Abstract
The nuclear envelope protein lamin A is encoded by the lamin A/C (LMNA) gene, which can contain missense mutations that cause Emery-Dreifuss muscular dystrophy (EDMD) (p.R453W). We fused mutated forms of the lamin A protein to bacterial DNA adenine methyltransferase (Dam) to define euchromatic-heterochromatin (epigenomic) transitions at the nuclear envelope during myogenesis (using DamID-seq). Lamin A missense mutations disrupted appropriate formation of lamin A–associated heterochromatin domains in an allele-specific manner—findings that were confirmed by chromatin immunoprecipitation–DNA sequencing (ChIP-seq) in murine H2K cells and DNA methylation studies in fibroblasts from muscular dystrophy patient who carried a distinct LMNA mutation (p.H222P). Observed perturbations of the epigenomic transitions included exit from pluripotency and cell cycle programs [euchromatin (open, transcribed) to heterochromatin (closed, silent)], as well as induction of myogenic loci (heterochromatin to euchromatin). In muscle biopsies from patients with either a gain- or change-of-function LMNA gene mutation or a loss-of-function mutation in the emerin gene, both of which cause EDMD, we observed inappropriate loss of heterochromatin formation at the Sox2 pluripotency locus, which was associated with persistent mRNA expression of Sox2. Overexpression of Sox2 inhibited myogenic differentiation in human immortalized myoblasts. Our findings suggest that nuclear envelopathies are disorders of developmental epigenetic programming that result from altered formation of lamina-associated domains.
INTRODUCTION
Genetic and biochemical perturbations of components of the nuclear envelope cause a broad range of clinical phenotypes, including muscular dystrophies, neuropathies, cardiomyopathies, and lipodystrophies (1). Dominant missense mutations in the lamin A/C gene (LMNA), which encodes the lamin A and lamin C intermediate filament proteins—scaffolding components that strengthen the nuclear envelope—cause the broadest range of phenotypes, and even neighboring mutations can show quite distinct tissue-restricted clinical disease. Skeletal muscle appears particularly susceptible to abnormalities of the nuclear envelope, with LMNA mutations causing both Emery-Dreifuss muscular dystrophy (EDMD) and limb-girdle muscular dystrophy (LGMD). Additional muscular dystrophy phenocopies are caused by mutations in genes that encode other nuclear envelope proteins, including emerin (encoded by the EMD gene), nesprin 1 (SYNE1), nesprin 2 (SYNE2), and four and a half LIM domain protein 1 (FHL1) (2).
Myogenic differentiation requires the coordinated execution of three key cellular programs: inactivation of pluripotency programs (that is, the Oct4-Nanog-Sox2 program), exit from the cell cycle [cell cycle regulatory proteins cyclin-dependent kinase 1 (CDK1) and the tumor-suppressor retinoblastoma 1 (Rb1)], and induction of myogenesis (by master transcriptional regulators MyoD and myogenin). In differentiating skeletal muscle cells, the LMNA gene is strongly induced at the onset of terminal differentiation of myoblasts (myogenic cells that either divide or differentiate to form muscle fibers) into syncytial myotubes (multinucleated fibers formed from fused myoblasts) (3). The lamin A/C protein has also been shown to play a key role in the myoblast-myotube transition (4, 5). Missense mutations that form the aberrant lamin A protein associated with muscular dystrophy cause impaired activation of the gene (MYOG) encoding myogenic factor 4, which drives inappropriate maintenance of heterochromatin at the MYOG promoter and downstream effects of the encoded protein (myogenin) on myogenesis in culture (6). The importance of wild-type lamin A/C for terminal differentiation is also seen in adipogenic cells, in which a lamin A protein with a familial partial lipodystrophy (FPLD) missense mutation disrupts adipocyte differentiation (7). The ability of exogenously expressed FPLD lamin A in 3T3-L1 cells (an adipose-like cell line) to inhibit adipogenesis in culture is controversial (8) and needs additional study. Together, these data suggest that laminopathies disrupt cell differentiation, but the mechanism of this disruption is not understood.
Some dominant gain- or change-of-function mutations in the lamin A/C proteins show similar muscular dystrophy phenotypes as those observed with loss of function of other nuclear envelope components (emerin, nesprin 1, and FHL1); thus, molecular pathogenesis might also be shared between these genetically distinct nuclear envelopathies. Consistent with this hypothesis, we previously showed shared mRNA expression fingerprints in muscle biopsies from EDMD patients with both LMNA [autosomal dominant form of EDMD (EDMD-AD)] and EMD (emerin) mutations [X-linked recessive form of EDMD (EDMD-XR)], suggesting a failure of appropriate induction of the myogenic terminal differentiation program (3). Mechanistic studies bolstering these findings were carried out in a mouse knockout model of emerin deficiency, in which the inappropriate timing of myogenic lineage–specific genes was confirmed (9). This study also showed evidence of failure to appropriately exit from the cell cycle (delayed suppression of the E2F transcription factor pathways) (9). Thus, exit from the cell cycle and induction of the myogenic cell fate program—two of three key programs that must be orchestrated during terminal differentiation and commitment to the myogenic lineage—are perturbed in emerin deficiency.
There is accumulating evidence that lamin A protein is directly involved in epigenomic regulation of chromatin through heterochromatic lamina-associated domains (LADs). The nuclear lamina is a fibrillar network of intermediate filaments and membrane-associated proteins that lies along and interacts with the inner nuclear membrane and DNA, and provides physical support and regulates DNA replication, transcription, and cell division. Genomic regions associated with nuclear lamina show enrichment in repressive heterochromatin marks and are associated with transcriptionally inactive domains (10). Transcriptional repositioning of silent genes to the nuclear periphery (11), together with sequestration of transcriptional factors by the interior of the nucleus (12), indicates that the nuclear lamina controls transcription and, likely, cell fate. Consistent with this hypothesis, murine Lmna-null cells fail to acquire appropriate heterochromatin region of the mouse genome at the nuclear envelope during myogenesis and show a loss of the myogenic program (13). Also, a human EDMD LMNA missense mutation expressed as a protein in Caenorhabditis elegans impairs tissue-specific reorganization of heterochromatin, with abnormal retention of a muscle-specific, transcriptionally silent gene at the nuclear periphery (14).
We hypothesized that different LMNA missense mutations alter the euchromatin-heterochromatin transitions during terminal differentiation of cells mediated by the nuclear envelope in a tissue-restricted manner. That is, nuclear envelopathies are epigenetic disorders. We assessed this at a genome-wide level by fusing the lamin A protein carrying various missense mutations to the bacterial DNA adenine methyltransferase (Dam) and used the DamID approach, which identifies DNA adenine methylation of the genomic regions that are transiently bound to or in close proximity to a fusion protein (15, 16). The resolution of the DamID approach is about 1 kb, or tens of nanometers from the tethered protein. In Drosophila, a DamID microarray approach was successfully used to assess euchromatin-heterochromatin transitions for the Escargot protein in intestinal cell development (17) and Hb9-lamina interactions in neuronal lineages (18). Here, we extended the DamID method to study gain- or change-of-function mutations in the lamin A protein, with next-generation DNA sequencing as the readout of the point of DNA binding (DamID-seq). Findings were validated with chromatin immunoprecipitation–DNA sequencing (ChIP-seq) and DNA methylation arrays.
RESULTS
LMNA mutations alter genomic LADs in human muscle cell nuclei
We generated lentiviral constructs that express the wild-type lamin A, the lamin A p.R453W mutation (which corresponds to EDMD) (19), or the lamin A p.R482W mutation [observed in FPLD (20)] fused to Dam (bacterial adenine methyltransferase) and transduced them into human myogenic cells. A DamOnly lentivirus was used as control. The Dam–lamin A fusion proteins showed the expected molecular weights and subcellular localization restricted to the nuclear envelope (fig. S1). Infections were done 24 hours after switching the cells to differentiation media, and genomic material was harvested 48 hours later (3 days after differentiation), when most of the cells had differentiated into multinucleate myotubes. DamID-seq experiments were performed in duplicate for each Dam-LMNA construct and the negative control (DamOnly). Library sizes and sequencing read depth were comparable between samples (~30 million reads), and >80% of reads started with the expected GATC sequence of the Dam adenine methylase recognition site. After sequencing, reads were aligned to a reference human genome (hg19), and domains of enrichment were called using SICER (spatial clustering approach for the identification of ChIP-enriched region) algorithm (31). Comparison of wild-type Dam-LMNA peaks with human myotube histone marks [ENCODE data (H3K9me3, GSM733730; H3K4me3, GSM733637)] showed a strong positive correlation with the heterochromatic histone mark H3K9me3 and anticorrelation with the euchromatic mark H3K4me3 (fig. S2). These data are consistent with the lamin A/C protein associated with heterochromatic regions, namely, LADs.
Genomic mapping of LADs showed wild-type lamin A associated with ~11,000 genomic domains, with an average domain length of 32 kb (~12% of the genome) (table S1). Both EDMD (p.R453W) and FPLD (p.R482W) lamin A mutations revealed a significant increase in the number of LADs, with nearly twice as many genomic loci labeled by DamID compared to wild type (table S1 and Fig. 1A). Comparative analysis of genes associated with LADs in human myogenic cells expressing one of the three lamin A variants showed that most of the LADs associated with wild-type lamin A were shared with one or both mutant constructs (1239 of 1960 LADs; 63%) (Fig. 1A). However, promiscuous binding of mutant lamin A to new chromatin sites not normally bound by wild-type lamin A led to a preponderance of mutation-specific LADs (promiscuous LADs). For p.R453W (EDMD), 69% of LADs (2397 of 3477) were not shared with wild type and 32% (1115 of 3477) were specific to the mutation. For p.R482W (FPLD), 80% (3996 of 5011) was not shared with wild type and 54% (2715 of 5011) was specific to the mutation (Fig. 1A). Wild-type LADs were larger (32.5-kb mean size) than mutant LADs (22.3 kb for lamin A–EDMD and 19.9 kb for lamin A–FPLD; P < 0.001 versus wild type; table S1). These data showed that disease-associated missense mutations cause allele-specific alterations of chromatin association with the nuclear envelope.
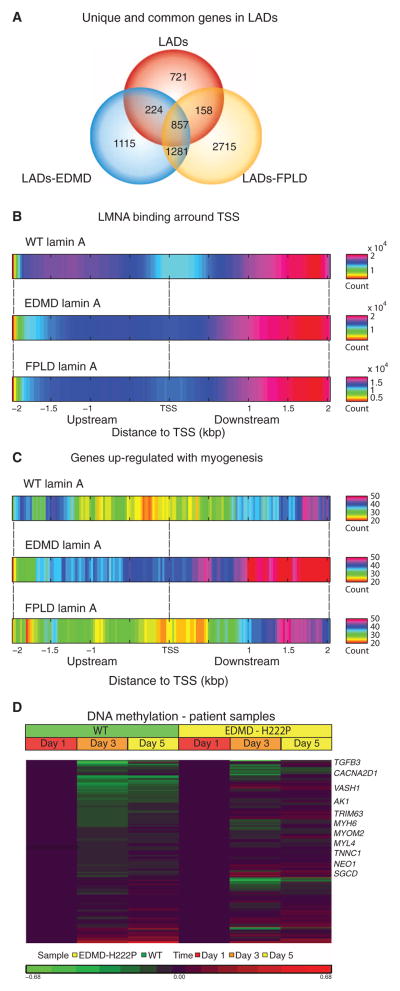
Fig. 1. Promiscuous formation of short LADs by mutation-bearing lamin A/C proteins is associated with allele-specific perturbation of myogenic programs.
Interactions between lamin A nuclear envelope protein and chromatin domains were assessed by Dam–lamin A fusion protein using DamID-seq [wild-type (WT), p.R453W EDMD, and p.R482W FPLD] in differentiating human myoblasts. LADs were scored and compared between the three lamin A/C variants. (A) Overlap in LADs (and genes found in LADs) among the three variants. Mutant lamin A proteins showed promiscuous formation of new LADs that were of shorter length compared to WT (fig. S1). (B) WT lamin A showed loss of LADs at TSS, whereas mutant lamin A showed abnormal increased association with start sites. WT and mutations in lamin A showed most binding downstream of TSS. (C) Restriction of analysis to transcript units associated with myogenic differentiation showed broad loss of lamin A binding with WT lamin A, consistent with euchromatinization of myogenic loci at the myotube stage. Lamin A containing an EDMD mutation (p.R453W) showed persistent heterochromatinization of myogenic loci, whereas p.R482W FPLD mutation retained appropriate loss of LADs similar to WT. (D) DNA methylation status of myogenesis-associated loci (n = 322) in fibroblasts from EDMD patients [carrying the LMNA mutation (p.H222P)] transfected with an MyoD expression vector shows expected loss in methylation similar to cells from control subjects. Genes involved in skeletal and cardiac muscle development that showed persistent methylation are marked at the left.
EDMD mutations show allele-specific alteration of epigenomic programming of myogenic differentiation
To focus on the potential effects of lamin A mutations on gene transcription, we evaluated lamin A gene interactions that occurred at regions encompassing the transcription start site (TSS) of genes associated with the LADs. Using 25–base pair (bp) windows, wild-type lamin A showed a selective lack of binding ~200 bp in length around the TSS and strong enrichment 1 to 2 kb downstream of the TSS (Fig. 1B). Mutant lamin A LADs did not show the same loss of binding within ~200 bp around the TSS but did show enrichment 1 to 2 kb downstream. This indicates that disease-causing lamin A/C mutations cause abnormally increased TSS-specific nuclear envelope–chromatin interactions.
We had previously shown delayed induction of some myogenesis-associated transcripts in emerin-null cells (9) and had hypothesized that this could result from a relative failure in heterochromatin-euchromatin conversion at these loci during myogenic development. To test this notion using the DamID-seq data, we selected mRNA transcripts that are significantly induced by the transition from myoblasts to myotubes [one-way analysis of variance (ANOVA), P <0.01; FC (fold change) > 1.2] (table S2) and then mapped these transcripts (n = 322). As hypothesized, wild-type lamin A showed marginal binding near the TSS of these myogenesis-related transcripts, consistent with euchromatinization of these loci and high expression in myotubes (Fig. 1C). In contrast, EDMD lamin A showed pervasive heterochromatinization of these loci. This effect was allele-specific; FPLD lamin A association with myogenic genes was more similar to wild type (Fig. 1C). These data are consistent with an allele-specific effect of LMNA gene mutations on the muscle developmental program and are in accordance with the specific clinical muscular phenotype associated with EDMD (p.R453W).
These data predicted that the genomic loci of genes involved in terminal differentiation might fail to move from the nuclear periphery (heterochromatin) to central nuclear locations (euchromatin). To test this, fluorescence in situ hybridization (FISH) analysis was carried out with the MYOG genomic locus in MyoD-converted human fibroblasts from EDMD-AD patients during myogenic differentiation induced in culture by serum starvation (fig. S3). Visualization of the MYOG locus showed retention at the nuclear periphery in p.H222P EDMD cells, whereas wild-type lamin A showed the expected central nuclear location consistent with euchromatinization [nuclear distance for wild type (n = 16 nuclei) versus p.H22P (n = 13 nuclei); P = 0.05, Student’s t test].
To extend our observations to patients affected by lamin A laminopathies, fibroblasts were obtained from an EDMD patient harboring a different heterozygous LMNA mutation (p.H222P) as well as control subjects. Cells were transfected with a doxycycline-inducible MyoD expression construct to force conversion to the myogenic lineage and induced the fibroblasts to differentiate into myotubes (fig. S4). EDMD and wild-type cells were harvested at 1, 3, and 5 days after differentiation, and genome-wide methylation patterns were detected by Illumina arrays (Infinium HumanMethylation450). The myogenesis-associated transcripts (n = 322; n = 210 were present in methylation set) were investigated for changes in methylation patterns as a function of myogenic differentiation normalized to day 1 (d1) (Fig. 1D). These data showed the predicted loss of DNA methylation (associated with transcriptional silencing) as a function of myogenic differentiation in wild-type cells, consistent with the DamID data presented above. In contrast, and similar to the DamID data, the EDMD cells showed persistent DNA methylation at many of these loci (81 of 210 genes in EDMD at d3 and 84 of 210 at d5 of differentiation; table S3), indicating a perturbation in the differentiation program, although cells still formed myotubes by visual inspection (Fig. 1D and fig. S4). These results were confirmed using human skeletal myoblasts/myotubes infected with LMNA constructs carrying the p. R453W (EDMD) and p.R482W (FPLD) mutations as well as the wild-type LMNA gene (fig. S5).
Delayed E2F cell cycle suppression is shared by lamin A (EDMD-AD) and emerin-null (EDMD-XR) myogenic cells
The EDMD phenotype results from AD missense mutations in LMNA (above; EDMD-AD) as well as loss of function of the lamin-associated emerin protein (EDMD-XR). We previously showed similarities in mRNA expression profiles (of genes related to the transition to myogenic terminal differentiation) between muscle biopsies from EDMD-AD and EDMD-XR patients (3). In emerin-null cells, we showed relative inability to exit from the cell cycle through the shutoff of E2F pathways (3, 9). We queried the DamID-seq data from wild-type and mutant lamin A (p.R453W EDMD-associated, p.482W FPLD-associated) proteins to determine whether key E2F pathway members, CDK1 and RB, showed abnormal regulation of chromatin (Fig. 2A). Consistent with our previously published mRNA and protein data in emerin-null cells (9), EDMD-mutated lamin A protein showed reduced association with the CDK1 and RB loci; FPLD lamin A data were more variable (Fig. 2C). To further investigate the effect of LMNA mutations on genes previously implicated in EDMD pathology, we focused our analysis on DNA methylation marks on cell cycle regulatory and myogenic differentiation genes. The RB1, HDAC1, CHRNG, and MCM3 (cell cycle genes) were differentially methylated in EDMD and showed decreased methylation levels compared to wild type and FPLD (Fig. 2H). Furthermore, in EDMD muscle cells, the gene encoding the master regulator of muscle differentiation, MYOD1, was persistently methylated and showed statistically higher levels of methylation relative to wild type and FPLD (Fig. 2H and table S4).
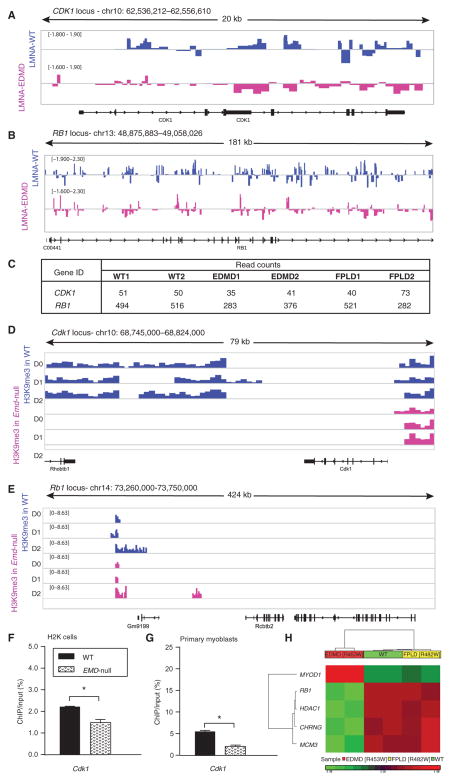
Fig. 2. Cell cycle loci show decreased association with EDMD lamin A and inadequate heterochromatin formation in differentiating EDMD myogenic cell.
Lamina association of cell cycle loci assessed by DamID-seq showed reduction in binding of mutated lamin A, which resulted in reduced heterochromatinization of these loci as assessed by ChIP-seq in emerin-null murine myogenic cells (H2K cells). (A and B) Wiggle track format (Wig) of lamin A enrichment on two cell cycle genes, (A) CDK1 and (B) RB1. Data are presented as log2 of DamLMNA-associated genomic regions/DamOnly genomic regions. (C) Normalized read counts of lamin A peaks at the CDK1 and RB1 loci. (D and E) Wig tracks of H3K9me3 enrichment at Cdk1 (D) and Rb1 (E) loci. (F and G) ChIP-qPCR validation of H3K9me3 enrichment at the Cdk1 locus in WT emerin and emerin-null (EMD-null) (F) murine H2K cells and (G) primary myoblasts from extensor digitorum longus (EDL) murine muscle. Data are means ± SEM. (H) DNA methylation analysis of human skeletal muscle cells infected with lentivirus encoding the WT LMNA gene or LMNA genes with one of two different mutations [WT, EDMD (p.R453W), and FPLD (p.R482W)] focused on genes previously implicated in EDMD pathology (3). Heat map of genes that show statistically significant differences (±1.2 FC; P < 0.05, FDR-corrected) between WT and EDMD and WT and FPLD cells. chr, chromosome.
To integrate the lamin A protein data (DamID) for additional epigenetic marks, we carried out ChIP-seq for H3K9me3 at three time points during myogenic differentiation of wild-type and emerin-null murine H2K cells. H3K9me3 peak mapping relative to known start sites of transcription units showed similar distribution patterns in wild-type and emerin-null cells, whereas most of the heterochromatic H3K9me3 peaks mapped to intergenic (noncoding) regions of the genome, 50 to 500 Mb away from the nearest gene TSS. Analysis of loci near TTS (±10 kb from TSS) showed that all samples displayed a relative decrease in sequence tag density compared to intergenic and intronic regions (fig. S6). We queried the Cdk1 and Rb1 loci to determine whether the DamID data correlated with heterochromatic marks. H3K9me3 was enriched in the vicinity of the Cdk1 and Rb1 loci in wild-type cells but was absent in emerin-null cells in the vicinity of Cdk1 or failed to show increased enrichment as a function of myogenic differentiation in the vicinity of Rb1, as seen in wild-type cells (Fig. 2B). However, these heterochromatic marks were quite far downstream of the transcript unit (10 kb from Cdk1 and 200 kb from Rb1). Thus, H3K9me3 and DamID marks were consistent but not conclusive of a direct effect of decreased heterochromatin enrichment on cell cycle genes in EDMD. To confirm these findings, ChIP polymerase chain reaction (PCR) was performed in both murine H2K and primary myoblast cells (Fig. 2C), which showed significant reduction in H3K9me3 enrichment at the Cdk1 promoter. These data were consistent with a delayed exit from the cell cycle, as seen in myogenic cells in muscle biopsies from patients with either EDMD-AD or EDMD-XR. This abnormal hetero-chromatinization of key cell cycle gene loci suggests that exit from the cell cycle is likely perturbed by nuclear envelop mutations.
Genome-wide prioritization of ChIP-seq alterations identifies aberrant persistence of pluripotency programs in emerin-null cells
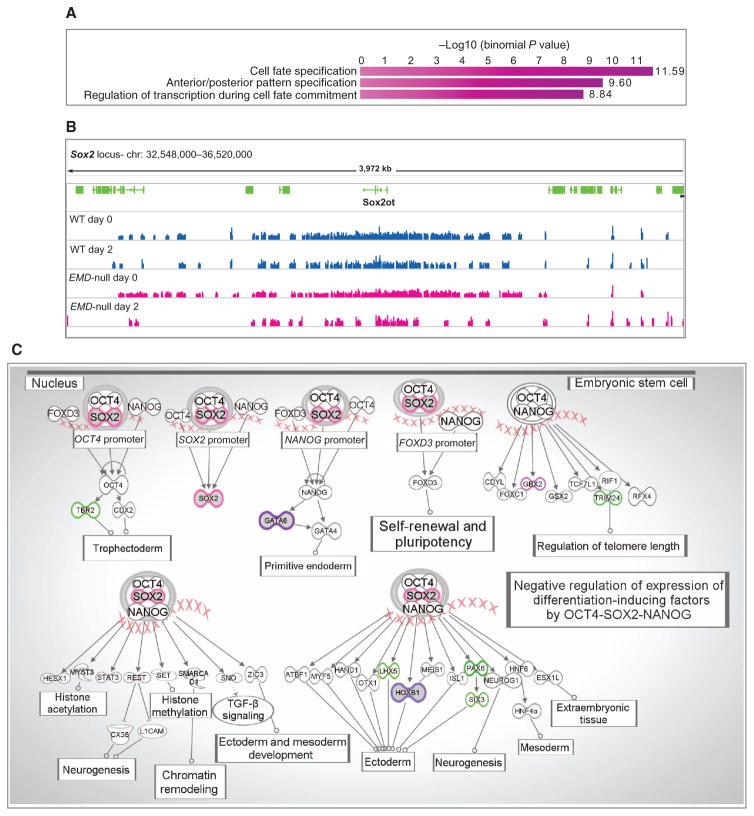
We then carried out a genome-wide assessment of changes in H3K9me3 heterochromatin marks during myogenic differentiation in emerin-null versus wild-type H2K cells. SICER-called peaks were intersected and annotated using BEDTools suite to obtain (i) normalized read counts of each peak for both conditions at all time points, (ii) cell type–specific peaks, and (iii) expanded heterochromatic islands during the d0 to d1 and d1 to d2 transitions. Using BEDTools suite, we generated a list of peaks that showed heterochromatic decrease in enrichment in emerin-null sample at d0 and d1 (myoblast-myotube transition). H3K9me3 ChIP-seq peaks were then analyzed using the GREAT web application (33) for genomic annotation of cis regulatory elements. The analysis revealed that loci involved in cell fate specification and transcriptional regulation of cell fate commitment, in particular Sox2 and its downstream targets (Gata6, Gbx2, Trim24, Hoxb1, Pax6, and others), showed loss of heterochromatin marks in emerin-null cells relative to wild type (Fig. 3 and table S5). Visualization of the enrichment around the Sox2 locus in emerin-null (pink) samples showed significant loss of heterochromatic H3K9me3 peaks at d2 relative to both emerin-null d0 and wild-type samples [d0: FC = −1.2 and FDR (false discovery rate) = 0.04: d1; FC = −1.3 and FDR = 0.02; d2: FC = −1.6 and FDR = 1.72 × 10−7).
Fig. 3. Decreased or absent H3K9m3 enrichment in murine emerin-null H2K cells is associated with genes known to be involved in cell fate regulation and transcriptional regulation in embryonic stem cells.
(A) Top 3 phase G0 biological processes associated with decreased H3K9me3 in emerin-null samples (GREAT web application). GREAT calculates the statistical enrichment (P value) for genomic regions based on ChIP-seq data and annotations from numerous ontologies. (B) Wig tracks of ChIP-seq data around the SOX2 locus. (C) Top canonical pathway generated using genes associated with a decrease (purple) or absence (green) of H3K9me3 enrichment in emerin-null H2K cells (Ingenuity Pathway Analysis). Pink x’s are a cartoon representation of gDNA. TGF-β, transforming growth factor–β.
Heterochromatic areas are increasingly recognized to expand along DNA during terminal differentiation of cells (21). We hypothesized that the nuclear envelope may serve a role in heterochromatic expansion along chromatin and that the process of loci expansion can be perturbed in emerin-null H2K cells. To test this hypothesis, we analyzed the expansion of heterochromatic loci as a function of myogenic differentiation (d0 to d2) using ChIP-seq data (Fig. 4). Proximal promoter analysis (−1000 bp and +200 bp) identified 1163 peaks in wild-type cells, 474 in emerin-null cells, and 390 peaks shared between the two conditions.
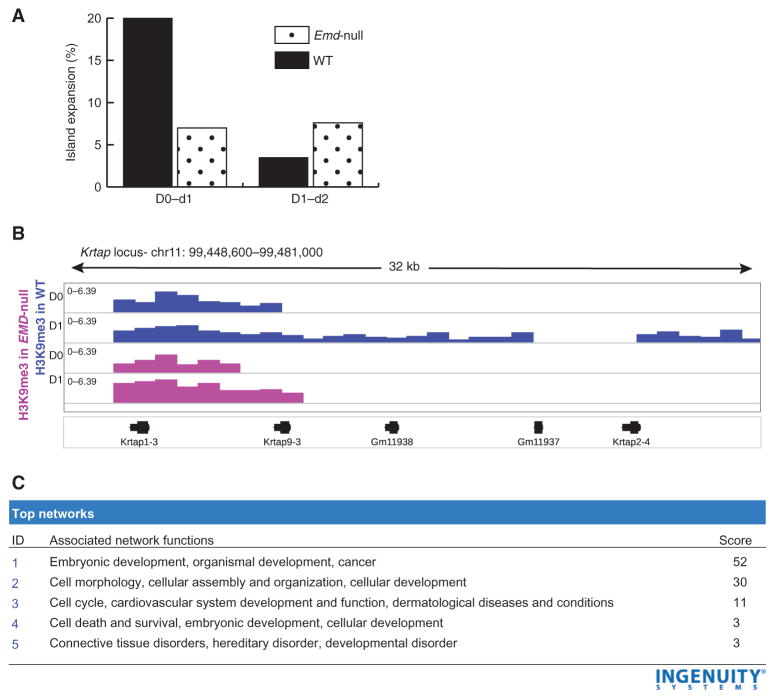
Fig. 4. Emerin-null cells show aberrant heterochromatin spreading during myogenic differentiation.
Heterochromatic expansion was studied by comparing ChIP-seq data between time points during myogenic differentiation in both WT and emerin-null H2K cells. (A) The percentage of heterochromatic gene promoter loci showing expansion was calculated between d0 to d1 and d1 to d2. In WT cells, 20% of loci showed expansion early in differentiation (d0 to d1); however, this rate slowed quickly to 4% (d1 to d2). Emerin-null cells showed a loss of these differentiation-specified transitions in heterochromatic expansion. (B) Visualization of ChIP-seq data for an exemplar locus (Krtap, keratin-associated proteins; Gm11938, predicted gene 11938; Gm11937, predicted gene 11937). H3K9me3 peaks in WT cells are shown in blue, and Emerin-null cells are shown in pink. Emerin-null cells show only marginal expansion (2 kb) between d0 and d1, whereas WT cells show a 12-kb expansion. (C) Gene ontology analysis of differentially expanded loci in WT versus emerin-null cells showed enrichment for genes involved in embryonic development.
Next, we sought to identify regions in which heterochromatin islands increased in length during d0 to d1 or d1 to d2 transition (twofold or more). In wild-type H2K cells, 20% of heterochromatic peaks at gene promoters showed heterochromatic expansion from d0 to d1, whereas only 7% of emerin-null peaks showed this expansion (Fig. 4A). Comparing d1 to d2 data, the expansion of heterochromatic loci in wild-type cells showed a marked slowing (fivefold) from 20 to 4% of loci. On the other hand, emerin-null cells maintained a relatively constant rate of 7% of loci with expansions.
An example of this differential heterochromatic expansion is visualized in Fig. 4B. Wild-type cells showed a 12-kb H3K9me3 expansion of a locus during the first day of differentiation, whereas emerin-null cells showed only a 2-kb expansion. We extracted the differentially expanded loci and entered the corresponding genes into Ingenuity Pathway Analysis. This showed that the differentially expanded heterochromatic loci were predominantly at gene loci involved in embryonic development (Fig. 4C).
Targeted validation shows inappropriate transcriptional up-regulation of Sox2 pluripotency pathways in emerin-null myogenic cells
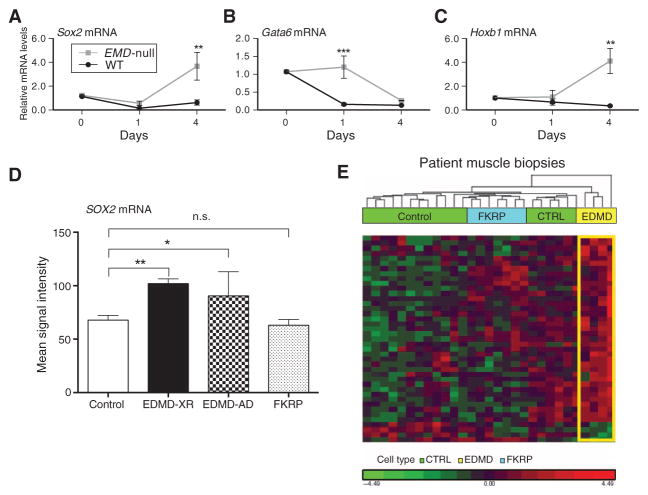
The genome-wide H3K9me3 ChIP-seq data suggested an abnormal gain of function of cell fate pathways in emerin-null cells due to lack of heterochromatin initiation, heterochromatin expansion, or both. One key pathway appeared to be Sox2-Oct4-Nanog (Fig. 3), a well-studied cell fate pathway induced in self-renewal and pluripotency (22). ChIP-seq results were validated in differentiating wild-type and emerin-null murine H2K myogenic cells using quantitative real-time PCR (qRT-PCR) (d0, d1, and d4). Sox2 gene expression showed a marked drop from d0 to d1 in wild-type cells, consistent with greater heterochromatin at the respective gene promoter (Fig. 5A). In contrast, emerin-null cells showed a smaller decrease in Sox2 mRNA expression from d0 to d1, with a fourfold increase from d1 to d4. We also studied two downstream targets of Sox2, Gata6, and Hoxb1, which are important for early development and differentiation of germ layers (Fig. 5, B and C). Gene expression data similarly confirmed ChIP-seq data, in which both Gata6 and Hoxb1 showed inappropriately high expression in emerin-null cells and abnormal patterns of expression change during myogenic differentiation (although not at exactly the same time point). To validate the mRNA findings at the protein level, wild-type and emerin-null H2K cells were studied at three time points after induction of myogenic differentiation (d1, d2, and d4), and Sox2 protein was measured by immunoblot (fig. S7A). Consistent with the mRNA findings, Sox2 protein decreased during differentiation of wild-type cells but increased in emerin-null cells.
Fig. 5. Sox2 and downstream target genes show abnormal gain of function in differentiating emerin-null myogenic cells and in muscle biopsies from EDMD patients.
(A to C) Differentiating WT and emerin-null H2K cells were studied at d0, d1, and d4 after induction of myogenic differentiation. qRT-PCR of (A) Sox2 and two downstream transcriptional target genes [Gata6 (B) and Hoxb1 (C)] showed down-regulation in WT cells, whereas emerin-null cells show abnormally high expression of all three genes. Two-way ANOVA with repeated measures, followed by Bonferroni’s multiple comparisons test, ***P < 0.0005 and **P < 0.005; n = 3. Error bars indicate ± SEM. (D and E) mRNA profiles were accessed (3) by Affymetrix microarrays in muscle biopsies taken from control, EDMD, and FKRP patients [control (CTRL), n = 13; EDMD-XR, n = 4; EDMD-AD, n = 3; FKRP, n = 7]. (D) Affymetrix signal intensity query of Sox2 mRNA showed abnormal up-regulation in both EDMD-XR and EDMD-AD patient muscle relative to control samples. (E) Hierarchical clustering of mRNA transcripts corresponding to H3K9me3 ChIP-seq loci that displayed specific decreases, relative to WT, in heterochromatin in emerin-null H2K myogenic cells (n = 42). The large majority of these murine ChIP-seq loci showed up-regulation of the corresponding mRNA transcripts in EDMD patient muscle biopsies (yellow) relative to control (green) and disease control FKRP (blue). n.s., not significant.
Our data in emerin-null murine myogenic cells suggested that human patients with EDMD would also show aberrant regulation of SOX2 in their muscle. To test this, we accessed our previously published 125 human muscle biopsy mRNA profiling data set, including muscle biopsies from EDMD (both emerin-null and lamin A/C dominant mutations) (table S6). We used RT-PCR of patient muscle biopsy mRNA to compare the steady-state mRNA levels of SOX2 in the EDMD biopsies relative to normal volunteer muscle and a different muscular dystrophy used as disease control [LGMD type 2I (LGMD2I); FKRP missense mutations] (Fig. 5D). SOX2 mRNA showed similar steady-state levels between normal volunteers and disease controls (LGMD2I) but significantly elevated mRNA levels in both lamin (EDMD-AD) and emerin (EDMD-XR) patient muscle.
We then used unsupervised hierarchical clustering to test for the association of additional transcripts with decreased heterochromatin in the human muscle biopsy data set, comparing AD (lamin A/C gain of function) forms with the FKRP/LGMD2I disease control (Fig. 5E and table S7). The results showed that significant up-regulation of multiple SOX2 pathway members together with other ChIP-seq targets was specific for AD lamin A/C patients. These data suggest that the gain of function of SOX2 pathways seen in murine emerin-null myogenic cells is likely shared with human EDMD patients.
Gain of function of SOX2 cell fate perturbations is specific to the disease type of lamin A/C mutation
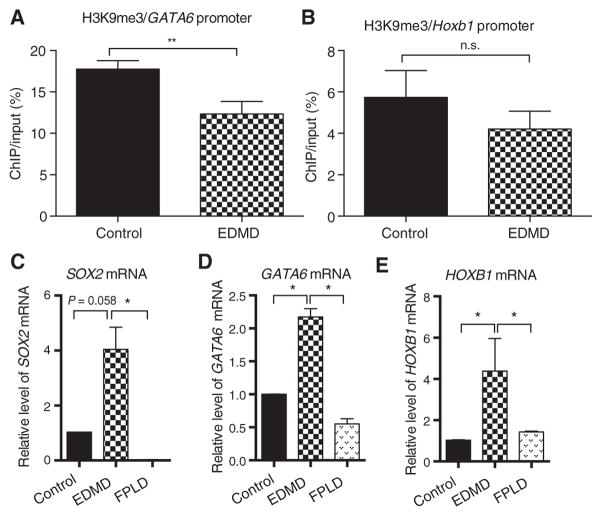
We used the same patient-derived MyoD-converted cells (wild-type, EDMD LMNA p.H222P, and FPLD LMNA p.R482W mutation) described above to carry out targeted validations of SOX2 pluripotency pathway persistence 4 days after triggering myogenesis in vitro. Human SOX2, GATA6, and HOXB1 loci were studied using H3K9me3 ChIP to assess heterochromatin levels at gene promoters, as well as mRNA qRT-PCR for geneexpression (Fig. 6). Cells carrying the EDMD LMNA p.H222P missense mutation showed reductions in heterochromatin at GATA6 and HOXB1 relative to the normal control (Fig. 6, A and B). SOX2 was not tested (failure of qPCR). mRNA transcript levels showed up-regulation relative to the normal control and FPLD cells (Fig. 6, C to E). The mRNA studies were validated at the protein level; SOX2 protein showed abnormal persistent expression in EDMD p.H222P myotubes compared to normal controls (fig. S7B). Data suggest that both EDMD-AD (LMNA missense gain of function) and EDMD-XR (emerin loss of function) share persistence of the SOX2-associated pluripotency pathways resulting from alterations in epigenetic programming. Last, these data are consistent with specific mutations in lamin A/C that cause a differential gain of function of abnormal initiation and spreading of heterochromatin marks, thus altering cell fate in a mutation-specific manner.
Fig. 6. Various lamin A/C mutations that cause either EDMD or FPLD give rise to differential regulation of Sox2 pathways.
Dermal fibroblasts from an EDMD patient carrying a lamin A/C p.H222P dominant mutation, an FPLD p.R482W lamin A/C mutation, and a control volunteer were converted to myogenic lineages, differentiated, and studied for Sox2 pathway gene regulation. (A and B) ChIP for the H3K9me3 heterochromatin mark showed mutation-specific loss of heterochromatin in EDMD for both Gata6 and Hoxb1. (C to E) Steady-state mRNA levels for the (C) Sox2, (D) Gata6, and (E) Hoxb1 genes in differentiated control, EDMD, and FPLD myogenic cells measured by qRT-PCR. For experimental versus control, *P = 0.05 and **P = 0.01. Error bars indicate ±SEM.
Wild-type lamin A protein directly interacts with the SOX2 locus, and this interaction is altered by LMNA missense mutations
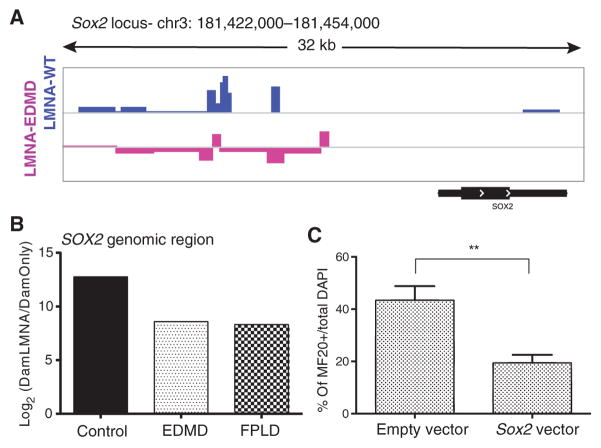
The ChIP-seq and ChIP-qPCR imply that lamin A directly interacts with the SOX2 gene locus and that mutations of lamin A or emerin disrupt this interaction, but this interaction could be mediated by other proteins. To test for direct protein-DNA interactions between wild-type and mutant lamin A proteins with the SOX2 genomic locus, we returned to the DamID method. First, we queried the DamID-seq data presented earlier for associations of lamin A protein with the SOX2 gene locus (Fig. 7A). This revealed wild-type lamin A to show association with the SOX2 upstream region and relative loss of this association with the p.R453W mutation. We then used the DamID methyladenine chromatin from the same myogenic time points to carry out targeted analysis of SOX2 locus enrichment (Fig. 7B). The results showed relative loss of lamin A protein association with the SOX2 locus for both the EDMD (p.R453W) and FPLD (p.R482W) LMNA mutations. Collectively, these data reveal that tethering of the SOX2 genomic region to the nuclear lamina is impaired when LMNA is mutated, as well as in emerin-null cells.
Fig. 7. The LMNA protein causes allele-specific perturbation by direct interaction with the SOX2 genomic locus, and SOX2 overexpression causes inhibitory effects on muscle differentiation.
The direct link between lamin A mutations and the SOX2 locus was studied by DamID-seq and DamID-qPCR using primers specific for SOX2. Effects of SOX2 on myogenic differentiation were assessed by overexpression experiments. (A) Wig tracks of lamin A enrichment at the SOX2 locus. (B) Validation experiment of lamin A enrichment at the SOX2 locus using DamID-qPCR. The analysis compared binding abilities of WT and mutated lamin A proteins at the SOX2 locus and showed decreased enrichment potential for the mutated lamins. (C) SOX2 overexpression significantly (P = 0.02, Student’s t test) reduced differentiation potential of human immortalized myoblasts. Differentiation potential was assessed by the ability of transfected cells [green fluorescent protein (GFP+) to form myosin heavy chain (MF20+)–positive myotubes and was calculated as the ratio of MF20+/total number of transfected myoblasts [GFP+ DAPI (4′,6-diamidino-2-phenylindole) cells]. Error bars indicate ± SEM.
We also confirmed the effects of LMNA mutations on myogenic differentiation. First, we infected human skeletal myogenic cells with various LMNA mutations [wild-type, p.R453W (EDMD), and p.R482W (FPLD)] and differentiated the cells into myotubes. Using a qRT-PCR assay, we assessed the myogenin mRNA levels and showed that the EDMD mutation (LMNA p.R453W) reduced the myogenin levels in differentiating myotubes, whereas the FPLD mutation (LMNA p.R482W) showed reduction in myogenin levels that was not statistically significant when compared to wild-type levels (fig. S8). These data show direct effects of LMNA mutations on muscle differentiation and, more importantly, show allele-specific effects (EDMD versus FPLD mutations).
Overexpression of SOX2 perturbs myogenesis
The epigenetic studies above suggest that three pathways are perturbed by both EDMD LMNA mutations and emerin loss-of-function mutations: exit from pluripotency programs (SOX2), exit from the cell cycle (E2F and Rb1), and induction of myogenesis. These three pathways are likely interrelated and coordinated during myogenic differentiation. To determine whether overexpression of SOX2 was sufficient to perturb myogenesis, SOX2 overexpression in normal human myoblasts was enhanced by transfection with a cytomegalovirus (CMV)–driven SOX2 expression vector, and a marker for commitment to the myogenic lineage MF20+ (myosin) was scored. Cells transfected with the SOX2 vector failed to enter the myogenic lineage compared to those transfected with empty vector (Fig. 7C).
DISCUSSION
We report the genome-wide assessment of physical interactions between the lamin A protein and regions of the genome during myogenesis using the DamID-seq approach. The mapping of LADs was done for wild-type lamin A protein and two human disease-causing missense mutations, p.R453W (EDMD) and p.R482W (FPLD). The data showed gain-of-function properties of mutant lamins, consistent with the observed dominant inheritance pattern of these mutations (and most laminopathies). Wild-type lamin A showed direct interactions with about 11,000 regions of the genome, whereas EDMD p.R453W increased interactions to 19,000 and FPLD p.R482W to 21,000 loci. The mutant lamins also showed dominant-negative features, with loss of the euchromatin transition of myogenic loci, and this finding was mutation-specific (seen by the EDMD mutation, but not FPLD) (Fig. 1).
Disease-specific disruptions of both the cell cycle and myogenesis pathways at the mRNA and protein levels had been previously observed in EDMD patient muscle biopsies (carrying either lamin A/C missense or emerin-null mutations) (3) and had been validated in emerin-null mice by studies of in vivo myogenesis (9). Our data show that these perturbations were likely the result of mutation-specific altered LADs and downstream consequences on DNA methylation at the genomic loci for members of these pathways (Fig. 2). For example, the genomic loci of both CDK1 and RB1 showed LAD sites in wild type consistent with exit from cell cycle, but these LADs were lost with EDMD p.R453W LMNA. Myogenic loci should show increasing euchromatin formation and mRNA expression during myogenesis, and this was seen by both DamID-seq and DNA methylation data. In contrast, there was broad loss of this transition in EDMD cells, as shown by both DamID-seq (p.R453W) and in genome-wide methylation studies of MyoD-converted muscle cells from an EDMD patient with a second mutation (p.H222P).
EDMD has features of a syndrome, with muscular dystrophy and marked muscle wasting as well as cardiac conduction block. The disease shows genetic heterogeneity caused by both dominant LMNA mutations and X-linked recessive EMD mutations, in which the encoded lamin A and emerin are interacting biochemical partners at the nuclear envelope. To integrate the epigenetic model across both disorders, genome-wide H3K9me3 (heterochromatin mark) ChIP-seq was carried out at three time points during the in vitro myogenic differentiation of emerin-null and wild-type murine myogenic cells. These data showed similar alterations in the epigenetic transitions of both cell cycle and myogenesis loci, as seen with the gain-of-function lamin A (p.R453W) DamID data. The increased time series permitted an additional assessment of epigenetic function, namely, spreading of heterochromatin laterally as a function of time (Fig. 4). For example, the initiation of heterochromatin at the Cdk1 locus appeared to fail entirely in emerin-null cells, whereas Rb1 heterochromatin initiated but showed relative inability to spread laterally (Figs. 2 and 4).
In carrying out a genome-wide prioritization of cellular functions altered in the emerin-null ChIP-seq time series, we noted significant alterations of Sox2-associated pluripotency pathways that we had not recognized in our previous studies (3, 9). Sox2 loci showed progressive gain of heterochromatin during myogenesis by both DamID-seq and ChIP-seq, but lamin A mutations and emerin loss of function both caused persistent expression of Sox2 pathway loci as a result of alterations in epigenetic programming. This change added abnormal persistence of pluripotency programs to myogenesis and cell cycle functions, all driven by alterations of LADs, histone associations, and DNA methylation patterns caused by lamin A missense and emerin loss of function. Overexpression of Sox2 protein in differentiating myogenic cells delayed their differentiation (Fig. 7), suggesting that persistence of pluripotency might be upstream of the myogenesis loci alterations.
We believe that the epigenetic effects of nuclear envelope disorders provide a unifying molecular model that explains the marked range of clinical phenotypes seen with different lamin A/C missense mutations as well as the clinical phenocopies seen with other nuclear envelop proteins, including emerin deficiency. Specific lamin A mutations have gain-of-function (promiscuous LADs) and dominant-negative (inability to initiate and spread LADs) consequences during the development of cell fate. Different mutations affect different cell lineages and cell-specific LADs. We provided evidence for allele specificity, where the epigenetics of myogenic loci was altered by a muscular dystrophy lamin A mutation (EDMD p.R453W) but not an adipose tissue disorder (FPLD p.R482W). It is important to speculate why geographically distant mutations such as R453W and H222P cause similar EDMD-specific epigenetic changes, whereas mutations in close proximity R453W (EDMD) and R482W (FPLD) do not. Our data suggest that an orchestrated association of lineage-specific (for example, MyoD) and nonlineage-specific (for example, LMNA) proteins is responsible for the appropriate developmental programming of distinct cell lineages. It is likely that there are multiple points of contact on the lamin A protein for lineage-specific proteins, and our data suggest that the R453W and H222P mutations alter different contact points for the same myogenic proteins disrupted in EDMD, whereas R482W does not. Krimm et al. (23) have carried out structure determination and mutant analyses of various EDMD and FPLD LMNA mutations to show that disease-specific mutations have structural consequences that differ at the molecular level. A limitation of our study is that we focused on EDMD (a muscle disease) using FPLD mutations as a disease control. Future studies are needed for other LMNA mutations that affect other cell lineages (for example, FPLD LMNA mutations in differentiating adipogenic cells; peripheral neuropathy LMNA mutations during neuronal development) before our models can be generalized to other laminopathies.
Although lineage-committed cells share an identical genome with embryonic stem cells, the epigenomic landscape differs markedly, with most changes arising from redistribution of transcriptional repressor marks H3K9me3 and H3K27me3 (21). Our model suggests that many of these changes are driven by the increased expression of lamin A during terminal differentiation of myoblasts and other cells (3, 4). This model is consistent with more general findings in pluripotent stem cells, in which epigenetic changes appear to drive cell fate memory, with large fractions of the genome associated with heterochromatin (H3K9m2), which is facilitated by chromatin nuclear lamina reassociation after mitosis in lineage-committed cells (24). The model is also supported by the findings of Towbin et al. (25) who have shown stepwise formation of H3K9me3 heterochromatic foci at the nuclear periphery using H3K9me1/2 as a substrate, suggesting that nuclear envelope plays an active role in epigenetic reprogramming.
Together, our data suggest that, in the presence of LMNA and EMD mutations and, by extension, all nuclear envelop disorders, there is inappropriate association of heterochromatin with nuclear lamina upon differentiation. This mishap results in three parallel events that may have a cumulative effect: slowing of exit from cell cycle (heterochromatinization of Cdk1 locus), slowing of exit from pluripotency programs (heterochromatinization of Sox2 locus), and poorly coordinated induction of terminal differentiation programs.
MATERIALS AND METHODS
Study design
Study design included discovery experiments (genome-wide) and validation experiments (preselected end points). No statistical outliers were excluded from analyses. The study rationale was to test the hypothesis that dominant missense mutations of the lamin A protein and loss of function of the emerin protein led to disruptions of interactions of DNA chromatin with the myogenic cell nuclear envelop during cell differentiation (myogenesis). Genome-wide discovery experiments (DamID, ChIP-seq, and genomic DNA methylation) were used to build molecular models that were then validated through targeted experiments using patient cells, patient muscle biopsies, mouse models, and in vitro–engineered cells.
Cell cultures
Conditionally immortalized H2K myoblasts
For the following experiments, conditionally immortal satellite cell-derived cell line H2K was used. H2K cell line was generated from the H2Kb-tsA58 immortomouse (26). Both emerin-null and wild-type mice are on the Bl6 background. The transgenic mice harbor a temperature-sensitive immortalizing T antigen gene (tsA58) under the control of an interferon-γ (IFN-γ)–inducible major histocompatibility complex class I promoter. Depending on culture conditions, the cells either exhibit continuous mitosis or alternatively terminally differentiate into myotubes. Emerin-null H2K mice were generated in our research center by T. Cohen and T. Partridge by breeding emerin-null (9) and H2Kb-tsA58 mice on the Bl6 background. Cells were harvested at the following time points: contact-inhibited myoblasts—80 to 90% confluent myogenic cells grown under permissive conditions; differentiating myotubes for 24, 48, 72, and 96 hours (d1 to d4)—80 to 90% confluent cells grown under permissive conditions for 24 hours, and induced to differentiate (serum withdrawal, nonpermissive temperature, and absence of IFN-γ).
Single fiber-derived satellite cell isolation and primary myotube differentiation
Emerin-null and wild-type mice aged 9 weeks were sacrificed and the EDL was dissected. Muscles were digested in 0.2% collagenase type 1/Dulbecco’s modified Eagle’s medium (DMEM) (Life Technologies), and individual myofibers were dissociated and washed, as described previously (27). To induce satellite cell activation, 6 to 12 myofibers were cultured on Matrigel-coated six-well plate in DMEM (Life Technologies) containing 10% (v/v) horse serum (Life Technologies), 10% (v/v) fetal bovine serum (Life Technologies), 1% (v/v) chick embryo extract (US Biological), 2% L-glutamine (Sigma-Aldrich), and 1% (v/v) penicillin/streptomycin solution (Sigma-Aldrich) at 37°C in 5% CO2. After myofiber removal, activated myoblasts were proliferated for 5 days. To induce differentiation, myoblasts were plated on Matrigel-coated dishes and kept in DMEM containing 5% horse serum (Life Technologies), 2% L-glutamine (Sigma-Aldrich), and 1% (v/v) penicillin/streptomycin solution (Sigma-Aldrich) at 37°C in 5% CO2. Cells were differentiated for 24 hours and harvested for downstream applications.
Immortalized MyoD-converted human myoblasts
EDMD and control patient fibroblasts were obtained from skin biopsies. Cells were immortalized using a lentiviral vector containing the sequence encoding the catalytic subunit of human telomerase as previously described (28). Inducible myogenic conversion was obtained using a lentiviral vector containing a murine MyoD under the control of the Tet-On inducible construct as described previously (29). Immortalized EDMD, FPLD, and control fibroblasts were proliferated to 90% confluence followed by MyoD induction by doxycycline. Four days after MyoD induction, myotubes were harvested for qRT-PCR and ChIP-qPCR experiments.
Immortalized human skeletal myoblasts
Human skeletal myoblasts were proliferated in Skeletal Muscle Cell Growth Medium (PromoCell) according to the manufacturer’s instructions except for fetal bovine serum (Invitrogen) that was adjusted to 20%. Differentiation was induced at high cell density by serum starvation [DMEM (Life Technologies) in the presence of gentamicin (50 μg/ml) and insulin (10 μg/ml)]. For DNA methylation analysis and myogenin qRT-PCR experiments, myogenic cells were infected with LMNA variants [wild-type, EDMD (p.R453W), FPLD (p.R482W)]. Myogenic cells were affected at myoblast stage and harvested the next day (myoblasts) or differentiated into myotubes and harvested 4 days after differentiation (myotubes).
Human embryonic kidney 293T cells
Human embryonic kidney (HEK) 293T cells were grown according to standard protocol in DMEM supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen), 1% penicillin/streptomycin (Invitrogen), and 2% L-glutamine (Invitrogen).
DamID-seq assay
Plasmid construction
pLgw V5-EcoDam (negative control), pLgw RFC1-V5-EcoDam, and pLgw CBX1-V5-EcoDam (positive control) vectors were obtained from the Bas van Steensel laboratory. We used Gateway cloning system (D-TOPO cloning) (Invitrogen) to generate entry clone (pENTR) for human LMNA ORF synthesized from RNA sample of a normal volunteer. LMNA entry clones harboring p.R453W and p.R482W mutations were generated using QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies). To clone these three versions of LMNA ORF into pLgw RFC1-V5-EcoDam, we used Gateway cloning system (LR clonase) (Invitrogen).
Lentiviral production and infection
HEK293T cells were grown in 10-cm dishes until 60 to 80% confluence. We used Lipofectamine 2000 (Invitrogen) for lentiviral production. We mixed 67.7 μl of Lipofectamine 2000 with 500 μl of Opti-MEM (minimum essential medium) and added it to the following mixture: 3.5 μg of pMD-G (envelope plasmid), 6.5 μg of pCMV-ΔR8.2 (packaging construct), 2.5 μg of pRSV-Rev (Rev-encoding plasmid), 10 μg of specific pLgw LMNA-V5-EcoDam, and 500 μl of Opti-MEM. The mixture was incubated at room temperature for 20 min and added dropwise to the HEK293T cells. After 8 hours, the medium was replaced with 6 ml of fresh medium. Virus containing medium was harvested on three consecutive days, and supernatants were pulled (18 μl of infectious medium), filtered (0.45 μm), and aliquoted (1-ml aliquots). Virus aliquots were stored at −80°C.
Human skeletal myoblasts were plated on six-well plates at high density. Differentiation was induced the next day by serum starvation [DMEM with insulin (10 μg/μl) and gentamicin (50 μg/μl)]. Myoblasts were differentiated for 24 hours; after which, they were infected with infectious medium diluted 2:1 in DMEM in the presence of 1.5 μl of Polybrene (10 mg/ml). The cells were then returned to 37°C for overnight incubation.
The following day, the medium was replaced with 3 ml of fresh differentiation medium. After 48 hours later, the cells were harvested and the genomic DNA (gDNA) was isolated using the Qiagen DNA Micro Kit, “Isolation of gDNA from Small Volumes of Blood” protocol. The DNA was eluted in 200 μl of buffer AE.
DamID library preparation and sequencing
The gDNA was ethanol-precipitated overnight at −20°C and dissolved in tris-EDTA (pH 7.5) to a concentration of 1 μg/μl. gDNA (2.5 μl) was digested with 0.5 μl of Dpn I (New England Biolabs; 20 U/μl) at 37°C overnight, and Dpn I was inactivated by heating to 80°C for 20 min. Dpn I–digested gDNA was ligated to the adaptor AdR [slowly annealed AdR-top (5′-CTAATACGACTCACTATAGGGCAGCGTGGTCGCGGCCGAGGA-3′) and AdR-bottom (5′-TCCTCGGCCG-3′)]. The ligation reaction was performed at 16°C for 2 hours using T4 Ligase (Roche; 5 U/μl). The enzyme was inactivated by heating to 65°C for 10 min. To remove the fragments that have unmethylated GATCs, we performed Dpn II digestion at 37°C for 1 hour.
PCR reaction was performed to amplify the regions flanked by adaptors. Reaction consisted of 10-μl Dpn II–digested DNA, 5-μl 10× cDNA (complementary DNA) PCR reaction buffer (Clontech), 0.625-μl primer bio-Adr-PCR (5′ bio-GGTCGCGGCCGAGGATC 3′; 100 μM), 1-μl deoxynucleotide triphosphates (10 mM), 1-μl PCR Advantage enzyme mix (Clontech, 50×), 32.375-μl double-distilled (ddH20). We used the following amplification reaction program: 1 cycle of 68°C (10 min), 94°C (1 min), 65°C (5 min), 68°C (15 min); 3 cycles of 94°C (1 min), 65°C (1 min), 68°C (10 min); 17 cycles of 94°C (1 min), 65°C (1 min), 68°C (2 min). The PCR products were cleaned with the Qiagen MinElute PCR Purification Kit and eluted in 20-μl ddH2O. The PCR products (2 μl) were run on a gel to verify successful digestion and amplification.
Each sample (3 μg) was diluted in 100-μl ddH2O and sonicated with Covaris using the following settings: 10% duty cycle, 175 Peak Incident Power, 200 cycles per burst for a total of 4.5 min. Biotinylated ends of the PCR products were pulled down using Dynabeads MyOne Streptavidin T1 (Invitrogen). These beads (50 μl) were washed three times with 1 ml of 1× Binding and Washing (B&W) buffer [5 mM tris-HCl (pH 7.5), 0.5 mM EDTA, 1 M NaCl]. Washed beads were resuspended in 75 μl of Covaris-sonicated DNA, 100 μl of 2× B&W buffer, and 25 μl H2O. This mixture was incubated at 4°C for 30 min on a rotator. The beads were washed three times with 200 μl of 1× B&W buffer.
DNA was released off the beads by Dpn II digestion at 37°C for 1 hour. The supernatant was saved and cleaned using the Qiagen MinElute Reaction Cleanup Kit. DNA was eluted in 20-μl H2O and quantified with Qubit. qPCR analysis was performed to validate the DamID DNA before submission for sequencing.
Bioinformatic analysis
Demultiplexed .fastq files were aligned to reference genome (hg19) using Bowtie (30) and uniquely mapped reads were kept for further analysis. Peaks were called using SICER algorithm (31) with parameter set as follows: window size (2000 bp), gap size (6000 bp), and FDR1e-3. Distribution of peaks was determined using HOMER (annotatePeaks.pl, GenomeOntology and Venn). Lamin A binding 2 kbp upstream/downstream of the TSS was queried using HOMER (annotatePeaks.pl, ghist). Similar analysis was performed for genes induced by myogenic induction. To obtain genes induced by myogenic induction in human skeletal myotubes, we downloaded ENCODE Affymetrix Exon 1.0 ST array data for skeletal myoblasts and myotubes (GSE15805). Data were normalized using Affymetrix Expression Console followed by Partek Genomics Suite analysis to obtain genes induced in myotubes (one-way ANOVA, P < 0.01; FC > 1.2). Alignment files (.bam) for the histone peaks were downloaded from the ENCODE (H3K9me3, GSM733730; H3K4me3, GSM733637). Lamin A, H3K9me3, and H3K4me3 binding around TSS was determined using HOMER (annotatePeaks.pl, ghist).
Methylation analysis
DNA methylation analysis was performed on three time points in two different patient cell types (normal and EDMD) using Illumina 450K methylation bead array. Normalization was performed using SWAN algorithm in R studio (32). Methylation pattern of genes induced by myogenesis was plotted using Partek Genomics Suite.
Second DNA methylation analysis was performed on human skeletal myoblasts and myotubes (day 4 of differentiation) infected with LMNA variants [wild-type, EDMD (p.R453W), and FPLD (p.R482W)] using Illumina 450K methylation bead array. Normalization and analysis were performed as described above.
Real-time PCR
Total RNA was isolated using TRIzol according to the manufacturer’s instructions. Two hundred nanograms of total RNA was reverse-transcribed into single-stranded cDNA and processed for RT-PCR (SYBR Green). Primers used can be found in table S8.
ChIP assay and ChIP-seq
ChIP-qPCR and ChIP-seq were performed with modifications following Myers laboratory protocol at http://myers.hudsonalpha.org/documents/Myers%20Lab%20ChIP-seq%20Protocol%20v041610.pdf. Briefly, cells were lysed in Farnham buffer [5 mM Pipes (pH 8.0), 85 mM KCl, 0.5% NP-40] and subsequently in radioimmunoprecipitation assay buffer [1× phosphate-buffered saline (PBS), 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS]. Chromatin was sheared by sonication (12 × 15 s) with 15-s break in between intervals in 1.5-ml microcentrifuge tubes using Sonifier Cell Disruptor 350. Fragmentation of chromatin was inspected on 1% agarose gel. Chromatin/DNA (50 μg) was immunoprecipitated overnight at 4°C with 3 μg of antibody against H3K9me3 (Abcam, ab8898). As a control, we used 5 μg of sheared, non-precipitated chromatin (input). Samples were incubated with magnetic beads (Dynabeads, Invitrogen) for 6 hours and washed successively in buffer I [20 mM tris-HCl (pH 8.0), 150 mM NaCl, 2 mM EDTA, 0.1% SDS, 1% Triton X-100], buffer II [20 mM tris-HCl (pH 8.0), 500 mM NaCl, 2 mM EDTA, 0.1% SDS, 1% Triton X-100], buffer III [10 mM tris-HCl (pH 8.0), 250 mM LiCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA], and tris-EDTA (pH 8.0). All washes were performed at 4°C for 10 min. qPCR primers used can be found in table S8. The results were reported as qPCR values normalized to input chromatin (gDNA) and presented as percentage of input. To ensure specific PCR amplification, every RT-PCR run was followed by a dissociation phase analysis (denaturation curve) to confirm the presence of a single dissociation peak. For ChIP-seq, chromatin was precipitated as described above, and libraries were prepared using Mondrian SP system and run on Illumina HiScan platform with multiplexed four samples per lane.
ChIP-seq data analysis
Demultiplexed .fastq files were aligned to reference genome (mm9) using Bowtie (30), and uniquely mapped reads were kept for further analysis. Peaks were called using SICER algorithm (31) with parameter set as follows: window size (1000 bp), gap size (3000 bp), and FDR1e-2. Genomic distribution of ChIP peaks and tag densities around TSS were determined with HOMER platform using the following commands: perl annotatePeaks.pl wt.bed mm9 > output.txt and annotatePeaks.pl tss.txt mm9 -size 10000 -hist 100 -d wt.bed emd.bed > outputfile.txt. Peaks specific to cell type were determined using BEDTools suite: bed-tools intersect -a A.bed -b B.bed –v. Differentially enriched peaks were studied by intersecting individual peak files for all conditions to generate union file using BEDTools intersect and annotated with read counts from individual .bed files using bedtools annotate counts –i union.bed -files F1.bed F2.bam FN.bed. The read counts were normalized to corresponding library sizes. Gene ontology of biological processes of peaks showing reduction/absence in H3K9me3 enrichment at the point of myogenic induction (d0 to d1) was identified using GREAT cis regulatory region prediction tool (33), whereas top canonical pathways were generated by Ingenuity Pathway Analysis. Heterochromatic expansion in proximal promoter region was defined as at least two times increase in peak length during d0 to d1 and d1 to d2 transition. Initially, sequencing and ChIP quality analysis were performed using FastQC and strand cross-correlation analysis (34). ChIP data for human myogenic cells were obtained from ENCODE (H3K9me3 – GSM733730, H3K4me3, GSM733637).
Expression data
Human skeletal myotubes and myoblasts
Affymetrix expression data for human skeletal myotubes and myoblasts (35, 36) were downloaded from Gene Expression Omnibus (GEO) (accession no. GSE15805).
Muscle biopsies
Affymetrix expression data for muscle tissue of control, EDMD-AD, EDMD-XR, and FKRP patients (3) were obtained from GEO (accession no. GSE3307). Sets were normalized using Expression Console from Affymetrix using PLIER (Probe Logarithmic Intensity Error) algorithm. Annotation, FC, and P values, as well as hierarchical clustering, were calculated and plotted using Partek Genomics Suite.
Western blot
Murine emerin-null and wild-type H2K cells were harvested at 24 (d1), 48 (d2), and 96 hours (d4) after differentiation. Patient cells were harvested 72 hours after differentiation. For each sample, 30 μg of total proteins was used for SDS–polyacrylamide gel electrophoresis and Western blotting. Sox2 levels were detected using anti-Sox2 antibody (Abcam, ab97959). Antibody against H3 was used as loading control (Abcam, ab1791). For detection of Dam-V5-LMNA proteins, antibody against V5 was used (Abcam, ab9116).
Myogenin FISH
FISH was performed as described previously (12). Briefly, EDMD patient and control myotubes (4 days of differentiation) were fixed with 4% paraformaldehyde in 1× PBS and permeabilized with 0.5% Triton X-100 in 1× PBS. gDNA was denatured by placing the slide in an 80°C dry bath for 5 min. DNA FISH probe containing myogenin locus was purchased from Empire Genomics (BAC clone, RP11-335O13; dye color, Orange 5-TAMRA dUTR). Slides were washed in a prehybridization buffer (2× SSC, 50% formamide) at 37°C for 1 hour before proceeding with hybridization step. FISH probe was denatured according to the manufacturer’s instruction. Hybridization was performed in a humidified chamber overnight 37°C in the dark. After hybridization, slides were washed in a prehybridization buffer at 37°C for 1 hour. Slides were then washed three times with PBST (1× PBS + 0.05% Tween 20) and mounted with VECTASHIELD (Vector Laboratories) and visualized with an Olympus FV-1000 confocal laser scanning microscope. Distances between each of the two MYOG loci and the nuclear envelop were measured from images.
Overexpression and myogenic differentiation experiments
Human immortalized myoblasts were transfected with SOX2 vector (EX-T2547-M61; GeneCopoeia) and corresponding empty vector (EX-T2547-NEG; GeneCopoeia). Both vectors carried information for GFP gene as well. Medium was changed the next day to induce myogenic differentiation. Cells were fixed 72 hours after differentiation induction and stained for myosin heavy chain (MF20, Developmental Studies Hybridoma Bank) and DAPI (ProLong Antifade Reagents with DAPI from Invitrogen). Cells that received the vector were scored on the basis of GFP expression, and myogenic potential was estimated as a ratio of MF20+ GFP+ cells over the total number of GFP+ cells. Four image fields containing 300 to 500 cells were scored for each transfection experiment.
Statistical analyses
The test for statistical significance was the two-tailed Student’s t test. Prediction of DamLMNA binding and H3K9me3 ChIP-seq enrichment was performed with the SICER algorithm. DNA methylation analysis was conducted with SWAN algorithm in R and Partek Genomics Suite. Multiple hypothesis testing was performed with the statistical data analysis software Partek Genomics Suite using P value adjustment.
Supplementary Material
Acknowledgments
We thank C. Stewart, T. Partridge, and T. Cohen for emerin-null mice and for emerin-null and wild-type H2K cell lines; B. van Steensel for Dam vectors; N. Markovic for the help with MATLAB scripting; M. Giri for help with microarray data analysis; P. Scacheri and A. Saiakhova for valuable input on ChIP-seq analysis; S. Bhuvanendran for help with acquiring confocal microscopy data; and the Myology Center of Research in Paris for their platform for immortalization of human cells. J.P. is a student in the Molecular Medicine Program of the Institute for Biomedical Sciences at The George Washington University. This work is from a dissertation to be presented to the above program in partial fulfillment of the requirements for the Ph.D. degree.
Funding: This work was supported by the U.S. NIH (grant 3R01 NS29525), the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin Diseases, and the Association Française contre les Myopathies (A.F.M.).
Footnotes
Author contributions: J.P. designed and performed experiments, analyzed data (DamID-seq, ChIP-seq, FISH, ChIP-qPCR, Western blot, qRT-PCR, DNA methylation, and overexpression experiment), and contributed to the writing and preparation of the manuscript. S.D. and V.S. helped design and perform the ChIP-seq experiment. V.F.G. isolated primary myoblast from single myofibers from EDL muscle from wild-type and emerin-null mice. J.K.J. helped develop the approach for acquiring and analyzing the imaging data. K.M., V.M., C.V., and G.B. provided MyoD-converted myogenic cells issued from patients’ fibroblasts harboring lamin A/C p.H222P (EDMD) and p.R482W (FPLD) mutations. E.P.H. designed experiments, analyzed data, and contributed to preparation of the manuscript.
Competing interests: The authors declare that they have no competing financial interests.
REFERENCES AND NOTES
- 1.Maraldi NM, Capanni C, Cenni V, Fini M, Lattanzi G. Laminopathies and lamin-associated signaling pathways. J Cell Biochem. 2011;112:979–992. doi: 10.1002/jcb.22992. [DOI] [PubMed] [Google Scholar]
- 2.Camozzi D, Capanni C, Cenni V, Mattioli E, Columbaro M, Squarzoni S, Lattanzi G. Diverse lamin-dependent mechanisms interact to control chromatin dynamics: Focus on laminopathies. Nucleus. 2014;5:427–440. doi: 10.4161/nucl.36289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bakay M, Wang Z, Melcon G, Schiltz L, Xuan J, Zhao P, Sartorelli V, Seo J, Pegoraro E, Angelini C, Shneiderman B, Escolar D, Chen YW, Winokur ST, Pachman LM, Fan C, Mandler R, Nevo Y, Gordon E, Zhu Y, Dong Y, Wang Y, Hoffman EP. Nuclear envelope dystrophies show a transcriptional fingerprint suggesting disruption of Rb–MyoD pathways in muscle regeneration. Brain. 2006;129:996–1013. doi: 10.1093/brain/awl023. [DOI] [PubMed] [Google Scholar]
- 4.Constantinescu D, Gray HL, Sammak PJ, Schatten GP, Csoka AB. Lamin A/C expression is a marker of mouse and human embryonic stem cell differentiation. Stem Cells. 2006;24:177–185. doi: 10.1634/stemcells.2004-0159. [DOI] [PubMed] [Google Scholar]
- 5.Rober RA, Weber K, Osborn M. Differential timing of nuclear lamin A/C expression in the various organs of the mouse embryo and the young animal: A developmental study. Development. 1989;105:365–378. doi: 10.1242/dev.105.2.365. [DOI] [PubMed] [Google Scholar]
- 6.Håkelien AM, Delbarre E, Gaustad KG, Buendia B, Collas P. Expression of the myo-dystrophic R453W mutation of lamin A in C2C12 myoblasts causes promoter-specific and global epigenetic defects. Exp Cell Res. 2008;314:1869–1880. doi: 10.1016/j.yexcr.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 7.Boguslavsky RL, Stewart CL, Worman HJ. Nuclear lamin A inhibits adipocyte differentiation: Implications for Dunnigan-type familial partial lipodystrophy. Hum Mol Genet. 2006;15:653–663. doi: 10.1093/hmg/ddi480. [DOI] [PubMed] [Google Scholar]
- 8.Kudlow BA, Jameson SA, Kennedy BK. HIV protease inhibitors block adipocyte differentiation independently of lamin A/C. AIDS. 2005;19:1565–1573. doi: 10.1097/01.aids.0000186827.91408.90. [DOI] [PubMed] [Google Scholar]
- 9.Melcon G, Kozlov S, Cutler DA, Sullivan T, Hernandez L, Zhao P, Mitchell S, Nader G, Bakay M, Rottman JN, Hoffman EP, Stewart CL. Loss of emerin at the nuclear envelope disrupts the Rb1/E2F and MyoD pathways during muscle regeneration. Hum Mol Genet. 2006;15:637–651. doi: 10.1093/hmg/ddi479. [DOI] [PubMed] [Google Scholar]
- 10.Kind J, van Steensel B. Genome–nuclear lamina interactions and gene regulation. Curr Opin Cell Biol. 2010;22:320–325. doi: 10.1016/j.ceb.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 11.Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–247. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- 12.Yao J, Fetter RD, Hu P, Betzig E, Tjian R. Subnuclear segregation of genes and core promoter factors in myogenesis. Genes Dev. 2011;25:569–580. doi: 10.1101/gad.2021411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Solovei I, Wang AS, Thanisch K, Schmidt CS, Krebs S, Zwerger M, Cohen TV, Devys D, Foisner R, Peichl L, Herrmann H, Blum H, Engelkamp D, Stewart CL, Leonhardt H, Joffe B. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell. 2013;152:584–598. doi: 10.1016/j.cell.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 14.Mattout A, Pike BL, Towbin BD, Bank EM, Gonzalez-Sandoval A, Stadler MB, Meister P, Gruenbaum Y, Gasser SM. An EDMD mutation in C. elegans lamin blocks muscle-specific gene relocation and compromises muscle integrity. Curr Biol. 2011;21:1603–1614. doi: 10.1016/j.cub.2011.08.030. [DOI] [PubMed] [Google Scholar]
- 15.Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, van Steensel B. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 16.Rodriguez A, Bjerling P. The links between chromatin spatial organization and biological function. Biochem Soc Trans. 2013;41:1634–1639. doi: 10.1042/BST20130213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korzelius J, Naumann SK, Loza-Coll MA, Chan JSK, Dutta D, Oberheim J, Gläßer C, Southall TD, Brand AH, Jones DL, Edgar BA. Escargot maintains stemness and suppresses differentiation in Drosophila intestinal stem cells. EMBO J. 2014;33:2967–2982. doi: 10.15252/embj.201489072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lacin H, Rusch J, Yeh RT, Fujioka M, Wilson BA, Zhu Y, Robie AA, Mistry H, Wang T, Jaynes JB, Skeath JB. Genome-wide identification of Drosophila Hb9 targets reveals a pivotal role in directing the transcriptome within eight neuronal lineages, including activation of nitric oxide synthase and Fd59a/Fox-D. Dev Biol. 2014;388:117–133. doi: 10.1016/j.ydbio.2014.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonne G, Di Barletta MR, Varnous S, Bécane HM, Hammouda EH, Merlini L, Muntoni F, Greenberg CR, Gary F, Urtizberea JA, Duboc D, Fardeau M, Toniolo D, Schwartz K. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat Genet. 1999;21:285–288. doi: 10.1038/6799. [DOI] [PubMed] [Google Scholar]
- 20.Shackleton S, Lloyd DJ, Jackson SNJ, Evans R, Niermeijer MF, Singh BM, Schmidt H, Brabant G, Kumar S, Durrington PN, Gregory S, O’Rahilly S, Trembath RC. LMNA, encoding lamin A/C, is mutated in partial lipodystrophy. Nat Genet. 2000;24:153–156. doi: 10.1038/72807. [DOI] [PubMed] [Google Scholar]
- 21.Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, Antosiewicz-Bourget J, Ye Z, Espinoza C, Agarwahl S, Shen L, Ruotti V, Wang W, Stewart R, Thomson JA, Ecker JR, Ren B. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, Wong KY, Sung KW, Lee CWH, Zhao XD, Chiu KP, Lipovich L, Kuznetsov VA, Robson P, Stanton LW, Wei CL, Ruan Y, Lim B, Ng HH. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 23.Krimm I, Östlund C, Gilquin B, Couprie J, Hossenlopp P, Mornon J-P, Bonne G, Courvalin J-C, Worman HJ, Zinn-Justin S. The Ig-like structure of the C-terminal domain of lamin A/C, mutated in muscular dystrophies, cardiomyopathy, and partial lipodystrophy. Structure. 2002;10:811–823. doi: 10.1016/s0969-2126(02)00777-3. [DOI] [PubMed] [Google Scholar]
- 24.Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat Genet. 2009;41:246–250. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Towbin BD, González-Aguilera C, Sack R, Gaidatzis D, Kalck V, Meister P, Askjaer P, Gasser SM. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell. 2012;150:934–947. doi: 10.1016/j.cell.2012.06.051. [DOI] [PubMed] [Google Scholar]
- 26.Morgan JE, Beauchamp JR, Pagel CN, Peckham M, Ataliotis P, Jat PS, Noble MD, Farmer K, Partridge TA. Myogenic cell lines derived from transgenic mice carrying a thermolabile T antigen: A model system for the derivation of tissue-specific and mutation-specific cell lines. Dev Biol. 1994;162:486–498. doi: 10.1006/dbio.1994.1103. [DOI] [PubMed] [Google Scholar]
- 27.Gnocchi VF, White RB, Ono Y, Ellis JA, Zammit PS. Further characterisation of the molecular signature of quiescent and activated mouse muscle satellite cells. PLOS One. 2009;4:e5205. doi: 10.1371/journal.pone.0005205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Auré K, Mamchaoui K, Frachon P, Butler-Browne GS, Lombès A, Mouly V. Impact on oxidative phosphorylation of immortalization with the telomerase gene. Neuromuscul Disord. 2007;17:368–375. doi: 10.1016/j.nmd.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 29.Chaouch S, Mouly V, Goyenvalle A, Vulin A, Mamchaoui K, Negroni E, Di Santo J, Butler-Browne G, Torrente Y, Garcia L, Furling D. Immortalized skin fibroblasts expressing conditional MyoD as a renewable and reliable source of converted human muscle cells to assess therapeutic strategies for muscular dystrophies: Validation of an exon-skipping approach to restore dystrophin in Duchenne muscular dystrophy cells. Hum Gene Ther. 2009;20:784–790. doi: 10.1089/hum.2008.163. [DOI] [PubMed] [Google Scholar]
- 30.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maksimovic J, Gordon L, Oshlack A. SWAN: Subset-quantile within array normalization for Illumina Infinium HumanMethylation450 BeadChips. Genome Biol. 2012;13:R44. doi: 10.1186/gb-2012-13-6-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol. 2008;26:1351–1359. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McDaniell R, Lee BK, Song L, Liu Z, Boyle AP, Erdos MR, Scott LJ, Morken MA, Kucera KS, Battenhouse A, Keefe D, Collins FS, Willard HF, Lieb JD, Furey TS, Crawford GE, Iyer VR, Birney E. Heritable individual-specific and allele-specific chromatin signatures in humans. Science. 2010;328:235–239. doi: 10.1126/science.1184655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, Garg K, John S, Sandstrom R, Bates D, Boatman L, Canfield TK, Diegel M, Dunn D, Ebersol AK, Frum T, Giste E, Johnson AK, Johnson EM, Kutyavin T, Lajoie B, Lee BK, Lee K, London D, Lotakis D, Neph S, Neri F, Nguyen ED, Qu H, Reynolds AP, Roach V, Safi A, Sanchez ME, Sanyal A, Shafer A, Simon JM, Song L, Vong S, Weaver M, Yan Y, Zhang Z, Zhang Z, Lenhard B, Tewari M, Dorschner MO, Hansen RS, Navas PA, Stamatoyannopoulos G, Iyer VR, Lieb JD, Sunyaev SR, Akey JM, Sabo PJ, Kaul R, Furey TS, Dekker J, Crawford GE, Stamatoyannopoulos JA. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.