Summary
Generation of induced pluripotent stem cells (iPSCs) from human urine-derived cells (hUCs) provides a convenient and non-invasive way to obtain patient-specific iPSCs. However, many isolated hUCs exhibit very poor proliferation and are difficult to reprogram. In this study, we optimized reprogramming approaches for hUCs with very poor proliferation. We report here that a compound cocktail containing cyclic pifithrin-a (a P53 inhibitor), A-83-01, CHIR99021, thiazovivin, NaB, and PD0325901 significantly improves the reprogramming efficiency (170-fold more) for hUCs. In addition, we showed that replacement of Matrigel with autologous hUC feeders can overcome the reprogramming failure due to the massive cell death that occurs during delivery of reprogramming factors. In summary, we describe improved approaches to enable iPSC generation from hUCs that were otherwise difficult to reprogram, a valuable asset for banking patient-specific iPSCs.
Keywords: CPFT-a, P53, small molecules, autologous UC feeders, hUCs, iPSCs
Graphical Abstract
Highlights
-
•
SM treatment significantly enhances the reprogramming of hUCs
-
•
Replacement of Matrigel with autologous hUCs as feeder facilitates reprogramming
-
•
Selection of cell-dependent reprogramming strategy is useful for banking iPSC lines
In this article, Pan G, Zhang H, Li Y, and colleagues show that poorly proliferating hUCs could be reprogrammed with the aid of small molecules (A-83-01, Chir, Tzv, CPFT-a, NaB, PD) and autologous UC feeders. The approaches using a small-molecule cocktail and autologous feeder cells significantly improves the reprogramming efficiency (170-fold more) and enable iPSC generation from hUCs with different proliferation states.
Introduction
Human induced pluripotent stem cell (iPSC) technology holds great potential for personalized regenerative medicine, drug discovery, and disease modeling without concerns about ethical issues associated with the human embryo (Yamanaka, 2009). For the achievement of this goal it is essential to generate patient-specific iPSCs in a way that is safe enough for clinical applications, for example, serum free, integration free, and animal components free. To date, several approaches have been published on the generation of virus-free human iPSCs, for example, using small molecules (SMs) (Esteban et al., 2010, Hou et al., 2013, Li et al., 2009, Lin et al., 2009, Mali et al., 2010, Shi et al., 2008, Yu et al., 2011, Zhu et al., 2010), non-viral minicircles (Jia et al., 2010), or non-integrating episomal vectors (Yu et al., 2009). We previously reported that human urine-derived cells (hUCs) could be reprogrammed into iPSCs with higher efficiencies than the widely used skin fibroblast, thus providing an easy and non-invasive way to obtain somatic cells for iPSC generation (Zhou et al., 2011). Moreover, we have shown that hUCs served as a good source to generate human iPSCs using a non-viral integration approach from different genetic backgrounds (Xue et al., 2013). However, in our experience culturing hUCs from different donors, we found that a significant number of hUCs exhibited very poor proliferation (Tables 1 and 2), and thus were problematic for further iPSC generation. However, reprogramming these poorly proliferating hUCs is usually necessary for banking human iPSC lines from different donors.
Table 1.
Isolation UCs from Healthy Adults
| No. of Samples Collected | No. of Samples with Successful Isolation | % UC Isolation | No. of UC Samples with Low Growth Rate | % of UC Samples with Low Growth Rate | |
|---|---|---|---|---|---|
| Male | 24 | 8 | 33 | 4 | 50 |
| Female | 21 | 5 | 24 | 2 | 40 |
| Total | 45 | 13 | 29 | 6 | 46 |
Table 2.
Isolation of UCs from Patients with Diabetes and Blood Disorders
| Disease | Sex | No. of Samples Collected | No. of Samples with Successful Isolation | % UC Isolation | No. of UC Samples with Low Growth Rate | % of UC Samples with Low Growth Rate |
|---|---|---|---|---|---|---|
| T1D | male | 6 | 4 | 67 | 3 | 75 |
| female | 3 | 2 | 67 | 0 | 0 | |
| total | 9 | 6 | 67 | 3 | 50 | |
| T2D | male | 38 | 14 | 37 | 9 | 64 |
| female | 12 | 1 | 8 | 1 | 100 | |
| total | 50 | 15 | 30 | 10 | 67 | |
| Blood disorders | male | 8 | 2 | 25 | 0 | 0 |
| female | 2 | 0 | 0 | 0 | 0 | |
| total | 10 | 2 | 20 | 0 | 0 | |
| Overall total | 69 | 23 | 33 | 13 | 57 |
T1D, type 1 diabetes; T2D, type 2 diabetes.
Many SMs have been reported to facilitate reprogramming. For example, sodium butyrate (N, NaB), a histone deacetylase (HDAC) inhibitor, greatly improved reprogramming efficiency by upregulating epigenetic remodeling and pluripotency-associated genes (Mali et al., 2010, Zhang and Wu, 2013). In another study, Yu et al. (2011) reported that an SM cocktail containing MEK inhibitor PD0325901 (PD), GSK3β inhibitor CHIR99021 (C, Chir), transforming growth factor β (TGF-β)/Activin/Nodal receptor inhibitor A-83-01 (A), ROCK inhibitor HA-100, and human leukemia inhibitory factor (hLIF), facilitates iPSC generation from human neonatal foreskin fibroblasts. Also, Chir was used to promote iPSC generation from human primary keratinocytes (Li et al., 2009). A summary of these SMs is presented in Table 3.
Table 3.
Six Small Molecules Used for Reprogramming
| Abbreviation | Small Molecule | Function in Reprogramming | Target | References |
|---|---|---|---|---|
| A | A-83-01 | promotes reprogramming of human epidermal keratinocytes using OCT4 and KLF4 | (−)TGF-β(smad2) | Yu et al., 2011, Zhu et al., 2010 |
| C | CHIR99021 | combined with parnate results in the reprogramming of human primary keratinocyte transduced with OCT4 and KLF4 | (−)GSK-3β | Yu et al., 2011, Zhu et al., 2010 |
| T | thiazovivin | ROCK inhibitor which dramatically improves the reprogramming efficiency in the presence of PD, Chir, A-83-01, and hLIF | (−)ROCK | Yu et al., 2011 |
| P | cyclic pifithrin-α | suppression or silencing of P53 significantly increased the reprogramming efficiency of human somatic cells | (−)P53 | Hong et al., 2009, Kawamura et al., 2009, Okita et al., 2011 |
| N | sodium butyrate | enhanced miR302/367 cluster, histone H3 acetylation, promoter DNA demethylation, and the expression of endogenous pluripotency-associated genes | (−)HDAC | Mali et al., 2010, Zhu et al., 2010, Zhang and Wu, 2013; |
| PD | PD0325901 | PD serves as a selection strategy when generating iPSCs from virally transduced neural progenitor cells, and contributes to stabilization of the iPSC state. Selectively binds and inhibits MEK, which may result in the inhibition of phosphorylation and activation of MAPK/ERK and the inhibition of tumor cell proliferation | (−)MEK-(+)MAPK/ERK | Yu et al., 2011, Lin et al., 2009, Zhu et al., 2010 |
See also Figure S2.
In addition, reprogramming factors OSKM have been shown to trigger cell senescence by upregulating P53, P16INK4a, and P21CIP1 (Banito et al., 2009). P53-mediated cell senescence has been known as a barrier to reprogramming (Hong et al., 2009, Kawamura et al., 2009) and downregulation of P53 could promote reprogramming and iPSC generation (Okita et al., 2011). Cyclic pifithrin-a (P, CPFT-a) is a transcriptional inhibitor of P53. Compared with P53 short hairpin RNA or dominant-negative P53 protein, inhibition of P53 by CPFT-a is efficient and easy to control. Because P53 plays an important role in safeguarding genome stability, it is ideal to release its inhibition in newly generated iPSCs after reprogramming. Here, to enable efficient iPSC generation from hUCs with various proliferation capabilities, we optimized non-viral approaches for hUC reprogramming. Firstly, we selected CPFT-a, a P53 inhibitor, and other SMs known to promote reprogramming such as A-83-01, Chir, thiazovivin (T, Tzv), and NaB, to form an SM cocktail. Aided by this SM cocktail and another SM, PD0325901 introduced only at later stage of reprogramming, the hUC reprogramming efficiency was significantly enhanced, up to 170-fold. The highly efficient reprogramming could be achieved in 2 weeks with the aid of these SMs. Furthermore, we found that non-transfected autologous hUCs can be used as feeder to overcome the cell death caused by electroporation for the transfection of reprogramming genes. Upon these optimizations, successful rates of generating viral-free iPSCs from hUCs with different states were greatly improved.
Results
SM Treatment Enables iPSC Generation from Poorly Proliferating hUCs
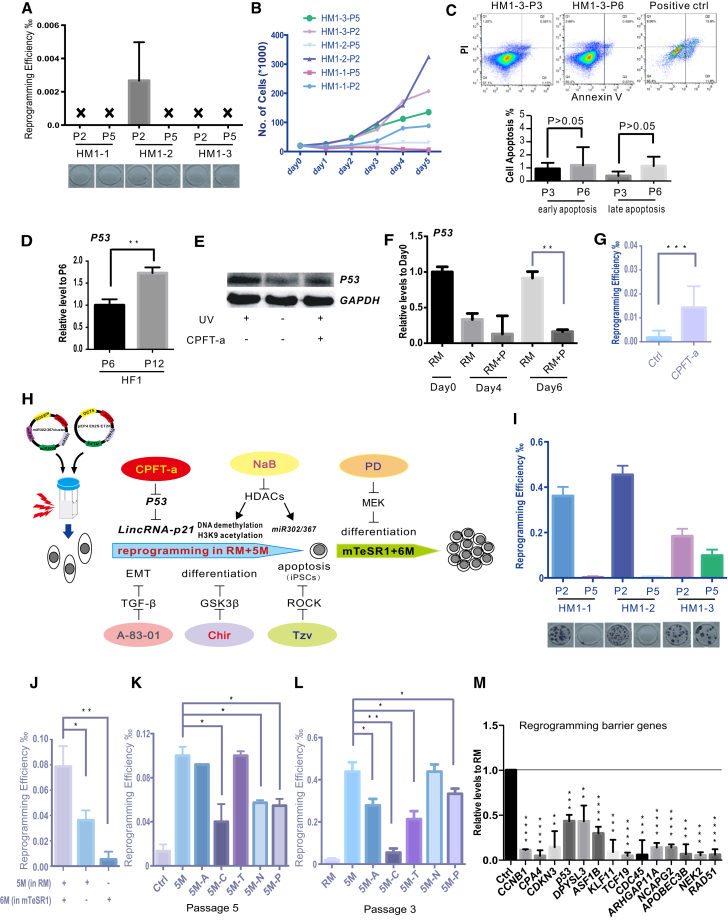
In our attempt to isolate hUCs from healthy adults or different patients with disease such as type 1 diabetes, type 2 diabetes, and other blood disorders, the successful rate for obtaining expandable hUCs is about 30% (Tables 1 and 2). Osmolality (OSM) of urine samples is an important index for successful hUCs isolation. We found that it is usually difficult to obtain cells if the OSM of urine is more than 598 mmol/l or less than 241 mmol/l. Furthermore, 50% of the successfully isolated hUCs are very poor in proliferation and the efficiencies for iPSC generation vary widely between different batches. For example, three different batches of hUC cell lines (HM1-1, HM1-2, and HM1-3) derived from the same person showed dramatically different reprogramming efficiencies using episomal vectors and the previously reported approaches (Yu et al., 2009, Xue et al., 2013) (Figure 1A). During this reprogramming process, RM medium (REGM + DMEM/high glucose containing minimal essential medium [MEM] non-essential amino acids [NEAA], glutaMAX-1, and 10% fetal bovine serum [FBS] 1:1; the combination of the three is referred to as RM used for hUC culture) was used at the initial induction stage (days 2–9), while mTeSR1 (human embryonic stem cell [hESC] culture medium) was used at later stage to expand the reprogrammed cells (days 10–13). To observe the morphological changes more clearly, we seeded an appropriate number of transfected hUCs onto 24-well plates at a density of 2.5 × 104 cells/well. As shown in Figure 1A, only HM1-2 at passage 2 (HM1-2-P2) produced alkaline phosphatase (AP)-positive iPSC colonies. Consistently, HM1-2-P2 showed the highest proliferation among all examined cell lines (Figure 1B), suggesting that cell proliferation is a critical factor for a successful iPSC generation. The apoptosis rates were not significantly increased in hUCs with higher passage number (Figure 1C), indicating that the apoptosis of hUCs might not be the rate-limiting event during reprogramming. It has been reported that P53 was activated and served as a critical barrier during reprogramming (Banito et al., 2009). We showed that the P53 level increased with the passages of hUCs (Figure 1D), suggesting that P53 might also be a negative factor for hUC reprogramming. To promote iPSC generation from poorly proliferating hUCs we chose CPFT-a, an SM inhibitor for P53, to suppress P53 during hUC reprogramming. We firstly confirmed the effective suppression of P53 by CPFT-a using western blot (Figure 1E). Then we showed that with CPFT-a added in reprogramming medium, P53 was downregulated while the reprogramming efficiency significantly improved (Figures 1F and 1G). In addition, we tested other SMs that are known to promote reprogramming in our hUC reprogramming system. To this end, we finalized five SMs (A-83-01, CHIR99021, Tzv, NaB, PD0325901) that could work with CPFT-a to promote hUC reprogramming. During iPSC generation, we added five SMs (A-83-01, Chir, Tzv, CPFT-a, NaB; i.e., ACTPN) at the early stage (first 7 days) and added one more compound, PD, at the later stage to expand the reprogrammed iPSCs (Figure 1H). We named this strategy as plan B (Figure 3A). Based on this strategy, all hUCs described in Figure 1A could be reprogrammed into iPSCs (Figure 1I). These data indicate that a combination of P53 inhibitor with other SMs in reprogramming medium enables iPSC generation from hUCs that are otherwise difficult to be reprogrammed.
Figure 1.
Cocktail of P53 Inhibitor CPFT-a and Other Small Molecules Improves Reprogramming Efficiency of UCs Isolated from Individual Healthy Adults
(A) UCs reprogramming using RM-mTeSR1 without small-molecule cocktail. UCs HM1-1, HM1-2, and HM1-3 represent UCs collected from a healthy male donor at passages (P) 2 and 5 and used for reprogramming (n = 3).
(B) Growth curves of UCs at passages 2 and 5 (n = 3).
(C) FACS analysis on cell apoptosis through the expression of annexin V and PI. The percentage of annexin V-positive cells was analyzed. HM1 includes HM1-1, HM1-2, and HM1-3. Positive control was treated with 800 μM H2O2 for 4 hr.
(D) The expression level of P53 tested by western blot. HF1 represents UCs collected from a healthy female donor at passages 6 and 12 and used for qPCR. ∗∗p < 0.01 (n = 3).
(E) Effect of cyclic pifithrin-a, a P53 inhibitor, on P53 expression. UV, 8 hr after UV ray treatment (15 s, 6000 μW/cm2). −UV, RM functions as the negative control without UV treatment. CPFT-a, incubated for 8 hr with 10 μM CPFT-a.
(F) The expression level of P53 of UCs in the process of reprogramming. UCs, HF14-P5. ∗∗p < 0.01 (n = 3).
(G) UCs reprogramming treated with CPFT-a. UCs, HM1-P3. ∗∗∗p < 0.001 (n = 3).
(H) Scheme of reprogramming plan B (R5 +5 M-mTeSR1 + 6M, ACTNP was added in RM for induction stage and ACTNP + PD was added in mTeSR1 for proliferation stage).
(I) Reprogramming efficiency using plan B. UCs HM1-1, HM1-2, and HM1-3, at passages 2 and 5, were used for reprogramming (n = 3).
(J) Reprogramming efficiency with small-molecule cocktail added at different stages. UCs, HM9-P3. ∗p < 0.05, ∗∗p < 0.01 (n = 3).
(K) Role of single small molecule in RM + 5M for UCs with poor proliferation ability. UCs, HF14-P5. ∗p < 0.05 (n = 3).
(L) Role of single small molecule in RM + 5M for UCs with good proliferation. UCs, HF14-P3. ∗p < 0.05, ∗∗p < 0.01. (n = 6).
(M) The expression level of 14 known reprogramming barriers in UCs treated by RM + 5M normalized. UCs, HM1. ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 (n = 3).
Data were analyzed using a two-tailed Student's t test. Error bars indicate the SD. Replicates were biological independent experiments. For a summary of small molecules used for reprogramming, see Table 3. For the role of PD in reprogramming, see Figure S3.
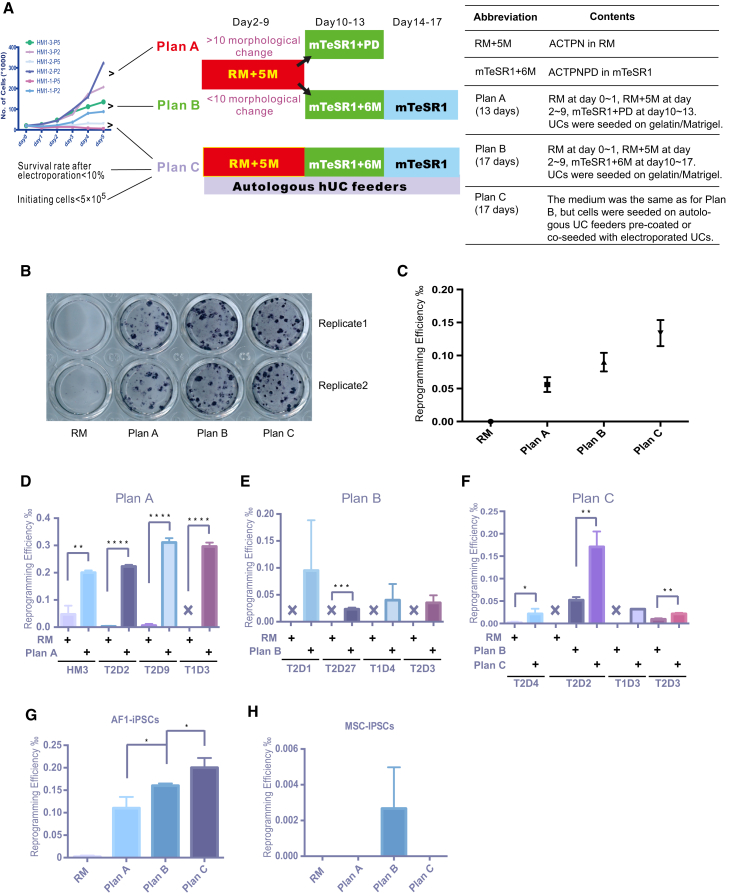
Figure 3.
Comparison of Plans A, B, and C on Reprogramming of UCs, Amniotic Fluid Cells, and Mesenchymal Stem Cells Collected from Umbilical Cord Blood
(A) Scheme of plans A, B, and C for reprogramming. Growth curves of UCs were the same as shown in Figure 1B.
(B) AP staining assay of iPSCs induced from HM1-3-P3 used RM, plans A, B, and C (n = 2).
(C) The bona fide iPS clones induced by RM, plans A, B, and C. UCs, HM1-3-P3 (n = 2).
(D–F) Plans A, B, and C were used to construct an iPSC bank. UCs, HM3, T2D2, T2D9, T1D3, T2D1, T2D27, T1D4, T2D3, T2D4. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 (n = 3).
(G) Amniotic fluid cells (AF1-P5) reprogramming using plans A, B, and C. AF1 was used as feeder in plan C. ∗p < 0.05 (n = 3).
(H) Mesenchymal stem cells collected from umbilical cord blood (MSC-P5) were reprogrammed by plans A, B, and C. HF1 was used as feeder in plan C (n = 3).
Data were analyzed using a two-tailed Student's t test. Error bars indicate the SD. Replicates were biological, independent experiments.
We further characterized iPSCs generated with the reprogramming strategy plan B. Two iPSCs, HM1-iPS and HF1-iPS, generated from healthy male and female adult UCs were selected for further characterization. These two iPSCs showed typical hESC morphology (Figures S1A and S1B) and could be well expanded on Matrigel in mTeSR1 medium. They maintained normal karyotype and AP positivity during multiple passaging (Figures S1C–S1F). PCR analysis confirmed that these iPSC colonies had no transgene expression or genomic integration (Figure S1G). Typical pluripotency genes such as OCT4, SSEA4, TRA-1-60, and TRA-1-81 were activated and detected by fluorescence-activated cell sorting (FACS) (Figure S1H). Moreover, both OCT4 and NANOG promoters were demethylated in these iPSCs based on the bisulfate sequencing analysis (Figure S1I). Upon injection into NOD/SCID mice, they formed typical teratomas containing tissues from all three germ layers (Figure S1J). In addition, we performed whole-genome transcriptome analysis on iPSCs generated with SMs. Four iPSCs from different batches of hUCs generated with SMs showed little difference to hESCs, H1, in terms of whole-genome transcriptome as well as expression of selected pluripotent genes (Figures S1N–S1P). Taking all the data together, we developed an optimized approach to enable iPSC generation from poorly proliferating hUCs and low reprogramming efficiency.
Minimizing SM Treatment during hUC Reprogramming
Although adding SMs dramatically promotes hUC reprogramming, too much SM treatment raised the concern about generating more unwanted genetic or epigenetic mutations (Wu et al., 2013). Thus, we sought to examine whether some SMs could be removed during the whole reprogramming process. HM9, an hUC line isolated from a healthy male, was used for the testing. The reprogramming efficiency went down by about half when all six SMs were withdrawn at later expansion stage (RM + 5M [ACTPN in RM]-mTeSR1) and was more dramatically reduced upon withdrawal at the early induction stage (RM-mTeSR1 + 6M [ACTPN + PD in mTeSR1]) (Figure 1J). These data indicate that SM exposure at the early stage is much more critical for reprogramming. Therefore, for highly proliferating hUCs, SMs could be removed at the later stage (RM + 5M-mTeSR1, referred to as plan A) for iPSC generation (Figure 3A). We then examined whether we could further remove some individual SMs at the early induction stage to reprogram those highly proliferating hUCs. Two hUCs with different proliferation rates, HF14-P3 (high) and HF14-P5 (low), were used for further analysis. For HF14-P5 with low proliferation, the reprogramming efficiency was significantly reduced when C, N, or P was withdrawn, while removal of A or T had little effect. These data suggested that C, N, and P played important roles in reprogramming poorly proliferating hUCs (HF14-P5) (Figure 1K). For HF14-P3 with higher proliferation, the reprogramming efficiency was significantly reduced when C was withdrawn, but not A, T, or P (Figure 1L). Based on these findings, we suggest that to reprogram highly proliferating hUCs, N could be omitted, while for poorly proliferating hUCs, A and T could be removed. In addition, we found that SM treatment inhibited the genes reported to be barriers to reprogramming (Qin et al., 2014) (Figure 1M), which could be part of the mechanism for these SMs in promoting hUC reprogramming.
Autologous UC Feeder Facilitates iPSC Generation from hUCs
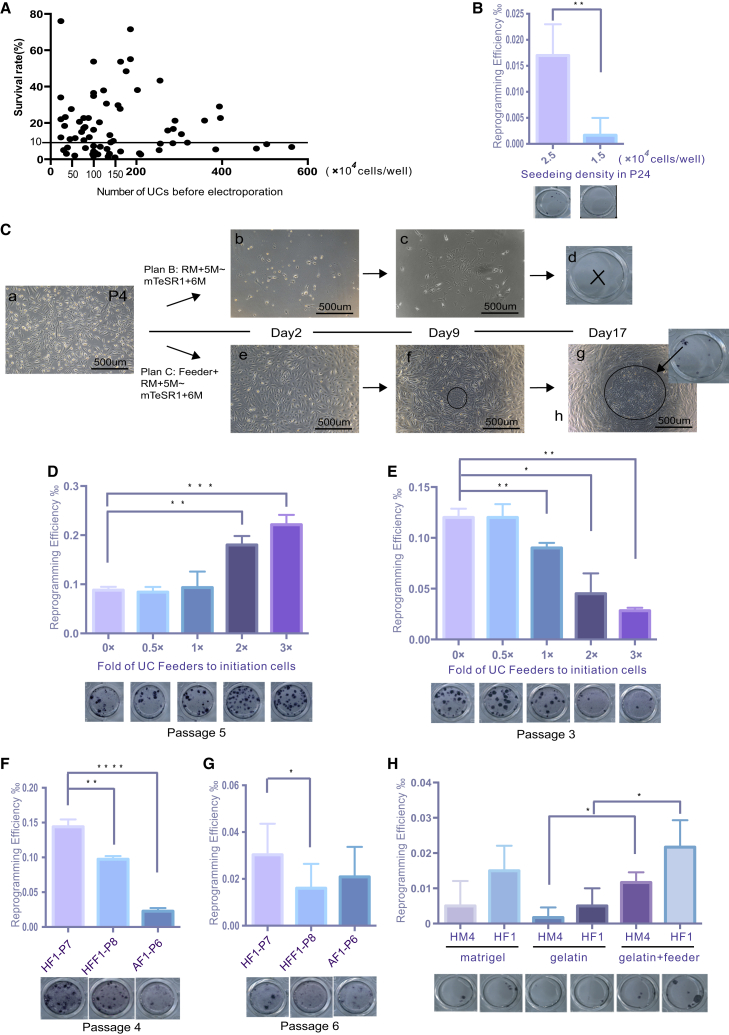
Another technical problem that usually occurs during iPSC generation is cell death caused by electroporation for the transfection of reprogramming factors. On many occasions, the survival rate of hUCs after electroporation was less than 10% (Figure 2A), which is problematic for subsequent iPSC generation. In our experience, iPSC colonies can be successfully generated if the number of surviving cells is more than 2.5 × 104 cells/well of a 24-well plate, while it usually fails if the surviving cells amount to fewer than 1.5 × 104 cells/well (Figure 2B). To overcome the reprogramming failure caused by low cell density, we replated the transfected hUCs onto the untransfected autologous hUCs for reprogramming and iPSC generation. T1D3, a hUC line isolated from a type 1 diabetes patient, could not be reprogrammed to generate iPSCs using plan B described above, due to low survival rate after electroporation (Figure 2C, upper panel). However, typical iPSC clones appeared upon plating the transfected hUCs onto the autologous untransfected hUCs as feeders for reprogramming (Figure 2C, lower panel and Figures S1K–S1P). We termed this approach plan C (Figure 3A). Moreover, we found that application of autologous hUC feeder also dramatically enhanced the reprogramming efficiency of hUCs, with higher passage number and poor proliferation (Figure 2D). However, for hUCs with lower passage numbers and good proliferation, including the hUC feeder was not beneficial and even inhibited iPSC generation (Figure 2E), indicating that too many hUCs may compete with the growth of reprogrammed cells. We also showed that employing other cell lineages as feeders, such as human foreskin cell and amniotic fluid cells, showed little effect on hUC reprogramming (Figures 2F and 2G). In addition, including the appropriate number of autologous hUC feeders could substitute Matrigel during reprogramming, which is helpful in developing a totally defined condition for iPSC generation (Figure 2H).
Figure 2.
Autologous UC Feeders Facilitate Reprogramming of UCs with Poor Proliferation Ability
(A) Survival rate of different UC samples after nucleofection. Each dot represents a UC sample for reprogramming. The cell number before and after nucleofection was calculated by cell count. UCs were collected from healthy adults and patients (n = 73).
(B) Poorly proliferating UCs at low cell density (1.5 × 104 cells in one well of a 24-well plate) displayed low reprogramming efficiency. ∗∗p < 0.01 (n = 3).
(C) UC T1D3-P4 with a low proliferation rate generated iPSCs with plan C. The iPSCs are indicated by a black circle. Scale bar, 500 μm.
(D) Optimization of the density of UC feeders for optimal reprogramming of UCs with poor proliferation ability. The fold was calculated based on UCs as initiating cells. UC, HM6-P5. ∗∗p < 0.01, ∗∗∗p < 0.001 (n = 3).
(E) Optimization the density of UC feeders for optimal reprogramming of UCs with good proliferation ability. The fold was calculated based on UCs as initiating cells. UCs, HM1-P3. ∗p < 0.05, ∗∗p < 0.01 (n = 3).
(F) Human foreskin fibroblast cell (HFF1-P8) and amniotic fluid cell (AF1-P6) as feeders for reprogramming of UCs at early passage. UCs, HF1-P4. ∗∗p < 0.01, ∗∗∗∗p < 0.0001 compared with HF1-P7 (n = 3).
(G) Human foreskin fibroblast cell (HFF1-P8) and amniotic fluid cell (AF1-P6) as feeders for reprogramming of UCs at late passage. UCs, HF1-P6. ∗p < 0.05 compared with HF1-P7 (n = 3).
(H) Substitution of Matrigel with autologous UC feeders on gelatin-coated plate. UCs, HM4, HF1. ∗p < 0.05 (n = 3).
Data were analyzed using a two-tailed Student's t test. Error bars indicate the SD. Replicates were biological, independent experiments. For characteristics of iPSCs generated with reprogramming strategies plan B and plan C, see Figure S1.
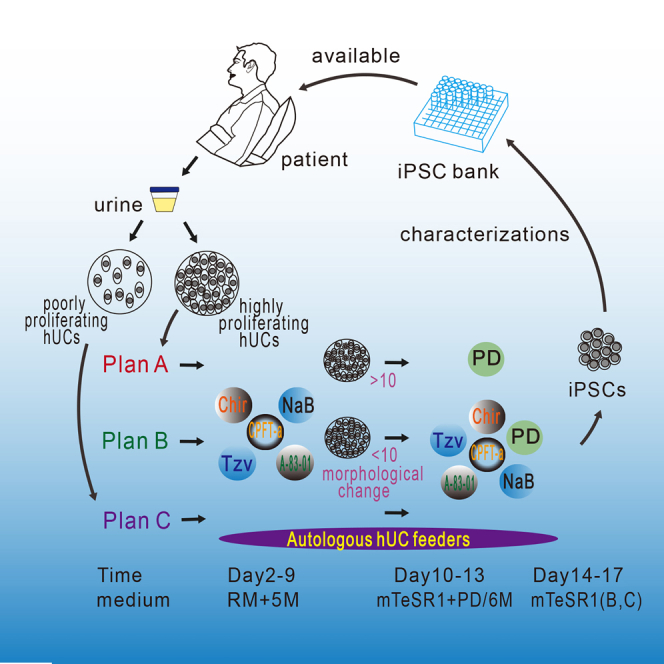
Cell-Dependent Reprogramming Strategy for Banking iPSC Lines
Banking human iPSCs from different genetic backgrounds would be valuable for further studies on disease modeling and drug screening as well as regenerative medicine. Here, we summarized three reprogramming strategies for reprogramming hUCs with various properties (Figure 3A). For banking iPSCs, we selected a particular strategy to generate hUC-derived iPSCs based on the proliferation capability of the isolated hUCs. For example, we chose plan A for hUCs with high proliferation and plan B for those with moderate proliferation. Plan C can be used if the survival rate of hUCs after electroporation is very low (Figure 3A). To test these strategies, we chose a hUC line with moderate proliferation to generate iPSCs using plans A, B, and C, respectively, and found that plan C was the best while plan B was better than plan A (Figures 3B and 3C). Based on the selected strategy, we were able to generate iPSCs from most batches of the isolated hUCs that was otherwise difficult to achieve using previously reported methods (Figures 3D–3F). We also examined these strategies in reprogramming other cell types, such as amniotic fluid cells (AF1-P5) and mesenchymal stem cells (MSCs). Our described approaches also enhanced iPSC generation from AF cells or MSCs, albeit the overall reprogramming efficiency of MSCs was much lower than that of AF cells (Figures 3G and 3H). In summary, we developed a panel of approaches to reprogram hUCs with different proliferation capabilities, potentially of value in banking human iPSC lines from different genetic backgrounds.
Discussion
Generation of human iPSCs through a non-virus integration approach is usually slow and less efficient and thus hampers its wider applications, particularly for banking iPSCs from different genetic backgrounds and patients. Cells isolated from human urine provide a convenient and non-invasive way to obtain patient cells for iPSC generation. However, we found that a high number of isolated hUCs showed limited proliferation and were difficult to reprogram for iPSC generation using the non-viral approach. To generate iPSCs from hUCs with low proliferation rates, we developed an optimized reprogramming process. Through application of SMs and autologous untransfected hUC feeders to enhance reprogramming, we were able to generate iPSCs from most batches of hUCs, particularly for those poorly proliferating hUCs that were difficult to reprogram using other methods. Our approaches would be valuable in banking iPSC lines from individuals with different genetic backgrounds.
The role of SMs in aiding iPSC generation has been extensively examined in different somatic cells, such as human adult/fetal fibroblasts, mouse embryonic fibroblasts, and human epidermal keratinocytes (Esteban et al., 2010, Hou et al., 2013, Li et al., 2009, Lin et al., 2009, Mali et al., 2010, Shi et al., 2008, Yu et al., 2011, Zhu et al., 2010). Here, we re-examined the role of different SMs in hUC reprogramming. We found that Chir and CPFT-a significantly improved hUC reprogramming (p < 0.001), while NaB showed moderate improvement (p < 0.05). In contrast, A-83-01 and Tzv had a minimal effect when they were used alone (Figure S2). PD0325901 has been shown to suppress the growth of non-reprogrammed cells and promote reprogramming when added at a later stage (Shi et al., 2008). Consistently our results show that during hUC reprogramming, adding PD0325901 at an early stage inhibits reprogramming but at a later stage promotes reprogramming (Figures S3A–S3D). Among these six tested SM, CPFT-a, a P53 inhibitor, has not been previously reported to be used for reprogramming. P53 has been reported to be a critical barrier during somatic cell reprogramming (Marion et al., 2009) and also an important tumor suppressor (Levine and Oren, 2009). For efficient reprogramming, P53 is usually a considered target for suppression during iPSC generation. In fact one of the reprogramming factors, SV40LT, is known as a negative regulator of P53. In addition, suppressing P53 through other biological approaches such as RNAi or even gene knockout promotes reprogramming, but raises safety concerns regarding genome instability due to the low activity of P53. Therefore, considering that SV40LT, another negative factor for P53, was already included for reprogramming, we minimized the application of CPFT-a during hUC reprogramming. Adding a low concentration of CPFT-a indeed suppressed P53 expression and enhanced the final efficiencies of iPSC generation during hUC reprogramming (Figures 1F and 1G), indicating a synergistic suppression of P53 with SV40LT. Another SM reported for enhancing iPSC generation, NaB, did not show a significant effect in promoting hUC reprogramming (Figure 1L). NaB was known to activate the expression of miR302/367, which further promotes reprogramming (Zhang and Wu, 2013). Liang et al. (2010) reported that NaB facilitated reprogramming only in the presence of exogenous c-Myc. For hUC reprogramming, we replaced C-Myc with miR302/367 as reprogramming factor, which might explain the less significant effect of NaB on hUC-derived iPSC generation. Nevertheless, due to the variation in different batches of hUCs (Figure S2), it is still beneficial to include NaB in the SM cocktail when reprogramming hUCs with poor proliferation.
Another improvement of our optimized strategy is the application of autologous hUC feeders to support iPSC generation. In most cases we found that if the number of surviving cells after factor transfection was not sufficient, it usually failed to generate iPSCs. To overcome this problem, we plated the transfected hUCs onto the autologous non-transfected cells for further reprogramming. Aided by the feeder, we were able to generate iPSCs from a very low number of surviving hUCs. Based on our experience, in the absence of Matrigel, if the cells reach confluence of between 80% and 100% at day 5 or 6 of reprogramming, we will definitely obtain bona fide iPSC clones. However, we found that during reprogramming, if the total number of UCs is too high, the reprogramming efficiency decreases. Thus, to reprogram UCs with a high growth rate, we need to optimize the number of autologous UCs as feeder. Application of a feeder to replace Matrigel as the extracellular matrix for reprogramming is valuable in generating iPSCs under conditions with no animal components. In summary, we described here optimized approaches for the generation of iPSCs from hUCs, particularly for those poorly proliferating hUCs that are otherwise difficult to reprogram. Using these strategies, we were able to generate iPSCs from almost 100% of different batches of isolated hUCs, which are valuable for banking iPSCs from patients with different genetic backgrounds.
Experimental Procedures
Ethical Statement
The individuals in this study have signed written informed consent for donating UCs for stem cell generation. The experiments involving human subject and animal research had been reviewed and approved by the Institutional Review Board at Guangzhou Institutes of Biomedicine and Health (GIBH) (no. 2010012). The studies using human cells and mice have been approved by the Human or Animal Subjects Oversight Committee of GIBH.
Collection and Expansion of UCs
One hundred and fourteen donors were recruited for urine samples with informed consent based on IRB approval (no. GIBH-IRB02-2009002) of GIBH. The purposes and procedures for isolating urine cells and generating stem cells were explained to donors in detail, and questions, if any, were answered in full. We then obtained a formal signed consent form and collected a total of 50–500 ml of urine from each donor. UCs were collected as described previously (Zhou et al., 2011, Zhou et al., 2012). In brief, urine samples were collected at the mid-stream from 114 individuals. Osmolarity pressure (OSM) of each urine sample was tested and urine samples' OSM between 241 mmol/l and 598 mmol/l were centrifuged to collect the exfoliated cells. The primary UCs were then processed and cultured and passaged in medium consisting of REBM (renal epithelial basal medium; Lonza) medium containing SingleQuot Kit CC-4127 REGM (Lonza)/DMEM/high glucose (1×) (Hyclone) containing 100× MEM NEAA (Gibco), 100× glutaMAX-1 (Gibco), and 10% FBS (Hyclone) 1:1 (the combination of the three is referred to as RM).
Cell Proliferation Curves
UCs (20,000) were seeded per well (24-well plate). Three wells were harvested every 24 hr to count the number of cells. The cell populations were analyzed using a two-tailed Student's t test. Error bars indicate the SD (n = 3).
iPSC Generation
UCs (5 × 105 to 1 × 106) were individualized by trypsin treatment (0.05% trypsin/0.5 mM EDTA, Gibco) and electroporated with indicated episomal plasmids using an Amaxa Basic Nucleofector Kit for primary mammalian epithelial cells, program T-020 (Lonza). The electroporated UCs were seeded onto P6 wells, and the next day passaged to Matrigel (354277, Becton Dickinson) pre-coated P24 wells. In each nucleofection, 6 μg of pEP4EO2SET2K (Yu et al., 2009) (contains OCT4, SOX2, SV40LT, and KLF4) and 4 μg of pCEP4-miR-302-367 cluster (Liao et al., 2011) (contains miR-302b, c, a, d, and miR-367) were used. The induced media during reprogramming and proliferation were named plan A, plan B, and plan C. The concentration of SMs was shown as follows: 0.5 μM A-83-01 (SML0788-5MG, Sigma), 3 μM CHIR99021 (252917-06-9, Guangzhou Laura Biotech), 0.5 μM Tzv (1226056-71-8, Guangzhou Laura Biotech), 250 μM sodium butyrate (303410-100G, Sigma), 0.3 μM cyclic pifithrin-a (04-0040, Stemgent), and 0.5 μM PD0325901 (Guangzhou Laura Biotech). plan A was RM at days 0–1, RM + 5M at days 2–9, and defined medium mTeSR1 (STEMCELL) at days 10–13. plan B was RM at days 0–1, RM + 5M at days 2–9, and mTeSR1 + 6M at days 10–17. The medium of plan C was the same as for plan B, but cells were seeded on autologous UC feeders pre-coated or co-seeded with electroporated UCs. Each medium was changed every other day. The iPSC colonies were picked at around days 13–18 and cultured in mTeSR1 on Matrigel. The culture medium was changed daily. The iPSCs were passaged with 0.5 mM EDTA (25200-056, Gibco).
iPSC Characterization
PCR analysis of exogenous reprogramming factors and episomal backbone integration, FACS analysis (OCT-3/4 antibody, Santa Cruz Biotechnology sc-5279; anti-SSEA4 antibody, Abcam AB16287; anti-TRA-1-81, Millipore MAB 4381; and anti-TRA-1-60, Millipore MAB 4360). AP staining, karyotyping, and bisulfate sequencing were performed as we described previously (Qin et al., 2008, Wang et al., 2013). Specifically, we used NBT and BCIP (GenView) for AP staining. The primers used for cloning, PCR analysis, qPCR, and bisulfate sequencing were used according to a previous report (Xue et al., 2013). Total RNA was extracted using Trizol (Invitrogen) and reverse transcribed by oligo(dT) or specific primers. qPCR was performed in triplicate with a CFX96 machine (Bio-Rad) and SYBR Green Premix EX Taq Kit (Takara) following the instructions by the manufacturer. Teratoma formation was done to examine the in vivo differentiation potential of human iPSCs derived with the 6-SMs method. iPSCs grown on Matrigel in mTeSR1 were collected with EDTA treatment, then resuspended by Matrigel and injected into hindlimb muscles of 6-week-old immunocompromised SCID mice (one 6-cm dish with 50%–80% confluence per injection per mouse). Two injections were performed for each iPSC clone. After 6–8 weeks, teratomas were obtained from all injections. The teratomas were dissected and fixed in 4% paraformaldehyde (Jingxin Biotechnology). Samples were embedded in paraffin and processed with H&E staining in the Experimental Pathology Department of GIBH.
Flow Cytometry Analysis
About 1 × 106 iPSCs were used for each staining. iPSCs were trypsinized and fixed with 1% paraformaldehyde dissolved in PBS for 10 min at 37°C. The cells were then washed with FACS buffer (PBS containing 2% FBS) and resuspended in 90% ethanol for permeabilization by incubating 30 min on ice. After washing, cells were sequentially incubated with primary and secondary antibody for 30 min at 37°C. Control samples were stained with isotype-matched control antibodies. The iPSCs were washed and resuspended in FACS buffer and then processed for analysis on FACSCalibur (BD).
RNA-Seq
Total mRNA was isolated from hUCs, iPSCs, and ESCs. RNA-seq libraries were constructed using the TruSeq RNA Sample Preparation Kit v2 (Illumina). Libraries were sequenced using MiSeq Reagent Kit v2. Hierarchical clustering of the RNA-seq data, heatmaps, and scatterplots were generated using glbase (Hutchins et al., 2014).
Author Contributions
D.L., L.L.W., H.Z., Y.H.L., and G.J.P. designed the experiments. L.L.W. and Q.Y.C. carried out vector construction. Y.L.S. carried out the immunoassays. H.Z. and Y.H.L. provided clinical samples. J.D.H., D.L., Q.S., and X.S. collected clinical samples. L.L.W., D.L., J.D.H., X.J.C., and T.Z. purified iPSCs. D.L., J.D., H.Z., Y.H.L., and G.J.P. interpreted of the data and drafted the manuscript. X.S.W. analyzed RNA-seq data. All of the authors revised and approved the final manuscript.
Acknowledgments
This work is supported by the National Basic Research Program of China (grant 2012CB966503, 2015CB964901, 2015CB964902), Cooperation Grant of the Natural Science Foundation of Guangdong Province (2014A030312012), the Frontier and Key Technology Innovation special grant from the Department of Science and Technology of Guangdong province (2014B020225006, 201508020258), International Science & Technology Cooperation Program of China (no. 2014DFA30180), the National Natural Science Foundation of China (grants 31371514, 81372249, and 81300431), the Science and Information Technology of Guangzhou Key Project (201508020258), “Hundred Talents Program” of the Chinese Academy of Science (to G.P.), Science and Technology Planning Project of Guangdong Province, China (grant 2013B091500072) and Project of Department of Education of Guangdong Province, China (grant 2014GKXM029), Science and Technology Project of Guangzhou (2014J4100129, 2015A020212023 to H.Z.). We thank Dr. Xiaodong Shu for critical comments on the manuscript.
Published: April 28, 2016
Footnotes
Supplemental Information includes three figures and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.04.001.
Contributor Information
Yuhua Li, Email: liyuhua2011gz@163.com.
Hua Zhang, Email: jimzhua@163.com.
Guangjin Pan, Email: pan_guangjin@gibh.ac.cn.
Accession Numbers
The accession number for the RNA-seq data of iPSC and hUC reported in this article is GEO: GSE79134. RNA-Seq data of ESCs are available in Gene Expression Omnibus (GEO: GSE69797) (Huang et al., 2015).
Supplemental Information
References
- Banito A., Rashid S.T., Acosta J.C., Li S., Pereira C.F., Geti I., Pinho S., Silva J.C., Azuara V., Walsh M. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev. 2009;23:2134–2139. doi: 10.1101/gad.1811609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban M.A., Wang T., Qin B., Yang J., Qin D., Cai J., Li W., Weng Z., Chen J., Ni S. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell. 2010;6:71–79. doi: 10.1016/j.stem.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Hong H., Takahashi K., Ichisaka T., Aoi T., Kanagawa O., Nakagawa M., Okita K., Yamanaka S. Suppression of induced pluripotent stem cell generation by the P53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou P., Li Y., Zhang X., Liu C., Guan J., Li H., Zhao T., Ye J., Yang W., Liu K. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science. 2013;341:651–654. doi: 10.1126/science.1239278. [DOI] [PubMed] [Google Scholar]
- Huang K., Du J., Ma N., Liu J., Wu P., Dong X., Meng M., Wang W., Chen X., Shi X. GATA2(-/-) human ESCs undergo attenuated endothelial to hematopoietic transition and thereafter granulocyte commitment. Cell Regen. 2015;4:4. doi: 10.1186/s13619-015-0018-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins A.P., Jauch R., Dyla M., Miranda-Saavedra D. glbase: a framework for combining, analyzing and displaying heterogeneous genomic and high-throughput sequencing data. Cell Regen. 2014;3:1. doi: 10.1186/2045-9769-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia F., Wilson K.D., Sun N., Gupta D.M., Huang M., Li Z., Panetta N.J., Chen Z.Y., Robbins R.C., Kay M.A. A nonviral minicircle vector for deriving human iPS cells. Nat. Methods. 2010;7:197–199. doi: 10.1038/nmeth.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura T., Suzuki J., Wang Y.V., Menendez S., Morera L.B., Raya A., Wahl G.M., Izpisua Belmonte J.C. Linking the P53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A.J., Oren M. The first 30 years of P53: growing ever more complex. Nat. Rev. Cancer. 2009;9:749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Zhou H., Abujarour R., Zhu S., Young Joo J., Lin T., Hao E., Scholer H.R., Hayek A., Ding S. Generation of human-induced pluripotent stem cells in the absence of exogenous Sox2. Stem Cells. 2009;27:2992–3000. doi: 10.1002/stem.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G., Taranova O., Xia K., Zhang Y. Butyrate promotes induced pluripotent stem cell generation. J. Biol. Chem. 2010;285:25516–25521. doi: 10.1074/jbc.M110.142059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao B., Bao X., Liu L., Feng S., Zovoilis A., Liu W., Xue Y., Cai J., Guo X., Qin B. MicroRNA cluster 302-367 enhances somatic cell reprogramming by accelerating a mesenchymal-to-epithelial transition. J. Biol. Chem. 2011;286:17359–17364. doi: 10.1074/jbc.C111.235960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T., Ambasudhan R., Yuan X., Li W., Hilcove S., Abujarour R., Lin X., Hahm H.S., Hao E., Hayek A. A chemical platform for improved induction of human iPSCs. Nat. Methods. 2009;6:805–808. doi: 10.1038/nmeth.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Chou B.K., Yen J., Ye Z., Zou J., Dowey S., Brodsky R.A., Ohm J.E., Yu W., Baylin S.B. Butyrate greatly enhances derivation of human induced pluripotent stem cells by promoting epigenetic remodeling and the expression of pluripotency-associated genes. Stem Cells. 2010;28:713–720. doi: 10.1002/stem.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion R.M., Strati K., Li H., Murga M., Blanco R., Ortega S., Fernandez-Capetillo O., Serrano M., Blasco M.A. A P53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K., Matsumura Y., Sato Y., Okada A., Morizane A., Okamoto S., Hong H., Nakagawa M., Tanabe K., Tezuka K. A more efficient method to generate integration-free human iPS cells. Nat. Methods. 2011;8:409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- Qin D., Gan Y., Shao K., Wang H., Li W., Wang T., He W., Xu J., Zhang Y., Kou Z. Mouse meningiocytes express Sox2 and yield high efficiency of chimeras after nuclear reprogramming with exogenous factors. J. Biol. Chem. 2008;283:33730–33735. doi: 10.1074/jbc.M806788200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H., Diaz A., Blouin L., Lebbink R.J., Patena W., Tanbun P., LeProust E.M., McManus M.T., Song J.S., Ramalho-Santos M. Systematic identification of barriers to human iPSC generation. Cell. 2014;158:449–461. doi: 10.1016/j.cell.2014.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Do J.T., Desponts C., Hahm H.S., Scholer H.R., Ding S. A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell. 2008;2:525–528. doi: 10.1016/j.stem.2008.05.011. [DOI] [PubMed] [Google Scholar]
- Wang L., Wang L., Huang W., Su H., Xue Y., Su Z., Liao B., Wang H., Bao X., Qin D. Generation of integration-free neural progenitor cells from cells in human urine. Nat. Methods. 2013;10:84–89. doi: 10.1038/nmeth.2283. [DOI] [PubMed] [Google Scholar]
- Wu Y.L., Pandian G.N., Ding Y.P., Zhang W., Tanaka Y., Sugiyama H. Clinical grade iPS cells: need for versatile small molecules and optimal cell sources. Chem. Biol. 2013;20:1311–1322. doi: 10.1016/j.chembiol.2013.09.016. [DOI] [PubMed] [Google Scholar]
- Xue Y., Cai X., Wang L., Liao B., Zhang H., Shan Y., Chen Q., Zhou T., Li X., Hou J. Generating a non-integrating human induced pluripotent stem cell bank from urine-derived cells. PLoS One. 2013;8:e70573. doi: 10.1371/journal.pone.0070573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S. A fresh look at iPS cells. Cell. 2009;137:13–17. doi: 10.1016/j.cell.2009.03.034. [DOI] [PubMed] [Google Scholar]
- Yu J., Hu K., Smuga-Otto K., Tian S., Stewart R., Slukvin I.I., Thomson J.A. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Chau K.F., Vodyanik M.A., Jiang J., Jiang Y. Efficient feeder-free episomal reprogramming with small molecules. PLoS One. 2011;6:e17557. doi: 10.1371/journal.pone.0017557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Wu W.S. Sodium butyrate promotes generation of human induced pluripotent stem cells through induction of the miR302/367 cluster. Stem Cells Dev. 2013;22:2268–2277. doi: 10.1089/scd.2012.0650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Benda C., Duzinger S., Huang Y., Li X., Li Y., Guo X., Cao G., Chen S., Hao L. Generation of induced pluripotent stem cells from urine. J. Am. Soc. Nephrol. 2011;22:1221–1228. doi: 10.1681/ASN.2011010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Benda C., Dunzinger S., Huang Y., Ho J.C., Yang J., Wang Y., Zhang Y., Zhuang Q., Li Y. Generation of human induced pluripotent stem cells from urine samples. Nat. Protoc. 2012;7:2080–2089. doi: 10.1038/nprot.2012.115. [DOI] [PubMed] [Google Scholar]
- Zhu S., Li W., Zhou H., Wei W., Ambasudhan R., Lin T., Kim J., Zhang K., Ding S. Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell. 2010;7:651–655. doi: 10.1016/j.stem.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.