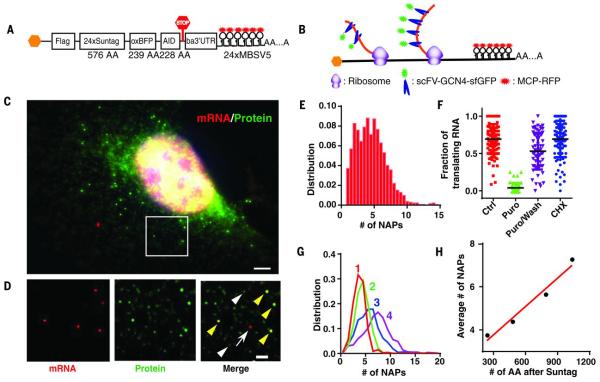
There are two obstacles to visualize the translation site (TLS): (i) the weak signal of the nascent chain and (ii) the background due to preexisting labeled proteins. To address the weak signal, we used the recently developed SunTag technology for signal amplification (16), which uses a genetically encoded antibody fragment, the single-chain variable fragment (scFV) of GCN4 antibody fused to a super folder green fluorescent protein (sfGFP) (scFV-sfGFP) (17). When the epitope peptide for the scFV is multimerized, multiple scFV-sfGFPs bind to a single reporter protein, rendering it visible above the fluorescence background, similar to MS2 stem loops that make the mRNA visible (Fig. 1, A and B) (18). Because the sfGFP is already mature and fluorescent, the NAP could be detected as soon as it emerges from the ribosome (Fig. 1B). In a polysome, multiple NAPs accumulate at the site of mRNA and form a bright TLS. To address the background fluorescence, we removed completed proteins by fusing the auxin-induced degron (AID) to the C terminus of the reporter in order to regulate its stability (19, 20). We inserted the protein of interest [blue fluorescent protein (oxBFP) (21)] between the SunTag and the AID (20) with a flag tag in the N terminus. The reporter mRNA included the 3′ untranslated region (3’UTR) of β-actin mRNA for cytoplasmic localization and the MS2 binding sites (MBS) for its visualization (22). The reporter (flag-SINAPS) was cloned into a lentiviral backbone for stable integration into the target cells (Fig. 1A).
Fig. 1.
System for single-molecule imaging of nascent peptides in live cells.
(A) Schematic of the SINAPS construct. Flag, flag tag; AID, auxin induced degron; ba3’UTR, β-actin 3′ untranslated region; AA, amino acid. (B) Schematic of SINAPS. scFV-sfGFP binds to and labels NAPs containing SunTag epitope emerging from ribosome. Polysome assembling on mRNA results in multiple NAPs. (C) smFISH and IF experiments on flag-SINAPS constructs. Green, IF against GFP; red, smFISH against mRNA. The bright green spots colocalizing with red mRNAs are TLSs. Scale bar, 5 μm. (D) The deconvolved image of the box in (C). (Left) smFISH. (Middle) IF. (Right) Merged. Yellow arrowheads indicate TLS; white arrowheads indicate single flag-SINAPS protein; and the white arrow indicates untranslating mRNA. Scale bar, 2 μm. (E) The integrated intensity of TLS is normalized with that of single proteins, giving the number of NAPs (full-length equivalent). (F) The fraction of translating mRNA under treatments. Each symbol represents a single cell. Ctrl, control; Puro, 100 μg/mL puromycin for 10 min; Puro/Wash, 100 μg/mL puromycin treatment for 10 min, then incubation in normal medium for 20 min; CHX, 2 μg/mL CHX treatment for 30 min. (G) The number of NAPs for different constructs at steady state. The longer the mRNA, the more NAPs at the TLS. (H) The mean number of NAPs scales linearly with the length of the coding region. Red line describes a linear fit with slope 0.0044 per amino acid.
The SINAPS reporter was expressed by viral infection in a U2OS human osteosarcoma cell line stably expressing two accessory proteins: the antibody scFV-sfGFP for labeling and the thermally stable rice Oryza Sativa transport inhibitor response 1 (OsTIR1) for degrading AID-containing protein (19, 23). A protein of the correct size was synthesized but became undetectable with Western blot in presence of 500 μM auxin Indole-3-acetic acid (IAA) (fig. S1). We performed single-molecule fluorescence in situ hybridization (smFISH) to detect mRNA and immunofluorescence (IF) against GFP to measure the protein (Fig. 1, C and D). The bright spots in the IF channel (green) colocalized with mRNA spots (red). The numerous dim green spots were single proteins labeled with SunTag (Fig. 1D, white arrow heads). They were used to convert the TLS intensity into the number of NAPs (maximum, 14; mean, 4.5) (Fig. 1E). We used puromycin, a tRNA analog that dissociates NAPs from the ribosomes, to verify that TLS disappeared (Fig. 1F and fig. S2, A and B); its removal (Puro/Wash) restored the TLS to steady-state levels (Fig. 1F and fig. S2, C to E). Low concentrations of the elongation inhibitor cycloheximide (CHX) increased NAPs on mRNAs (Fig. 1F and fig. S2F). We inserted varying lengths of the protein domains between SunTag and the AID to determine whether the number of NAPs scaled linearly with the length of the coding region (Fig. 1G and fig. S2G). A linear fit described the data with a slope of 0.0044 per amino acid (Fig. 1H). With the translation elongation speed determined as v = 4.7 amino acids/s, this reveals an average initiation rate of 1.3 per min (supplementary materials, Eq. 4).
Diffusion dynamics of translating RNA
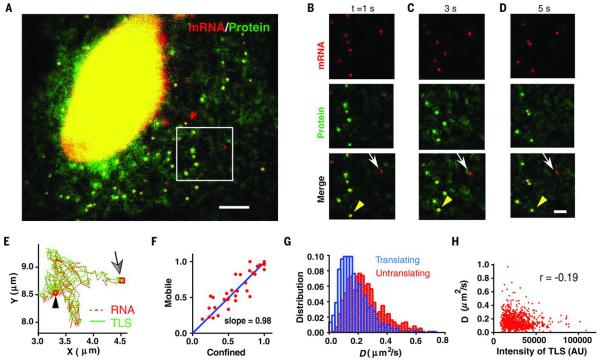
To describe the dynamics of translation, we performed live cell imaging. We labeled the mRNA with MS2 coat protein (MCP) fused with Halotag (24) and applied Halo-JF549 dyes for its superior brightness and photostability (23, 25). We imaged mRNAs and TLS simultaneously using their distinct fluorescent tags (stdMCP-Halo-JF549 and scFV-sfGFP) (26) on two precisely aligned cameras (27). We observed bright TLS moving together with mRNAs (Fig. 2, A to D, and movies S1 and S2). As a control, in puromycin-treated cells there were no TLSs detected (movie S3). The brightness of the mRNA and TLS allowed accurate determination of their positions with Matlab software (28). We tracked the positions of mRNA and TLS (29) and identified TLS moving together with mRNAs (Fig. 2E) (23). We classified the tracks of mRNAs into two categories: confined and freely diffusing. For each category, we determined the fraction of mRNA tracks with TLS in each cell and defined them as translating mRNAs. Translation was independent of whether the mRNAs were confined or freely diffusing (Fig. 2F). For freely diffusing mRNAs, the diffusion coefficients (D) were measured through mean-square displacements. The translating mRNAs diffused slower on average but overlapped in their distribution with untranslating ones (Fig. 2G). The D of translating mRNA was weakly anticorrelated with TLS intensity, which is consistent with larger polysomes slowing mRNA mobility (Fig. 2H). Therefore, the mRNA mobility correlates with its translation level on average but is not an exact predictor of translation status of individual mRNAs coding for cytosolic proteins, which is different from membrane-targeted mRNAs.
Fig. 2.
Dynamics of mRNAs in translation.
(A) Snapshot of live cell imaging of cells stably expressing flag-SINAPS (movie S1). Green, scFV-sfGFP; red, stdMCP-Halotag-JF549. Scale bar, 5 μm. (B to D) Montage of movie S2 at time (t) = 1, 3 and 5 s. (Top) mRNA. (Middle) TLS and free proteins. (Bottom) Merged. Yellow arrowheads inidcate TLS, and white arrows indicate untranslating mRNAs. Scale bar, 2 μm. (E) The mRNA (red) and TLS (green) move together. Arrow (square) indicates the start of the track. The black arrowhead (circle) indicates the end of the track. (F) The motion of mRNA is classified either as confined or mobile. The fraction of translating mRNA in each category is almost identical in each cell. Each symbol represents a cell. (G) The histogram of diffusion coefficients (D) of freely diffusing mRNAs. Blue, translating mRNA; red, untranslating mRNA. (H) The diffusion coefficient of the translating mRNA is only weakly anticorrelated with the integrated intensity of TLS; correlation coefficient (r) = −0.19.
Local translation on the ER
Local protein synthesis on the ER and other membrane-bound cellular compartments plays a critical role in protein trafficking to establish and maintain membrane function (30). Introducing an ER-targeting signal in the N terminus of the SINAPS reporter will target the reporter protein to the ER (Fig. 3A). Because the scFV antibody is cytoplasmic, the SunTag motif needs to be exposed to cytosol to be labeled (Fig. 3B). We fused the first 29 amino acids of cytochrome p450 to the N terminus of the reporter [cytoplasmic end of an ER signal-anchor membrane protein (CytERM)] (21). CytERM-SINAPS labeled the tubular ER structure when expressed in U2OS cells stably infected with scFV-sfGFP and OsTIR1 (fig. S3, A and B). At low expression, individual CytERM-SINAPS proteins were visible owing to their slower mobility on the membrane (fig. S3, C to E, and movie S4). Unlike mRNAs coding for cytoplasmic proteins, the majority of CytERM-SINAPS mRNAs showed confined motion (Fig. 3, C to G, and movies S5 to S7). Consistent with this, all mRNAs colocalizing with TLSs were confined and showed low mobility (Fig. 3, H to J). Confined mRNAs without bright translation sites might have recently initiated translation. In contrast, none of the freely diffusing mRNAs moved together with TLS (Fig. 3, C to J). This confirmed that the reporter proteins were inserted into the ER cotranslationally, and the mRNAs were tethered to the ER by NAPs. To further support this interpretation, when treated with puromycin the mRNAs immediately dissociated from the ER (within 2 min) and became freely diffusing particles without TLS (Fig. 3, G and K, and movies S5 and S8). These experiments demonstrate that the mRNAs are tethered to the ER only when translated.
Fig. 3.
Local translation on the ER.
(A) The schematic of the ER translation reporter. (B) The CytERM peptide is inserted into the ER, while the rest protein domain is kept in the cytosol. The mRNA is tethered to the ER by CytERM, and TLS is labeled with scFV-sfGFP. Although only one ribosome is shown, it is likely that mRNAs are translated by polysomes. The items are described similarly as in Fig. 2. (C) Snapshot of a live cell expressing CytERM-SINAPS (movie S6). Green, scFV-sfGFP; red: stdMCP-Halotag-JF549. Scale bar, 5 μm. (D toF) Montage of movie S7 at t = 2, 6 and 10 s. (Top) mRNAs. (Middle) TLS and free proteins. (Bottom) Merged. Yellow arrowheads indicate TLS; white arrows indicate untranslating mRNAs. Scale bar, 2 μm. (G) The fraction of mobile mRNAs for cytoplasmic mRNAs (cyt), Flag-SINAPS; the ER-targeted mRNAs (ER), CytERM-SINAPS; and CytERM-SINAPS mRNAs in the presence of 100 μg/mL puromycin (ER+Puro). Unpaired t test, ***P < 0.001. NS, not significant. See also movie S5. (H) (Top) A translating mRNA in (C) to (F) shows a confined motion. (Bottom) An untranslating mRNA is freely diffusing. (I) Most translating CytERM-SINAPS mRNAs were confined with very small diffusion coefficient (blue). The untranslating mRNA had higher diffusion coefficient (red). (J) The confined mRNAs had a significantly higher fraction in translation (average 69%) than that of freely diffusing ones (average 3%; all fast moving mRNAs are not translating). (K) The diffusion coefficient of CytERM-SINAPS mRNA when treated with 100 μg/mL puromycin. mRNAs were released from the ER and freely diffuse.
Translation kinetics in live cells
Because the translating CytERM-SINAPS mRNA were confined and could be tracked for many minutes, we performed fluorescence recovery after photobleaching (FRAP) experiments on single TLSs (Fig. 4A). We applied a two-second pulse of diffraction-limited 491-nm laser light to bleach the selected single TLS (23). The integrated fluorescence intensity was monitored while tracking the TLS (movies S9 and S10). The fluorescence recovered over 90% of the pre-bleach value within 4 min (Fig. 4, B and E). As a control, neighboring TLS not bleached showed consistent signal throughout the experiment (Fig. 4C). In contrast, the fluorescence only recovered to ~20% of pre-bleach value when ribosomes were stalled by high concentrations of CHX (Fig. 4, D and E). To derive the recovery curve, we assumed that the scFV binds to each epitope segment with equal probability at the TLS or on mature proteins. We modeled the translation by ribosomes as a stochastic process, with constant initiation and elongation rates. The normalized recovery curve depended only on one parameter: the elongation rate of ribosomes [the gene length is a constant for a given reporter (supplementary text)]. The modeling correctly predicted the FRAP curve with Monte Carlo simulation (fig. S4). Fitting the theoretical curve to the experimentally observed data yielded the average translation elongation rate v = 4.7 ± 0.6 amino acids/s (Fig. 4E). The translation rate was in agreement with previous measurements by using ribosome profiling (v = 5.6 amino acids/s) (31). Experiments on mRNAs with different lengths of coding regions yielded similar translation rates (v = 5.6 ± 1.4 amino acids/s and v = 5.0 ± 1.1 amino acids/s) (fig. S5). These results indicate that the SINAPS reporter mRNAs undergo constitutive translation in U2OS cells.
Fig. 4.
Translation kinetics in live cells measured with single-molecule FRAP of TLS.
(A) Schematics of FRAP. The existing NAPs on the TLS were bleached with a focused 491-nm laser at t = 0 (black). The fluorescence recovers as existing ribosomes synthesize the SunTag motifs and new ribosomes arrive and make new NAPs (green). At t = ∞, the fluorescence should recover to the beginning value at steady state. (B toD) Montage of FRAP experiments at different time points for (B) control, (C) nonbleached TLS, and (D) CHX treatment. (E) There was little recovery for CHX treatment (D) (black squares) compared with control (B) (blue circles). Fitting the theory (red line) to the experimental data (n = 31 movies) yielded the translation elongation speed v = 4.7 ± 0.6 amino acids/s.
Translation in neurons
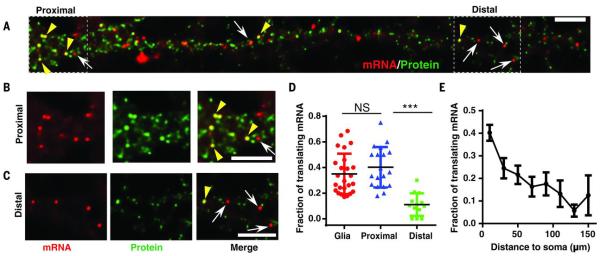
We applied SINAPS to study local translation in neurons, where local regulation of translation is particularly important owing to polarized neuronal processes (32). We combined the OsTIR1 and scFV-sfGFP into a polycistronic plasmid with an internal ribosome entry site (IRES) (23). We expressed flag-SINAPS and OsTIR1-IRES-scFV-sfGFP in cultured primary hippocampal neurons by dual lentiviral infection. We performed smFISH and IF experiments to investigate the translation of individual mRNAs. Bright puncta in the SunTag channel colocalizing with the mRNA indicated that the mRNA was being translated (Fig. 5, A to C, and fig. S6). Although the numbers of NAPs on mRNAs in primary neurons and glia were similar to that in U2OS (Fig. 1E and fig. S6, B and E), the fraction of translating mRNAs was less (Figs. 1G and 5D), suggesting a more stringent regulation of mRNA translatability in primary cells. In addition, translation in neurons was spatially modulated. In proximal dendrites (<30 μm from soma), the percentage of translating mRNAs (~40%) was similar to that of the glial cells cultured in the same dish. However, the percentage gradually decreased as a function of distance to the soma (Fig. 5E), as low as ~10% for distal dendrites (>100 μm) (Fig. 5D). This suggests translation repression in distal dendrites (33).
Fig. 5.
Spatial distribution of translation sites in neurons.
(A) smFISH and IF experiment. Flag-SINAPS was coexpressed with OsTIR1-IRES-scFV-sfGFP in dissociated primary hippocampal neuron with lentiviral infection at 7 days in vitro. The neurons were fixed at 14 to 21 days in vitro, and smFISH and IF experiments were performed. The dendrites of the neuron were straightened with ImageJ. The whole neuron is shown in fig. S6. Red, mRNA; green, TLS and free protein. Scale bar, 5 μm. (B and C) Segments in the (B) proximal and (C) distal dendrite were enlarged. (Left) mRNA. (Middle) TLS and free protein. (Right) Merged. Yellow arrowheads indicate TLS; white arrows indicate untranslating mRNAs. Scale bar, 5 μm. (D) The fraction of translating mRNA was similar for proximal dendrites (<30 μm to soma) and glial cells in the same culture dish (fig. S6), but significantly less in distal dendrites (>100 μm from soma). Unpaired t test, ***P < 0.001. (E) The fraction of translating mRNA in dendrite as a function of distance to soma (53 dendrites, 19 neurons).
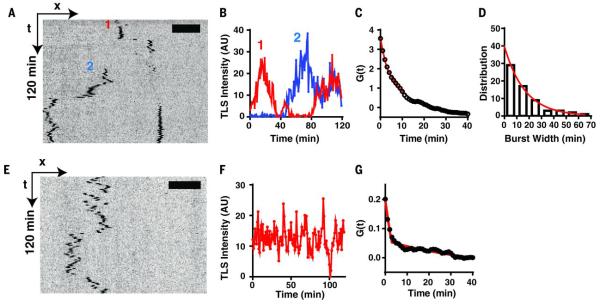
To investigate the dynamics of translation, we imaged the SINAPS reporter in live neurons. We observed bright green particles, mostly in proximal dendrites, but some also distributed in distal dendrites (movie S11). They were much brighter than the fast diffusing single proteins (visible at high power). These were bona fide TLSs because puromycin treatment abolished them within 2 min (fig. S7). We focused on RNA in the distal dendrites because they showed dynamic behavior and were sparsely distributed. We tracked the TLS for 2 hours. The position and integrated intensity of TLS were determined with a tracking algorithm in order to generate the spatial trajectory and intensity trace of the sites (23). TLSs were mobile in a time scale of minutes (Fig. 6A and movies S11 to S13). The mRNA sampled several anchoring sites in dendrites while undergoing active translation. This is in contrast to the hypothesis that the RNA was translationally repressed during transport (34). The TLS intensity over time encoded the translation dynamics of the mRNA (Fig. 6B). To extract the kinetic parameters from these observations, it was necessary to deconvolve the effect of multiple ribosomes generating NAPs. We adopted a fluctuation analysis approach that was developed for transcription (5). The translation dynamics was described by an autocorrelation function G(τ) (Fig. 6C and supplementary text). We observed that many translation events occurred in a bursting fashion (movies S11 to S13): The translation was “on” or “off” with interspersed long periods of no translation at all. We modeled translation as the random telegraph model used for describing transcription kinetics. When the RNA is on, the translation is described by a stochastic process, with constant initiation and elongation rate. The autocorrelation function for this scenario has been worked out previously for modeling transcription (5). The autocorrelation function depends on the initiation rate c and a total dwell time T of NAP at TLS determined by elongation speed and the length of the coding region. When the mRNA is in the off state, there is no translation. The two states interchange randomly and have lifetimes τon and τoff, respectively. We further assumed that translations were correlated in the same on state but statistically independent between successive on states. Fitting the theoretical function to the experimental data yielded a dwell time of an individual NAP (T = 170 ± 50 s, which is consistent with the length of the reporter) and an average translation initiation rate of c = 2.1/min. The average lifetime of the on state was τon = 13 min (Fig. 6C). Alternatively, we measured the width of the translation bursts directly from intensity traces (only from the full off-on-off intensity traces), which yielded the average lifetime of bursts τon = 17 min, agreeing with the fluctuation analysis (Fig. 6D). About 20% of mRNAs underwent constitutive translation during the 2 hours of imaging (Fig. 6, E to G, and movie S14). By singling out those TLSs and analyzing them separately, we obtained a similar dwell time (T = 164 ± 24 s) but a much longer τon > 120 min, which is consistent with a constitutive translation kinetics for these mRNAs. Therefore, the elongation speed was independent of whether translation was bursting or constitutive.
Fig. 6.
Translation dynamics in live neurons.
Flag-SINAPS reporter was expressed in primary hippocampal neuron and imaged live. (A) The kymograph of two flag-SINAPS TLSs in dendrites from time-lapse imaging (movies S11 to S13). (B) The integrated intensities of the two TLSs (labeled as 1 and 2, respectively) showing bursting behaviors. (C) Fitting the autocorrelation function of the integrated intensity (average of 61 TLSs) yielded the residence time T = 170 ± 50 s, initiation rate = 2.1/min, and the average length of translation bursts τon = 13 min. (D) The translation burst size was directly measured for tracks showing complete off-on-off cycles. Exponential fit of the histogram yielded the average length of the burst τon = 17 min. (E) Kymograph, (F) integrated intensity trace, and (G) autocorrelation function of constitutive TLS (defined as translating more than 90% of the time during the 2-hour imaging window, average of 13 TLSs). Fitting of correlation function yielded T = 164 ± 24 s, initiation rate = 2.9/min. The length of translation burst was τon >120 min, which is consistent with a constitutive translation.
Discussion
We studied translation dynamics of single mRNAs in live cells by directly imaging the “newly born” nascent peptide emerging from the ribosome. This was possible because the scFV-GCN4 had high affinity to its epitope [the dissociation constant Kd is less than 4.4 × 10−11 M for wild-type scFV-GCN4 (35)]. FRAP experiments of scFV-GCN4 bound to mitochondria showed that the off time was in the 10-min range (16). Therefore, the on rate (kon) for the scFV-GCN4 is fast (~107/M/s). The concentration of scFV-GCN4 in the cytoplasm is typically >100 nM. Therefore, the time between the NAP segment appearing from the ribosome and being bound by scFV is in the sub-second range. Each epitope peptide has 24 amino acids and takes ~5 s to synthesize. Therefore, the binding of the scFV is fast enough to track translation kinetics. The measured translation speed is consistent with the literature (31), further demonstrating the validity of the technology and that the binding of the scFV does not interfere with the translation kinetics.
Our results using single-particle tracking of mRNA and TLS support the widespread view of translation dynamics on the ER. For secretory and membrane proteins, their mRNAs show the following characteristics: (i) Translating mRNAs were tethered to the ER and show confined motion (Fig. 3, G to J). The translation-dependent tethering is universal for all signal peptides we have tested (CytERM, prolactin, and immunoglobulin heavy chain). (ii) mRNAs were translated multiple rounds on the ER because mRNA were tethered to the same location much longer than the time needed for a single round of translation. (iii) Multiple ribosomes translated the same mRNA tethered to the ER at the same time because TLS was much brighter than a single protein. And (iv) the tethering of the mRNA to the ER depended on the NAP because puromycin treatment releases mRNA from restricted motion to freely diffusing motion (Fig. 3, G and K). Therefore, our data are consistent with a “cotranslational translocation” model (7, 36) that the mRNA that contains ER-targeting signal peptide was tethered to the ER by the NAP during the initial translation. The mRNA remained on the ER because ribosomes continually initiated translation. For cytosolic proteins, their mRNAs were mainly freely diffusing. Therefore, they were mostly translated by cytosolic ribosomes, not by the ER-bound ribosomes (37, 38).
In neurons, it has been proposed that localizing mRNAs are packaged into granules and transported long distances (33). It is hypothesized that mRNAs are suppressed for translation during this transport (34). By visualizing translation of single mRNAs, we demonstrated that mRNAs with the β-actin 3’UTR are actively translated in proximal dendrites within 30 μm of the soma. Many fewer mRNAs in distal dendrites are actively translating, possibly in a repressed granule state (33). These mRNAs show a bursting translation behavior, similar to that seen for nascent mRNA during transcription (5). In addition, we found that mRNA can scan while actively translating and possibly provide newly synthesized proteins immediately for visited spines. This single-molecule methodology provides a powerful tool with which to determine the mechanism of local translation of mRNA and particularly its relationship to local neuronal activity.
The SINAPS technology provides a general approach to measure translation kinetics of single mRNAs. Because all components of the technology are genetically encoded, it can elucidate translation regulation and dynamics with unprecedented spatiotemporal precision without perturbation directly in live cells or animals. For low-abundance proteins, the degron in the reporter is not needed. Further development will be needed to study translation of endogenous genes. This work is complementary to a companion manuscript (39) that demonstrates single-molecule translation by the use of a multi-epitope reporter paired with exogenous fragment antigen-binding (FAB) antibody to detect nascent chains. The measurements for initiation and elongation agree within a factor of two, indicating that the general approach is reproducible between laboratories and different cell types.
Supplementary Material
Movie S1
Translation sites and mRNAs of flag-SINAPS were moving together U2OS Cell (Fig. 2A) stably expressing scFV-sfGFP, OsTIR1, stdMCP-Halo and flag-SINAPS was streamed at 50ms for 200 frames. Green: scFV-sfGFP, Red: stdMCP-Halotag-JF549. Scale Bar: 5 μm
Movie S5
Comparing the diffusion of flag-SINAPS and CytERM-SINAPS mRNAs U2OS cells stably expressing stdMCP-stdGFP were transiently transfected with flag-SINAPS (left panel) or CytERM-SINAPS (middle and right panels) constructs. All mRNAs were labeled with stdMCP-stdGFP (22). Most of cytoplasmic flag-SINAPS were freely diffusing (left). In contrast, CytERM-SINAPS mRNAs were mostly confined (middle). However, when treated with puromycin, most CytERM-SINAPS mRNAs became freely diffusing (right), very similar to the flag-SINAPS mRNAs (left). Note the mRNAs were labeled by stdMCP-stdGFP and scFV-sfGFP was not expressed in the cells. The white ovals were draw to isolate nuclei regions. The yellow boxes drawn in each panel were arbitrary regions in the cytoplasm and meant to focus the attention. Scale bar: 5μm.
Movie S6
CytERM-SINAPS TLS were confined, while non-translating mRNAs were freely diffusion U2OS Cell (Fig. 3) stably expressing scFV-sfGFP, OsTIR1, stdMCP-Halo and CytERM-SINAPS was streamed at 50ms for 200 frames. Green: scFV-sfGFP, Red: stdMCP-Halotag-JF549. Scale Bar: 5 μm
Movie S7
A selected portion of the cell shown in Movie S6 and Figs. 3D-F Left: mRNA, middle: TLS and free proteins, right: merge. Scale Bar 2 μm. The red circles were tracked mRNAs. The green square was a tracked TLS. In the movie, we showed the tracks of one translating mRNA and one untranslating mRNA. The translating mRNA was confined and the untranslating mRNA was freely diffusing.
Movie S8
CytERM-SINAPS TLS disappeared and mRNAs freely diffused in the presence puromycin U2OS Cell stably expressing scFV-sfGFP, OsTIR1, stdMCP-Halo and CytERM-SINAPS was streamed at 50ms for 200 frames in the presence of 100μg/mL puromycin. There were no bright green translation sites moving together with mRNAs. In addition, all mRNAs were freely diffusing instead of confined. Green: scFV-sfGFP, Red: stdMCP-Halotag-JF549. Scale Bar: 2μm.
Movie S9
FRAP of a single TLS: The overview of the whole cell After two pre-bleaching images, a single selected translation site was bleached before frame 3 in U2OS cells stably expressing scFV-sfGFP, OsTIR1 and CytERM-SINAPS. The fluorescence recovery was monitored with time lapse imaging every 10s. The images were max-projection of z-stacks. Top: the FRAP movie with tracked TLS; bottom: the fluorescence intensity of the bleached TLS. Scale bar: 5 μm.
Movie S10
FRAP of single TLSs: comparison between different conditions Single translation sites were bleached in U2OS cells stably expressing scFV-sfGFP, OsTIR1 and CytERM-SINAPS. Left: control; middle: non-bleached; right: bleached in the presence of CHX. Scale bar: 2μm.
Movie S11
Flag-SINAPS in hippocampal neuron: overview of TLS in the neuron. Primary hippocampal neurons were infected with OsTIR1-IRES-scFV-sfGFP and flag-SINAPS. The neuron was imaged with time-lapse movie every one minute. The ROI was straightened and showed in Movie S12 and analyzed in Fig. 6A-C. Scale bar: 5 μm
Movie S12
Tracking of TLS in neuron Analysis of the TLSs in selected ROI shown in Movie S11. Top: original movie; middle: inverted and the TLS tracked; bottom: the integrated intensities of tracked translation sites in the middle panel. Scale bar: 5 μm
Movie S13
An example of TLS scanning along the dendrite Primary hippocampal neurons were infected with OsTIR1-IRES-scFV-sfGFP and flag-SINAPS. Top: original movie; middle: inverted and the TLS tracked; bottom: the integrated intensities of tracked translation sites in the middle panel. Scale bar: 5 μm
Movie S14
Constitutive translating mRNAs in neuron A constitutively translating mRNA in primary hippocampal neurons infected with OsTIR1-IRES-scFV-sfGFP and flag-SINAPS. Top: original movie; middle: inverted and the TLS tracked; bottom: the integrated intensity of tracked translation site in the middle panel. Scale bar: 5 μm
Movie S2
A selected portion of the cell shown in Movie S1 and Figs. 2B-D Left: mRNA, middle: TLS and free protein, right: merge. Scale Bar: 2 μm. The red circles were tracked mRNA. The green square was tracked TLS. In the movie, we showed the tracks of one translating mRNA and one untranslating mRNA.
Movie S3
Translation sites disappeared in the presence of puromycin U2OS Cell stably expressing scFV-sfGFP, OsTIR1, stdMCP-Halo and flag-SINAPS was streamed at 50ms for 200 frames in the presence of 100μg/mL puromycin. There were no bright green TLS moving together with mRNAs. Green: scFV-sfGFP, Red: stdMCP-Halotag-JF549. Scale Bar: 5μm.
Movie S4
Single CytERM-SINAPS proteins could be tracked U2OS Cell (Fig. S3) stably expressing scFV-sfGFP, OsTIR1 and CytERM-SINAPS was streamed at 50ms for 60 frames. Single CytERM-SINAPS protein (green circle) on the ER membrane could be tracked. The translation site (Green square) was confined and much brighter. Scale bar: 2 μm.
ACKNOWLEDGMENTS
We thank X. Meng for cloning some of the constructs and Western blotting, E. L. Snapp for providing the oxBFP plasmid and the sequence of CytERM, E. Tutucci for constructive discussion and suggestions, and L. Lavis for providing JF dyes Halotag. The authors thank T. Morisaki et al. for sharing their preliminary data and the Transcription Imaging Consortium at the Janelia Research Campus of the HHMI for creating the environment for our interactions and discussions. This work was supported by NIH grant NS083085 to R.H.S.
REFERENCES AND NOTES
- 1.Schwanhäusser B, et al. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 2.Sonenberg N, Hinnebusch AG. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holt CE, Schuman EM. Neuron. 2013;80:648–657. doi: 10.1016/j.neuron.2013.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buxbaum AR, Haimovich G, Singer RH. Nat. Rev. Mol. Cell Biol. 2015;16:95–109. doi: 10.1038/nrm3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larson DR, Zenklusen D, Wu B, Chao JA, Singer RH. Science. 2011;332:475–478. doi: 10.1126/science.1202142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jan CH, Williams CC, Weissman JS. Science. 2014;346:1257521. doi: 10.1126/science.1257521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu B, Buxbaum AR, Katz ZB, Yoon YJ, Singer RH. Cell. 2015;162:211–220. doi: 10.1016/j.cell.2015.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dieterich DC, et al. Nat. Neurosci. 2010;13:897–905. doi: 10.1038/nn.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Starck SR, Green HM, Alberola-Ila J, Roberts RW. Chem. Biol. 2004;11:999–1008. doi: 10.1016/j.chembiol.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 11.Aakalu G, Smith WB, Nguyen N, Jiang C, Schuman EM. Neuron. 2001;30:489–502. doi: 10.1016/s0896-6273(01)00295-1. [DOI] [PubMed] [Google Scholar]
- 12.Wang DO, et al. Science. 2009;324:1536–1540. doi: 10.1126/science.1173205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butko MT, et al. Nat. Neurosci. 2012;15:1742–1751. doi: 10.1038/nn.3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taniguchi Y, et al. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halstead JM, et al. Science. 2015;347:1367–1671. doi: 10.1126/science.aaa3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD. Cell. 2014;159:635–646. doi: 10.1016/j.cell.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pédelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Nat. Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 18.Bertrand E, et al. Mol. Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 19.Holland AJ, Fachinetti D, Han JS, Cleveland DW. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E3350–E3357. doi: 10.1073/pnas.1216880109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. Nat. Methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- 21.Costantini LM, Fossati M, Francolini M, Snapp EL. Traffic. 2012;13:643–649. doi: 10.1111/j.1600-0854.2012.01336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu B, et al. Genes Dev. 2015;29:876–886. doi: 10.1101/gad.259358.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Materials and methods are available as supplementary materials on Science Online.
- 24.Los GV, et al. ACS Chem. Biol. 2008;3:373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 25.Grimm JB, et al. Nat. Methods. 12:244–250. doi: 10.1038/nmeth.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]; 2015;3:250. [Google Scholar]
- 26.Wu B, Chao JA, Singer RH. Biophys. J. 2012;102:2936–2944. doi: 10.1016/j.bpj.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grünwald D, Singer RH. Nature. 2010;467:604–607. doi: 10.1038/nature09438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lionnet T, et al. Nat. Methods. 2011;8:165–170. doi: 10.1038/nmeth.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jaqaman K, et al. Nat. Methods. 2008;5:695–702. doi: 10.1038/nmeth.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rapoport TA. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- 31.Ingolia NT, Lareau LF, Weissman JS. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jung H, Gkogkas CG, Sonenberg N, Holt CE. Cell. 2014;157:26–40. doi: 10.1016/j.cell.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buxbaum AR, Wu B, Singer RH. Science. 2014;343:419–422. doi: 10.1126/science.1242939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doyle M, Kiebler MA. EMBO J. 2011;30:3540–3552. doi: 10.1038/emboj.2011.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wörn A, et al. J. Biol. Chem. 2000;275:2795–2803. doi: 10.1074/jbc.275.4.2795. [DOI] [PubMed] [Google Scholar]
- 36.Jan CH, Williams CC, Weissman JS. Science. 2015;348:1217. doi: 10.1126/science.aaa8299. [DOI] [PubMed] [Google Scholar]
- 37.Reid DW, Nicchitta CV. Nat. Rev. Mol. Cell Biol. 2015;16:221–231. doi: 10.1038/nrm3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reid DW, Nicchitta CV. Science. 2015;348:1217. doi: 10.1126/science.aaa7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morisaki T, et al. Science. 2016;352:1425–1429. doi: 10.1126/science.aaf0899. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1
Translation sites and mRNAs of flag-SINAPS were moving together U2OS Cell (Fig. 2A) stably expressing scFV-sfGFP, OsTIR1, stdMCP-Halo and flag-SINAPS was streamed at 50ms for 200 frames. Green: scFV-sfGFP, Red: stdMCP-Halotag-JF549. Scale Bar: 5 μm
Movie S5
Comparing the diffusion of flag-SINAPS and CytERM-SINAPS mRNAs U2OS cells stably expressing stdMCP-stdGFP were transiently transfected with flag-SINAPS (left panel) or CytERM-SINAPS (middle and right panels) constructs. All mRNAs were labeled with stdMCP-stdGFP (22). Most of cytoplasmic flag-SINAPS were freely diffusing (left). In contrast, CytERM-SINAPS mRNAs were mostly confined (middle). However, when treated with puromycin, most CytERM-SINAPS mRNAs became freely diffusing (right), very similar to the flag-SINAPS mRNAs (left). Note the mRNAs were labeled by stdMCP-stdGFP and scFV-sfGFP was not expressed in the cells. The white ovals were draw to isolate nuclei regions. The yellow boxes drawn in each panel were arbitrary regions in the cytoplasm and meant to focus the attention. Scale bar: 5μm.
Movie S6
CytERM-SINAPS TLS were confined, while non-translating mRNAs were freely diffusion U2OS Cell (Fig. 3) stably expressing scFV-sfGFP, OsTIR1, stdMCP-Halo and CytERM-SINAPS was streamed at 50ms for 200 frames. Green: scFV-sfGFP, Red: stdMCP-Halotag-JF549. Scale Bar: 5 μm
Movie S7
A selected portion of the cell shown in Movie S6 and Figs. 3D-F Left: mRNA, middle: TLS and free proteins, right: merge. Scale Bar 2 μm. The red circles were tracked mRNAs. The green square was a tracked TLS. In the movie, we showed the tracks of one translating mRNA and one untranslating mRNA. The translating mRNA was confined and the untranslating mRNA was freely diffusing.
Movie S8
CytERM-SINAPS TLS disappeared and mRNAs freely diffused in the presence puromycin U2OS Cell stably expressing scFV-sfGFP, OsTIR1, stdMCP-Halo and CytERM-SINAPS was streamed at 50ms for 200 frames in the presence of 100μg/mL puromycin. There were no bright green translation sites moving together with mRNAs. In addition, all mRNAs were freely diffusing instead of confined. Green: scFV-sfGFP, Red: stdMCP-Halotag-JF549. Scale Bar: 2μm.
Movie S9
FRAP of a single TLS: The overview of the whole cell After two pre-bleaching images, a single selected translation site was bleached before frame 3 in U2OS cells stably expressing scFV-sfGFP, OsTIR1 and CytERM-SINAPS. The fluorescence recovery was monitored with time lapse imaging every 10s. The images were max-projection of z-stacks. Top: the FRAP movie with tracked TLS; bottom: the fluorescence intensity of the bleached TLS. Scale bar: 5 μm.
Movie S10
FRAP of single TLSs: comparison between different conditions Single translation sites were bleached in U2OS cells stably expressing scFV-sfGFP, OsTIR1 and CytERM-SINAPS. Left: control; middle: non-bleached; right: bleached in the presence of CHX. Scale bar: 2μm.
Movie S11
Flag-SINAPS in hippocampal neuron: overview of TLS in the neuron. Primary hippocampal neurons were infected with OsTIR1-IRES-scFV-sfGFP and flag-SINAPS. The neuron was imaged with time-lapse movie every one minute. The ROI was straightened and showed in Movie S12 and analyzed in Fig. 6A-C. Scale bar: 5 μm
Movie S12
Tracking of TLS in neuron Analysis of the TLSs in selected ROI shown in Movie S11. Top: original movie; middle: inverted and the TLS tracked; bottom: the integrated intensities of tracked translation sites in the middle panel. Scale bar: 5 μm
Movie S13
An example of TLS scanning along the dendrite Primary hippocampal neurons were infected with OsTIR1-IRES-scFV-sfGFP and flag-SINAPS. Top: original movie; middle: inverted and the TLS tracked; bottom: the integrated intensities of tracked translation sites in the middle panel. Scale bar: 5 μm
Movie S14
Constitutive translating mRNAs in neuron A constitutively translating mRNA in primary hippocampal neurons infected with OsTIR1-IRES-scFV-sfGFP and flag-SINAPS. Top: original movie; middle: inverted and the TLS tracked; bottom: the integrated intensity of tracked translation site in the middle panel. Scale bar: 5 μm
Movie S2
A selected portion of the cell shown in Movie S1 and Figs. 2B-D Left: mRNA, middle: TLS and free protein, right: merge. Scale Bar: 2 μm. The red circles were tracked mRNA. The green square was tracked TLS. In the movie, we showed the tracks of one translating mRNA and one untranslating mRNA.
Movie S3
Translation sites disappeared in the presence of puromycin U2OS Cell stably expressing scFV-sfGFP, OsTIR1, stdMCP-Halo and flag-SINAPS was streamed at 50ms for 200 frames in the presence of 100μg/mL puromycin. There were no bright green TLS moving together with mRNAs. Green: scFV-sfGFP, Red: stdMCP-Halotag-JF549. Scale Bar: 5μm.
Movie S4
Single CytERM-SINAPS proteins could be tracked U2OS Cell (Fig. S3) stably expressing scFV-sfGFP, OsTIR1 and CytERM-SINAPS was streamed at 50ms for 60 frames. Single CytERM-SINAPS protein (green circle) on the ER membrane could be tracked. The translation site (Green square) was confined and much brighter. Scale bar: 2 μm.