Abstract
Background:
In most cases, prostatitis can be caused by a bacterial agent such as Ureaplasma urealyticum. Considering to the cumbersome of the culture method for the detection of Ureaplasma species in clinical samples such as prostate; PCR method that is faster and more appropriate than the cultivation methods, can be utilized for the detection of U. urealyticum and U. parvum. PCR-RFLP method can differentiate both biovars and assist in studies of the clinical diagnosis, epidemiology and pathology of this species in human. The aim of this study was to molecular detection of U. urealyticumin in prostate tissue samples based on PCR- RFLP.
Methods:
Two hundred prostate tissue samples were collected from patient suffering from prostatitis. The PCR assay was used to amplify a 559 bp fragment of 16S-23SRNA interspace region of Ureaplasma. After sequencing, PCR products from positive samples were digested with TaqI restriction enzyme.
Results:
Seven cases (3.5%) out of 200 prostate tissue samples were positive for U. urealyticum. Results of PCR products sequencing demonstrated that all isolates were U. parvum biovar. PCR-RFLP results shown that there was not any differentiation in pattern of enzymatic digestion, in addition, all isolates were U. parvum, serovar 3.
Discussion:
U. urealyticum can be one of the causing agents of prostatitis. Using PCR-RFLP with specific primer and restriction enzyme is a rapid and cost-effect method for detection and differentiation of Ureaplasma from clinical samples.
Key Words: Ureaplasma urealyticum, Ureaplasma parvum, PCR-RFLP, Prostatitis, Prostate tissue
Introduction
Prostatitis is an inflammation of the prostate gland, which can be varied from an acute to chronic medical condition (1). It is mostly caused by a bacterial infection (2). In recent years, the role of genital Mycoplasma especially U. urealyticum has been evaluated (3).
The genus Ureaplasma belongs to Mycoplasmataceae family and mollicutes class. Like other members of this class, Ureaplasma is lacking a cell wall and the smallest free-living organisms known. U. urealyticum is considered as agent of nongonococcal urethritis (NGU), acquired arthritis and prostatitis in men (4). It is subdivided to U. parvum, including serotypes 1, 3, 6 and 14 and is designated as biovar 1. The other biovar of U. urealyticum include the remaining 10 of established serotypes and designated as biovar 2 (5).
Less than 60% DNA homology are between biovars (6). Using culture for detection of Ureaplasma species in patient samples such as prostate tissues is laborious, taking several days and requiring a high degree of technical skill (7). PCR method is specific, sensitive and provides results in short time. Due to its equivalent and fast performance than to culture, it can be replaced to conventional culture for detection of U.urealyticum and U. parvum (8, 9). In addition, culture alone does not differentiate U. urealyticum from U. parvum. In several molecular typing methods introduced for U.urealyticum, PCR-RFLP has some advantages such as easy interpretation, implementation and using for large quantities of samples (10, 11).
Both biovars can be differentiated by this method, which assists in studies of the clinical significance, epidemiology and pathology of this species in human. Ryo et al. examined the 16sRNA-23sRNA intergenic spacer regions of U.urealyticum and showed that this region also can be used for biovar identification (12). For the diagnosis of bacterial prostatitis, samples such as voided urine, urethral swabs, and expressed prostatic secretions can be used. The normal bacterial flora in the area of genitourinary causes contamination of samples and makes it difficult to interpret the results. To avoid contamination with flora, the appropriate sample is direct using of prostate tissue (1). Recently, a study was done especially on prostate samples for detection of Ureaplasma in cancer cases but there is not any report of the role of U. urealyticum in prostatitis in Iran (13).
The aim of this study was to molecular detection of U. urealyticum in human prostate tissue samples in prostatitis cases based on PCR-RFLP.
Materials and Methods:
In this project, a cross sectional study was conducted and 200 prostate tissues were collected from prostatitis patients referred to Tehran hospitals during 2008 to 2010 for the preparation and examination. Only men with prostatitis were enrolled in this study and all of them subjected to prostate biopsy by a physician.
The prostatitis tissues were embedded with paraffin by pathologist. In the laboratory, using a microtome with disposable blades, 5-10 µ sections were prepared from blocks and transferred into sterile micro-tubes. To prevention cross contamination between the samples, the microtome blade and gloves were changed and the microtome were washed with 70% ethanol between samples. DNA was extracted from paraffin-embedded tissue samples using a special kit (manufactured by QiaGen Co). Briefly, 25 mg of cutting prostate tissue mixed with 20-µg proteinase K and incubated at 56 ˚C until the tissue was completely lysed. Two hundred µl of first buffer was added to the sample and incubated for 10 min at 70 ˚C. Then 200 µl of ethanol were mixed with sample. Mixture was applied to the column and centrifuged at 600-x g for 1 min. Another Buffer was added in a sequence as protocol. At the end of process, DNA was collected in a clean micro-tube by Elution Buffer and stored at 4 °C for PCR or at -20 °C for long-term storage.
A PCR assay with primers UuF5’-(TGGAGTTAAGTCGTAACAAG)-3’ and UuR5’-(CTGAGATGTTTCACTTCACC)-3’ was used and amplified a 559-bp sequence. Primers for amplification of 16S-23SRNA interspace region of Ureaplasma were synthesized by CinnaGen Company. The PCR reaction was performed in a total volume of 30 µl. Each reaction contained 15 µl Master Mix (2X), 1.5 mM MgCl2, 1µg DNA template, 20 pmol of each reverse and forward primer, and sterile distilled water. PCR reaction mixture with Ureaplasma DNA and PCR reaction mixture without DNA were used as positive and negative control, respectively. The initial denaturation performed for 5 min at 95 ˚C. Total of 35 cycles was carried out as follows: denaturation for 30 sec at 94 ºC, annealing for 60 sec at 56 ºC and extension for 60 sec at 72 ºC. The final extension was completed for 5 min at 72 ºC. PCR product electrophoresed on 1.8% gel agarose- TBE buffer (Boric acid 27.5 gr, Tris base 54 gr, EDTA 20 ml (pH=8). For confirming of U. urealyticum, the PCR product was send for sequencing.
After sequencing, the BLAST program was used for alignment the sequence with other sequences available in database. The sequences were studied with Web Cutter software. Therefore, a list of restriction enzyme cutting sites was prepared. To identify polymorphism, the PCR product obtained from Ureaplasma DNA was digested with EcoRI, AluI, TaqI, CacI8 and BbsI. For enzymatic digestion, 15 µl reactions was prepared in the following ingredient; 6.3 µl sterile distilled water, 1.5 µl enzyme buffer, 0.2 µl restriction enzyme and 7µl PCR product. After providing the reactions, micro-tubes containing all enzyme except TaqI were put in the water bath at 37 ºC for 2 h and micro-tubes containing the enzyme TaqI were put at 65 ºC for 2 h. After 2 h, products of RFLP were electrophoresed on 3% agarose gel. Finally, the size of the fragment resulted from enzymatic digestion of all positive PCR samples, was compared with each other and the biovars were identified.
Statistical analysis was conducted to determine how many samples were positive for U. urealyticum. Perspective analyses were performed and data with rounded-up numerical values (percentage) were documented.
Results
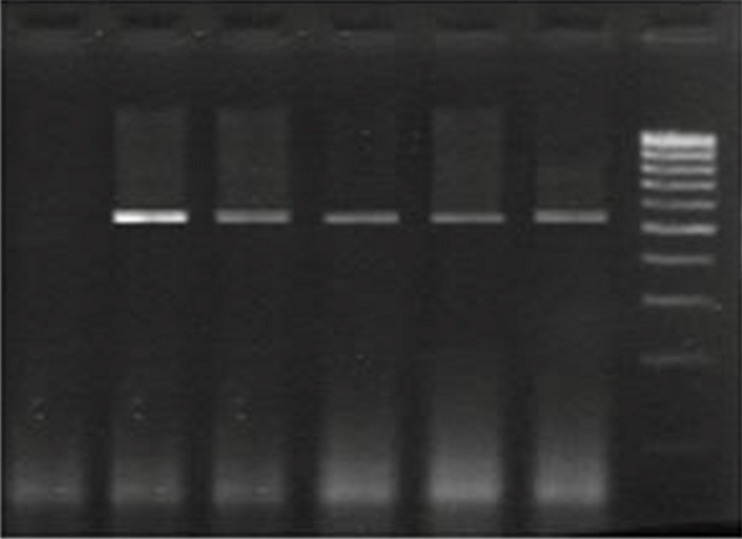
In 200 prostate tissue samples that their DNA was extracted and examined by PCR method, seven (3.5%) cases were positive for U. urealyticum and a fragment with 559 bp size was obtained (Fig. 1). Patients ranged in terms of age from 41 to 90 yr. Two patients were diagnosed with cancer which their samples contained U. urealyticum.
Fig.1.
Agarose gel electrophoresis of PCR amplified products. Lane 1. Negative control. Lane 2. Positive control. Lane 3,4,5,6. Positive samples containing of Ureaplasma urealyticum (559 bp band). Lane 7. Size marker (100bp DNA ladder
Obtaining results of PCR product sequencing and using of BLAST software shown that all of positive prostate samples contained Parvum biovar. Urealyticum biovar was not in samples.
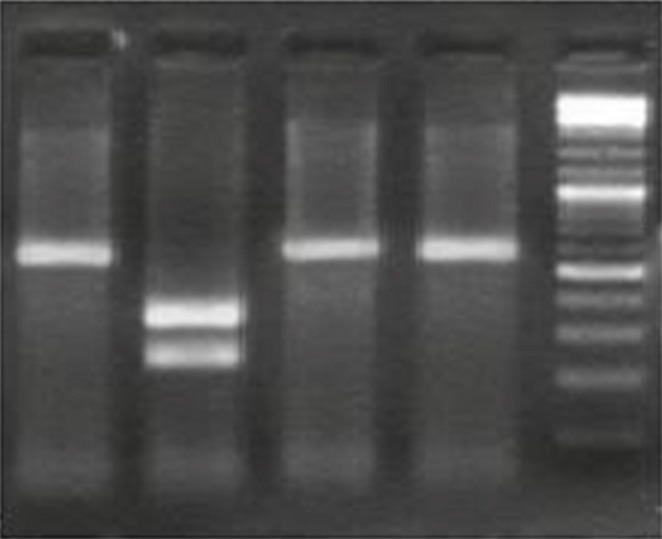
The PCR products were confirmed by RFLP. The restriction enzyme selection was based on PCR product sequencing results. The PCR products of U.urealyticum were digested by TaqI, which yielded 227 and 332 bp fragments (Fig. 2). Results of PCR-RFLP shown that there was not any differentiation in pattern of enzymatic digestion on PCR product and all isolates were from one type of U. parvum biovar belonging to serovar 3.
Fig. 2.
Enzymatic digestion of PCR product obtained from positive samples. Lane 1. 559bp-long PCR product Ureaplasma urealyticum as positive control, Lane 2. PCR-RFLP pattern of Ureaplasma urealyticum after digestion with TaqI, specific enzyme that made two pieces of 227 bp and 332 bp; Lane 3, 4. PCR-RFLP pattern of U. urealyticum after digestion with Cac8I and BpsI (it is not digested); Lane 5. Size marker (100bp DNA ladder
Discussion
Prostatitis is a common disorder of men and it is estimated that half of men suffer from symptoms at same stage of their live (14, 15). In many cases, urologists often find themselves, incapable to diagnosis and treat. Different bacteria causing the infection and the role of Ureuplasma as an etiologic agent of prostatitis are controversial (16). Ureaplasma one of the most common pathogens causing sexually transmitted disease that is important to determine its prevalence in men with prostatitis. Although Ureaplasma species have ability to visibly grow on the media but using molecular methods for identification of species and serovar for interpretation of pathogenicity is important (17, 18).
In this study, U.urealyticum was detected in 3.5% of prostate tissue samples using conventional PCR. In a study, multiplex PCR was used for detection of U.urealyticum in 92 specimens including prostate secretion and urine from patients. Amplifying a fragment of 16S rRNA gene by multiplex PCR method, detected U.urealyticum in 4 (4.3%) samples (19). Lee et al. used multiplex PCR for detection of U.urealyticum and other bacteria in one-step in 96 urine samples. U. urealyticum was not detected in urine samples (20). However, present study differs from most of studies in type and size of sample and using of method. In these previous studies, urine samples and prostate secretions were used for detection of bacteria while we used prostate tissue for direct detection of Ureaplasma in prostate gland tissue.
PCR in the detection of pathogens in bacterial prostatitis is very effective. Because of many patients taking several antibiotics before an accurate diagnosis of bacterial, this will interfere with the growth of bacteria in vitro. Therefore, using PCR we can identify the causative agent directly and rapidly. Because the gene sequence obtained from PCR method has a specific restriction site for Ureaplasma species, using this method is possible to distinguish the different species. In this study, RFLP method was used for typing to better understand the pathology, epidemiology and clinical significant of Ureaplasma species in human prostatitis. In the first step, result of PCR product sequencing showed that U. parvum (biovar 1) was predominant isolate and U. urealyticum (biovar 2) was not detected. In other hand, there was no difference between obtained patterns of Taq1 enzyme digestion on PCR product from positive samples. These results state that all of U. parvum biovar belong to serovar 3. The findings are consistent with previous studies (21, 22). In a study, U. parvum was the most frequently occurring species (23). Eventually, U. parvum serovar 3 has a possible photogenic role.
Conclusion
U. urealyticum could be one of the causing agents of prostatitis. Since all of biovars belong to U. parvum, therefore it can be said that the causative agents of prostatitis in men are due to play a single biovar that makes it easier to follow up and treatment of infection origin. Using of PCR-RFLP with specific primer and restriction enzyme is a rapid and cost-effect method for detection and differentiation of Ureaplasama from clinical samples.
Acknowledgments
We specially thank the Deputy of Research Affairs of Iran University of Medical Sciences for their continuous support.
Conflict of interest
The authors declare that there is no conflict of interests.
References
- 1.Domingue G, Hellstrom WP. Prostatistis. Clinical Microbiology Reviewes. 1998:604–13. doi: 10.1128/cmr.11.4.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Škerk V. Azithromycin in the treatment of chronic bacterial prostatitis caused by traditional and unusual pathogens. 20th Europea Congress of Clinical Microbiology and Infectious Diseases.2010. [Google Scholar]
- 3.Brunner H, Weidner W, Schiefer HG. Studies on the role of Ureaplasma urealyticum and Mycoplasma hominis in prostatitis. J Infect Dis. 1983;147(5):807–13. doi: 10.1093/infdis/147.5.807. [DOI] [PubMed] [Google Scholar]
- 4.Domingues D, Tavira LT, Duarte A, Sanca A, Prieto E, Exposto F. Ureaplasma urealyticum Biovar Determination in Women Attending a Family Planning Clinic in Guiné-Bissau, Using Polymerase Chain Reaction of the Multiple-Banded Antigen Gene. J Clin Lab Anal. 2002;16:71–5. doi: 10.1002/jcla.10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yi J, Yoon B, Kim EC. Detection and biovar discrimination of Ureaplasmaurealyticum by real-time PCR. Mol Cell Probes. 2005;19(4):225–60. doi: 10.1016/j.mcp.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 6.Pitcher D, Sillis M, Robertson J. Simple Method for Determining Biovar and Serovar Types of Ureaplasma urealyticum Clinical Isolates Using PCR–Single-Strand Conformation Polymorphism Analysis. J Clin Microbiol. 2002;l39(5):1840–4. doi: 10.1128/JCM.39.5.1840-1844.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stellrecht KA, WoronAM , Mishrik NG, Venezia RA. Comparison of multiplex PCR assay with culture detection of genital mycoplasmas. J Clin Microbiol. 2004;42:1528–33. doi: 10.1128/JCM.42.4.1528-1533.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farrell JJ, Larson JA, Akeson JW, Lowery KS, Rounds MA, Sampath R, et al. Ureaplasmaparvum prosthetic joint infection detected by PCR. J Clin Microbiol. 2014;52:2248–50. doi: 10.1128/JCM.00432-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petrikkos G, Hadjisoteriou M, Daikos G. PCR versus culture in the detection of vaginal Ureaplasmaurealyticum and Mycoplasma hominis. Int J Gynecol Obstetr. 2007;97(3):202–3. doi: 10.1016/j.ijgo.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 10.Ota M, Asamura H, Oki T, Sada M. Restriction enzyme analysis of PCR products. Methods Mol Biol. 2009;578:405–14. doi: 10.1007/978-1-60327-411-1_25. [DOI] [PubMed] [Google Scholar]
- 11.Rasmussen H. In: Restriction Fragment Length Polymorphism Analysis of PCR-Amplified Fragments (PCR-RFLP) and Gel Electrophoresis – Valuable Tool for Genotyping and Genetic Fingerprinting. Magdeldin , editor. InTech Publication; 2012. [Google Scholar]
- 12.Harasawa R, Kanamoto Y. Differentiation of two biovars of Ureaplasma urealyticum based on the 16S-23S rRNA intergenic spacer region. J Clin Microbiol. 1999;37(12):4135–8. doi: 10.1128/jcm.37.12.4135-4138.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eslami G, Goudarzi H, Baseri N, Ghalavand Z, Taherpour A, Zhaam H, et al. The Prevalence of Ureaplasma Urealyticum and Mycoplasma Genitalium in Patients with Prostate Cancer in Shohada Hospital in Tehran, Iran. NBM. 2015;2:73–8. [Google Scholar]
- 14.Viscardi R. Ureaplasma species: Role in Diseases of Prematurity. Clin Perinatol. 2010;37(2):393–409. doi: 10.1016/j.clp.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi YS, Kim KS, Choi SW, Kim S, Bae WJ, Cho HJ, et al. Microbiological etiology of bacterial prostatistis in general hospital and primary care clinic in korea. Prostate Int. 2013;1(3):133–8. doi: 10.12954/PI.13023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vaidyanathan R, Mishra V. Chronic Prostatitis: Current Concept. Indian J Urol. 2008;24(1):22–7. doi: 10.4103/0970-1591.38598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naher HS, Said IH. Culturing and PCR Methods for Detection of Mycoplasma hominis and Ureaplasma urealyticum in Women with Genitourinary Tract Infections. Int Res J Med Sci. 2013;1(3):25–9. [Google Scholar]
- 18.Waites KB, Xiao L, ParalanovV , Viscardi RM, Glass JI. Molecular methods for the detection of Mycoplasma and Ureaplasma infection in humans. J Mol Diagn. 2012;14(5):437–50. doi: 10.1016/j.jmoldx.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee MK, Kim TH, Lee M. Detection of Cryptic Microorganisms in Patients with Chronic Prostatitis by Multiplex Polymerase Chain Reaction. Korean J Urol. 2007;48(3):304–9. doi: 10.4111/kju.2007.48.3.304. [Google Scholar]
- 20.Lee SR, Chung JM, Kim YJ. Rapid one step detection of pathogenic bacteria in urine with sexually transmitted disease (STD) and prostatitis patient by multiplex PCR assay (mPCR) J Microbiol. 2007;45(5):453–9. [PubMed] [Google Scholar]
- 21.Kong F, Zhenfang MA, James G, Gordon S, Gilbert GL. Species Identification and Subtyping of Ureaplasma parvum and Ureaplasma urealyticum Using PCR-Based Assays. J Clin Microbiol. 2000;38(3):1175–79. doi: 10.1128/jcm.38.3.1175-1179.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Povlsen K, Jensen JS, Lind I. Detection of Ureaplasma urealyticum by PCR and Biovar Determination by Liquid Hybridization. J Clin Microbiol. 1998;36(11):3211–216. doi: 10.1128/jcm.36.11.3211-3216.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mander R, Raukas E, Turk S, Korrovits P, Punab M. Mycoplasmas in semen of chronic prostatistis patients. Scand J Urol Nephrol. 2005;39(6):479–82. doi: 10.1080/00365590500199822. [DOI] [PubMed] [Google Scholar]