Abstract
Computational, genomic, and proteomic approaches have been used to discover non-annotated protein-coding small open reading frames (smORFs). Some novel smORFs have crucial biological roles in cells and organisms, which motivates the search for additional smORFs. Proteomic smORF discovery methods are advantageous because they detect smORF-encoded polypeptides (SEPs) to validate smORF translation and SEP stability. Because SEPs are shorter and less abundant than average proteins, SEP detection using proteomics faces unique challenges. Here, we optimize several steps in the SEP discovery workflow to improve SEP isolation and identification. These changes have led to the detection of several new human SEPs (novel human genes), improved confidence in the SEP assignments, and enabled quantification of SEPs under different cellular conditions. These improvements will allow faster detection and characterization of new SEPs and smORFs.
TOC Graphic
INTRODUCTION
An expression screen for genes that prevent neuronal cell death revealed a novel class of human bioactive peptides1. In this screen, a neuronal cell line was engineered to express the Alzheimer’s disease protein V642I-APP. Transfection of these engineered cells with a cDNA library identified neuroprotective genes that prevented cell death. One of the protective genes was identified as a 16S ribosomal RNA, which was shown to contain a previously unknown 75-bp protein-coding short open reading frame (smORF). smORFs are defined as protein-coding sORF of less than 100 amino acids. The 16S ribosomal smORF produces a 24-amino acid peptide called humanin, which prevents cell death by inhibiting pro-apoptotic BCL-2 proteins2,3.
Humanin differs from traditional bioactive peptides, peptide hormones and neuropeptides, in two ways. First, peptide hormones and neuropeptides are generated from proteolysis of longer proteins called prohormones4–8. By contrast, humanin is translated from a smORF as a peptide and does not require further proteolysis for activation. Second, peptide hormones and neuropeptides bind through cell surface receptors, receptor tyrosine kinases (RTKs) and G protein-coupled receptors (GPCRs), while humanin binds an intracellular protein. These differences indicate that humanin is part of a distinct class of bioactive peptides.
Additional work has revealed that genomes harbor many non-annotated smORFs, and some of these smORFs are biologically active9–11. In flies, for example, deletion of tal/pri gene, which encodes several smORFs, results in loss of segmentation of the embryo, and a truncated limb and a missing tarsus in the adult fly12,13. Functional smORFs have also been identified in bacteria14–16, plants 17, and other eukaryotes17–24.
The biological activity of these novel genes has led to emerging strategies for smORF discovery. smORFs have been discovered by computational9,18,19,25, genomic (Ribo-Seq)18,26,27, and proteomic methods28,29. While computational and genomics methods infer protein-coding genes, proteomics provides direct evidence for smORF translation and demonstrates that the resulting smORF-encoded polypeptides (SEPs) are stable enough to be detected. We use a cutoff of 150 amino acids for SEPs because we found a substantial fraction of non-annotated protein-coding ORFs between 100–150 amino acids (about 10% of our total)21.
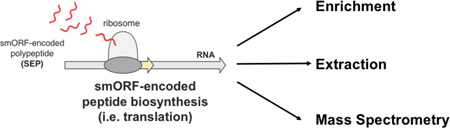
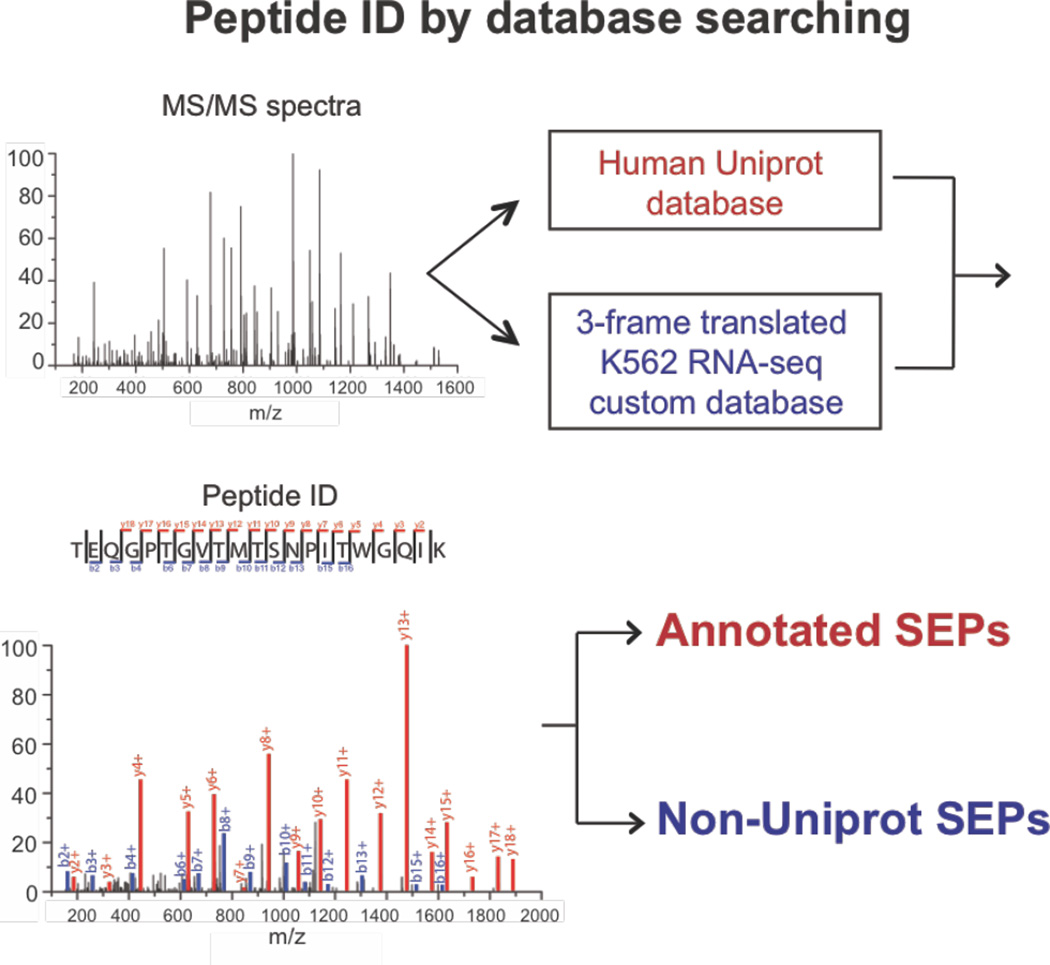
Proteomic discovery of SEPs and smORFs requires the combination of proteomics and genomics (i.e. RNA-Seq), referred to as proteogenomics28,29. Novel SEP discovery begins by enriching proteome for low molecular weight peptides and small proteins (<30 kilodaltons (kDa)). This fraction is proteolytically digested and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) proteomics21,23,24,28. The resulting LC-MS/MS dataset is then interrogated using a protein database from the three-frame translation of the RNA-Seq data21–23 (Figure 1). Removal of known proteins identifies non-annotated SEPs and smORFs. To identify known (i.e. annotated) SEPs, the human UNIPROT database is used in this workflow instead (Figure 1).
Figure 1.
Overview of the SEP discovery workflow. To identify known and novel SEPs MS/MS spectra are searched against the Human UNIPROT database (known SEPs) and a 3-frame translated RNA-Seq custom database (novel SEPs). Peptides that uniquely match to a UNIPROT protein entry that is less than 150 amino acids in length are annotated as known SEPs. Peptides that match to an entry in the form RNA-Seq 3-frame translated database that is less than 150 amino acids in length and do not overlap with any UNIPROT proteins are novel SEPs (i.e. non-annotated, non-UNIPROT).
The small size of SEPs compared to proteins make smORF/SEP discovery using proteomics challenging. We typically have to identify a smORF/SEP from a single tryptic peptide because they are shorter than normal proteins. We previously improved proteome fractionation methods to identify more SEPs21. Here, we examine the impact of different isolation, enrichment, and mass spectrometry approaches to improve the workflow further. These efforts led to a more confident identification of SEPs and the discovery of 37 non-annotated human SEPs (i.e. 37 novel human genes).
MATERIALS AND METHODS
Cell Culture
K562 and A549 cells were maintained in RPMI and F-12K media, respectively. HeLa and HEK293 cells were cultured using DMEM. The media contained 10% fetal bovine serum (FBS). Cells were grown under an atmosphere of 5% CO2 at 37°C until confluent. Before cells lysis and enrichment of SEPs, the media was removed from adherent cells by aspiration (A549, HeLa, HEK293) or non-adherent cells (K562) by centrifugation. HEPES-buffered saline (pH 7.5) was used to wash the cells to remove residual media and FBS.
SEP Enrichment Methods
We tested three conditions for SEP enrichment: (1) acid precipitation, (2) 30-kDa molecular weight cut off (MWCO) filter, and (3) reverse-phase (C8) cartridge enrichment. Cellular proteomes from 4 × 107 cells were extracted by lysis with boiling water. After cooling the samples on ice, the cells were sonicated for 20 bursts at output level 2 with a 30% duty cycle (Branson Sonifier 250; Ultrasonic Convertor). For the acid precipitation, the addition of acetic acid (to a final concentration of 0.25% by volume) was followed by centrifugation at 14,000 × g for 20 min at 4 °C. This step precipitates larger proteins to reduce the complexity of the supernatant and enriches lower molecular weight proteins that are then analyzed by LC-MS/MS proteomics for SEPs. For the 30-kDa MWCO, the addition of acetic acid (to a final concentration of 0.25% by volume) was followed by centrifugation at 14,000 × g for 20 min at 4 °C. The supernatant is then passed through a 30-kDa MWCO filter and the flow through is analyzed for SEPs. Lastly, the reverse phase enrichment, the cellular extracts are centrifuged at 25,000 × g for 30 min and supernatants removed and filtered through 5 µM syringe filters followed by enrichment of SEPs using Bond Elute C8 silica cartridges (Agilent Technologies, Santa Clara, CA). Approximately 100 mg sorbent was used per 10 mg total lysate protein. Cartridges were prepared with one column volume methanol and then equilibrated with two-column volumes triethylammonium formate (TEAF) buffer, pH 3.0 before the sample was applied. The cartridges were then washed with two column volumes TEAF and the SEP enriched fraction eluted by the addition of acetonitrile:TEAF pH 3.0 (3:1) and lyophilized using a Savant Speed-Vac concentrator. BCA protein assay (Thermo Scientific) was used to measure protein concentration of each sample after extraction and enrichment.
SEP Extraction Methods
Four different methods were compared for extraction of SEPs from 4 × 107 total cells: (1) 50 mM HCl, 0.1% β-mercaptoethanol (β-ME); 0.05% Triton X-100 at room temperature (lysis buffer); (2) 1 N acetic acid/0.1 N HCl at room temperature; (3) boiling in water; or (4) boiling in lysis buffer. After extraction using these four methods, the extracts were centrifuged at 25,000 × g for 30 min, and supernatants filtered through 5 µM syringe filters. The flow through was then enriched for SEPs by binding and elution using Bond Elute C8 silica cartridges (Agilent Technologies, Santa Clara, CA). Approximately 100 mg sorbent was used per 10 mg total lysate protein. Cartridges were prepared with one column volume methanol and equilibrated with two-column volumes triethylammonium formate (TEAF) buffer, pH 3.0 before the sample was applied. The cartridges were then washed with two column volumes TEAF and the SEP enriched fraction eluted by the addition of acetonitrile:TEAF pH 3.0 (3:1) and lyophilized using a Savant Speed-Vac concentrator. BCA protein assay (Thermo Scientific) was used to measure protein concentration of each sample after extraction and enrichment.
Digestion and Sample Preparation for LC-MS/MS
An aliquot of 100 µg of enriched samples was precipitated with chloroform/methanol extraction. Dried pellets were dissolved in 8 M urea/100 mM TEAB, pH 8.5. Proteins were reduced with 5 mM tris 2-carboxyethylphosphine hydrochloride (TCEP, Sigma-Aldrich) and alkylated with 10 mM iodoacetamide (Sigma-Aldrich). Proteins were digested overnight at 37 °C in 2 M urea/100 mM TEAB, pH 8.5, with trypsin (Promega). Digestion was stopped with formic acid, 5 % final concentration.
Q Exactive LC-MS/MS analysis
Digests were analyzed by LC-MS using an Easy-nLC1000 (Proxeon) and a Q Exactive mass spectrometer (Thermo Scientific). An EASY-Spray column (Thermo Scientific) 25 cm by 75um packed with PepMap C18 2um particles was used. Electrospray was performed directly from the tip of the analytical column. Buffer A and B were 0.1 % formic acid in water and acetonitrile, respectively, and the solvent flow rate was 300 nl/min. Each sample was run in triplicate. The digested samples were loaded onto the column using an autosampler, and the samples were desalted online using a trapping column. Peptide separation was performed with 6-hour reverse phase gradient. The gradient increases from 5–22% B over 280 min, 22–32% B over 60 min, 32–90% B over 10 min, followed by a hold at 90% B for 10 min. The column was re-equilibrated with buffer A before injection.
The Q Exactive was operated in a data-dependent mode. Full MS1 scans were collected with a mass range of 400 to 1800 m/z at 70k resolution. The 10 most abundant ions per scan were selected for MS/MS with an isolation window of 2 m/z and HCD energy of 25 and resolution of 17.5k. Maximum fill times were 60 and 120 ms for MS and MS/MS scans, respectively. An underfill ratio of 0.1% was utilized for peak selection, dynamic exclusion was enabled for 15s and unassigned and singly charge ions were excluded. Data was collected with default values for AGC target of 1e6 and 5e5 and maximum injection times of 60 and 120ms for MS and MS/MS scans respectively. Data was also collected with sensitive settings for comparison. AGC of MS and MS/MS scans were increased to 5e6 and 5e6 respectively and maximum fill times were increased to 120 ms and 500 ms. All other parameters remained unchanged.
Orbitrap Fusion Tribrid LC-MS/MS Analysis
C8 SPE enriched samples were analyzed on an Orbitrap Fusion Tribrid mass spectrometer (Thermo Scientific). The digest was injected directly onto a 50cm, 75um ID column packed with BEH 1.7um C18 resin (Waters). Samples were separated at a flow rate of 200 nl/min on an nLC 1000 (Thermo Scientific). Buffers A and B were 0.1% formic acid in water and acetonitrile, respectively. A gradient of 1–22%B over 160 min, an increase to 32%B over 60 min, an increase to 90%B over another 10 min and held at 90%B for a final 10 min of washing was used. The column was re-equilibrated with 20 µl of buffer A before the injection of sample. Peptides were eluted directly from the tip of the column and nanosprayed directly into the mass spectrometer by application of 2.5 kV at the back of the column. The Orbitrap Fusion was operated in a data-dependent mode. Full MS scans were collected in the Orbitrap at 120K resolution with a mass range of 400 to 1500 m/z and an AGC target of 4e5 and maximum fill time of 50 ms. The cycle time was set to 3 sec. Within this 3 sec widow the most abundant ions per scan were selected for fragmentation by either CID in the ion trap with an AGC target of 1e4 and maximum fill time of 35 ms or HCD and detection in the Orbitrap with an AGC target of 5e5 and max fill time of 250 ms. Collision energy was set to 35 for both CID and HCD, and a minimum intensity of 5000 was required for selection. Quadrupole isolation at 1.6 m/z was used, monoisotopic precursor selection was enabled, and dynamic exclusion was used with exclusion duration of 10 sec.
Data Analysis to Identify Annotated and Non-annotated SEPs
Tandem mass spectra were extracted from raw files using RawExtract 1.9.9.2 and searched with ProLuCID30 using Integrated Proteomics Pipeline – IP2 (Integrated Proteomics Applications). We used two databases in these searches, a custom database created from the in silico 3-frame translation of RNA-Seq data from K562 cells (RNA-Seq database), and the UNIPROT Human database. The transcriptome data is deposited on GEO (GSE34740). The search space included all fullytryptic and half-tryptic peptide candidates. Carbamidomethylation on cysteine was considered as a static modification.
To determine annotated and non-annotated SEPs, data files from technical replicates were combined and searched by ProLuCID. For HCD, data was searched with 50-ppm precursor ion tolerance then filtered to 10-ppm, and 50-ppm fragment ion tolerance with a maximum of two internal missed cleavages using either the custom database or UNIPROT Human database. For CID, data was searched with 500-ppm precursor ion tolerance then filtered to 10-ppm, and 50-ppm fragment ion tolerance with a maximum of two internal missed cleavages using either the custom database or UNIPROT Human database. Identified spectra were filtered and grouped into proteins using DTASelect31,32. Proteins and SEPs required, at least, one peptide to be identified with a setting of less than 1% FDR for all searches. Unique peptides identified by searching the UNIPROT database that belonged to smORFs of fewer than a 150 codons were kept and were referred to as ‘annotated SEPs’.
To identify non-annotated SEPs, data files from technical duplicates were combined and searched by ProLu-CID. Data was searched with 50-ppm precursor ion tolerance then filtered to 10-ppm, and 50-ppm fragment ion tolerance with a maximum of two internal missed cleavages using only the custom database. The results from the custom database search were then filtered against the UNIPROT human database using a string-searching algorithm to remove any annotated peptides. We visually inspect the MS2 spectra for all of the smORF/SEP peptides to validate the assignment. In particular, we required that any critical amino acid residues that uniquely distinguish the peptide detected in the MS2 data.
The next step is to determine whether the non-annotated peptides are from smORFs or not. The non-annotated peptides are searched against NCBI Human Reference Sequence Database (RefSeq) using tBLASTn, which identifies an RNA that could have produced the SEP. After identifying an RNA and sequence that encodes the peptide, we annotate the downstream in-frame stop codon, and then try to identify the upstream in-frame start codon.
We assign start codons to any in-frame ATG. If there is no in-frame ATG, we look for an in-frame near-cognate codon (i.e. ACG, AAG, CUG, etc.) in a Kozak sequence33 to assign as the start codon. Lastly, if an in-frame ATG or near-cognate start codon cannot be found, we identify the upstream in-frame stop codon, and if the distance between the upstream and downstream in-frame stop codons is less than 150 codons, we annotated the gene as a smORF. If the peptides did not match to any RNA sequences with the RefSeq RNA database, it means that they were derived from RNAs that were present in the RNA-Seq data but not in the RefSeq database. For these peptides, we repeat these steps for assigning the smORF using RNAs from the RNA-Seq database.
Arsenite Treatment Experiments
HEK293 cells were grown to ~70% confluence and then treated with 10 µM sodium arsenite for 24 hours. Cellular proteins were extracted using the lysis buffer followed by centrifugation 20,000 × g for 20 min at 4 °C to remove any insoluble particulates. The concentrations were determined using a Bradford assay and 100 µg was taken forward for digestion and sample preparation (see above) and LC-MS/MS using the Q Exactive. After collection of the data, LC-MS peaks corresponding to two SEPs and two proteins were identified and quantified using Skyline. XICs were extracted with Skyline and peak identity was confirmed by correlating retention time to the identified spectra from the database search results. The AUC (area under the curve) for the peptide ions was used to determine the relative quantity of each peptide between control and arsenite-treated samples. The extraction of the isotopic peaks for each peptide and comparison to the theoretical isotopic distribution at a resolution of 60k validated the selected peptide ion we used for quantitation.
Raising SLC35A4-SEP Antibody
Antisera against SLC35A4 was raised in rabbits against a synthetic peptide fragment encoding Cys34SLC35A4(2–34) coupled to maleimide activated keyhole limpet hemocyanin (ThermoFisher, Waltham MA). The peptide, ADDKDSLPKLKDLAFLKNQLESLQRRVEDEVNC, was synthesized and C18 HPLC purified by RS Synthesis (Louisville, KY); purity was 99.0%. Immunogen was prepared by emulsification of Freund's complete adjuvant-modified Mycobacterium butyricum (EMD Millipore, Billerica MA) with an equal volume of phosphate buffered saline (PBS) containing 1.0 mg conjugate/ml for initial injections. For booster injections, incomplete Freund's adjuvant was mixed with an equal of PBS containing 0.5 mg conjugate/ml. For each immunization, an animal received a total of 1 ml emulsion in 20 intradermal sites in the lumbar region. Three individual rabbits were injected every three weeks and were bled one week following booster injections. Bleeds were screened for titer and specificity; antiserum PBL #7383, 6/25/15 bleed, was used for these studies. All animal procedures were approved by the Institutional Animal Care and Use Committee of the Salk Institute and were conducted in accordance with the National Institutes of Health guidelines.
Western Blot Analysis
Control and sodium arsenite-treated HEK293 cells were extracted by lysis buffer. Protein concentration was measured using Bradford assay (BioRad). 30 µg of total protein from each sample was loaded on a 4–12 % BisTris gel, 10-well (Bolt, Life Technologies) and run in MES running buffer at 200V for 20 min. Proteins were transferred to PVDF membrane and then blocked at room temperature for 1 hour using LiCor Blocking Buffer. The membrane was then blotted with primary antibody; rabbit anti-beta actin (LiCor) 1:1000 for 1 hour at room temperature; rabbit anti-HO-1 (Cell Signaling) overnight at 4 °C; or rabbit anti-SLC35A4 SEP at 1:5000 dilution overnight at 4°C. Washed membrane three time with TBS-T, then blotted with secondary antibody: goat anti-rabbit IRDye 800CW (LiCor) at 1:10000 dilution, rocked 1 hour at room temperature. Washed membrane three times with TBS-T then scanned the membrane using LiCor Odyssey CLx at IR700 and IR800. The built-in tool in Odyssey CLx was used to quantify the intensity of the bands of interest.
RESULTS AND DISCUSSION
Enrichment optimization
Identifying all the SEPs in cells and tissues is required to characterize smORF biology. In a complex mixture such as total cell lysate, detecting small and low abundant proteins is challenging as detection is naturally biased towards the detection of more abundant proteins34. Therefore, SEP detection will likely benefit from an enrichment step, but we have yet to test this assumption. Here, we compare different enrichment methods for their ability to identify the greatest number of known and unknown SEPs from cells.
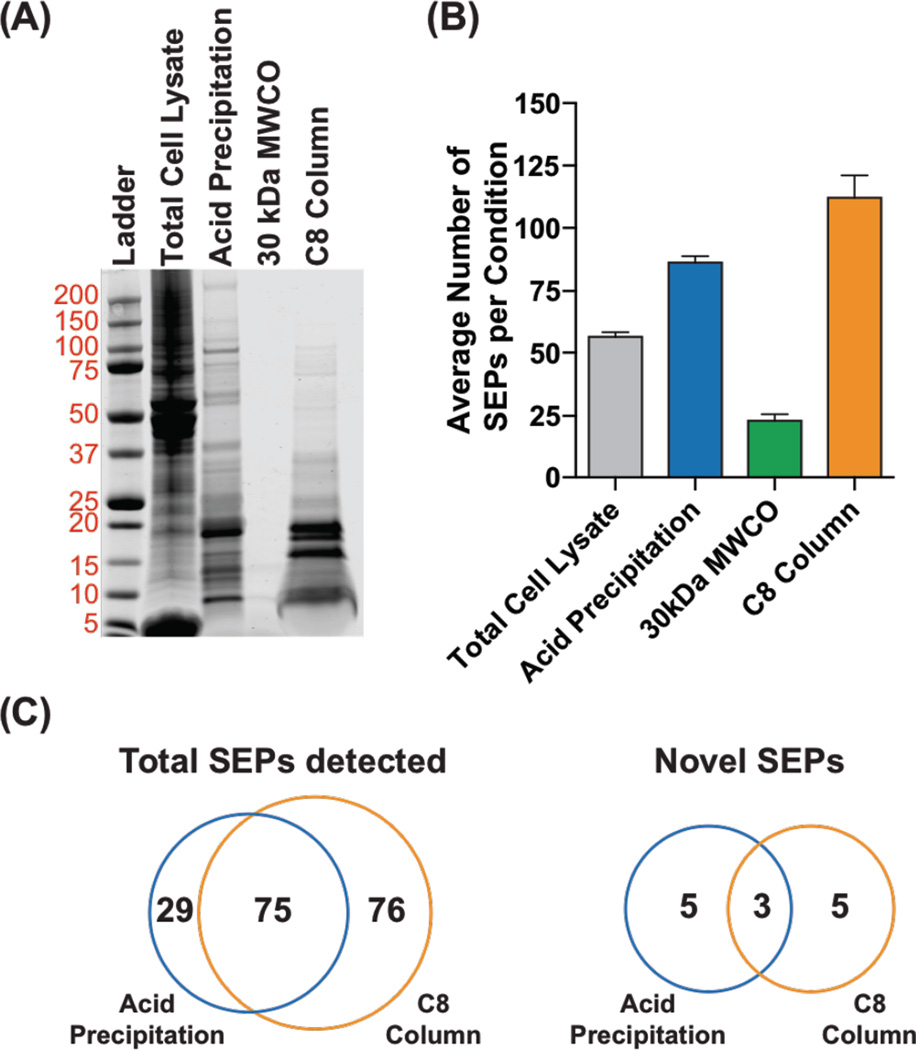
We began these experiments using K562 cells, which we chose because the first SEPs were discovered using this cell line22. The total proteome is prepared by boiling K562 cells to inactivate all proteolytic activity and then lysing the cells by sonication. We used three methods to enrich the < 30 kDa proteome: 1) acetic acid precipitation; 2) molecular weight cutoff (MWCO) filtration (30 kDa); or 3) solid-phase extraction (SPE). A BCA assay quantified the protein concentrations in each of these enriched samples, and an equal amount of total protein was analyzed by SDS-PAGE gel (Figure 2A). The results are clear. The 30-kDa MWCO resulted in poor recovery compared to the acid precipitation and SPE.
Figure 2.
Comparison of different methods for SEP enrichment using K562 cells. (A) Cells lysates were prepared by boiling in water followed by sonication. SEPs were enriched from this lysate by acid precipitation, a 30-kDa MWCO filter, or C8 SPE (i.e. C8 column). The results from these enrichments were analyzed by SDS-PAGE (30 µg total protein per lane, Coomassie stain). (B) Analysis of these samples by proteomics identified the average number of SEPs in each sample. (C) Venn diagrams of the total SEPs (known and novel) and novel SEPs in the acid precipitation and C8 column samples detected by proteomics.
Analysis of total lysate by SDS-PAGE reveals that a majority of the proteome is larger than 30 kDa. Acetic acid precipitation aggregates larger proteins leaving behind lower molecular weight proteins solution. SDS-PAGE of the solution after acetic acid precipitation led to the majority of the signal coming from proteins less than 30 kDa (Figure 2). Previously, we had relied on MWCO filtration to enrich the lower molecular weight proteome, but this method results in significantly less protein by SDS-PAGE, which hurts our ability to detect SEPs (Figure 2). The solid phase extraction method using selective carbon groups (C8) bonded to silica-based sorbents was originally developed to enrich plasma and tissue extracts for peptide hormones by removing larger molecular weight proteins before measurement by radioimmunoassay 35,36. Applying this method to enrich the lower molecular weight proteins gave excellent results by SDS-PAGE (Figure 2).
We then determined whether the results we measured by SDS-PAGE correlated with the number of known and unknown SEPs that we could detect using proteomics. Enriched and non-enriched proteome samples were reduced, alkylated, and trypsin digested followed by LC-MS/MS analysis. Samples were analyzed using a 6-hour gradient on a Q-Exactive mass spectrometer set to a top 10-mode. The decoy database searching was used to identify the acquired MS/MS spectra using two databases (Figure 1), the RNA-Seq database and the UNIPROT database.
Analysis of the LC-MS/MS datasets using the human UNIPROT database revealed 70, 96, 35, 143 known SEPs from the non-enriched, acetic acid precipitated, MWCO and SPE enriched samples, respectively. We analyzed the LC-MS/MS datasets using a custom database made from the three-frame translation of RNA-Seq data from K562 cells, which contains all potential translated proteins in K562 cells. A search of our proteomics data against the RNA-Seq database enabled us to identify several non-annotated SEPs.
From the non-enriched, acetic acid precipitated, MWCO and SPE enriched samples, we identified 4, 8, 1, and 8 non-annotated SEPs, respectively. The average number of SEPs detected, annotated or novel, correlate with the protein recovery we observed by SDS-PAGE (Figure 2B, and Figure S2). The data indicates that the acetic acid precipitation and C8 SPE methods are better than the 30 kDa MWCO filter we have used in the past. Many SEPs that were only identified using the acetic acid precipitation or C8 SPE method (Figure 2C). This was consistent in several other cell lines that we tested. (Figure S1). Also, all the methods provide SEP of similar lengths and hydrophobicity (Figure S3 and Figure S4). Therefore, we recommend using both enrichment methods moving forward to maximize the total number of SEPs detected.
Different methods for SEP extraction
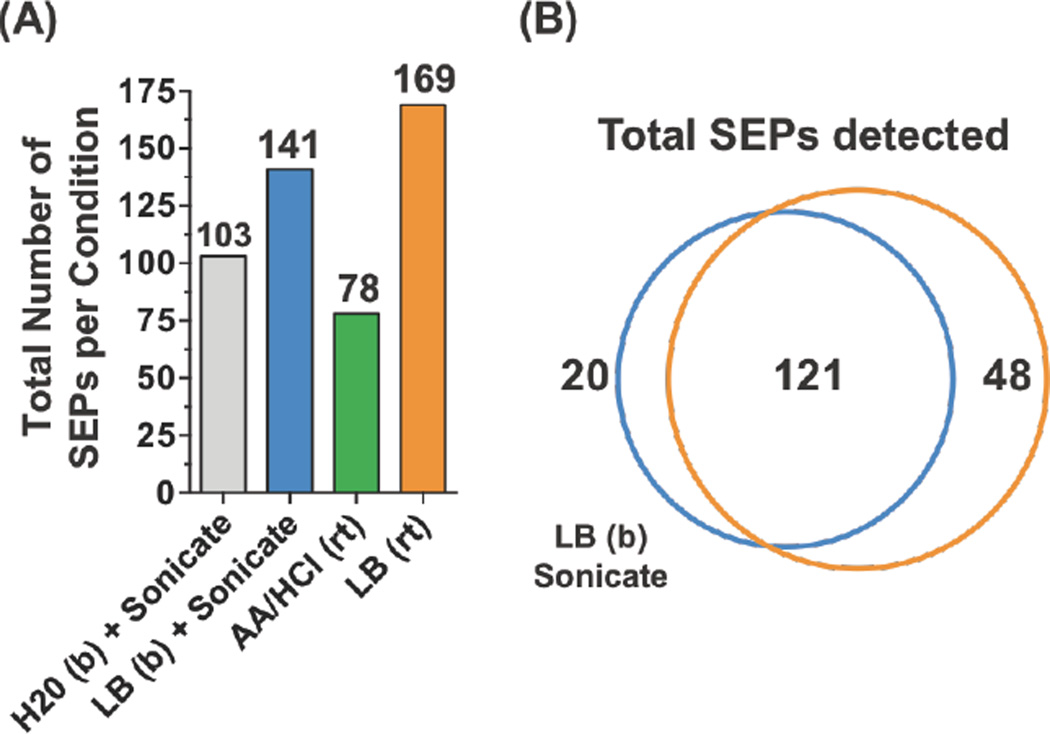
We compared several distinct methods for isolating SEPs from the lung cancer cell line A549 (i.e. extraction methods) (Figure 3A). We selected another cell line to ensure that our methods translated to the more conventional adherent cells. We tested four different extraction methods: 1) water + sonication; 2) lysis buffer + sonication; 3) acetic acid (1N) + HCl (0.1N); or 4) lysis buffer. After extraction, we used SPE to prepare the sample for LC-MS/MS. We searched the proteomics data against the Human UNIPROT database and three-frame translated RNA-Seq custom database for peptide identification. Samples extracted in the lysis buffer detected the most SEPs while acid extraction resulted in fewest SEPs detected. Overall, the lysis buffer performs better than water or acid alone, while boiling did not seem to have a strong effect (Figure S5). The number and identity of SEPs detected with or without boiling are similar. Overall, the combination of extracting cell lysate in the lysis buffer and enriched with C8 column provided the highest recovery of small peptidome and the largest number of SEPs detected (Figure 3B). LC-MS/MS Optimization. For SEP discovery, good spectral quality is essential because SEPs are low abundant, with a single peptide detected per SEP in most cases. The confidence of the peptide identification depends on good quality MS/MS spectra—i.e. good sequence coverage and a low background are necessary. Previously, we used an Orbitrap Velos hybrid ion trap mass spectrometer (Thermo Fisher Scientific) with Collision Induced Dissociation (CID) and low-resolution MS/MS spectra acquisition. Low-resolution spectra detected in the linear ion trap can often have high background noise, especially for low abundant species such as SEPs. High-resolution MS/MS data, obtained using an Orbitrap, can solve this problem but leads to less sensitivity since more ions are required for detection.
Figure 3.
Different extraction methods have a minimal impact on total number of SEPs detected. (A) Total number of SEPs identified from A549 cells using four different SEP extraction methods: boiling (b) water and sonication; boiling lysis buffer (LB, 50 mM HCl, 0.1% beta-ME, 0.05% Triton X-100) and sonication; acetic acid (AA) and hydrochloric acid (HCl) at room temperature (rt); and lysis buffer at room temperature. (B) Comparison of the extraction methods demonstrated good overlap between the methods with lysis buffer at room temperature capturing the most SEPs.
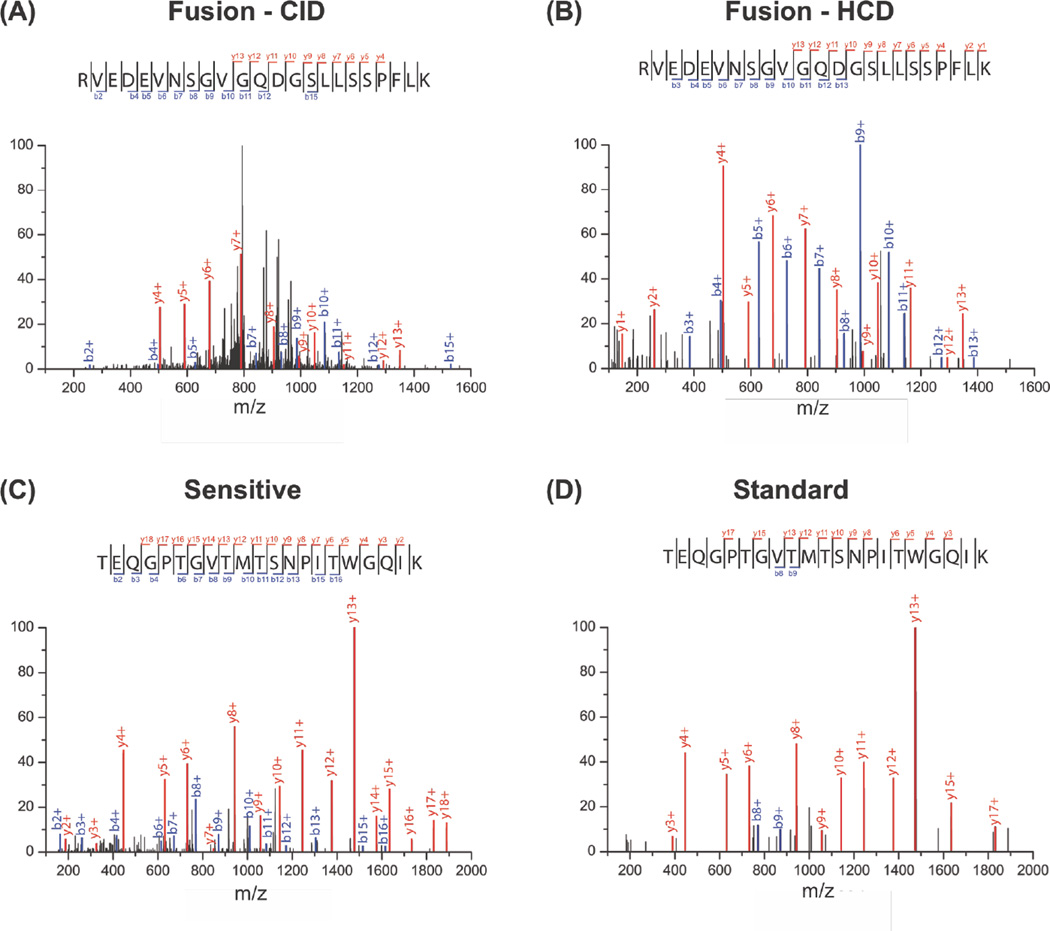
High-energy Collisional Dissociation (HCD) is reported to provide better sequence coverage than CID, provided the HCD energy is adequate for the peptide37. Improved sequence coverage can benefit SEP detection by providing more confidence in the SEP peptide detected. We tested whether HCD would improve SEP peptide characterization. For example, MS/MS of a SEP peptide by low-resolution CID and high-resolution HCD on the Fusion Tribrid MS reveals increased sequence coverage using HCD (Figure 4A, B). We found modest improvements in peptide coverage using HCD. For instance, CID identified 11 b-ions and 10 y-ions, while HCD detected 11 b-ions and 12 y-ions. Qualitatively, the HCD spectrum is less noisy, and major peaks in the CID spectra are not assigned (Figure 4A, B). A similar improvement in coverage was observed using HCD with the QE mass spectrometer (Figure S6). These results indicate that HCD provides a slight improvement in sequence coverage of peptides and much lower background, but doesn’t effect the total number of SEPs we detect.
Figure 4.
Comparison of MS/MS spectra acquired using different fragmentation methods and automatic gain control. (A) MS/MS spectrum of the same SEP peptide acquired by low resolution CID or (B) high resolution HCD (Fusion Tribrid MS). (C) MS/MS spectrum of the same SEP peptide acquired with sensitive or (D) standard setting (QExactive MS).
We also optimized the Automatic Gain Control (AGC) and fill time of the Q Exactive to increase coverage in the MS2 spectra. The higher AGC setting and longer max fill times (sensitive) identified 13 b-ions and 17 y-ions, while the default AGC and fill time (standard) settings detected 2 b-ions and 12 y-ions (Figure. 4C, D and Figure S7). All data presented herein was collected under the “sensitive” settings to ensure good spectral quality. With the sensitive setting, we observe a marked improvement in the number of detected ions to provide significantly better sequence coverage. Therefore, increased fill times and high AGC settings should be used for SEP discovery.
Label-free SEP quantitation
We do not obtain many spectral counts for SEPs due to their overall short length, which has prevented us from using spectral counting to quantify SEP levels. Here, we look at using the area under the curve in the MS1 spectra to quantitate SEP levels. We decided to compare SEP levels in control and arsenite-treated HEK293 cells. This system is ideal for these experiments because known increases in heme oxygenase 1 (HO-1) expression can be used as a positive control. Moreover, SCL35A4 mRNA38, which includes the SLC35A4 smORF, was reported to be elevated under these conditions, which suggests that arsenite treatment might regulate SEP levels.
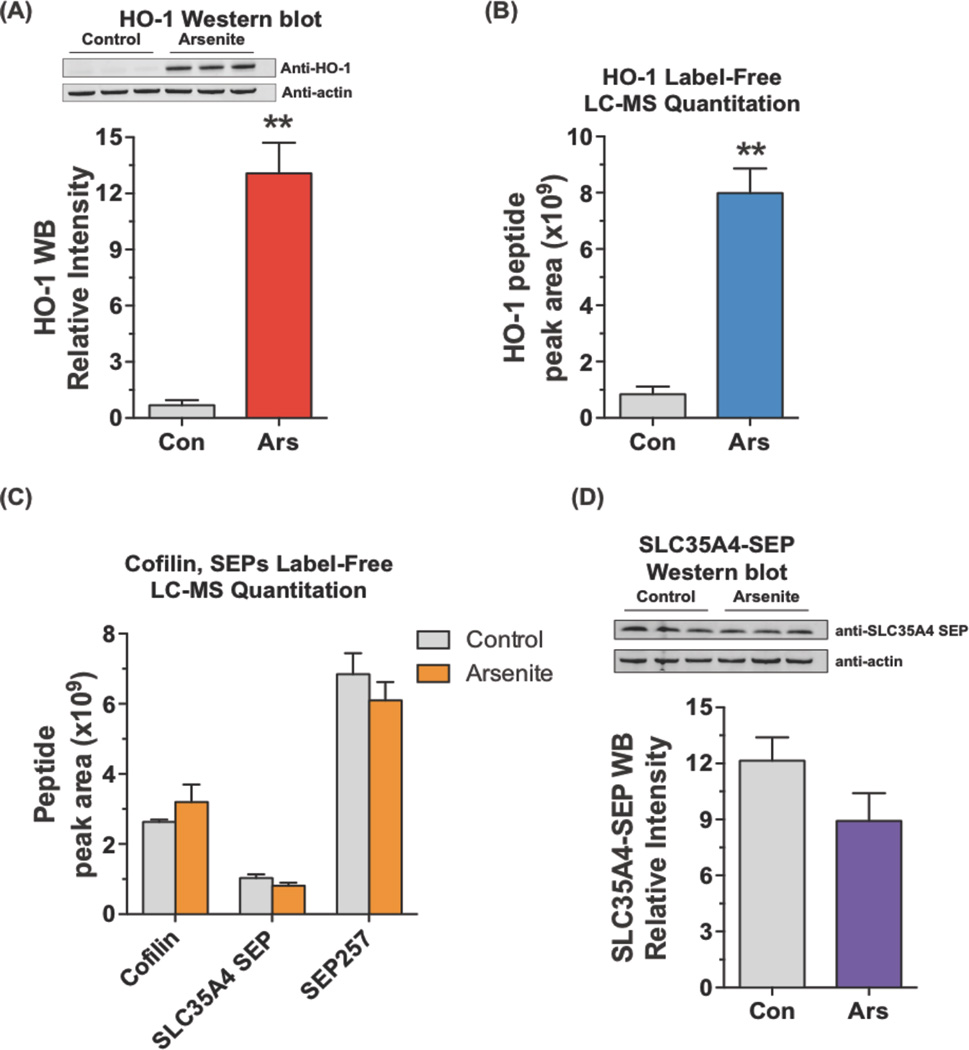
Sodium arsenite-treated (10 µM) and untreated HEK293 cells were extracted and analyzed by LC-MS/MS. Heme oxygenase 1 (HO-1) was reported to be up-regulated by arsenite treatment of HEK293 cells in a previous proteomics study39, and we validated this change by Western blot showing HO-1 was highly expressed in arsenite-treated samples (p <0.01) (Figure 5A). We looked at HO-1 levels by label-free LC-MS, by quantitating the area under the LC-MS peak for an HO-1 peptide in the MS1 data. We performed label-free quantitative analysis using Skyline software40,41 that extracts peak area of the detected peptides from MS1 by retention time and accurate mass. Using peak areas allows us to quantitate relative protein or SEP expression level between two conditions. This analysis showed a strong increase in HO-1 peptide levels in the arsenite-treated sample demonstrating that the label-free quantitation is similar to a Western blot (Figure 5A, B).
Figure 5.
Quantitation of SEPs upon arsenite treatment. (A) HEK293 cells were treated with 10 µM sodium arsenite for 24 hours. Western blot analysis revealed increased HO-1 expression upon on arsenite treatment (10 µM, 24 hours). The intensity of the bands on the Western blot was quantified by LiCor Odyssey CLx and normalized by beta-actin. (B) Peak area (MS1) of the HO-1 peptide agrees with Western blot. (C) Peak areas (MS1) of cofilin, SLC35A4-SEP, and SEP257 were unchanged upon arsenite treatment. (D) SLC35A4-SEP levels were also measured by Western blot, which agreed with the proteomics quantitation. The intensity of the bands on the blot was quantified by LiCor Odyssey CLx and normalized by beta-actin. (Student’s t-test, **, p < 0.01)
We measured the levels of three peptides to determine what effect, if any, arsenite has on SEP levels. The peptides included two SEPs, SLC35A4-SEP and SEP257, and cofilin, which was the negative control. As expected, analysis of the area under the curve for a cofilin peptide revealed that cofilin levels were unchanged between the arsenite- and control-treated samples. A similar analysis of SLC35A-SEP and SEP257 demonstrated that these two peptides were unchanged between the control and arsenite-treated samples (Figure 5C, Figure S8). Furthermore, most SEPs have similar ion intensities such that this label-free quantitation method should be general (Figure S9).
We wanted to confirm that SLC35A4-SEP is unchanged, so we generated an antibody against SLC35A4-SEP, which we used for Western blot analysis. We tested this antibody by overexpressing SLC35A4 and demonstrated that it efficiently detects SLC35A4-SEP (Figure S10). Using this antibody against control and arsenite-treated samples shows that SLC35A4-SEP levels are unchanged (Figure 5D), supporting our quantitative label-free mass spectrometry results. The label-free quantitative measures SEP levels between two different conditions will be of tremendous use in distinguishing SEPs that are changing under different biological conditions, even though we did not find any changes in this example.
Analysis of Novel Human SEPs
In this study, we detected 37 novel human SEPs (Table S1), which come from smORFs that are not annotated in the RefSeq database. Each of these smORFs represents a novel human gene. The new SEPs are translated from smORFs in the 5’UTR (5 SEPs), 3’UTR (2 SEPs), non-coding RNAs (6 SEPs) and 24 RNAs that were not in the RefSeq database but are present in our RNA-Seq data. Most of the new smORFs (21 in total (55%)) have an AUG start codon, while the remaining 16 SEPs (45%) do not. This observation is in agreement with previous studies21,23,24,27, indicating that a significant portion of SEPs can be translated from non-canonical AUG start codon.
A few of the SEPs are unknown isoforms of known proteins. For instance, one of the SEP peptides we detected, GYFDSGDYNMAK, is derived from a 119 amino acid SEP from a non-annotated smORF with an ATG start. When we align this SEP to non-redundant human proteins using pBLAST it has >85% sequence homology to several alpha-endosulfine protein isoforms (Figure 6A and Figure S11). Thus, we conclude that SEP252 is a novel alpha-endosulfine protein isoform, and we demonstrate how SEP discovery can help find additional, non-annotated, isoforms of known small proteins.
Figure 6.
Some novel SEPs are new isoforms of known proteins or fragments of longer proteins. (A) SEP252 is a new isoform of the protein alpha-endosulfine (ENSA) and this connection were discovered because the SEP peptide (red) is homologous to ENSA but different enough to realize that this peptide is from a non-annotated smORF. Alignment of the entire smORF demonstrates high sequence homology (>80%) to various ENSA isoforms indicating that this SEP is a member of the ENSA family of proteins. (B) SEP266 was identified through a peptide that is homologous (red) to another peptide from Elongation factor 1-alpha 1 (EEF1A1) but differs by one amino acid (blue) indicating that it belongs to a non-annotated smORF. Alignment of the entire smORF shows high sequence homology (>80%) to part of EEF1A1.
Another group of newly discovered SEPs have sequence homology to known proteins but the SEP and the known protein are different lengths, a part of much long proteins (Figure 6B and Figure S11). For example, one of the SEPs peptides, NMITETSQADCAVLIVAAGVGEFEAGISK, belongs to a 123 amino acid long SEP with ATG start. pBLAST of this sequence demonstrated strong sequence homology of this SEP to a 462 amino acid long eukaryotic translation elongation factor 1 from residue 49 to 169. Truncated variants of EEF1A1 have previously been shown to promote42 or suppress43 cancer cell growth suggesting that this SEP266 might be an interesting candidate for downstream cell biological studies. The discovery of truncated forms of known proteins, such as EEF1A1, might provide new insight into the biological regulation of these proteins.
CONCLUSIONS
By testing and optimizing several different parameters in the SEP workflow, we have improved the number of SEPs detect, and enhanced the confidence in those assignments. The identification of smORFs and SEPs becomes increasingly important as new biological functions are emerging. For example, new mammalian SEPs that regulate muscle endurance44 and metabolism10 have recently been discovered. As a potential pool of molecules with roles in fundamental biology, the discovery of smORFs and SEPs is of paramount importance. Here, we highlight the power of proteomics in contributing to this field by defining a new workflow that improves on the enrichment, mass spectrometry, and quantitation of human SEPs.
Supplementary Material
Acknowledgments
This study was supported by the NIH (R01 GM102491, A.S), the NCI Cancer Center Support Grant P30 (CA014195 MASS core, A.S.), The Leona M. and Harry B. Helmsley Charitable Trust grant (#2012-PG-MED002, A.S.), and Dr. Frederick Paulsen Chair/Ferring Pharmaceuticals (A.S.), and NIH (P41 GM103533 and R01 MH067880, J.K.D., J.R.Y.), and NIH (R01 HG004037, I.J., M.K.) and GENCODE Welcome Trust grant (U41 HG007234, I.J., M.K.).
ABBREVIATIONS
- SEP
smORF-Encoded Polypeptide
- kDa
kilodalton
- MWCO
molecular weight cutoff
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- MS
mass spectrometry
- SPE
solid phase extraction
- PAGE
polyacrylamide gel electrophoresis
- CDS
coding sequence
- UTR
untranslated region
- CID
collision induced dissociation
- HCD
high-energy collisional dissociation
- AGC
automatic gain control
- HO-1
heme oxygenase 1
Footnotes
ASSOCIATED CONTENT
Supporting Information
This material is available free of charge via the Internet at http://pubs.acs.org
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The Authors declare no competing financial interest.
REFERENCES
- 1.Hashimoto Y, Niikura T, Tajima H, Yasukawa T, Sudo H, Ito Y, Kita Y, Kawasumi M, Kouyama K, Doyu M, Sobue G, Koide T, Tsuji S, Lang J, Kurokawa K, Nishimoto I. Proc Natl Acad Sci U S A. 2001;98:6336–6341. doi: 10.1073/pnas.101133498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo B, Zhai D, Cabezas E, Welsh K, Nouraini S, Satterthwait AC, Reed JC. Nature. 2003;423:456–461. doi: 10.1038/nature01627. [DOI] [PubMed] [Google Scholar]
- 3.Zhai D, Luciano F, Zhu X, Guo B, Satterthwait AC, Reed JC. J Biol Chem. 2005;280:15815–15824. doi: 10.1074/jbc.M411902200. [DOI] [PubMed] [Google Scholar]
- 4.Bliss M. The Discovery of Insulin. United States of America: University of Chicago Press; 2013. [Google Scholar]
- 5.Bliss M, Purkis R. The discovery of insulin. Chicago: University of Chicago Press; 1982. [Google Scholar]
- 6.De Lecea , Kilduff T, Peyron C, Gao X-B, Foye P, Danielson P, Fukuhara C, Battenberg E, Gautvik V, Bartlett Fn. Proc. Natl. Acad. Sci. 1998;95:322–327. doi: 10.1073/pnas.95.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sakurai T, Amemiya A, Ishii M, Matsuzaki I, Chemelli RM, Tanaka H, Williams SC, Richardson JA, Kozlowski GP, Wilson S. Cell. 1998;92:573–585. doi: 10.1016/s0092-8674(00)80949-6. [DOI] [PubMed] [Google Scholar]
- 8.Vale W, Spiess J, Rivier C, Rivier J. Science. 1981;213:1394–1397. doi: 10.1126/science.6267699. [DOI] [PubMed] [Google Scholar]
- 9.Ladoukakis E, Pereira V, Magny EG, Eyre-Walker A, Couso JP. Genome Biol. 2011;12:R118. doi: 10.1186/gb-2011-12-11-r118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee C, Zeng J, Drew BG, Sallam T, Martin-Montalvo A, Wan J, Kim SJ, Mehta H, Hevener AL, de Cabo R, Cohen P. Cell Metab. 2015;21:443–454. doi: 10.1016/j.cmet.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Slavoff SA, Heo J, Budnik BA, Hanakahi LA, Saghatelian A. J Biol Chem. 2014;289:10950–10957. doi: 10.1074/jbc.C113.533968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galindo MI, Pueyo JI, Fouix S, Bishop SA, Couso JP. PLoS Biol. 2007;5:e106. doi: 10.1371/journal.pbio.0050106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kondo T, Hashimoto Y, Kato K, Inagaki S, Hayashi S, Kageyama Y. Nat. Cell Biol. 2007;9:660–665. doi: 10.1038/ncb1595. [DOI] [PubMed] [Google Scholar]
- 14.Hemm MR, Paul BJ, Miranda-Ríos J, Zhang A, Soltanzad N, Storz G. J. Bacteriol. 2010;192:46–58. doi: 10.1128/JB.00872-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hemm MR, Paul BJ, Schneider TD, Storz G, Rudd KE. Mol. Microbiol. 2008;70:1487–1501. doi: 10.1111/j.1365-2958.2008.06495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wadler CS, Vanderpool CK. Proc. Natl. Acad. Sci. 2007;104:20454–20459. doi: 10.1073/pnas.0708102104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanada K, Higuchi-Takeuchi M, Okamoto M, Yoshizumi T, Shimizu M, Nakaminami K, Nishi R, Ohashi C, Iida K, Tanaka M. Proc. Natl. Acad. Sci. 2013;110:2395–2400. doi: 10.1073/pnas.1213958110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aspden JL, Eyre-Walker YC, Phillips RJ, Amin U, Mumtaz MAS, Brocard M, Couso J-P. Elife. 2014;3:e03528. doi: 10.7554/eLife.03528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frith MC, Forrest AR, Nourbakhsh E, Pang KC, Kai C, Kawai J, Carninci P, Hayashizaki Y, Bailey TL, Grimmond SM. PLoS Genet. 2006;2:e52. doi: 10.1371/journal.pgen.0020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kastenmayer JP, Ni L, Chu A, Kitchen LE, Au WC, Yang H, Carter CD, Wheeler D, Davis RW, Boeke JD, Snyder MA, Basrai MA. Genome Res. 2006;16:365–373. doi: 10.1101/gr.4355406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma J, Ward CC, Jungreis I, Slavoff SA, Schwaid AG, Neveu J, Budnik BA, Kellis M, Saghatelian A. J. Proteome. Res. 2014;13:1757–1765. doi: 10.1021/pr401280w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oyama M, Kozuka-Hata H, Suzuki Y, Semba K, Yamamoto T, Sugano S. Mol. Cell. Proteomics. 2007;6:1000–1006. doi: 10.1074/mcp.M600297-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Slavoff SA, Mitchell AJ, Schwaid AG, Cabili MN, Ma J, Levin JZ, Karger AD, Budnik BA, Rinn JL, Saghatelian A. Nat. Chem. Biol. 2013;9:59–64. doi: 10.1038/nchembio.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vanderperre B, Lucier JF, Bissonnette C, Motard J, Tremblay G, Vanderperre S, Wisztorski M, Salzet M, Boisvert FM, Roucou X. PLoS One. 2013;8:e70698. doi: 10.1371/journal.pone.0070698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vanderperre B, Lucier JF, Roucou X. Database (Oxford) 2012;2012:bas025. doi: 10.1093/database/bas025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bazzini AA, Johnstone TG, Christiano R, Mackowiak SD, Obermayer B, Fleming ES, Vejnar CE, Lee MT, Rajewsky N, Walther TC, Giraldez AJ. EMBO J. 2014;33:981–993. doi: 10.1002/embj.201488411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ingolia NT, Lareau LF, Weissman JS. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Branca RM, Orre LM, Johansson HJ, Granholm V, Huss M, Pérez-Bercoff Å, Forshed J, Käll L, Lehtiö J. Nat. Methods. 2014;11:59–62. doi: 10.1038/nmeth.2732. [DOI] [PubMed] [Google Scholar]
- 29.Castellana N, Bafna V. J Proteomics. 2010;73:2124–2135. doi: 10.1016/j.jprot.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu T, Park SK, Venable JD, Wohlschlegel JA, Diedrich JK, Cociorva D, Lu B, Liao L, Hewel J, Han X, Wong CC, Fonslow B, Delahunty C, Gao Y, Shah H, Yates JR., 3rd J Proteomics. 2015;129:16–24. doi: 10.1016/j.jprot.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cociorva D, D LT, Yates JR. Curr Protoc Bioinformatics. 2007;16 doi: 10.1002/0471250953.bi1304s16. 13.4:13.4.1–13.4.14. [DOI] [PubMed] [Google Scholar]
- 32.Tabb DL, McDonald WH, Yates JR., 3rd J Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kozak M. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 34.Liu H, Sadygov RG, Yates JR., 3rd Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 35.Vale W, Vaughan J, Jolley D, Yamamoto G, Bruhn T, Seifert H, Perrin M, Thorner M, Rivier J. Methods Enzymol. 1986;124:389–401. doi: 10.1016/0076-6879(86)24030-6. [DOI] [PubMed] [Google Scholar]
- 36.Vale W, Vaughan J, Yamamoto G, Bruhn T, Douglas C, Dalton D, Rivier C, Rivier J. Methods Enzymol. 1983;103:565–577. doi: 10.1016/s0076-6879(83)03040-2. [DOI] [PubMed] [Google Scholar]
- 37.Diedrich JK, Pinto AF, Yates JR., 3rd J Am Soc Mass Spectrom. 2013;24:1690–1699. doi: 10.1007/s13361-013-0709-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andreev DE, O'Connor PB, Fahey C, Kenny EM, Terenin IM, Dmitriev SE, Cormican P, Morris DW, Shatsky IN, Baranov PV. Elife. 2015;4:e03971. doi: 10.7554/eLife.03971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lau AT, He QY, Chiu JF. Biochem J. 2004;382:641–650. doi: 10.1042/BJ20040224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schilling B, Rardin MJ, MacLean BX, Zawadzka AM, Frewen BE, Cusack MP, Sorensen DJ, Bereman MS, Jing E, Wu CC, Verdin E, Kahn CR, Maccoss MJ, Gibson BW. Mol Cell Proteomics. 2012;11:202–214. doi: 10.1074/mcp.M112.017707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dahl LD, Corydon TJ, Ränkel L, Nielsen KM, Füchtbauer E-M, Knudsen CR. Cancer Cell Int. 2014;14:17. doi: 10.1186/1475-2867-14-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rho SB, Park YG, Park K, Lee S-H, Lee J-H. FEBS Letters. 2006;580:4073–4080. doi: 10.1016/j.febslet.2006.06.047. [DOI] [PubMed] [Google Scholar]
- 44.Anderson DM, Anderson KM, Chang CL, Makarewich CA, Nelson BR, McAnally JR, Kasaragod P, Shelton JM, Liou J, Bassel-Duby R, Olson EN. Cell. 2015;160:595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.