Abstract
Glutathione S-transferase pi has been shown to reactivate 1-cysteine peroxiredoxin (1-Cys Prx) by formation of a complex. A model of the complex was proposed based on the crystal structures of the two enzymes. We have now characterized the complex of GST pi/1-Cys Prx by determining the Mw of the complex, by measuring the catalytic activity of the GST pi monomer, and by identifying the interaction sites between GST pi and 1-Cys Prx. The Mw of the purified GST pi/1-Cys Prx complex is 50,200 at pH 8.0 in the presence of 2.5 mM glutathione, as measured by light scattering, providing direct evidence that the active complex is a heterodimer composed of equimolar amounts of the two proteins. In the presence of 4 M KBr, GST pi is dissociated to monomer and retains catalytic activity, but the Km value for GSH is increased substantially. To identify the peptides of GST pi that interact with 1-Cys Prx, GST pi was digested with V8 protease and the peptides were purified. The binding by 1-Cys Prx of each of four pure GST pi peptides (residues 41–85, 115–124, 131–163, and 164–197) was investigated by protein fluorescence titration. An apparent stoichiometry of 1 mol/subunit 1-Cys Prx was measured for each peptide and the formation of the heterodimer is decreased when these peptides are included in the incubation mixture. These results support our proposed model of the heterodimer.
Keywords: Glutathione S-transferase pi, 1-Cys peroxiredoxin, Heterodimer
Glutathione S-transferases (GSTs1), which catalyze the nucleophilic attack by the thiol of glutathione on electrophilic substrates, constitute a family of enzymes important in the detoxification of xenobiotics, endogenous compounds, and the products of oxidative stress [1,2]. The pi isozyme (GST pi), crystallized as a homodimer with a subunit molecular weight of 23,500, is of particular interest because it exhibits diverse roles in mammalian cells: it provides a defense against carcinogenesis, since it catalyzes the inactivation of known carcinogens [3]; it contributes significantly to the development of resistance to cancer chemotherapy, since GST pi levels increase in tumors and the enzyme metabolizes key anticancer drugs [4–7]; and it promotes the cellular response to oxidative stress, since GST pi has recently been reported to activate the anti-oxidant enzyme 1-Cys peroxiredoxin [8,9].
1-Cys peroxiredoxin (1-Cys Prx, Prdx 6, Prx VI, and AOP2), a homodimer with a subunit molecular weight of 25,000, is an anti-oxidant enzyme that catalyzes the reduction of hydroperoxides to alcohols using a strictly conserved cysteine [10]. The oxidized 1-Cys Prx intermediate must react with another thiol compound to regenerate the sulfhydryl cysteine of active 1-Cys Prx. However, that thiol had not been identified.
Recently, we demonstrated that reactivation of human 1-Cys Prx occurs by formation of a complex between 1-Cys Prx and human GST pi in the presence of glutathione (GSH), in which the two proteins are present in equimolar mixtures [9]. The complex is dissociated when GSH is removed by dialysis [9]. Complex production in the presence of GSH is followed by glutathionylation of 1-Cys Prx, and subsequently by the slow formation of an intermolecular disulfide between 1-Cys Prx and GST pi [9]. The disulfide is then reduced by GSH, regenerating an active 1-Cys Prx [8,9]. A plausible energy-minimized model of the complex was proposed; this ‘in silico’ model was constructed by docking one subunit from the crystal structure of GST pi with one subunit from the crystal structure of 1-Cys Prx using the program ZDOCKpro 1.0. In the present paper, we evaluate the validity of our proposed model by measuring the molecular weight of the complex, by determining whether GST pi retains catalytic activity as a monomer, and by identifying the contact sites between GST pi and 1-Cys Prx in the active complex. A preliminary version of this study has been presented [11].
Materials and methods
Materials
Reduced GSH, 1-chloro-2,4-dinitrobenzene (CDNB), S-hexylglutathione Sepharose, S-hexylglutathione, imidazole, and chemicals for the preparation of buffers were obtained from Sigma Chemical Co. Nickel–nitrilotriacetic acid agarose (Ni–NTA) was purchased from Qiagen, Inc. 1,6-Hexanediol was supplied by ACROS Organics. Synthetic GST pi peptide fragments were obtained from EZBiolab. Centriplus YM-10 concentrators were from Millipore. Staphylococcal Protease V8 was purchased from Worthington Biochemical Corporation. All chemicals were of reagent grade.
Expression and purification of GST pi and 1-Cys Prx
For the expression of human GST pi, the WT plasmid was transformed into Escherichia coli JM105 and the cells were grown and induced for expression of GST pi [12]. For the expression of human 1-Cys Prx (Prdx 6), the WT plasmid was transformed in E. coli BL21 (DE3) cells [13]. Because GST pi does not have a His6-tag, purification of the enzyme was performed using a S-hexylglutathione agarose affinity column, as described previously [11]. In all cases, GST pi was eluted using a buffer containing 2.5 mM S-hexylglutathione. 1-Cys Prx contains a His6-tag at the C-terminus; thus, purification of the protein was performed using a Ni–NTA column, as described previously [13]. All of the enzymes were purified to homogeneity, yielding a single peptide by N-terminal sequencing on an Applied Biosystems gas-phase sequencer (Model Procise) equipped with an on-line Microgradient Delivery System (Model 140 C) and a computer (Model 610 Macintosh). Each of the enzymes exhibited a single band by SDS–PAGE. All purified GSTs were stored in aliquots at −80 °C, and all purified 1-Cys Prx proteins were stored in aliquots at 4 °C, both enzymes in 0.05 M Tris–Cl buffer at pH 8.0.
Determination of the molecular mass of the enzymes
Light scattering measurements with a miniDAWN laser photometer (Wyatt Technology Corp., Santa Barbara, CA) was used to determine the molecular mass of GST pi, 1-Cys Prx, and the complex. All samples (0.1–0.3 mg/mL) were in 0.05 M Tris–Cl, pH 8.0 and filtered through a 0.02-μm filter before being used and, when indicated, a saturating concentration of GSH was included. The addition of GSH to the enzyme samples was used to determine if this substrate would promote dimerization. Data were collected at a laser wavelength of 690 nm and analyzed using ASTRA software for Windows. The A280 nm was used to determine the protein concentration (E0.1%280 nm = 1.24 for GST pi [14], 1.07 for 1-Cys Prx, and 1.16 for the complex [9]).
Light scattering was also conducted for GST pi samples (0.1–0.4 mg/mL) in 0.05 M sodium–MES buffer (pH 6.5) containing various concentrations of KBr (0–4 M). The average molecular mass of each sample was also determined in the presence of a saturating GSH concentration.
Standard assay for GST activity
Enzymatic activity toward 1-chloro-2,4-dinitrobenzene (CDNB) was measured in a total volume of 1.0 mL using a Hewlett–Packard 8453 spectrophotometer by monitoring the formation of the conjugate of CDNB (3 mM) and GSH (2.5 mM) at 340 nm (Δε = 9600 M−1 cm−1) in 0.1 M potassium phosphate buffer (pH 6.5), containing 1 mM EDTA and 2.5% ethanol at 25 °C, according to the method of Habig et al. [15]. An enzyme unit is defined as the amount of enzyme that converts 1 μmol/min of CDNB to the conjugate. All measurements were corrected for the spontaneous nonenzymatic rate of formation of the conjugate of GSH and CDNB.
The apparent Km value of GST pi for glutathione was determined at 25 °C by varying the glutathione concentrations (0.01–2 mM) while keeping a constant CDNB concentration (3 mM). Similarly, the apparent Km value for CDNB was determined from a range of concentrations of CDNB (0.01–4.0 mM) at a constant glutathione concentration (2.5 mM) in 0.1 M potassium phosphate buffer containing 1 mM EDTA and 2.5% ethanol (pH 6.5).
The kinetic parameters were also determined for GST pi in 0.05 M sodium–MES (pH 6.5) containing the various concentrations of KBr. For these measurements, the enzyme was stored in 0.05 M sodium–MES (pH 6.5) and was diluted into the assay buffer of 0.05 M sodium–MES (pH 6.5) and KBr immediately before the activity measurements. In determining the Km for CDNB in the presence of KBr, the constant [glutathione] was maintained at 25 mM.
Formation of GST pi/1-Cys Prx complex under standard conditions
Heterodimers were generated in which one subunit was a non-His-tagged GST pi and the other subunit was a His-tagged 1-Cys Prx. The enzymes were incubated together in 20% 1,6-hexanediol in 0.05 M Tris–Cl at pH 8.0 for 2 h at 25 °C to facilitate complete dissociation to monomers [16,9]. A total of 1 mg of each enzyme in 1 mL of buffer was used. This mixture was then dialyzed overnight (molecular weight cutoff = 10 kDa) against 50 mM Tris–Cl buffer containing 2.5 mM reduced GSH at pH 8.0 and 4 °C to remove 1,6-hexanediol and allow the reformation of dimers. The mixture of two proteins was then loaded onto a Ni–NTA column (1.5 mL) equilibrated in 0.05 M Tris–Cl buffer at pH 8.0 containing 2.5 mM reduced GSH at 4 °C. The enzymes separate because 1-Cys Prx has a His-tag on each subunit and binds tightly to the Ni–NTA column, while the GST pi has no His-tag and does not bind to the column. The heterodimer (in which only one of the two subunits has a His-tag) binds less tightly to the Ni–NTA column than does 1-Cys Prx alone (in which every subunit contains a His-tag). The column was washed with 0.05 M Tris–Cl buffer at pH 8.0, containing 2.5 mM GSH, to remove unbound GST pi. The heterodimer and 1-Cys Prx were separated using a linear gradient from 0.05 M Tris–Cl at pH 8.0 containing 2.5 mM reduced GSH to the same buffer with the addition of 0.3 M imidazole (20 mL of each buffer). (If the column is eluted, with buffer lacking GSH, the heterodimer complex dissociates.) The column eluate was monitored at A280 nm. Fractions of 1 mL were collected, and the GST activity was determined under standard conditions. The three peaks were pooled separately and concentrated to approximately 1 mL using the Centriplus YM-10 centrifugal filter device (molecular weight cutoff = 10 kDa). To remove the imidazole, the heterodimer and 1-Cys Prx were dialyzed against 0.05 M Tris–Cl buffer at pH 8.0 containing 2.5 mM reduced GSH. The activity of the recovered GST pi, as well as the purified heterodimer, was determined under standard conditions. The A280 nm was used to determine the protein concentration. All three enzymes were stored at 4 °C in 0.05 M Tris–Cl buffer at pH 8.0 containing 2.5 mM reduced GSH. The N-terminal sequences of the recovered proteins were determined to confirm the composition and purity of the enzymes.
Proteolytic digestion of GST pi by Staphylococcal protease V8
GST pi (1.0–2.0 mg/mL) was incubated for 30 min in 0.05 M Tris–Cl buffer (pH 8.0) with 6 M Urea and 10 mM N-ethylmaleimide at 25 °C to block the free −SH groups. The enzyme solution was then dialyzed overnight (molecular weight cutoff = 10 kDa) against 4 L of 50 mM ammonium bicarbonate (pH 7.8). The enzyme solution was lyophilized and then resolubilized in 1.0 mL of 2 M urea in 50 mM ammonium bicarbonate (pH 7.8) and incubated at 37 °C for 2 h. V8 protease [5.0% (w/w)] and the enzyme sample were incubated for 2.5 h at 37 °C. A second aliquot of the V8 protease solution was added, and incubation was continued for another 2.5 h at 37 °C.
HPLC separation of peptides
The proteolytic digest was injected onto a Varian 5000 LC HPLC (Varian, Walnut Creek, CA) equipped with a Phenomenex C18 reverse-phase column (0.46 × 25 cm) equilibrated with solvent A (0.1% trifluoroacetic acid in water). After elution with solvent A (0% solvent B) for 10 min, a linear gradient was run to 10% solvent B (0.075% trifluoroacetic acid in acetonitrile) at 30 min followed by a linear gradient to 25% solvent B at 105 min. The linear gradient was continued to 35% solvent B at 115 min, followed by 45% solvent B at 180 min, and finally a 30-min gradient to 100% solvent B. The column was eluted for 10 min with 100% solvent B. The flow rate was 1 mL/min. The eluate was monitored at 220 nm with 1 mL fractions collected. The peptide peaks resulting from the HPLC separation were lyophilized and analyzed by N-terminal sequencing as described previously.
Formation of GST pi/1-Cys Prx complex in the presence of GST pi peptide fragments
GST pi/1-Cys Prx complexes were formed under standard conditions (2 h of incubation with 20% 1,6-hexanediol at 25 °C) as described above except that either a fraction from the separated GST pi proteolytic digest or 100 μM of a pure synthetic GST-derived peptide was included. The GST pi, heterodimer complex and 1-Cys Prx were separated using Ni–NTA chromatography. The conditions were the same as described above, except that dialysis was conducted using a membrane with a molecular weight cutoff of 500 to remove 1,6-hexanediol while retaining the proteins and the test peptides. The activity of the recovered GST pi homodimer, as well as the purified complex, was determined under standard conditions as described above. The A280 nm was used to determine the protein concentration. The N-terminal sequences of the recovered GST pi, 1-Cys Prx, and purified heterodimers were determined to confirm the purity of the enzymes.
Fluorescence spectrophotometry
The steady-state fluorescence spectra of 1-Cys Prx (2 μM enzyme subunits) in 0.05 M Tris–Cl buffer, pH 8.0 were measured on a Perkin-Elmer MPF-3 fluorescence spectrophotometer at 25 °C. The enzyme solutions were excited at 280 nm and the emission spectrum of each sample was scanned and recorded in the range of 290–420 nm. The bandwidth for excitation and emission was 10 nm. The spectra were corrected for the background contributed by the buffer.
The stoichiometry of binding for each synthetic GST peptide fragment was determined by fluorescence titration. The decrease in protein fluorescence of 1-Cys Prx (2 μM) in 0.05 M Tris–Cl buffer, pH 8.0, was monitored as a function of peptide concentration (0.04–4 μM). For all measurements, the excitation wavelength was 280 nm and the emission was monitored at 330 nm at 25 °C. In a typical experiment, two parallel samples containing a 1 mL solution of 1-Cys Prx or buffer alone were titrated with 2 μL aliquots of a concentrated GST pi peptide solution. The final dilution did not exceed 2–3% of the initial volume. Fluorescence titration data were corrected for dilutions.
Protein–protein docking of the GST pi and 1-Cys Prx complex
We have used a two stage procedure called ZDOCKpro 1.0, as described previously [9]. The ZDOCKpro package is based on the ZDOCK and RDOCK programs within Insight II molecular modeling software (Molecular Simulations, Inc.). The Protein Data Bank structures used were PDB: 1PRX (1-Cys Prx) and 19GS (GST pi). The model of the heterodimer complex is the same one presented previously [9].
Results
The complex between GST pi and 1-Cys Prx is a heterodimer
Previously, we isolated and characterized a complex between GST pi and 1-Cys Prx that had a molar ratio of 1:1 based on N-terminal sequencing, but the molecular mass of this complex had not been determined [9]. In order to assess the composition of the complex, molecular masses for wild-type human GST pi, 1-Cys Prx, and the complex were determined using light scattering (Table 1). First, the molecular mass of wild-type human GST pi was examined over a range of protein concentrations (0.1–0.4 mg/mL) in the absence and presence of glutathione. The monomeric mass of GST pi is 23,500 Da; thus, if the enzyme were completely dimeric in solution, the molecular mass should be approximately 47,000 Da. In contrast, the results shown in Table 1 (columns 2 and 3) indicate that the molecular mass of GST pi is significantly lower than that expected for a fully dimeric species, suggesting that in solution, GST pi exists in a dimer–monomer equilibrium with the monomer predominating.2 The percentage of dimer and monomer in the protein solutions can be determined from the measured average mass at a known protein concentration; the fraction of the total concentration that is monomer and dimer can be calculated using the known mass values of the monomer and the dimer. For example, at a concentration of 0.1 mg/mL of GST pi, the average molecular mass is 29,300 Da in the presence of GSH as compared to 47,000 Da if the protein was 100% dimer, suggesting that the enzyme is only 25% dimeric in solution. The existence of appreciable amounts of GST pi monomer in solution may account, in part, for the ability of this isozyme of GST to form a complex with 1-Cys Prx. For comparison, under similar conditions, the Mw is 50,100 Da for the alpha-GST [17] and 46,900 Da for the mu-GST [18], indicating that these enzymes are, respectively, 88% and 83% dimeric in solution. These results may explain, in part, the observation that of the glutathione S-transferases tested for the ability to form a complex with 1-Cys Prx, alpha class GST had no effect and mu class was only 20% as effective as GST pi [9].
Table 1.
Molecular mass of human GST pi, 1-Cys Prx, and the complex in 0.05 M Tris-chloride buffer (pH 8.0), determined by light scattering in the absence and presence of GSH
| [Protein] (mg/mL) | Mass of GST pi (Da) |
Mass of 1-Cys Prx (Da) |
Mass of GST pi/1-Cys Prx complex (Da) | ||
|---|---|---|---|---|---|
| –GSH | +GSHa | –GSH | +GSHb | +GSHa | |
| 0.1 | 33,200 ± 1800 | 29,300 ± 2100 | 37,400 ± 1000 | 38,700 ± 1100 | 49,300 ± 300 |
| 0.2 | 32,900 ± 1000 | 34,100 ± 2000 | 40,500 ± 400 | 36,900 ± 700 | 49,400 ± 900 |
| 0.3 | 34,300 ± 1400 | 36,300 ± 1800 | 38,800 ± 100 | 37,600 ± 400 | 51,800 ± 4700 |
| 0.4 | 35,100 ± 1300 | 38,700 ± 1800 | 43,000 ± 200 | 35,200 ± 200 | – |
The concentration of GSH (2.5 mM) is saturating.
The concentration of GSH is 2.5 mM.
Since GSH has been shown to be an important component in the formation and activation of the complex between GST pi and 1-Cys Prx [9], the molecular mass of GST pi was also determined in the presence of the same concentration of GSH used to form the complex (2.5 mM). Table 1 (columns 2 and 3) shows that the addition of GSH does not appreciably change the molecular mass of GST pi indicating that GSH does not promote complex formation by shifting the dimer–monomer equilibrium of GST pi.
It was important to ascertain the molecular mass of 1-Cys Prx alone. The 1-Cys Prx subunit (with 6 His) has a molecular mass of 25,900; in this case, if 1-Cys Prx is completely dimeric in solution, the molecular mass should be approximately 52,000. The results of Table 1 (columns 4 and 5) suggest that 1-Cys Prx also exists in an equilibrium mixture of dimer and monomer, as shown by an average molecular mass of about 37,000 Da, significantly lower than the value expected for a dimer. Furthermore, the presence of GSH did not have an effect on the equilibrium. For 1-Cys Prx, the average molecular mass suggests that in solution this protein is 53% dimeric, which also supports the proposal that the observed equilibrium may be partly responsible for its ability to form a complex with another protein.
The molecular weight of the purified GST pi/1-Cys Prx complex was determined only in the presence of GSH because of the instability of the complex in the absence of GSH [9]. Under these conditions, the average molecular mass of the complex is equal to that of a dimer (~50 kDa; Table 1); the complex cannot be explained by a mixture of 1-Cys Prx + GST pi since the molecular mass is greater than that for either single protein when measured alone. For the first time, we have direct evidence that the active GST pi/1-Cys Prx complex is indeed a dimer.
GST pi is active as a monomer
In our previous studies [9], we showed that the complex is active with respect to both peroxidase and glutathione S-transferase activity. However, the GST activity in the complex (when considered per mg of GST present) was only 25% of that of GST pi alone. Since we now know that the complex exists as a heterodimer in which one subunit of GST pi interacts with one subunit of 1-Cys Prx, it is likely that GST pi retains some catalytic activity as a monomer. Furthermore, the glutathione S-transferase activity should properly be compared with that of a monomer of GST. Until now the question of whether the GST pi monomer has activity has not been addressed.
Monovalent anions have been used to disrupt the electrostatic interactions between subunits of multimeric proteins [19]. KBr was selected for this study since, in accordance with the Hofmeister series of monovalent anions, bromide is an effective protein destabilizing and dissociating reagent [19]. Moreover, Hearne and Colman [18] established that 0.05 M MES (pH 6.5) containing a range of KBr concentrations was a satisfactory system for examining the dimer–monomer equilibrium of the mu-class GST. Therefore, 0.5–4.0 M KBr was added to GST pi in 0.05 M MES (pH 6.5). Table 2 (column 2) shows that the average molecular mass of GST pi in 0.05 M MES (pH 6.5) is dependent on the concentration of KBr present. Appreciable amounts of dimer remain at concentrations up to 2.0 M KBr (Table 2). Above 2.0 M KBr, the average molecular mass decreases as the concentration of KBr increases. The enzyme becomes predominantly monomeric (~94%) in the presence of 4 M KBr (Table 2). The Mw was evaluated in the absence and presence of saturating GSH concentrations to determine whether this substrate promotes dimerization. The results (Table 2, column 3) show that GSH has little effect on the average molecular mass of the enzyme in the presence of KBr.
Table 2.
Molecular mass and specific activities of GST pi in 0.05 M MES (pH 6.5) containing various concentrations of KBr, determined by light scattering in the absence and presence of GSH
| Concentration of KBra (M) | Mw without GSH (Da) | Mw with GSHb (Da) | Specific activityb,c (μmol/min/mg) |
|---|---|---|---|
| 0 | 32,400 ± 1000 | 33,800 ± 1000 | 71.6 ± 4.0 |
| 0.5 | 31,500 ± 400 | 32,500 ± 200 | 70.2 ± 4.1 |
| 1.0 | 31,000 ± 1400 | 33,900 ± 700 | 48.0 ± 5.5 |
| 1.5 | 30,700 ± 1900 | 31,400 ± 300 | 32.3 ± 1.0 |
| 2.0 | 30,600 ± 1700 | 30,600 ± 700 | 17.7 ± 1.6 |
| 2.5 | 28,300 ± 1400 | 31,600 ± 1300 | 17.6 ± 2.2 |
| 3.0 | 27,500 ± 900 | 26,100 ± 500 | 13.4 ± 1.3 |
| 3.5 | 25,500 ± 600 | 25,700 ± 2500 | 19.4 ± 1.0 |
| 4.0 | 24,800 ± 2800 | 24,300 ± 2000 | 12.8 ± 1.3 |
In all cases, the enzyme (0.1-0.4 mg/mL) was in 0.05 M MES (pH 6.5) containing various concentrations of KBr.
The concentration of GSH (25 mM) is saturating.
GST activity, in the presence of various concentrations of KBr, was measured using CDNB as the electrophilic substrate.
GST pi was also assayed for activity in the presence of various concentrations of KBr using CDNB as the electrophilic substrate. As shown in Table 2 (column 4), the specific activity between 0 and 0.5 M KBr is essentially the same, but sharply decreases between 0.5 and 1.0 M KBr. At concentrations greater than 1.0 M KBr the specific activity gradually decreases until the values plateau at 2.0 M KBr (~15 lmol/min/mg). In contrast, most of the decrease in molecular mass of GST pi occurs between 2.0 and 4.0 M KBr (Table 2).
Incubation of GST pi over 5 h in 0.05 M sodium–MES (pH 6.5) containing 4 M KBr indicates that the enzyme, when assayed under the same salt conditions as those in the incubation solution, exhibits the same activity over the entire time period. Since there is no observable time dependence of change in enzymatic activity, we conclude that the activity of the wild-type enzyme in 0.05 M MES (pH 6.5) containing KBr is established rapidly and does not change over at least 5 h.
The kinetic parameters of GST pi were determined in the presence of increasing concentrations of KBr to evaluate whether the electrophilic substrate, the glutathione substrate, or both are affected by the KBr and the dissociation of the dimer. Table 3 shows the Km value for GSH and the Km and Vmax values for the xenobiotic substrate CDNB. For CDNB, the Km values do not change appreciably. In contrast, the Km for GSH exhibits a ~28-fold increase. Some of this Km effect is likely due to the dissociation of GST pi and some due to the effect of KBr on the catalytic reaction, because there is little change in molecular weight up to 2.0 M KBr while the Km value for GSH increases 16-fold. Similar to the specific activity values, the Vmax decreases to approximately 13 μmol/min/mg, clearly indicating that the monomer of GST pi retains appreciable activity.
Table 3.
CDNB and GSH kinetic parameters of GST pi in 0.05 M MES (pH 6.5) containing various concentrations of KBr
| Concentration of KBr (M) | aVmaxCDNB (μmol/min/mg) | bKmCDNB (mM) | cKmGSH (mM) |
|---|---|---|---|
| 0 | 71.6 ± 4.0 | 1.0 ± 0.04 | 0.1 ± 0.04 |
| 0.5 | 70.2 ± 4.1 | 1.0 ± 0.1 | 0.4 ± 0.02 |
| 1.0 | 48.0 ± 5.5 | 0.9 ± 0.1 | 0.6 ± 0.1 |
| 1.5 | 32.3 ± 1.0 | 0.6 ± 0.03 | 1.2 ± 0.2 |
| 2.0 | 17.7 ± 1.6 | 0.8 ± 0.01 | 1.6 ± 0.02 |
| 3.0 | 13.4 ± 1.3 | 0.6 ± 0.04 | 2.2 ± 0.1 |
| 4.0 | 12.8 ± 1.3 | 0.6 ± 0.03 | 2.8 ± 0.3 |
The Km values were generally determined at 25 °C under saturating conditions of the invariant substrate and the Vmax values were determined by an extrapolation of the velocity vs. the concentration of CDNB to infinite concentrations using SigmaPlot for data analysis. The data presented are the averages of at least two trials.
The concentration of GSH used for the determination of the KmCDNB was 25 mM.
The concentration of CDNB used in the determination of the KmGSH was 3 mM.
To determine whether the monomer of GST pi has activity, we assumed that the measured Vmax is the sum of the contributions from the dimer and the monomer:
Vmax = x (fraction of total that is dimer) + y (fraction of total that is monomer), where x is the specific activity of the dimer and y is the specific activity of the monomer. We can calculate the fraction of the total that is dimer and monomer from the measured average molecular mass. We selected molecular mass values for the enzyme in the presence of 2.0 M KBr because this activity reflects the effect of KBr on the activity of the enzyme in the absence of any appreciable change in the dimer–monomer equilibrium. Assuming that the activity for the monomer in 2.0 M KBr is the same as the activity of the monomer in the presence of 4.0 M KBr (where GST pi is 94% monomeric), one has two equations and two unknowns; thus a Vmax value can be calculated for the monomer and the dimer of GST pi of 12.0 and 31.1 μmol/min/mg, respectively. The monomer of GST pi is catalytically competent and contributes 28% of the overall specific activity. We have previously reported that the activity of the GST pi subunit within the GST pi/1-Cys Prx heterodimer is 28% that of GST pi alone. Thus, the GST activity in the complex is comparable to that of a separate GST pi monomer suggesting that the GST subunit functions independently in the heterodimer.
Effect of V8 proteolytic fragments of GST pi on the formation of the complex
The heterodimer complex is formed by incubation of purified GST pi (without a His6-tag) and 1-Cys Prx (with a C-terminal His6-tag) at pH 8.0 in buffer containing 20% 1,6-hexanediol to dissociate the homodimers, followed by dialysis against a buffer containing 2.5 mM glutathione (GSH) but lacking l,6-hexanediol [9]. The heterodimer can be purified by chromatography on a nickel–nitriloacetic acid-agarose column in the presence of GSH as described in Materials and methods. Fig. 1 (open circles) illustrates the separation of the enzymes. As proved by N-terminal sequencing in our earlier paper [9], Peak I consists of GST pi and has no His6-tag; Peak II is composed of GST pi/1-Cys Prx heterodimer and it has only one His6-tag; and Peak III consists of 1-Cys Prx, which has two His6-tags/dimer.
Fig. 1.
A representative plot comparing the separation of dimeric species of GST pi and 1-Cys Prx for the incubation mixture alone (○) and the incubation mixture in the presence of a complete, unfractionated digest of GST pi (●) using a Ni–NTA column as monitored by absorbance at 280 nm. The column was initially (fractions 1–15) eluted with 2.5 mM glutathione in 50 mM Tris–Cl, pH 8.0 (Buffer A). At fraction 15, a linear gradient was started from Buffer A (20 mL) to 0.3 M imidazole in Buffer A (20 mL). After the gradient, elution was continued with 0.3 M imidazole in Buffer A.
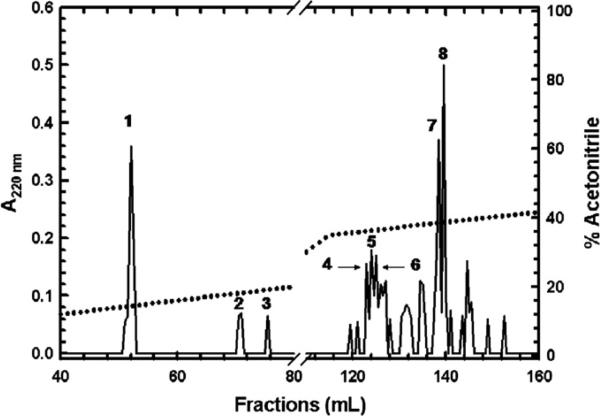
We sought to identify the contact sites between GST pi and 1-Cys Prx in the heterodimer. So that we were not restricted to the model we have suggested of the complex [9, Fig. 6], we have subjected GST pi to proteolytic digestion by Staphylococcal protease V8 in ammonium bicarbonate buffer, pH 7.8, and tested the peptide peaks for their ability to inhibit the formation of the heterodimer (i.e., Peak II of Fig. 1). Fig. 2 shows the HPLC separation with the peptide peaks numbered. Peaks 1–8 account for the entire sequence of GST pi. The additional peaks likely represent alternate proteolytic cleavages. The molecular weights of most of the peptides range from 650 to 3500. By using a dialysis membrane with a molecular weight cutoff of 500, we have removed the hexanediol (molecular weight ~110) to allow the two proteins to form homo- and heterodimers, yet retained most of the test peptides. The extent of formation of the heterodimer in the presence of the peptide was measured by chromatography on a Ni–NTA column, as in Fig. 1, solid circles.
Fig. 6.
Model of the GST pi/1-Cys Prx heterodimer illustrating the relative locations of the four GST pi peptide fragments that bind to 1-Cys Prx and that inhibit the formation of the complex. Ribbon representation of 1-Cys Prx (PDB 1PRX) complexed with GST pi (PDB 19GS). The backbone of the subunit B of 1-Cys Prx is cyan with tryptophan 33, 82, and 181 highlighted in yellow, and the backbone of the subunit A of GST pi is pink with peptide 41–85 highlighted in blue, peptide 115–124 highlighted in green, peptide 131–163 highlighted in red, and peptide 164–197 highlighted in orange. Glutathione is shown in brown, cysteine 47 of GST pi is green, and cysteine 47 of 1-Cys Prx is yellow.
Fig. 2.
HPLC separation of GST pi peptides resulting from digestion with Staphylococcal V8 protease. The digest was fractionated on a C18 column, as described under Materials and methods, and the effluent was monitored at A220 nm. The gradient in acetonitrile is shown by the dotted line.
Table 4 reports the results of these experiments in terms of the area of the heterodimer peak, as measured by A280 nm and by GST pi activity. From the results, it was observed that the heterodimer complex between 1-Cys Prx and GST pi did not form when a complete, unfractionated digest of GST pi was included in the mixture, (compare B with A in Table 4 and the solid with the open circles in Fig. 1). These results indicated that one or more of the GST pi-derived peptides was competing with GST pi and inhibiting the formation of the heterodimer complex. Furthermore, including the mixture of peptides 86–97 + 113–130 (Table 4, Peak 4), peptide 41–85 (Table 4, Peak 6), peptide 131–163 (Table 4, Peak 7), or the mixture of peptides 86–97 and 164–197 (Table 4, Peak 8) in the incubation mixture of GST pi and 1-Cys Prx substantially decreased the formation of the heterodimer complex. In contrast, peptides 32–40 (Table 4, Peak 1), 98–112 + 198–209 (Table 4, Peak 2), 198–209 alone (Table 4, Peak 3) and 1–30 (Table 4, Peak 5) had little effect on the extent of formation of the heterodimer. We thus identified five GST pi peptide regions that may be involved in the interactions between GST pi and 1-Cys Prx complex: residues 41–85, 86–97, 113–130, 131–163, and 164–197. However, for conclusive results, each of these peptides had to be tested separately with defined concentrations. The peptide of residues 115–124 was substituted for 113–130 since, based on the molecular model, this was the region of the peptide making closest contact with 1-Cys Prx. In addition, peptide fragment 32–40 was added to test whether the failure to observe effects for residues 32–36 and 37–40 was due to the fact that the peptides were too small to be retained throughout the workup procedure. These peptides were synthesized for further examination; the sequences of these peptides are shown in Fig. 3. Each of the synthetic peptides was >90% pure.
Table 4.
Yield of GST pi/1-Cys Prx heterodimer in the presence of peptide fragments derived from various regions of GST pia
| GST pi peptide peaks examined | Peak II (heterodimer) |
|
|---|---|---|
| Total Abs280 nm units | Total GST activity unitsb (ΔOD340 nm/min) | |
| A. None | 1.18 | 1.36 |
| B. Complete, unfractionated digest | No peak | No peak |
| 1. Residues 32-36 and 37-40 | 1.05 | 1.17 |
| 2. Residues 98-112 and 198-209 | 0.87 | 0.91 |
| 3. Residues 198-209 | 1.09 | 1.21 |
| 4. Residues 86-97 and 113-130 | 0.21 | 0.33 |
| 5. Residues 1-30 | 1.12 | 1.29 |
| 6. Residues 41-85 and V8 peptide | No peak | No peak |
| 7. Residues 131-163 | 0.37 | 0.46 |
| 8. Residues 86-97 and 164-197 | 0.62 | 0.54 |
GST pi and 1-Cys Prx (40 μM of each) together with each GST pi peptide peak were treated with 20% 1,6-hexanediol, followed by dialysis against buffer containing 2.5 mM GSH and loaded onto a Ni-NTA column, as described in Materials and methods.
GST activity was measured using CDNB as the electrophilic substrate, as described in Materials and methods, and expressed as AOD340 nm/min.
Fig. 3.
Sequences of synthetic peptides of GST pi that were examined for interaction with 1-Cys Prx.
Effect of pure, synthetic fragments of GST pi on the formation of the complex
The influence of the synthetic peptides of GST pi on heterodimer formation was investigated by including 100 μM of each peptide in the incubation mixture of GST pi and 1-Cys Prx. The extent of formation of the heterodimer in the presence of the peptide was measured by chromatography on a Ni–NTA column, as in Fig. 1. The results are summarized in Table 5. Peptides 41–85 and 115–124 completely inhibited complex formation (Table 5, lines 2 and 3), peptides 131–163 and 164–197 partially inhibited the formation of the complex (Table 5, lines 4 and 5), and peptides 32–40 and 86–97 did not appreciably affect formation of the complex (Table 5, lines 6 and 7). These results agree with the model of the heterodimer complex [9] since the peptides that substantially decreased complex formation are close to the subunit interface and suggest they may bind to 1-Cys Prx.
Table 5.
Yield of heterodimer protein peak from the Ni-NTA column in the presence of various synthetic GST pi peptidesa
| GST pi peptides | Peak II (heterodimer) |
|
|---|---|---|
| Total Abs280 nm units | Total GST activity unitsb (ΔOD340 nm/min) | |
| 1. None | 1.18 | 1.36 |
| 2. Residues 41-85 | No peak | No peak |
| 3. Residues 115-124 | No peak | No peak |
| 4. Residues 131-163 | 0.49 | 0.62 |
| 5. Residues164-197 | 0.66 | 0.74 |
| 6. Residues 32-40 | 1.10 | 1.21 |
| 7. Residues 86-97 | 0.94 | 1.09 |
GST pi and 1-Cys Prx proteins (40 μM of each) together with 100 μM of synthetic GST pi peptide were treated with 20% 1,6-hexanediol followed by dialysis against buffer containing 2.5 mM GSH and loaded onto a Ni-NTA column, as described in Materials and methods.
GST activity was measured using CDNB as the electrophilic substrate, as described in Materials and methods, and expressed as ΔOD340 nm/min.
Effect of GST pi peptide fragments on the fluorescence of 1-Cys Prx
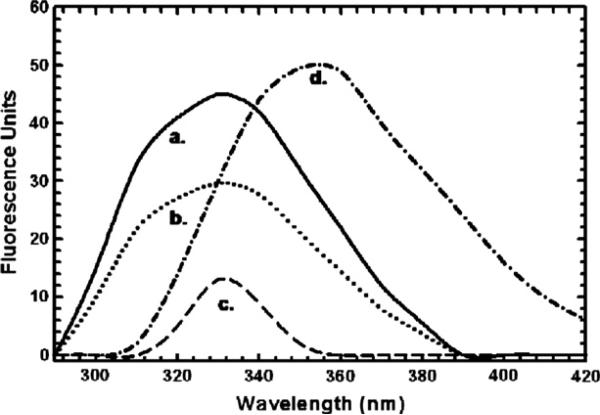
1-Cys Prx contains three tryptophans per monomer. Based on the proposed model of the complex, tryptophan 33, 82, and 181 are near the postulated interface; these tryptophans may be spectroscopically affected upon the binding of the GST pi fragments. When we compared the emission spectrum of 1-Cys Prx (Fig. 4, spectrum a) with that of free tryptophan (Fig. 4, spectrum d), we observed a shift in the maximum emission peak likely due to shielding of the tryptophan residues in the proteins from the aqueous phase [20].
Fig. 4.
The steady-state fluorescence spectra of 1-Cys Prx (2 μM enzyme subunits) in 0.05 Tris–chloride buffer (pH 8.0) was measured on a Perkin-Elmer MPF-3 fluorescence spectrophotometer. Emission spectrum (excitation at 280 nm, bandwidth of 10 nm) of 1-Cys Prx alone (a), 1-Cys Prx in the presence of 3.4 μM GST pi fragment 131–163 (b), 1-Cys Prx in the presence of 3.4 μM GST pi fragment 41–85 (c), free tryptophan (6 μM) (d).
A comparison of the emission spectrum (with excitation at 280 nm) of 2 μM of 1-Cys Prx alone (Fig. 4, spectrum a) with that of the same concentration of 1-Cys Prx in the presence of 3.4 μM GST pi peptide 41–85 (Fig. 4, spectrum c) reveals a decrease in the relative fluorescence intensity of 1-Cys Prx from 45 at its emission maximum (330 nm), to ~14 in the presence of fragment 41–85, a reduction to 31% of its original value of 1-Cys Prx caused by the addition of peptide 41–85. In contrast, when the emission spectrum of 1-Cys Prx is measured with the same concentration of GST pi fragment 131–163 (3.4 μM) (Fig. 4, spectrum b), the fluorescence intensity is reduced to only 29 fluorescence units, or to 65% of the fluorescence intensity of 1-Cys Prx alone. The decrease in the fluorescence intensity at the emission maximum of 1-Cys Prx in the presence of peptide 41–85 is perhaps due to a further change in solvent exposure of the tryptophan residues in the enzyme or to quenching by a proximate amino acid; but with fragment 131–163, a smaller change in tryptophan fluorescence is observed.
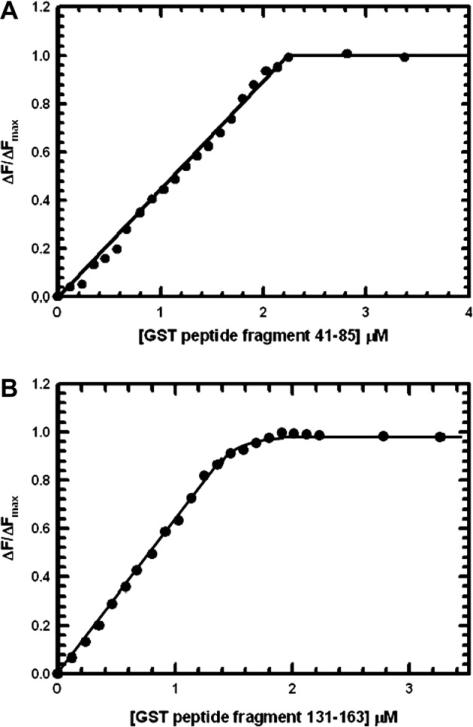
The binding of GST pi residues 32–40, 41–85, 86–97, 115–124, 131–163, and 164–197 to 1-Cys Prx in 0.05 M Tris–Cl, pH 8.0 was studied by monitoring the effect of increasing concentrations of each peptide on the fluorescence intensity at 330 nm of 1-Cys Prx (2 μM subunit concentration), as described in Materials and methods. Examples of these titrations are shown in Fig. 5A and B. From the concentration of the peptide at which the ΔF/ΔFmax first reaches 1.0 (see Fig. 5A), a stoichiometry was estimated of about 1 mol of peptide bound per mol of 1-Cys Prx subunit for GST pi residues 41–85 and 115–124 (Table 6, lines 1 and 2); in these cases ΔFmax is ~32. Furthermore, a stoichiometry of about 1.0 mol of peptide per enzyme subunit is obtained for GST pi residues 131–163 (see Fig. 5B) and 164–197 (Table 6, lines 3 and 4). However, in these cases ΔFmax is ~15. Under similar conditions, 1-Cys Prx does not bind peptides 32–40 and 86–97 (Table 6 lines 5 and 6), as demonstrated by their failure to change the fluorescence of 2 μM 1-Cys Prx when added at concentrations as high as 20 μM; these experiments indicate the specificity of the designated GST pi peptides in their interactions with 1-Cys Prx.
Fig. 5.
Fluorescence determination of the stoichiometry of binding for synthetic GST pi fragments 41–85 (A) and 131–163 to 1-Cys Prx (B). 1-Cys Prx (2 μM subunit concentration) in 0.05 M Tris–chloride buffer (pH 8.0) was titrated with the pure GST peptides at 25 °C. The results of the titration are plotted as fraction of maximum change (ΔF/ΔFmax) vs. the concentration of GST pi peptide 131–163. The decrease in protein fluorescence emission (ΔF) was recorded at 330 nm (excitation at 280 nm). The maximum change in fluorescence units (ΔFmax) is 32 for peptide 41–85 and 18 for peptide 131–163.
Table 6.
Determination of the stoichiometry of binding for synthetic GST pi peptide fragments and 1-Cys Prxa
| GST pi peptides | Maximum change in fluorescence units (ΔF) | Bound peptide/enzyme subunit |
|---|---|---|
| 1. Residues 41-85 | 32 | 1.10 |
| 2. Residues 115-124 | 35 | 0.95 |
| 3. Residues 131-163 | 18 | 0.95 |
| 4. Residues 164-197 | 11 | 0.73 |
| 5. Residues 32-40 | 0 | 0 |
| 6. Residues 86-97 | 0 | 0 |
The stoichiometry for the peptides was determined from the protein fluorescence titrations, as illustrated by Fig. 5. The titration experiments were conducted in 0.05 M Tris-Cl buffer, pH 8.0 at 25 °C at the concentration of enzyme subunits of 2 μM.
It is unclear whether the peptide binding sites for 1-Cys Prx are mutually exclusive or distinct. To distinguish between these possibilities, the fluorescence intensity at 330 nm of 1-Cys Prx (2 μM subunit concentration) was measured in the presence of 10 μM GST pi peptide 41–85 along with 10 μM of either peptides 115–124, 131–163, or 164–197. The combination of 41–85 with either 115–124 or 164–197 yields an average ΔFmax of ~31, similar to the ΔFmax of peptide 41–85 alone (Table 6, line 1). In contrast, the peptide mixture of 41–85 + 131–163 exhibits a slightly larger ΔFmax ~ 38. Moreover, the addition of 10 μM of all the peptides to 1-Cys Prx also yielded a value for ΔFmax ~ 38. These results suggest that peptide fragment 131–163 may bind at a different location from all the other fragments, consistent with the original model of the complex.
Discussion
In previous studies [9], we proposed that the mechanism of 1-Cys peroxiredoxin reactivation initially requires heterodimer formation between GST pi and 1-Cys Prx in the presence of glutathione. The formation of the complex allows GSH to have access to the oxidized cysteine 47 of 1-Cys Prx, facilitating its glutathionylation. The GST-bound GSH is thus functioning like the “resolving cysteine” residue in the 2-Cys peroxiredoxins [21]. Glutathionylation of 1-Cys Prx changes the conformation of the heterodimer, permitting a disulfide to form slowly between Cys-47 of GST pi and Cys-47 of 1-Cys Prx. (However, the intersubunit disulfide is not required for the formation or stabilization of the heterodimer since a heterodimer complex can be isolated chromographically even when S-methyl glutathione replaces reduced glutathione [9]; in that case, though, the processes of glutathionylation, inter-subunit disulfide formation and reactivation of 1-Cys peroxiredoxin do not occur). In contrast, in the presence of GSH, the intersubunit disulfide is reduced by GSH with regeneration of the peroxidative cysteine 47 of 1-Cys Prx [9].
We have determined that the GST pi/1-Cys Prx complex exists as a dimer. This conclusion comes from the observation that the molecular mass of the complex is ~50,000 Da in the presence of GSH [i.e., the sum of the mass of one subunit of GST pi (23,500) and one subunit of 1-Cys Prx (25,900)]. While GST pi and 1-Cys Prx have both been crystallized as dimers, our experiments show that, in solution, the individual proteins exist as mixtures of dimer and monomer since the average molecular mass of the individual proteins is significantly lower than that predicted for dimers. Indeed, the existence of insolution of appreciable amounts of monomeric GST pi and of monomeric 1-Cys Prx make it possible to form a heterodimic complex.
In our previous paper [9], we proposed that hexanediol was needed in the heterodimer incubation mixture to dissociate the enzymes to monomers. In light of the results of the present study, we suggest that hexanediol is actually needed to facilitate conformational changes that allow heterodimer formation. In past experiments involving the use of hexanediol with GST A1-1, the organic solvent was apparently needed mainly to dissociate the dimers.
The observation that the heterodimer complex is strictly dimeric in solution, while GST pi and 1-Cys Prx exist in dimer–monomer mixtures, also suggests that the stability of the complex is greater than that of either the dimer of GST pi or the dimer of 1-Cys Prx. This conclusion is further supported by the fact that Peak II is the largest peak in Fig. 1 after dissociation and reformation of the dimers. Although the reformation of dimers from a mixed pool of subunits is a random event, the greater stability of the heterodimer favors that species (i.e., Kd is lowest for the heterodimer).
There are several observations suggesting that GST pi is catalytically active as a monomer. For example, the active GST pi/1-Cys Prx complex, now identified as a heterodimer, has glutathione S-transferase activity. Additionally, although GST pi has been crystallized as a dimer, the sites for glutathione and for small electrophilic substrates (such as CDNB) are predominantly contained within each subunit; thus it is reasonable that monomers, produced under mild conditions, have activity. In a previous study, a monomeric species of a human GST pi was engineered by introducing 10 site specific mutations [22]. However, the absence of catalytic activity in this structurally stable monomer may not have general implications since it was so drastically altered.
We have here used 0.05 M MES, pH 6.5 containing a range of KBr concentrations to examine the dimer–monomer equilibrium of GST pi. Based on the crystal structure analyses of GSTs, three major interactions between the subunits in the dimer have been identified [23–25]. These interactions are the electrostatic and hydrogen-bonds between the polar amino acid residues of one subunit and that of the other subunit, the stacking interaction of two symmetrically equivalent arginines (one from each subunit), and the hydrophobic interactions, including a ball-and-socket motif. Electrostatic interactions of multimeric proteins can be disrupted by the addition of salts [26]. For example, the addition of salt resulted in the modulation of the dimer–monomer equilibrium of beta-lactoglobulin at pH 3 [27] and of GST mu at pH 6.5 [18] due to a shielding of the attractive charged groups in the proteins. We speculate that the addition of KBr also shields the electrostatic attractive forces at the GST pi subunit interface, thus, shifting the dimer–monomer equilibrium toward monomer. We found that the largest change in the specific activity was observed between 0.5 and 2.0 M KBr, while the largest change in the enzyme's average mass is observed between 2.0 and 4.0 M KBr. Therefore, the decrease in the enzyme's activity is primarily due to an effect of KBr on the catalytic reaction, and not to the dissociation of the subunits. The kinetic parameters of GST pi determined in the presence of increasing concentrations of KBr indicate that the binding site that is significantly affected by the separation of the subunits and the ionic interactions is the glutathione site rather than the electrophilic site. At the glutathione site, there is an extensive network of interactions between GSH and the amino acids of the enzyme, most of which involve residues from the same subunit binding the GSH [24]. However, there is a specific attractive interaction between the α-amino group of glutathione and the carboxylate of Asp98 from the neighboring subunit [24]. When the enzyme subunits are separated, Asp98 from the opposite subunit is unavailable and the Km for GSH increases. In contrast, the site for CDNB is defined entirely by the side chains of residues within the same subunit [28]; thus separation of the subunits does not affect the binding by GST pi of this xenobiotic substrate. Furthermore, as indicated in the results from the effect of KBr on Vmax, the GST pi monomer is catalytically competent; in fact, the Vmax of the dimer is only 2.6 times that of the monomer.
A major aim of this work was to identify segments of GST pi that belong to the interface of the complex. It has long been known that fluorescence spectroscopy can be a sensitive approach for investigating the interaction of proteins with target peptides. We have used this tool to probe the interaction of 1-Cys peroxiredoxin with several peptides which could encompass the interaction sites between GST pi and 1-Cys Prx. Tryptophan 33, 82, and 181, highlighted in the model of the GST pi/1-Cys Prx, are in the vicinity of the interface of the complex (Fig. 6), which enabled us to monitor the interaction between the GST peptides and 1-Cys Prx by means of the intrinsic fluorescence of these chromophores. These tryptophan residues of 1-Cys Prx exist in a solvent-restricted or hydrophobic environment as shown by the blue shift in the maximum emission peak observed with 1-Cys Prx when compared to free tryptophan [20]. In the presence of four GST pi peptides, a decrease in the maximum fluorescence of 1-Cys Prx at 330 nm is observed as compared to the 1-Cys Prx alone, which suggests that these fragments interact with 1-Cys Prx, further shielding it from the solvent or quenching the tryptophans [20]. Tyrosines and/or aspartates, for example, which are present in these GST pi fragments (see Fig. 3), can account for the quenching in fluorescence that is observed.
The identification of the GST pi segments that bind to 1-Cys Prx and inhibit the formation of the complex allows for the prediction of the types of interactions that could lead to subunit recognition and stabilization of the heterodimer. Based on the model of the complex (Figs. 6 and 7A) and analysis using the ZDock program, which thoroughly analyzes the most favorable protein–protein interaction sites, hydrophobic forces could be the major interactions between the two proteins. For example, GST pi fragment 115–124 is close to fragment 163–169 of 1-Cys Prx. Tyr118 of GST pi is thus only 3.5 Å away from Val163 of 1-Cys Prx, a distance sufficiently close for a hydrophobic interaction. Moreover, Val119 of GST pi is situated 4.1 Å from Val164 of 1-Cys Prx, reinforcing the likelihood of hydrophobic interactions between the two types of subunit. In the case of GST pi fragment 41–85, the aromatic ring of Tyr49 of GST pi is 3 Å from that of Tyr149 of 1-Cys Prx. This distance is sufficiently close for a pi–pi stacking interaction (less than 5.6 Å) [29]. Furthermore, the phenolic ring of Tyr63 of GST pi is 4.4 Å from the benzyl ring of Phe43 of 1-Cys Prx, also suggesting these two residues may interact through pi–pi stacking. Electrostatic interactions between the two enzymes may also be important. For example, the negatively charged carboxylate of Asp171 of GST pi is 4.2 Å from the positively charged amino group of Arg219 of 1-Cys Prx. Additionally, the imidazole group of His162 in GST pi is situated 4.9 Å from the carboxyl group of Asp180 of 1-Cys Prx. Therefore, if the pK of the Histidine is high enough, these two residues may interact electrostatically.
Fig. 7.
Model of the GST pi/1-Cys Prx heterodimer, and crystal structures of homodimeric 1-Cys Prx (PDB 1 PRX) and homodimeric human GST pi (PDB 19GS). (A) Heterodimer. The backbone of the 1-Cys Prx subunit is shown in cyan with peptides which includes regions close (<6 Å) to GST pi highlighted in white (left to right): 43–49, 149–155, 160–170 (helix), 176–184, and 218–221. The backbone of the GST pi subunit is shown in pink, with the peptides tested for interaction with 1-Cys Prx highlighted in blue (41–85), green (115–124), red (131–163), and orange (164–197), as in Fig. 6. Glutathione is shown in brown. (B) 1-Cys Prx homodimer. The backbone of subunit B is cyan with the peptides close to GST pi in the heterodimer highlighted in yellow: 43–49, 149–155, 160–170, 176–184, and 218–221. The backbone of subunit A is gold, with the peptides close to GST pi in the heterodimer pictured in white: 218–221, 176–184, 160–170, 149–155, and 43–49. (C) GST pi homodimer. The backbone of subunit B is yellow with the peptides tested for interaction with 1-Cys Prx highlighted in cyan (41–85), green (115–124), red (131–163), and orange (164–197). The backbone of subunit A is pink, with the peptides tested highlighted in blue (41–85), green (115–124), red (131–163), and orange (164–197).
Fig. 7 shows the crystal structures of homodimeric human 1-Cys Prx and GST pi as compared to the proposed model of the heterodimer (Fig. 7A); in this heterodimer, on the 1-Cys Prx subunit (cyan) we have highlighted in white the peptides considered close to GST pi. Fig. 7B depicts 1-Cys Prx with the two subunits shown in cyan and gold, respectively, and with peptides in close contact with GST pi in the heterodimer highlighted in yellow and white. Interestingly, the amino acids that appear to be part of the heterodimer interface are also part of the subunit interface of the 1-Cys Prx homodimer. In contrast, when we analyze the crystal structure of homodimeric human GST pi (Fig. 7C), with subunits shown in pink and yellow, and highlight the major peptides (shown in the same color as in Fig. 7A) that are in close contact with 1-Cys Prx in the heterodimer, most of the amino acids that appear to be part of heterodimer interface are not part of the subunit interface of the GST pi homodimer. The red, green and orange peptide regions are far from the interface of the GST pi subunits (Fig. 7C). Only a small section of the peptide 41–85 (highlighted in cyan on one subunit and blue on the other subunit) makes close contact in the GST pi homodimer (Fig. 7C). We suggest that GST pi predominantly utilizes a different interface for other protein–protein interactions that is distinguishable from the homodimeric interface. GST pi appears to form complexes with other proteins such as Jun N-terminal kinase [30–32] and tumor necrosis factor alpha [33]. If the other proteins interact entirely with the same site of GST pi occupied by 1-Cys Prx, complexes with these proteins seem likely to form with a GST monomer. In the case of the GST pi/1-Cys Prx, the subunit interface of 1-Cys Prx is the same for another subunit of 1-Cys Prx or for a GST pi subunit, thus allowing only one subunit of 1-Cys Prx to interact with one subunit of GST pi.
The involvement of GST pi in protein–protein interactions constitutes an important new role for GST pi and must affect the manner in which GST pi influences the cellular response to oxidative stress. Furthermore, since both enzymes are overexpressed in tumor cells, understanding this important interaction may lead to an innovative approach for the development of peptidomimetic agents that may inhibit the formation of the complex in patients undergoing cancer radiation therapy. Radiation therapy for cancer results in the local formation of peroxides that injure the tumor cells, to the benefit of the patients. On the other hand, active GST pi/1-Cys Prx complex may rapidly decompose the peroxides, thereby interfering with the effects of radiation therapy. In this case, inhibitors of complex formation could be a useful adjuvant to radiation therapy.
In summary, we have examined the interaction of glutathione S-transferase pi with 1-Cys peroxiredoxin in order to increase knowledge of the chemical and structural basis of the activation by GST pi of 1-Cys Prx. Three main characteristics were identified: first, the complex is a heterodimer. This led to the second characteristic: GST pi is active as a monomer. Finally, we identified GST pi residues 41–85 and 115–124 as the critical contact sites between GST pi and 1-Cys Prx.
Acknowledgments
We thank Chati Lum Zony for her contributions to the initial experiments and Dr. Yu Chu Huang for N-terminal sequencing of the peptides and proteins. We also thank Dr. Sheldon Feinstein for supplying the extracts of E. coli harboring recombinant human 1-Cys Prx (Prdx 6). The work was supported by NIH Grant R01-CA66561 (R.F.C.), by NIH fellowship 1F31 GM 75387 (L.A.R.) and by NIH Grant P01-HL 79063 (A.B.F.).
Footnotes
Abbreviations used: GST, glutathione S-transferase; GST pi, pi-class glutathione S-transferase; 1-Cys Prx, 1-Cysteine peroxiredoxin; Ni–NTA, nickel–nitriloacetic acid agarose; TRIS, tris (hydroxymethyl) aminomethane; EDTA, disodium ethylenediamine tetraacetate; MES, 2-(N-morpholino) ethane-sulfonate; WT, wild-type; GSH, glutathione; CDNB, 1-chloro-2,4-dinitrobenzene; SDS–PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis.
In a separate study, Y.-C. Huang, S. Misquitta, S.Y. Blond, R.F. Colman (manuscript in preparation) have shown that when the protein concentration range is extended from 0.008 to 1.2 mg/ml, the average mass of GST pi changes from 25,500 to 47,000. Furthermore, the substrate CDNB has little influence on the molecular mass.
References
- 1.Mannervik B, Danielson UH. CRC Crit. Rev. Biochem. 1988;23:283–337. doi: 10.3109/10409238809088226. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong RN. Chem. Res. Toxicol. 1997;10:2–18. doi: 10.1021/tx960072x. [DOI] [PubMed] [Google Scholar]
- 3.Coles B, Ketterer B. Crit. Rev. Biochem. Mol. Biol. 1990;25:47–70. doi: 10.3109/10409239009090605. [DOI] [PubMed] [Google Scholar]
- 4.Lee FY, Sciandra J, Siemann DW. Biochem. Pharmacol. 1989;38:3697–3705. doi: 10.1016/0006-2952(89)90575-3. [DOI] [PubMed] [Google Scholar]
- 5.Moscow JA, Fairchild CR, Madden MJ, Ransom DT, Wieand HS, O'Brien EE, Poplack DG, Cossman J, Myers CE, Cowan KH. Cancer Res. 1989;49:1422–1428. [PubMed] [Google Scholar]
- 6.Nakagawa K, Saijo N, Tsuchida S, Sakai M, Tsunokawa Y, Yokota J, Muramatsu M, Sato K, Terada M, Tew KD. J. Biol. Chem. 1990;265:4296–4301. [PubMed] [Google Scholar]
- 7.Tsuchida S, Sato K. Crit. Rev. Biochem. Mol. Biol. 1992;27:337–384. doi: 10.3109/10409239209082566. [DOI] [PubMed] [Google Scholar]
- 8.Manevich Y, Feinstein SI, Fisher AB. Proc. Natl. Acad. Sci. USA. 2004;101:3780–3785. doi: 10.1073/pnas.0400181101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ralat LA, Manevich Y, Fisher AB, Colman RF. Biochemistry. 2006;45:360–372. doi: 10.1021/bi0520737. [DOI] [PubMed] [Google Scholar]
- 10.Choi HJ, Kang SW, Yang CH, Rhee SG, Ryu SE. Nat. Struct. Biol. 1998;5:400–406. doi: 10.1038/nsb0598-400. [DOI] [PubMed] [Google Scholar]
- 11.Ralat LA, Misquitta SA, Zony CL, Manevich Y, Fisher AB, Colman RF. FASEB J. 2007;21:651–653. [Google Scholar]
- 12.Ralat LA, Colman RF. J. Biol. Chem. 2004;279:50204–50213. doi: 10.1074/jbc.M407445200. [DOI] [PubMed] [Google Scholar]
- 13.Chen JW, Dodia C, Feinstein SI, Jain MK, Fisher AB. J. Biol. Chem. 2000;275:28421–28427. doi: 10.1074/jbc.M005073200. [DOI] [PubMed] [Google Scholar]
- 14.Pettigrew NE, Brush EJ, Colman RF. Biochemistry. 2001;40:7549–7558. doi: 10.1021/bi002840w. [DOI] [PubMed] [Google Scholar]
- 15.Habig WH, Pabst MJ, Jakoby WB. J. Biol. Chem. 1974;249:7130–7139. [PubMed] [Google Scholar]
- 16.Misquitta SA, Colman RF. Biochemistry. 2005;44:8608–8619. doi: 10.1021/bi050449a. [DOI] [PubMed] [Google Scholar]
- 17.Vargo MA, Nguyen L, Colman RF. Biochemistry. 2004;43:3327–3335. doi: 10.1021/bi030245z. [DOI] [PubMed] [Google Scholar]
- 18.Hearne JL, Colman RF. Biochemistry. 2006;45:5974–5984. doi: 10.1021/bi060249e. [DOI] [PubMed] [Google Scholar]
- 19.Cacace MG, Landau EM, Ramsden JJ. Q. Rev. Biophys. 1997;30:241–277. doi: 10.1017/s0033583597003363. [DOI] [PubMed] [Google Scholar]
- 20.Matyus L, Szollosi J, Jenei A. J. Photochem. Photobiol. B. 2006;83:223–236. doi: 10.1016/j.jphotobiol.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 21.Wood ZA, Schroeder E, Harris JR, Poole LB. Trends Biochem. Sci. 2003;28:32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 22.Abdalla AM, Bruns CM, Trainer JA, Mannervik B, Stenberg G. Protein Eng. 2002;15:827–834. doi: 10.1093/protein/15.10.827. [DOI] [PubMed] [Google Scholar]
- 23.Ji X, Zhang P, Armstrong RN, Gilliland GL. Biochemistry. 1992;31:10169–10184. doi: 10.1021/bi00157a004. [DOI] [PubMed] [Google Scholar]
- 24.Reinemer P, Dirr HW, Ladenstein R, Huber R, Lo Bello M, Federici G, Parker MW. J. Mol. Biol. 1992;227:214–226. doi: 10.1016/0022-2836(92)90692-d. [DOI] [PubMed] [Google Scholar]
- 25.Sinning I, Kleywegt GJ, Cowan SW, Reinemer P, Dirr HW, Huber R, Gilliland GL, Armstrong RN, Ji X, Board PG, et al. J. Mol. Biol. 1993;232:192–212. doi: 10.1006/jmbi.1993.1376. [DOI] [PubMed] [Google Scholar]
- 26.Jencks WP. Catalysis in Chemistry and Enzymology. McGraw-Hill, Inc.; New York, NY: 1969. pp. 585–586. [Google Scholar]
- 27.Sakurai K, Oobatake M, Goto Y. Protein Sci. 2001;10:2325–2335. doi: 10.1110/ps.17001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ji X, Tordova M, O'Donnell R, Parsons JF, Hayden JB, Gilliland GL, Zimniak P. Biochemistry. 1997;36:9690–9702. doi: 10.1021/bi970805s. [DOI] [PubMed] [Google Scholar]
- 29.Mao L, Wang Y, Lui Y, Hu X. J. Mol. Biol. 2004;336:787–807. doi: 10.1016/j.jmb.2003.12.056. [DOI] [PubMed] [Google Scholar]
- 30.Adler V, Yin Z, Fuchs SY, Benezra M, Rosario L, Tew KD, Pincus MR, Sardana M, Henderson CJ, Wolf CR, Davis RJ, Ronai Z. EMBO J. 1999;18:1321–1334. doi: 10.1093/emboj/18.5.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang T, Arifoglu P, Ronai Z, Tew KD. J. Biol. Chem. 2001;276:20999–21003. doi: 10.1074/jbc.M101355200. [DOI] [PubMed] [Google Scholar]
- 32.Elsby R, Kitteringham NR, Goldring CE, Lovatt CA, Chamberlain M, Henderson CJ, Wolf CR, Park BK. J. Biol. Chem. 2003;278:22243–22249. doi: 10.1074/jbc.M301211200. [DOI] [PubMed] [Google Scholar]
- 33.Wu Y, Fan Y, Xue B, Luo L, Shen J, Zhang S, Jiang Y, Yin Z. Oncogene. 2006;25:5787–5800. doi: 10.1038/sj.onc.1209576. [DOI] [PubMed] [Google Scholar]