Abstract
Activation of oncogenes and loss of tumour suppressors promote metabolic reprogramming in cancer, resulting in enhanced nutrient uptake to supply energetic and biosynthetic pathways. However, nutrient limitations within solid tumours may require that malignant cells exhibit metabolic flexibility to sustain growth and survival. Here, we highlight these adaptive mechanisms and also discuss emerging approaches to probe tumour metabolism in vivo and their potential to expand the metabolic repertoire of malignant cells even further.
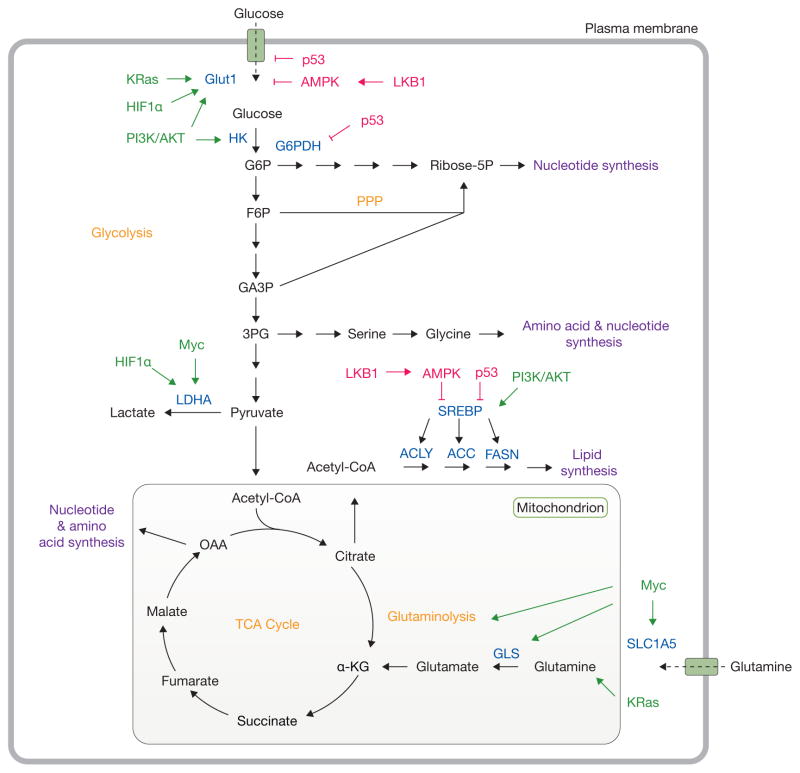
Metabolic reprogramming is considered a hallmark of cancer1, and has been an area of accelerated research over the last decade. A common theme emerging from this work is that when nutrients are abundant, oncogenic signalling pathways direct enhanced nutrient acquisition and facilitate assimilation of carbon into macromolecules such as lipids, proteins and nucleic acids. The net effect of these activities is to support cell growth and proliferation. Within the hierarchy of pathways altered in cancer, glucose and glutamine metabolism are consistently reprogrammed by mutations in MYC, TP53, the Ras-related oncogenes, and the LKB1-AMP kinase (AMPK) and PI3 kinase (PI3K) signalling pathways, among others (Fig. 1). Oncogenic Ras stimulates both glucose uptake via enhanced expression of GLUT1, and utilization of glucose by anabolic pathways2,3. Ras also regulates glutamine metabolism, specifically directing glutamine carbon into pathways that support biosynthesis, redox homeostasis and ultimately cell survival and growth4–6. Increased MYC elicits numerous metabolic effects through reprogrammed gene expression. These include enhanced glycolysis, in part through transcriptional activation of LDHA (ref. 7); enhanced mitochondrial biogenesis8; and enhanced glutamine catabolism9,10, culminating in biomass assimilation. The convergence of so many pathways on glucose and glutamine may reflect the fact that both nutrients are abundant, and both feed into multiple nodes of central metabolism. Glutamine has the added advantage of providing its two nitrogen atoms to synthesize hexosamines, nucleotides and amino acids, all of which are also required for growth11.
Figure 1.
Oncogenic signalling and nutrient availability influence cell metabolism. Oncogenic signalling regulates the acquisition of abundant nutrients including glucose and glutamine, and their utilization to support biosynthetic pathways (purple). Mutations in oncoproteins (green) lead to increased glucose uptake coupled to enhanced lactate production through the Warburg effect. Additionally, KRas and Myc promote glutamine metabolism to fuel the TCA cycle. Intermediates from glycolysis and the TCA cycle supply biosynthetic pathways to produce macromolecules necessary for cell proliferation. Tumour suppressors (red) such as LKB1, AMPK and p53 act at various nodes to oppose biosynthetic metabolism. Important nutrient transporters, transcription factors, and metabolic enzymes are highlighted in blue. Classic metabolic pathways are highlighted in orange. HK, hexokinase; G6P, glucose-6-phosphate; G6PDH, glucose-6-phosphate dehydrogenase; Ribose-5P, ribose-5-phosphate; F6P, fructose-6-phosphate; PPP, pentose phosphate pathway; GA3P, glyceraldehyde-3-phosphate; 3PG, 3-phosphoglycerate; LDHA, lactate dehydrogenase A; SREBP, sterol regulatory element-binding protein; ACLY, ATP citrate lyase; ACC, acetyl-CoA carboxylase; FASN, fatty acid synthase; GLS, glutaminase, α-KG, alpha-ketoglutarate; OAA, oxaloacetate; TCA, tricarboxylic acid.
However, in order to withstand the harsh environment of solid tumours, cancer cells must also optimize nutrient utilization when resources are scarce. Recent work has highlighted the importance of metabolic flexibility in both cultured cells and in vivo. For example, glucose deprivation, or growth in the harsh environment of the subcutaneous space in mice, elicits a selective pressure for KRAS mutations in colon cancer cells12. In this context, mutated KRAS rendered cells tolerant of low glucose conditions. Similarly, cancer cells in culture can restructure their metabolism to compensate for the loss of either glucose or glutamine, often using one nutrient to fill metabolite pools normally supplied by the other13–15. High-throughput screens revealed that cells chronically exposed to low glucose require oxidative phosphorylation as a means to maintain growth16. In a similar vein, subsets of lymphoma preferentially use, and can be highly dependent on, oxidative metabolism rather than the more classical glycolytic phenotype17.
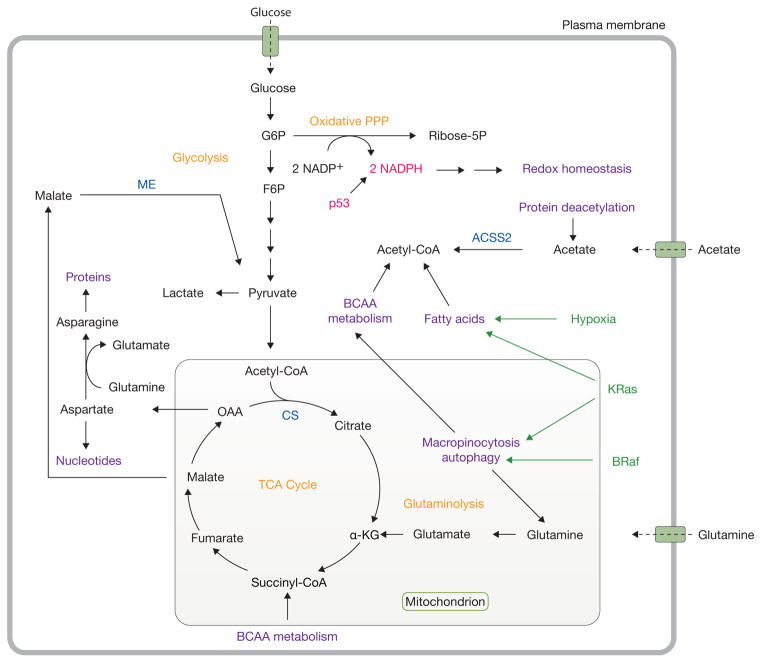
Overall, emerging evidence suggests that tumour cells have much more complex metabolic requirements than previously appreciated, and that numerous pathways complement glucose- and glutamine-dependent biomass production (Fig. 2). Here we discuss some of these pathways and how they contribute to cell survival and growth.
Figure 2.
Catabolic pathways support metabolism during nutrient stress. Under conditions of nutrient deprivation and other stresses, tumorigenic mutations also reprogram metabolism to support cell survival. In particular, KRas stimulates autophagy and macropinocytosis and facilitates fatty acid uptake. Mutations in BRaf also lead to enhanced autophagy. p53 promotes NADPH production through the pentose phosphate pathway to maintain redox homeostasis. Acetate and branched-chain amino acids serve as alternative substrates to support metabolism. Oncogenic drivers are highlighted in green, while tumour suppressors are shown in red. Important metabolic enzymes are highlighted in blue. Classic cancer metabolic pathways are shown in orange, while emerging pathways and activities supporting cell proliferation are shown in purple. G6P, glucose-6-phosphate; PPP, pentose phosphate pathway; ribose-5P, ribose-5-phosphate; F6P, fructose-6-phosphate; TCA, tricarboxylic acid; α-KG, alpha-ketoglutarate; OAA, oxaloacetate; BCAA, branched-chain amino acid; CS, citrate synthase; ME, malic enzyme; ACSS2, acetyl-CoA synthetase 2.
Protein and amino acid scavenging and catabolism in the face of nutrient deprivation
Although most cultured cancer cells use glutamine to supply the oxaloacetate (OAA) pool, complementing glucose-dependent acetyl-CoA formation (Fig. 1), glutamine’s ability to fuel alternative forms of metabolism has emerged as an important component of cell survival. Glucose deprivation in Myc-enhanced lymphoma cells stimulates a pathway whereby glutamine carbon is re-routed to acetyl-CoA13, and which can be mimicked by silencing the mitochondrial pyruvate carrier (MPC)18,19. This pathway is dispensable in glucose-replete cells with normal MPC function, but essential for survival and tumour growth when MPC is impaired19, indicating the importance of this mode of glutamine oxidation during nutrient limitation.
Glutamine deprivation also induces metabolic vulnerabilities. Citrate synthase loss was found to protect cells against apoptosis during glutamine deprivation20. Normally, citrate synthase condenses glutamine-derived OAA with acetyl-CoA to maintain TCA cycle function21. However, when glutamine is scarce, shunting OAA towards asparagine rather than citrate suppresses the unfolded protein response and supports cell survival20. Exogenous asparagine mimics citrate-synthase silencing during glutamine withdrawal. Although asparagine is normally considered a non-essential amino acid, rapidly proliferating cells need an abundant supply for protein synthesis, which is the basis of L-Asparaginase use in cancer therapy22,23. Because expression of asparagine synthetase correlates with poor prognosis in glioma and neuroblastoma20, these findings suggest that the ability to maintain an asparagine pool may provide an advantage to tumour cells in vivo.
Other mechanisms also enable cancer cells to deal with glutamine deprivation. Commisso et al. demonstrated that glutamine deprivation stimulates macropinocytosis in Ras-expressing cancer cells24. This process enables cells to scavenge fluid and macromolecules, using a system of membrane ruffling to capture and incorporate extracellular material. Extracellular proteins were identified as important components of the cargo captured and internalized in macropinosomes, allowing starved cells to generate pools of glutamine and other amino acids to supply the TCA cycle24. This mechanism relieved cells with oncogenic KRAS or Src from dependence on extracellular glutamine, and was required for maximal growth of KRAS tumours in vivo. Thus, macropinocytosis provides a mode of metabolic flexibility enabling some transformed cells to compensate for interruptions in the extracellular supply of free amino acids.
In addition to scavenging extracellular protein, cancer cells also activate autophagic degradation of macromolecules when deprived of nutrients or of the signals that stimulate nutrient uptake25–28. During autophagy, damaged organelles and their macromolecular components are degraded, providing recycled small molecule nutrients to feed intermediary metabolism27,29,30. Autophagy may also function to eliminate defective mitochondria, thereby reducing accumulation of reactive oxygen species (ROS) and improving cellular fitness. In Ras- or BRaf-driven mouse models of cancer, autophagy is crucial for tumour growth and/or progression. KRAS-driven pancreatic tumours in mice require autophagy for maximal growth31, and loss of essential autophagy genes impairs mitochondrial function in KRAS-driven lung tumours32,33. Interestingly, impaired autophagy results in the formation of oncocytomas, benign tumours filled with damaged mitochondria32. This implies that formation of aggressive KRAS-driven carcinomas requires both autophagy and effective mitochondrial function. Autophagy is also essential for maximal growth of BRafV600E lung tumours, and inhibiting autophagy extends the survival of mice bearing these tumours34. Furthermore, although chronic ablation of the autophagy gene Atg7 causes a number of systemic effects in mice, acute Atg7 deletion specifically impaired growth of pre-existing KRAS-driven lung tumours prior to the appearance of pathology in normal tissues35. Overall, these findings emphasize a role for autophagy in driving aggressive tumour formation and maintenance by providing an intracellular nutrient supply to support cell survival and growth.
Amino acids other than glutamine also contribute to bioenergetics. Branched-chain amino acids (BCAA) are important metabolic substrates because their degradation provides acetyl-CoA and/or anaplerotic substrates for the TCA cycle. Population-based human studies have demonstrated that elevated plasma BCAAs correlate with insulin resistance, type 2 diabetes and pancreatic cancer risk36–39. A cell-autonomous role for BCAA metabolism was uncovered in glioma, as the subset of these tumours lacking mutations in isocitrate dehydrogenases 1 and 2 (IDH1 and IDH2) express high levels of BCAA transaminase 1 (BCAT1), which initiates BCAA catabolism40. Silencing BCAT1 impairs cell proliferation and tumour growth, suggesting that BCAAs are functionally important metabolic substrates in these tumours. Future studies should provide insight into precisely how these amino acids support cancer and other diseases.
Emerging roles for lipid scavenging and fatty acid oxidation in tumour cell survival and growth
Lipids compose a substantial fraction of the dry weight of mammalian cells, and maintaining a supply of lipids is essential for cell proliferation41–43. In the presence of oxygen and abundant extracellular nutrients, most cultured cancer cells synthesize fatty acids de novo41,44. However, under conditions of metabolic stress, scavenging extracellular lipids has emerged as an important adaptive mechanism for cancer cells to maintain viability and/or growth45,46. Scavenging, rather than synthesizing lipids, spares cells from the need to supply carbon and reducing power (that is, NADPH) for this demanding pathway. Hypoxia and oncogenic HRAS or KRAS stimulate uptake and utilization of lysophospholipids (phospholipids missing one of the two acyl chains) to supply an intracellular lipid pool for growth45. As a result of their ability to acquire lipids in this fashion, KRAS-driven tumour cells are resistant to inhibition of stearoyl-CoA desaturase 1 (SCD1), the enzyme that normally desaturates fatty acids synthesized de novo prior to their incorporation into complex lipids45. Extracellular lipids, particularly desaturated fatty acids, are also important in cells with dysregulated mTORC1 activity subjected to hypoxia46. In this context, increased protein synthesis and reduced lipid desaturation leads to activation of the unfolded protein response and cell death, a phenotype which is rescued by unsaturated fatty acids. These findings were reproduced in cultures of clear cell renal cell carcinoma, glioblastoma and bladder cancer, suggesting that solid tumours may rely on the extracellular environment to provide fatty acids.
Fatty acids also supply energy, as mitochondrial fatty acid oxidation produces more than twice as much ATP per mole as oxidation of glucose or amino acids. A subset of diffuse large B-cell lymphomas appears to prefer fatty acid oxidation as a fuel and to express high levels of enzymes required to oxidize fatty acids, even under nutrient replete conditions17. Autophagy and related processes enable other cells to capitalize on fatty acids for fuel. In p53-deficient KRasG12D non-small cell lung cancers, impairing autophagy results in dysfunctional mitochondria, lipid accumulation, defective fatty acid oxidation and enhanced sensitivity to starvation32. Furthermore, fatty acid oxidation and other oxidative mitochondrial pathways seem to enable cancer cells to survive periods of tumour regression47. In an inducible model of KRAS-driven pancreatic cancer, tumour regression induced by kinase inhibitors or withdrawal of KRAS results in a dormant population of tumour cells, which are highly dependent on mitochondrial respiration for survival47. Inhibiting either autophagy or fatty acid oxidation reduces the tumour-initiating potential of this population, suggesting that these catabolic pathways are critical in enabling cancer cells to promote tumour formation after an initial round of treatment.
Other work demonstrates cooperative mechanisms by which stromal cells provide fatty acids to tumour cells as a fuel source, particularly in ovarian cancer. These tumour cells frequently metastasize to the omentum, a large fold of fatty tissue in the abdomen48,49. Co-culture of ovarian cancer cells with adipocytes revealed that adipocyte transfer of fatty acids activates AMPK and fatty acid oxidation in the cancer cells, enhancing cell proliferation48. These findings raise a number of questions regarding the role of the tumour microenvironment in promoting cell metabolism and the possibility of metabolite transfer between cells.
Acetate as an alternative metabolic substrate
Acetate, a 2-carbon fatty acid, is one of the smallest nutrients in mammals. It is readily converted to acetyl-CoA by acetyl-CoA synthetases, of which mammals contain at least three, ACSS1, ACSS2 and ACSS3. ACSS1 and ACSS3 encode mitochondrial enzymes whereas ACSS2 encodes an enzyme that is predominantly cytosolic50. Although circulating acetate is in the low-micromolar range51, recent work has demonstrated a role for acetate and acetate-metabolizing enzymes in tumours. First, infusion of 13C-glucose and 13C-acetate into mice bearing orthotopic glioblastomas demonstrated that both substrates could be oxidized52. However, when confronted with the same mixture of 13C-glucose and 13C-acetate in the plasma, the tumours oxidized acetate to a far greater extent than did the surrounding normal brain tissue. Acetate oxidation was also observed in metastatic brain tumours, suggesting that this pathway may be a general feature of tumour growth in the brain. Compellingly, infusion of human patients with a similar combination of 13C-glucose and 13C-acetate also revealed extensive acetate utilization in gliomas and brain metastases52.
A mouse model of ACSS2 deficiency was used to determine the role of this enzyme in tumour growth53. Although these mice have no overt phenotypes, ACSS2-deficient embryonic fibroblasts are defective in utilizing exogenous acetate for lipogenesis and histone acetylation. Moreover, in two models of hepatocellular carcinoma, ACSS2 knockout reduces tumour burden53. The development of selective ACSS2 inhibitors, coupled with the dispensability of ACSS2 for normal development, suggests that targeting acetate metabolism may have therapeutic potential in some forms of cancer53. Furthermore, cancer cells increase their dependence on acetate during conditions of hypoxia and nutrient stress, potentially adding to the therapeutic potential of ACSS2 inhibitors54. It remains to be seen whether the primary role of ACSS2-mediated production of acetyl-CoA from acetate is in energy metabolism, biosynthesis, acetylation of histones and other proteins, or a combination of these effects.
NADPH metabolism contributes to redox homeostasis
Beyond alternative carbon sources, there is growing interest in cofactor metabolism in cancer, particularly in the pathways that enable NADPH to participate in reductive biosynthesis and redox homeostasis. Aberrant oncogenic signalling and hypoxia induce ROS and increase the demand for robust systems to produce NADPH55,56. Although moderate ROS levels amplify tumorigenic signals, high levels induce death56. Thus, understanding redox regulation may prove useful in developing new therapies.
AMPK balances catabolic and anabolic pathways to match metabolic supply and demand57. Loss of either AMPK or its activator LKB1 enhances glycolysis58,59 and induces oxidative stress following glucose deprivation60. By phosphorylating the acetyl-CoA carboxylases ACC1 and ACC2, AMPK responds to glucose deprivation by limiting flux through the NADPH-consuming pathway of de novo fatty acid synthesis and promoting fatty acid oxidation; together, these effects conserve NADPH and allow cells to resist oxidative stress in culture60. Importantly, silencing ACC1 or ACC2 also promotes tumour growth in vivo60. Thus, in addition to its well-known role in energy balance, AMPK helps maintain redox homeostasis during nutrient deprivation.
Glutamine also impacts NADPH metabolism and redox control through a variety of mechanisms11. An unusual glutamine-dependent pathway allows pancreatic ductal adenocarcinoma (PDAC) to maintain a reduced glutathione pool to combat oxidative stress6. In KRAS-mutated PDAC cells, glutamine-dependent carbon flux progresses through GOT1, a cytosolic aspartate aminotransferase that produces OAA. Cytosolic malate dehydrogenase and malic enzyme then produce malate and pyruvate, respectively, with the latter reaction also producing NADPH. Suppression of any of several enzymes in this pathway results in oxidation of the glutathione pool, enhanced ROS levels and reduced PDAC tumour growth6.
One barrier to understanding redox control is the compartmentalization of distinct NADPH-producing pathways in the mitochondria or cytosol. Recent reports have sought to understand the importance of each pathway to overall NADPH metabolism. One study exploited the fact that a particular NADPH-utilizing reaction, synthesis of 2-hydroxyglutarate (2-HG) by oncogenic isoforms of IDH1 (cytosolic) or IDH2 (mitochondrial), is confined to a single compartment depending on which mutant is expressed61. Because 2-HG is scarce in most tissues, heterologous expression of mutant IDH1 or IDH2 coupled to metabolic labelling with 2H-labelled tracers supplying 2H to the NADPH pool allows 2-HG labelling to provide a read-out for NADPH metabolism in these two compartments. Labelling cells with [3-2H]glucose, which results in the production of labelled NADPH in the cytosol via the pentose phosphate pathway (PPP; Fig. 3), leads to significant labelling of 2-HG in cells expressing mutant IDH1, but not mutant IDH2 (ref. 61).
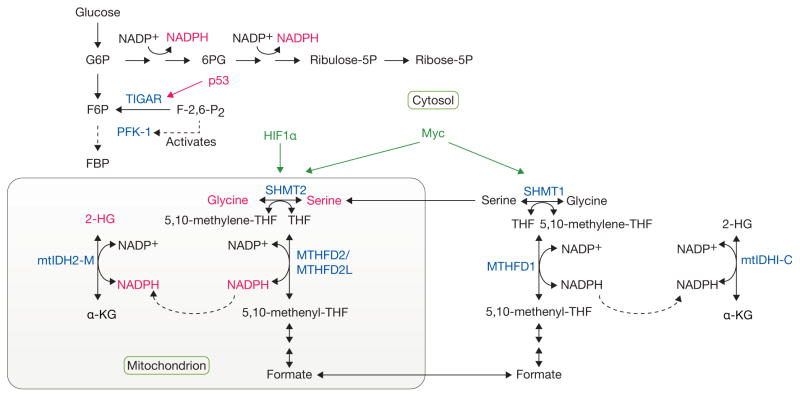
Figure 3.
Complex, compartmentalized pathways of NADPH production. Metabolic tracing experiments have provided tools to follow the cytosolic production of NADPH through the pentose phosphate pathway (PPP), as well as the contribution of serine/glycine metabolism to NADPH generation in the mitochondria. Given that mutant forms of isocitrate dehydrogenase (mtIDH1/2), localized to the cytosol (C) or mitochondria (M), respectively, give rise to increased 2-hydroxyglutarate (2-HG) levels, this reporter system was used to trace the production of NADPH in the mitochondria from serine/glycine through methylenetetrahydrofolate dehydrogenase 2 (like) (MTHFD2/MTHFD2L) enzymes. p53 regulates NADPH production by inducing the expression of TIGAR, a fructose-2,6-bisphosphatase, which reduces the levels of fructose-2,6-bisphosphate (F-2,6-P2), resulting in reduced activation of phosphofructokinase-1 (PFK-1). This reduction in the rate of glycolysis, allows for shunting of glycolytic intermediates into the PPP and the concomitant generation of NADPH. HIF1α and Myc regulate the expression of serine hydroxymethyltransferase (SHMT1/2) enzymes, thereby regulating serine/glycine metabolism. Tumor suppressors and oncogenes are shown in red and green, respectively. Important metabolic enzymes involved in these pathways are shown in blue. G6P, glucose-6-phosphate; 6PG, 6-phosphogluconate; Ribulose-5P, ribulose-5-phosphate; Ribose-5P, ribose-5-phosphate; F6P, fructose-6-phosphate; FBP, fructose-1,6-bisphosphate; THF, tetrahydrofolate; α-KG, alpha-ketoglutarate.
Similar logic was applied to the compartmentalization of serine-dependent one-carbon metabolism (Fig. 3). This pathway also generates NADPH, as serine hydroxymethyltransferases SHMT1 (cytosol) and SHMT2 (mitochondrial) transfer a methyl group from serine to tetrahydrofolate (THF) in the one-carbon pool, producing glycine and 5,10-methylene THF (ref. 62). The latter metabolite is oxidized to 5,10-methenyl-THF by the methylene THF dehydrogenases, MTHFD1 (cytosol) and MTHFD2/MTHFD2L (mitochondria) which generate NADPH62. Importantly, transfer of 2H from serine to 2-HG occurs only in cells expressing mutant IDH2, suggesting that serine/glycine-dependent NADPH production occurs primarily through a MTHFD2/MTHFD2L-dependent pathway in the mitochondria61. A separate study also used a combination of tracers to demonstrate that serine/one-carbon metabolism is a major source of cellular NADPH63.
Interestingly, SHMT2 expression is induced under hypoxia in a HIF1α- and Myc-dependent manner64. Silencing SHMT2 in Myc-expressing cells not only decreases the NADPH/NADP+ and reduced/oxidized glutathione ratios, but increases ROS and leads to hypoxia-induced cell death and tumour growth suppression. The fact that SHMT2 expression is elevated in several types of cancer and correlates with poor prognosis suggests that this enzyme may represent a therapeutic target65. Perhaps related to these findings, a number of studies have pinpointed consumption of either serine or glycine as factors related to cancer cell proliferation66,67. Given the importance of de novo serine synthesis through the PHGDH pathway68–70, more studies are likely to follow up on the role of serine/glycine and NADPH metabolism in promoting tumour growth, and whether these pathways can be exploited therapeutically.
Efforts to understand cancer metabolism in intact biological systems
With the increasing complexity of metabolism in cancer cell culture comes an increasing need to evaluate metabolism of intact tumours in situ. Within intact tumours, heterogeneous nutrient supply, oxygenation, and cell–cell interactions are all expected to have profound effects on metabolism. Metabolomic profiling is widely used to assess steady-state abundances of metabolites from many of the pathways described above, identifying differences between tumour tissue and normal tissue, or between individual tumours differing by driver mutation. As mass spectrometry systems and analytical software packages improve, it is becoming possible to assess increasingly large fractions of the metabolome. Metabolomics led to the discovery of massive elevations of 2-HG in tumours with mutated IDH1 or IDH271,72, and more recently the discovery of systematic alterations of glucose metabolism in renal cell carcinoma73. Discovery of large changes in metabolite pools can be translated into clinical approaches to monitor levels of that metabolite non-invasively, particularly by 1H magnetic resonance spectroscopy (MRS). MRS detects metabolites at high-micromolar to millimolar concentrations and maps them to an anatomic location. MRS is commonly used to assess metabolite pools in brain tumours, and recent MRS protocols have emerged to monitor 2-HG levels in gliomas74–77.
It is also possible to obtain dynamic views of tumour metabolism. Positron emission tomography (PET) detects and localizes gamma rays emitted by radionuclides like 18F and 11C. Labelling nutrients of interest with these tracers enables their uptake and retention to be observed within tumours. The most widely used PET tracer is 18FDG, a glucose analogue that is phosphorylated by hexokinase and trapped inside glycolytic tumours. 18FDG-PET is an important clinical technique to assess the distribution of tumour tissue in patients with metastatic disease and to monitor the effects of therapy78,79. New PET tracers that probe emerging aspects of cancer metabolism are being developed, and some are already used in clinical studies or practice. These tracers, which include 18F-glutamine, 18F-glutamate, and 11C-acetate80–86, are designed to detect tumours in which 18FDG uptake is non-diagnostic because the tumour displays relatively little glucose uptake or is in the vicinity of a glucose-avid organ like the brain.
Despite the utility of radioisotopes in imaging tumours, they provide little information about the metabolism of the labelled probe. For example, avid 18FDG uptake does not imply that the tumour cannot take up other nutrients, nor does it make any predictions about allocation of glucose carbon into any particular downstream pathway. To assess intermediary metabolism in live tumours, a growing number of studies52,87–91 have used infusions of 13C-labelled nutrients in tumour-bearing mice and humans to delineate bona fide tumour metabolism, as discussed above with respect to acetate52. The safety of 13C, coupled with the wide array of labelled nutrients available from commercial sources, makes in vivo isotope labelling a flexible and informative approach to study tumour metabolism.
Several studies have administered 13C-glucose to human subjects with solid tumours. A 2009 study provided a bolus of uniformly 13C-labelled glucose (that is, glucose in which all 6 carbons are labelled as 13C) to lung cancer patients several hours prior to resection of the tumour. Extraction of metabolites from tumour and lung, followed by assessment of 13C enrichment in glucose-derived metabolites, demonstrated enhanced labelling in a number of intermediates from glycolysis and the TCA cycle87. A larger follow-up study using a similar approach to introduce 13C-glucose provided additional qualitative evidence that glucose is used as an anaplerotic precursor to support a number of biosynthetic activities in lung tumours91. Pyruvate carboxylase, which enables carbon from glucose to supply an anaplerotic flux to the TCA cycle, is over-expressed in tumours relative to normal lung, consistent with a model in which glucose-dependent anaplerotic activity contributes to lung tumour growth in humans.
Other studies have used continuous infusions of 13C-labelled substrates throughout the operative procedure rather than bolus injections in an attempt to increase 13C enrichment in metabolite pools and to approach steady-state labelling in the tumour88–90. The latter consideration is important because establishing isotopic steady state may enable the application of quantitative models of metabolic flux. Infusions of 13C-glucose in orthotopic mouse models of human glioblastoma revealed that these 18FDG-avid tumours use both glycolysis and glucose oxidation in vivo89,90. These tumours also use glucose as a carbon source to synthesize a large glutamine pool, use pyruvate carboxylase as a source of anaplerotic flux, and display only marginal catabolism of infused 13C-glutamine90. Furthermore, primary cultures established from the tumours did not require glutamine to maintain viability, suggesting that glucose rather than glutamine is the key substrate to maintain TCA cycle function. This is quite different from the phenotypes observed in established glioma cell lines, most of which are highly dependent on glutamine to supply pools of TCA cycle intermediates and related molecules15,20,21. A similar 13C-glucose infusion study was performed in human patients with glioblastoma or brain metastases. As in the orthotopic tumours, these human tumours oxidize glucose extensively in the TCA cycle and synthesize glutamine from glucose88. Strikingly, comparing acetyl-CoA enrichment to glucose enrichment indicates that the tumours oxidize other substrates in addition to glucose, and in fact that glucose may be only a minor source of acetyl-CoA in the TCA cycle. This finding is potentially important, because oxidative metabolism in the normal brain is dominated by glucose92, suggesting that targeting alternative oxidative pathways may cut off the fuel supply to the tumour but be tolerated by the rest of the brain.
Conclusions and current challenges
Cancer cell metabolism appears more pleiotropic the more it is studied. The ability to engage pathways supplied by alternative nutrients, particularly during deprivation of glucose, glutamine and oxygen, may be a key feature selected during transformation or tumorigenesis. The extent to which these alternative pathways represent true vulnerabilities remains to be determined. However, there would appear to be numerous opportunities to test the impact of targeting some of these pathways in human cancer. Clinical trials examining the efficacy of autophagy inhibitors are already underway93–95. Targeting folate and nucleotide metabolism with methotrexate, pemetrexed and other agents is already known to provide clinical benefits96, and new insights into the integration of one-carbon metabolism with redox homeostasis may generate additional opportunities, perhaps in combination with drugs which enhance oxidative stress55,97. In vivo approaches to understand metabolism in intact tumours should shed light on the complexity of the metabolic pathways supporting tumour growth and provide insights into therapies which could be used to target these cancers.
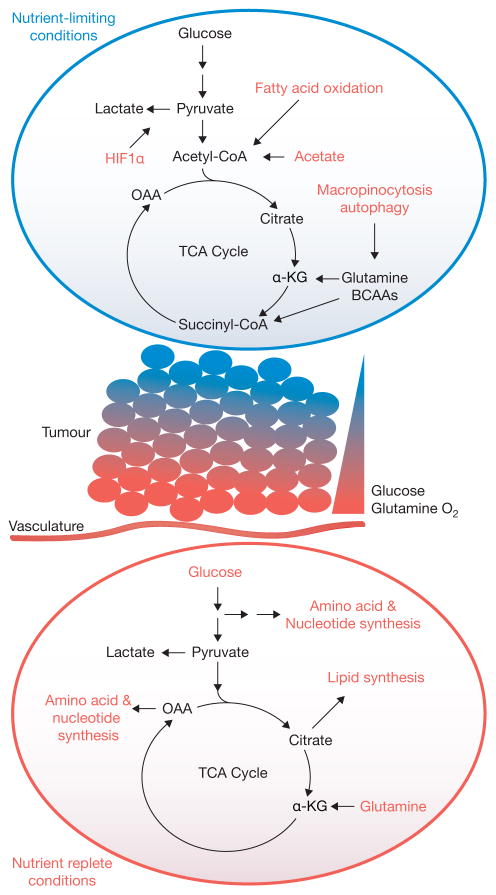
A looming challenge in cancer metabolism is to begin to understand metabolic heterogeneity within intact tumours. It is already clear from mouse models that both the driver mutation and tissue of origin influence metabolism when the tumour is considered as a single compartment98. Much less is known about heterogeneity within individual tumours, where regional differences in nutrient availability, localized effects of stromal and inflammatory cells, and cell-autonomous effects regulated by clonal expansion of mutants may all alter metabolic preferences and flexibility. Given the large repertoire of pathways available to transformed cells, one can envision a scenario where different parts of the tumour rely on starkly different metabolic platforms for cell survival and growth (Fig. 4). This picture is complicated further by metabolic effects within the microenvironment. For example, cancer cells secrete lactate, acidifying the microenvironment and triggering an inflammatory response resulting in the release of cytokines and other factors which promote tumour progression99. However, in some settings, lactate produced by stromal cells may provide a bioenergetic substrate for the cancer cells to support survival and growth100,101. Thus, metabolite exchanges and other aspects of cell–cell communications likely add an important dimension of metabolic heterogeneity within solid tumours. It is clear already that genetic heterogeneity plays an important role in promoting metastasis and contributes to clinical resistance102–104. It may be possible to merge metabolomics and/or in vivo isotope infusions with histological and molecular studies to map out distinct metabolic domains and correlate them with particular combinations of mutations or effects of the microenvironment.
Figure 4.
A model for metabolic heterogeneity in solid tumours. Within solid tumours, regional metabolic activities likely vary significantly according to influences of the microenvironment, particularly access to nutrients and oxygen. In the illustrated example, cells located close to vasculature use their favourable access to nutrients and oxygen to feed oncogene-stimulated anabolic pathways. However, gradients of nutrient availability demand that alternative pathways, including macromolecular degradation, are increasingly engaged at more remote sites to support cell viability. Oncogenes may also enhance the cell’s ability to activate these pathways. OAA, oxaloacetate; α-KG, alpha-ketoglutarate; TCA, tricarboxylic acid; BCAA, branched-chain amino acid.
Acknowledgments
We thank members of the DeBerardinis lab for their helpful comments on this review. R.J.D. is supported by grants from the N.I.H. (CA157996), Cancer Prevention and Research Institute of Texas (RP130272) and V Foundation. L.K.B. is supported by an N.I.H. Training Grant (5T32CA124334-08).
Footnotes
AUTHOR CONTRIBUTIONS
L.K.B. and R.J.D. wrote the paper and designed the illustrations.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Ying H, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flier JS, Mueckler MM, Usher P, Lodish HF. Elevated levels of glucose transport and transporter messenger RNA are induced by ras or src oncogenes. Science. 1987;235:1492–1495. doi: 10.1126/science.3103217. [DOI] [PubMed] [Google Scholar]
- 4.Gaglio D, et al. Oncogenic K-Ras decouples glucose and glutamine metabolism to support cancer cell growth. Mol Syst Biol. 2011;7:523. doi: 10.1038/msb.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weinberg F, et al. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci USA. 2010;107:8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Son J, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496:101–105. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shim H, et al. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc Natl Acad Sci USA. 1997;94:6658–6663. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li F, et al. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol. 2005;25:6225–6234. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wise DR, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci USA. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao P, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeBerardinis RJ, Cheng T. Q’s next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29:313–324. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yun J, et al. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science. 2009;325:1555–1559. doi: 10.1126/science.1174229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le A, et al. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 2012;15:110–121. doi: 10.1016/j.cmet.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang C, et al. Glioblastoma cells require glutamate dehydrogenase to survive impairments of glucose metabolism or Akt signaling. Cancer Res. 2009;69:7986–7993. doi: 10.1158/0008-5472.CAN-09-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng T, et al. Pyruvate carboxylase is required for glutamine-independent growth of tumor cells. Proc Natl Acad Sci USA. 2011;108:8674–8679. doi: 10.1073/pnas.1016627108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Birsoy K, et al. Metabolic determinants of cancer cell sensitivity to glucose limitation and biguanides. Nature. 2014;508:108–112. doi: 10.1038/nature13110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caro P, et al. Metabolic signatures uncover distinct targets in molecular subsets of diffuse large B cell lymphoma. Cancer Cell. 2012;22:547–560. doi: 10.1016/j.ccr.2012.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vacanti NM, et al. Regulation of substrate utilization by the mitochondrial pyruvate carrier. Mol Cell. 2014;56:425–435. doi: 10.1016/j.molcel.2014.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang C, et al. Glutamine oxidation maintains the TCA cycle and cell survival during impaired mitochondrial pyruvate transport. Mol Cell. 2014;56:414–424. doi: 10.1016/j.molcel.2014.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J, et al. Asparagine plays a critical role in regulating cellular adaptation to glutamine depletion. Mol Cell. 2014;56:205–218. doi: 10.1016/j.molcel.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeBerardinis RJ, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci USA. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Avramis VI. Asparaginases: biochemical pharmacology and modes of drug resistance. Anticancer Res. 2012;32:2423–2437. [PubMed] [Google Scholar]
- 23.Kawedia JD, Rytting ME. Asparaginase in acute lymphoblastic leukemia. Clin Lymphoma Myeloma Leuk. 2014;14:S14–S17. doi: 10.1016/j.clml.2014.06.017. [DOI] [PubMed] [Google Scholar]
- 24.Commisso C, et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 2013;497:633–637. doi: 10.1038/nature12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldsmith J, Levine B, Debnath J. Autophagy and cancer metabolism. Methods Enzymol. 2014;542:25–57. doi: 10.1016/B978-0-12-416618-9.00002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marino G, Niso-Santano M, Baehrecke EH, Kroemer G. Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol. 2014;15:81–94. doi: 10.1038/nrm3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lum JJ, et al. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 29.Deberardinis RJ, Lum JJ, Thompson CB. Phosphatidylinositol 3-kinase-dependent modulation of carnitine palmitoyltransferase 1A expression regulates lipid metabolism during hematopoietic cell growth. J Biol Chem. 2006;281:37372–37380. doi: 10.1074/jbc.M608372200. [DOI] [PubMed] [Google Scholar]
- 30.Lum JJ, DeBerardinis RJ, Thompson CB. Autophagy in metazoans: cell survival in the land of plenty. Nat Rev Mol Cell Biol. 2005;6:439–448. doi: 10.1038/nrm1660. [DOI] [PubMed] [Google Scholar]
- 31.Yang S, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–729. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo JY, et al. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev. 2013;27:1447–1461. doi: 10.1101/gad.219642.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo JY, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strohecker AM, et al. Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600E-driven lung tumors. Cancer Discov. 2013;3:1272–1285. doi: 10.1158/2159-8290.CD-13-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karsli-Uzunbas G, et al. Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov. 2014;4:914–927. doi: 10.1158/2159-8290.CD-14-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang TJ, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17:448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walford GA, et al. Branched chain and aromatic amino acids change acutely following two medical therapies for type 2 diabetes mellitus. Metabolism. 2013;62:1772–1778. doi: 10.1016/j.metabol.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Newgard CB, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9:311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayers JR, et al. Elevation of circulating branched-chain amino acids is an early event in human pancreatic adenocarcinoma development. Nat Med. 2014;20:1193–1198. doi: 10.1038/nm.3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tonjes M, et al. BCAT1 promotes cell proliferation through amino acid catabolism in gliomas carrying wild-type IDH1. Nat Med. 2013;19:901–908. doi: 10.1038/nm.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swinnen JV, Brusselmans K, Verhoeven G. Increased lipogenesis in cancer cells: new players, novel targets. Curr Opin Clin Nutr Metab Care. 2006;9:358–365. doi: 10.1097/01.mco.0000232894.28674.30. [DOI] [PubMed] [Google Scholar]
- 42.Currie E, Schulze A, Zechner R, Walther TC, Farese RV. Jr Cellular fatty acid metabolism and cancer. Cell Metab. 2013;18:153–161. doi: 10.1016/j.cmet.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 44.Bauer DE, Hatzivassiliou G, Zhao F, Andreadis C, Thompson CB. ATP citrate lyase is an important component of cell growth and transformation. Oncogene. 2005;24:6314–6322. doi: 10.1038/sj.onc.1208773. [DOI] [PubMed] [Google Scholar]
- 45.Kamphorst JJ, et al. Hypoxic and Ras-transformed cells support growth by scavenging unsaturated fatty acids from lysophospholipids. Proc Natl Acad Sci USA. 2013;110:8882–8887. doi: 10.1073/pnas.1307237110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Young RM, et al. Dysregulated mTORC1 renders cells critically dependent on desaturated lipids for survival under tumor-like stress. Genes Dev. 2013;27:1115–1131. doi: 10.1101/gad.198630.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Viale A, et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514:628–632. doi: 10.1038/nature13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nieman KM, et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat Med. 2011;17:1498–1503. doi: 10.1038/nm.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ackerman D, Simon MC. Hypoxia, lipids, and cancer: surviving the harsh tumor microenvironment. Trends Cell Biol. 2014;24:472–478. doi: 10.1016/j.tcb.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perez-Chacon G, Astudillo AM, Balgoma D, Balboa MA, Balsinde J. Control of free arachidonic acid levels by phospholipases A2 and lysophospholipid acyltransferases. Biochim Biophys Acta. 2009;1791:1103–1113. doi: 10.1016/j.bbalip.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 51.Psychogios N, et al. The human serum metabolome. PLoS ONE. 2011;6:e16957. doi: 10.1371/journal.pone.0016957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mashimo T, et al. Acetate is a bioenergetic substrate for human glioblastoma and brain metastases. Cell. 2014;159:1603–1614. doi: 10.1016/j.cell.2014.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Comerford SA, et al. Acetate dependence of tumors. Cell. 2014;159:1591–1602. doi: 10.1016/j.cell.2014.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schug ZT, et al. Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress. Cancer cell. 2015;27:57–71. doi: 10.1016/j.ccell.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gorrini C, Harris IS, Mak TW. Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov. 2013;12:931–947. doi: 10.1038/nrd4002. [DOI] [PubMed] [Google Scholar]
- 56.Schieber M, Chandel NS. ROS function in redox signaling and oxidative stress. Curr Biol. 2014;24:R453–R462. doi: 10.1016/j.cub.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer. 2009;9:563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Faubert B, et al. Loss of the tumor suppressor LKB1 promotes metabolic reprogramming of cancer cells via HIF-1alpha. Proc Natl Acad Sci USA. 2014;111:2554–2559. doi: 10.1073/pnas.1312570111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Faubert B, et al. AMPK is a negative regulator of the Warburg effect and suppresses tumor growth in vivo. Cell Metab. 2013;17:113–124. doi: 10.1016/j.cmet.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jeon SM, Chandel NS, Hay N. AMPK regulates NADPH homeostasis to promote tumour cell survival during energy stress. Nature. 2012;485:661–665. doi: 10.1038/nature11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lewis CA, et al. Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of Mammalian cells. Mol Cell. 2014;55:253–263. doi: 10.1016/j.molcel.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Christensen KE, MacKenzie RE. Mitochondrial one-carbon metabolism is adapted to the specific needs of yeast, plants and mammals. BioEssays. 2006;28:595–605. doi: 10.1002/bies.20420. [DOI] [PubMed] [Google Scholar]
- 63.Fan J, et al. Quantitative flux analysis reveals folate-dependent NADPH production. Nature. 2014;510:298–302. doi: 10.1038/nature13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ye J, et al. Serine catabolism regulates mitochondrial redox control during hypoxia. Cancer Discov. 2014;4:1406–1417. doi: 10.1158/2159-8290.CD-14-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee GY, et al. Comparative oncogenomics identifies PSMB4 and SHMT2 as potential cancer driver genes. Cancer Res. 2014;74:3114–3126. doi: 10.1158/0008-5472.CAN-13-2683. [DOI] [PubMed] [Google Scholar]
- 66.Jain M, et al. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336:1040–1044. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Labuschagne CF, van den Broek NJ, Mackay GM, Vousden KH, Maddocks OD. Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell Rep. 2014;7:1248–1258. doi: 10.1016/j.celrep.2014.04.045. [DOI] [PubMed] [Google Scholar]
- 68.Possemato R, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Locasale JW, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43:869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chaneton B, et al. Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature. 2012;491:458–462. doi: 10.1038/nature11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ward PS, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li B, et al. Fructose-1, 6-bisphosphatase opposes renal carcinoma progression. Nature. 2014;513:251–255. doi: 10.1038/nature13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Andronesi OC, et al. Detection of 2-hydroxyglutarate in IDH-mutated glioma patients by in vivo spectral-editing and 2D correlation magnetic resonance spectroscopy. Sci Transl Med. 2012;4:116ra4. doi: 10.1126/scitranslmed.3002693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Choi C, et al. 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat Med. 2012;18:624–629. doi: 10.1038/nm.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Elkhaled A, et al. Magnetic resonance of 2-hydroxyglutarate in IDH1-mutated low-grade gliomas. Sci Transl Med. 2012;4:116ra115. doi: 10.1126/scitranslmed.3002796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pope WB, et al. Non-invasive detection of 2-hydroxyglutarate and other metabolites in IDH1 mutant glioma patients using magnetic resonance spectroscopy. J Neuro-oncol. 2012;107:197–205. doi: 10.1007/s11060-011-0737-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gallamini A, Zwarthoed C, Borra A. Positron Emission tomography (PET) in oncology. Cancers. 2014;6:1821–1889. doi: 10.3390/cancers6041821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mankoff DA, et al. Tumor-specific positron emission tomography imaging in patients: [18F] fluorodeoxyglucose and beyond. Clin Cancer Res. 2007;13:3460–3469. doi: 10.1158/1078-0432.CCR-07-0074. [DOI] [PubMed] [Google Scholar]
- 80.Wu Z, et al. [(18)F](2S, 4S)-4-(3-Fluoropropyl)glutamine as a tumor imaging agent. Mol Pharm. 2014;11:3852–3866. doi: 10.1021/mp500236y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wachter S, et al. 11C-acetate positron emission tomography imaging and image fusion with computed tomography and magnetic resonance imaging in patients with recurrent prostate cancer. J Clin Oncol. 2006;24:2513–2519. doi: 10.1200/JCO.2005.03.5279. [DOI] [PubMed] [Google Scholar]
- 82.Ploessl K, Wang L, Lieberman BP, Qu W, Kung HF. Comparative evaluation of 18F-labeled glutamic acid and glutamine as tumor metabolic imaging agents. J Nucl Med. 2012;53:1616–1624. doi: 10.2967/jnumed.111.101279. [DOI] [PubMed] [Google Scholar]
- 83.Oyama N, et al. 11C-acetate PET imaging of prostate cancer: detection of recurrent disease at PSA relapse. J Nucl Med. 2003;44:549–555. [PubMed] [Google Scholar]
- 84.Mena E, et al. 11C-Acetate PET/CT in localized prostate cancer: a study with MRI and histopathologic correlation. J Nucl Med. 2012;53:538–545. doi: 10.2967/jnumed.111.096032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Qu W, et al. Synthesis of optically pure 4-fluoro-glutamines as potential metabolic imaging agents for tumors. J Am Chem Soc. 2011;133:1122–1133. doi: 10.1021/ja109203d. [DOI] [PubMed] [Google Scholar]
- 86.Venetti S, et al. Glutamine-based PET imaging facilitates enhanced metabolic evaluation of gliomas in vivo. Sci Trans Med. 2015;7:274ra17. doi: 10.1126/scitranslmed.aaa1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fan TW, et al. Altered regulation of metabolic pathways in human lung cancer discerned by 13C stable isotope-resolved metabolomics (SIRM) Mol Cancer. 2009;8:41. doi: 10.1186/1476-4598-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Maher EA, et al. Metabolism of [U-13C]glucose in human brain tumors in vivo. NMR Biomed. 2012;25:1234–1244. doi: 10.1002/nbm.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Marin-Valencia I, et al. Glucose metabolism via the pentose phosphate pathway, glycolysis and Krebs cycle in an orthotopic mouse model of human brain tumors. NMR Biomed. 2012;25:1177–1186. doi: 10.1002/nbm.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Marin-Valencia I, et al. Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell Metab. 2012;15:827–837. doi: 10.1016/j.cmet.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sellers K, et al. Pyruvate carboxylase is critical in non-small cell lung cancer. J Clin Invest. 2015;125:687–698. doi: 10.1172/JCI72873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Clarke DD, Sokoloff L. In: Basic Neurochemistry: Molecular, Cellular and Medical Aspects. 6. Siegel GJ, Agranoff BW, Albers RW, Fisher SK, Uhler MD, editors. Ch 31. Lippincott-Raven; 1999. [Google Scholar]
- 93.Yang ZJ, Chee CE, Huang S, Sinicrope FA. The role of autophagy in cancer: therapeutic implications. Mol Cancer Ther. 2011;10:1533–1541. doi: 10.1158/1535-7163.MCT-11-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Farrow JM, Yang JC, Evans CP. Autophagy as a modulator and target in prostate cancer. Nat Rev Urol. 2014;11:508–516. doi: 10.1038/nrurol.2014.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Amaravadi RK, et al. Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res. 2011;17:654–666. doi: 10.1158/1078-0432.CCR-10-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wilson PM, Danenberg PV, Johnston PG, Lenz HJ, Ladner RD. Standing the test of time: targeting thymidylate biosynthesis in cancer therapy. Nat Rev Clin Oncol. 2014;11:282–298. doi: 10.1038/nrclinonc.2014.51. [DOI] [PubMed] [Google Scholar]
- 97.Glasauer A, Chandel NS. Targeting antioxidants for cancer therapy. Biochem Pharmacol. 2014;92:90–101. doi: 10.1016/j.bcp.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 98.Yuneva MO, et al. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell Metabol. 2012;15:157–170. doi: 10.1016/j.cmet.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yabu M, et al. IL-23-dependent and -independent enhancement pathways of IL-17A production by lactic acid. Int Immunol. 2011;23:29–41. doi: 10.1093/intimm/dxq455. [DOI] [PubMed] [Google Scholar]
- 100.Lisanti MP, et al. Understanding the “lethal” drivers of tumor-stroma co-evolution: emerging role(s) for hypoxia, oxidative stress and autophagy/mitophagy in the tumor micro-environment. Cancer Biol Ther. 2010;10:537–542. doi: 10.4161/cbt.10.6.13370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Martinez-Outschoorn UE, et al. Oxidative stress in cancer associated fibroblasts drives tumor-stroma co-evolution: A new paradigm for understanding tumor metabolism, the field effect and genomic instability in cancer cells. Cell Cycle. 2010;9:3256–3276. doi: 10.4161/cc.9.16.12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yachida S, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ding L, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]