Abstract
Contrary to DNA double-helical structures, hydrogen bonds (H-bonds) involving nitrogen as the acceptor are not common in protein structures. We systematically searched N-H…N H-bonds in two different sets of protein structures. Data set I consists of neutron diffraction and ultrahigh-resolution x-ray structures (0.9 Å resolution or better) and the hydrogen atom positions in these structures were determined experimentally. Data set II contains structures determined using x-ray diffraction (resolution ≤ 1.8 Å) and the positions of hydrogen atoms were generated using a computational method. We identified 114 and 14,347 potential N-H…N H-bonds from these two data sets, respectively, and 56–66% of these were of the Ni+1-Hi+1…Ni type, with Ni being the proline backbone nitrogen. To further understand the nature of such unusual contacts, we performed quantum chemical calculations on the model compound N-acetyl-L-proline-N-methylamide (Ace-Pro-NMe) with coordinates taken from the experimentally determined structures. A potential energy profile generated by varying the ψ dihedral angle in Ace-Pro-NMe indicates that the conformation with the N-H…N H-bond is the most stable. An analysis of H-bond-forming proline residues reveals that more than 30% of the proline carbonyl groups are also involved in n → π∗ interactions with the carbonyl carbon of the preceding residue. Natural bond orbital analyses demonstrate that the strength of N-H…N H-bonds is less than half of that observed for a conventional H-bond. This study clearly establishes the H-bonding capability of proline nitrogen and its prevalence in protein structures. We found many proteins with multiple instances of H-bond-forming prolines. With more than 15% of all proline residues participating in N-H…N H-bonds, we suggest a new, to our knowledge, structural role for proline in providing stability to loops and capping regions of secondary structures in proteins.
Introduction
Hydrogen bonds (H-bonds) are one of the fundamental noncovalent interactions that provide stability to biomolecular structures (1). In proteins, the intrahelical and interstrand H-bonds stabilize the secondary structures, α-helices, and β-sheets, respectively. In addition to the main-chain (MC) amino and carbonyl functional groups, the side chains (SCs) of 10 naturally occurring amino acids can participate in H-bond interactions as the donor and/or acceptor. In proteins, H-bonds vary in strength and type depending upon the nature of the donor and acceptor atoms (2, 3, 4). With the most abundant elements of oxygen, nitrogen, and carbon, the frequently observed H-bonds in proteins include the O-H…O, N-H…O, and C-H…O types (5, 6, 7). The π-electron cloud of aromatic residues also takes part in interactions that can be considered similar to H-bonds, such as N-H…π, O-H…π, S-H…π, and C-H…π (8). The most frequently observed H-bonds involve the backbone amino and the carbonyl groups of the type N-H…O bonds that occur within α-helices and between β-strands. The weak C-H…O H-bond has gained attention in recent years (9, 10) and has been implicated in several biological processes. C-H…O H-bonds are one of the factors that may be responsible for providing stability to protein aggregates involving β-sheets (11). The majority of the observed H-bonds in proteins involve oxygen as the acceptor. With nitrogen as the acceptor, O-H…N, N-H…N, and C-H…N are the other possible H-bonds in proteins. Although the H-bonds of Watson-Crick basepairs in DNA double-helical structures involve nitrogen as the acceptor, there are fewer instances in which nitrogen acts as the H-bond acceptor in protein structures.
H-bonds of type N-H…N have been observed in structures of several small organic compounds (12, 13, 14, 15, 16, 17). Various reports have specifically identified H-bonds involving nitrogen as the acceptor in proteins and discussed the significance of such interactions with regard to structural stability and functional importance. For example, NMR studies have identified the Nδ nitrogen atom of a histidine residue as the H-bond acceptor in the H-bond network in ankyrin repeats (18). Ligands of proteins are known to interact through several noncovalent interactions (19, 20) and some of these molecules participate in H-bonds with nitrogen atoms as the acceptor (21, 22). Such H-bonds have been shown to be part of the pharmacophore and are important for binding to the target protein (23, 24). Perrin et al. (25) tested the capability of nitrogen atoms in the imidazole moiety to act as an H-bond acceptor or donor in cytochrome P450 using a series of compounds, and investigated their potential to be novel inhibitors.
Recently, Adhikary et al. (26) investigated possible H-bond interactions between the backbone N-H of the ith residue and the amide N of the previous residue. They investigated specific proline residues in the N-terminal region of Src homology domain 3 (SH3) protein. The infrared stretching frequencies and the blue shifts of the absorptions clearly indicated the presence of Ni+1-Hi+1…Ni H-bonds involving two proline residues. It would be interesting to identify, analyze, and characterize the H-bonds in proteins in which nitrogen is the acceptor. In this study, we systematically analyzed high-resolution protein crystal structures as well as protein structures determined by the neutron diffraction method. We considered six different types of H-bonds in which oxygen or nitrogen is the acceptor: N-H…O, O-H…O, C-H…O, N-H…N, O-H…N, and C-H…N. We constructed two different data sets of high-resolution protein structures to identify these H-bonds and found that <2.5% constitute the H-bonds in which nitrogen is the acceptor. The majority of N-H…N H-bonds are formed by MC amino groups, and >90% of such H-bonds involve proline backbone nitrogen. Further quantum chemical calculations on model compounds demonstrated that the conformation of proline with the Ni+1-Hi+1…Ni H-bond is energetically the most favorable. To our knowledge, a systematic analysis of N-H…N H-bonds in protein structures has not been performed heretofore. Our analysis helps to establish the H-bonding capability of proline imino nitrogen in protein structures, which may have further implications for the structural stability of proteins.
Materials and Methods
Selection of protein structure data sets
To identify H-bonds with oxygen or nitrogen as the acceptor, we constructed two different high-quality data sets of protein structures from the Protein Data Bank (PDB) (27). The first data set, data set I, contained structures determined by neutron diffraction or x-ray diffraction (resolution ≤ 0.9 Å) or by a neutron diffraction/x-ray diffraction hybrid method with a resolution of 0.9 Å or better. The hydrogen positions in all of these structures were experimentally determined. The second data set, data set II, comprised high-resolution x-ray structures determined as of March 31, 2015, with a resolution of 1.8 Å or better, R-value ≤ 0.20, and R-free ≤ 0.25. The positions of hydrogen atoms in these structures were fixed using the program REDUCE (28). In both data sets, the redundancy was removed at a 30% cutoff level using the program CD-HIT (29, 30).
Identification of H-bond interactions
We identified H-bonds of the D-H…A type (the donor atom D and the acceptor atom A are either nitrogen or oxygen) using the following distance and angle criteria: d(D…A) ≤ 3.5 Å; d(H…A) ≤ 2.5 Å; θ(D-H…A) ≥ 90°; θ(H…A-AA) ≥ 90°, where AA is the acceptor antecedent atom (31, 32). A slightly different criterion was applied for the weak C-H…O and C-H…N H-bonds (31). Although the distance criteria were the same (d(C…O/N) ≤ 3.5 Å; d(H…O/N) ≤ 2.5 Å), we considered only the angle θ(C-H…O/N) to identify C-H…O/N H-bonds. If the distance criteria were satisfied and if θ(C-H…O/N) ≥ 120°, this interaction was classified as a C-H…O or C-H…N H-bond.
Quantum chemical calculations on model compounds
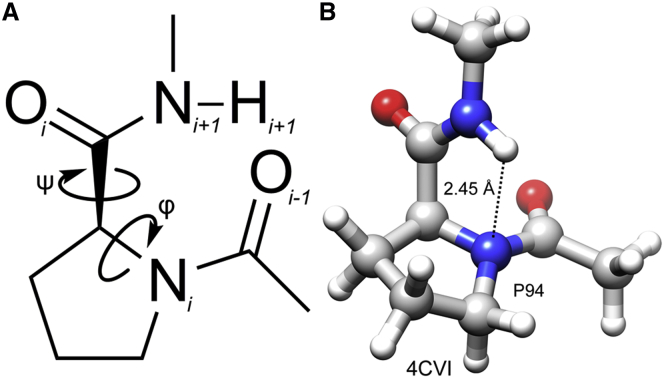
To understand the nature of N-H…N H-bonds involving the proline backbone nitrogen, we considered N-acetyl L-proline N′-methyl amide (Ace-Pro-NMe) as a model compound (Fig. 1 A). For quantum chemical calculations, we considered residue Pro-94 from the protein cytochrome c peroxidase, whose structure was determined using neutron diffraction (PDB: 4CVI) (33). The distance and angle criteria for an N-H…N H-bond between the backbone N-H of Ile-95 and the backbone N of Pro-94 were satisfied (Fig. 1 B). The coordinates of Pro-94, along with the backbone atoms of the preceding and succeeding residues, were directly imported from the experimental structure and used for the model compound. Methyl groups were introduced as capping groups to generate an Ace-Pro-NMe molecule using the program MOLDEN (34). First, the positions of hydrogen atoms were optimized using the model chemistry BP86 (35, 36) density functional theory and Ahlrich’s triple-ζ def2-TZVP (37, 38). The electronic structure program package ORCA v3.0.2 (39) was used for optimization.
Figure 1.
(A) Chemical structure of Ace-Pro-NMe, used for quantum chemical calculations. Proline is referred to as the ith residue and the subscripts for all of the atoms are labeled accordingly. Due to the rigid pyrrolidine ring, the dihedral angle φ is mostly close to −60°, whereas the other dihedral angle ψ is variable. The potential energy profile was created as described in the Materials and Methods. All geometrical parameters were fixed corresponding to the conformation of proline residue, Pro-94, from the protein cytochrome peroxidase (PDB: 4CVI) and the ψ angle was varied. (B) Example of a proline residue from the protein cytochrome peroxidase (PDB: 4CVI) that satisfies the geometrical criteria for the N-H…N H-bond. All of the molecular plots were rendered by Chimera software (40) and the standard colors are used to represent nitrogen, oxygen, carbon, and hydrogen atoms. The same convention is used for all molecular plots. To see this figure in color, go online.
To assess the stabilizing effect of the N-H…N H-bond on the conformation of the model compound, we performed single-point energy calculations by scanning different conformations of Ace-Pro-NMe, and created a potential energy surface. For this purpose, we first considered the two peptide bonds of the molecule as shown in Fig. 1 A in the trans conformation. Due to proline’s rigid pyrrolidine ring, the dihedral angle φ of proline was fixed at −69.3°, the value observed in the crystal structure of cytochrome peroxidase (PDB: 4CVI). Only the single bond defining the ψ dihedral angle was rotatable. All of the bond lengths and bond angles were fixed. With only ψ as a variable, starting from the crystal structure value, we rotated the ψ angle every 5° and performed a single-point energy calculation for each conformation. Thus, we generated 72 conformations and compared the single-point energy of each conformation with that corresponding to the crystal structure conformation. The potential energy surface thus created was plotted against ψ and other distance parameters. Details of the quantum chemical calculations performed on the model compound are provided below.
To calculate the single-point energy of each conformation of Ace-Pro-NMe molecule obtained by changing the dihedral angle ψ, we used the model chemistry M06-2X (41) density functional theory in conjunction with Dunning’s correlation-consistent, quadruple-ζ basis set augmented with diffuse functions, AUG-cc-pVQZ (42). Gaussian 09 (43) was used for this purpose. We also performed natural bond orbital (NBO) analyses (44, 45) on the model compound Ace-Pro-NMe at the M06-2X/AUG-cc-pVQZ level of theory for all 72 conformations. NBO version 3.1 (46) as implemented in Gaussian 09 was used for this purpose. We estimated the stabilization energies that resulted from the interaction between N-H…N bonding and lone pair NBOs using second-order perturbation theory.
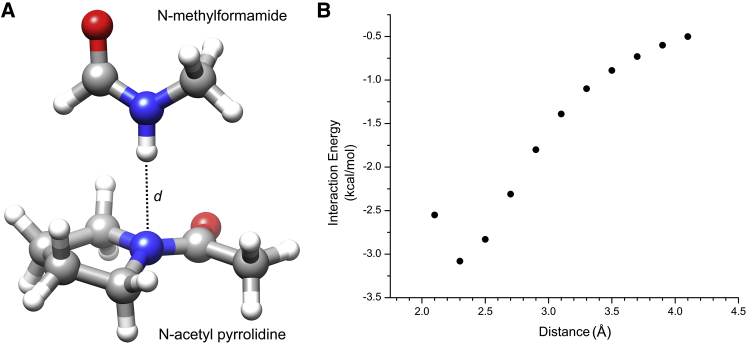
Since the N-H…N H-bond in Ace-Pro-NMe is an intramolecular interaction, to determine the contribution of this H-bond alone, we also carried out quantum chemical calculations on the model compounds N-methylformamide (NMF) and N-acetylpyrrolidine (NAP). In this system, we investigated the intermolecular H-bond interaction that formed between the N-H group of NMF and the nitrogen atom of NAP (Fig. 2 A). The angle between the N-H group and the donor nitrogen (θ(N-H…N)) was kept at 180°. We performed quantum chemical calculations by varying the distance d between the N-H of NMF and the nitrogen of NAP from 2.1 to 4.1 Å in steps of 0.2 Å. We calculated the interaction energy between the two molecules using the following equation:
| (1) |
where Eint is the interaction energy between NMF and NAP, EAB represents the single-point energy of the complex between NMF and NAP, and EA and EB correspond to the single-point energies of NMF and NAP, respectively. We calculated the single-point energies using the same level of theory and basis set as described above. We employed Boys and Bernardi’s (47) standard counterpoise correction method to account for the basis set superposition error.
Figure 2.
(A) The model compounds NMF and NAP are involved in the N-H…N H-bond interaction. The distance d between the N-H group of NMF and the nitrogen of NAP was varied from 2.1 to 4.1 Å in steps of 0.2 Å. Single-point energy calculations were carried out at each point and the basis set superposition error-corrected interaction energies between NMF and NAP were calculated as described in Materials and Methods. (B) Interaction energy profile between NMF and NAP as a function of distance d. To see this figure in color, go online.
Results
Data set I, with structures determined using the neutron diffraction method and/or x-ray diffraction with ultrahigh resolution, consists of 68 polypeptide chains from 64 structures after redundancy was removed at a 30% cutoff level. Data set II is comprised of 5542 polypeptide chains from 5336 x-ray structures with a resolution of 1.8 Å or better. The frequency of proline residues in both data sets is ∼5% (542 and 62,717 proline residues, respectively, out of a total number of 10,535 residues in data set I and 1,369,420 residues in data set II). To identify the N-H…N type of H-bonds, we considered all of the nitrogen atoms from the backbone and SCs of nitrogen-containing amino acids (His, Trp, Asn, Gln, Lys, and Arg) that could act as a potential donor or acceptor. We used the distance and angle criteria as described in the Materials and Methods section to identify N-H…N H-bonds. To determine the relative abundance of N-H…N H-bonds with respect to other types of H-bonds in which oxygen or nitrogen acts as an H-bond acceptor, we also identified five other types of H-bonds, namely, N-H…O, O-H…O, C-H…O, O-H…N, and C-H…N. The fractions of each type of H-bond determined for structures from data sets I and II are provided in Fig. S1 in the Supporting Material.
The trend is very similar for both data sets. As expected, the N-H…O H-bond is the most frequently observed H-bond, accounting for 60% and 76% of the total H-bonds in data sets I and II, respectively. This is mainly due to the secondary structures that give rise to either intrahelical or interstrand H-bonds. The H-bonds C-H…O (16–23%) and O-H…O (6–14%) are the next most predominantly observed H-bonds. The proportion of N-H…O H-bonds is higher and that of O-H…O and C-H…O H-bonds is lower in data set II compared with data set I. This perhaps could be due to a bias in the way hydrogens were built using the REDUCE program (28). Thus, overall, the three major H-bonds (N-H…O, O-H…O, and C-H…O) account for ∼98% of the six types of H-bonds analyzed in both data sets, and they involve oxygen as the acceptor. The remaining three types of H-bonds in which nitrogen is the acceptor constitute only ∼2%, with the N-H…N type accounting for more than 1.1%. In absolute numbers, we identified 114 and 14,347 potential N-H…N H-bonds from data set I and data set II, respectively. The acceptor and donor nitrogen atoms could be contributed by MC or SC atoms. In both data sets, in the majority of N-H…N H-bonds, both the donor and acceptor nitrogen atoms came from the MC-MC type. Among the total number of N-H…N H-bonds, 74% and 65% were of the MC-MC type in data sets I and II, respectively, and surprisingly, in almost 90% of them the acceptor nitrogen was contributed by the proline backbone in both data sets. Moreover, the N-H…N H-bonds were formed between the nitrogen of proline as the acceptor and the N-H amino group from the MC of the succeeding residue (the other major residue that participated in N-H…N H-bonds was histidine, and its imidazole group formed N-H…N H-bonds in almost 25–30% of cases; see Table S1). In this study, we focused our attention on further characterizing the Ni+1-Hi+1…Ni H-bonds in which proline is the ith residue.
Ni+1-Hi+1…Ni H-bonds formed by proline residues
We calculated several distance and angle parameters between the proline backbone nitrogen atoms and the backbone amino group of its succeeding residue for both data sets. We evaluated the same parameters for all other proline residues that did not satisfy the criteria for potential N-H…N H-bonds and compared them with those that did (Table 1). If we consider all proline residues, ∼25% and 15% of them participate in the Ni+1-Hi+1…Ni type of H-bond interactions in data set I and data set II, respectively. The average geometrical parameters are nearly identical for both data sets. When proline is involved in an H-bond interaction with the succeeding residue, the average distance between the Ni of proline and the Hi+1 atom of the succeeding residue is 2.4 Å, which is 1.0 Å less than the same parameters calculated for other proline residues. Similarly, the average distance between the two nitrogen atoms, Ni of proline and Ni+1 of the next residue, is ∼2.8 Å when there is an N-H…N H-bond, which is 0.7 Å smaller than that found for all other proline residues. The relevant angles that define the N-H…N H-bond, θ(Ni+1-Hi+1…Ni) and θ(Hi+1…Ni-AAi), also exhibit an ∼18–25° difference between H-bond-forming proline and other proline residues. We determined the statistical significance of the differences in distance and angle parameters calculated for H-bond-forming proline residues and all other proline residues using an unpaired t-test, and found that these differences are extremely statistically significant (p < 0.0001). We also calculated the (φ, ψ) values of the H-bond-forming proline residues and all other proline residues. Ramachandran plots of (φ, ψ) for both groups of proline residues belonging to data sets I and II clearly show that (φ, ψ) of H-bond-forming proline residues fall in the right-handed α-helical region and the bridge region connecting the helical and extended regions (see Fig. S2).
Table 1.
Average (Standard Deviation) of Geometrical Parameters for the H-Bond-Forming Proline Residues and All Other Proline Residues
| Data Set | Residuea | d(Ni…Hi+1)b (Å) | d(Ni…Ni+1) (Å) | θ(Ni…Hi+1-Ni+1) (°) | θ(AAi-Ni…Hi+1)c (°) |
|---|---|---|---|---|---|
| I | H-bond-forming prolines (75) | 2.41 (0.08) | 2.77 (0.05) | 105.0 (3.03) | 105.84 (6.25) |
| All other prolines (220) | 3.45 (0.59) | 3.38 (0.40) | 78.40 (16.21) | 125.0 (13.60) | |
| II | H-bond-forming prolines (8106) | 2.44 (0.05) | 2.76 (0.04) | 102.47 (1.70) | 105.94 (6.55) |
| All other prolines (46,497) | 3.41 (0.60) | 3.34 (0.36) | 77.76 (15.02) | 124.07 (14.74) |
The total number of residues from each data set for H-bond-forming proline and all other proline residues is given in brackets. The index i refers to the proline residue. The occupancy of H-bond-forming atoms should be 1. In data set I, only if the hydrogen positions are defined in the experimental structures, then the proline residues were considered for the analysis.
d(A…B) indicates the distance between atoms A and B; θ(A…B-C) denotes the angle formed by atoms A, B, and C.
Atom AA represents the acceptor antecedent atom.
n → π∗ interactions in proline residues with N-H…N H-bonds
In a series of studies, Bartlett et al. (48, 50) and Newberry et al. (49, 51) suggested that n → π∗ interactions between adjacent carbonyl groups of the polypeptide backbone can stabilize protein secondary and tertiary structures. In n → π∗ interactions, the lone pair electrons of the backbone carbonyl oxygen of the preceding residue i-1 overlaps with the π∗ antibonding orbital of the ith residue in protein structures, and this is especially prevalent in α-helical structures. Among different residues, proline seems to have a high preference for participating in n → π∗ interactions with its preceding residue (48). Hence, we were interested in determining whether the proline residues with an N-H…N H-bond interaction also have any preference for forming an n → π∗ interaction. The carbonyl oxygen of the preceding residue (Oi-1) and the carbonyl group of proline (Ci=Oi) were considered for this purpose. If the distance d(Oi-1…Ci) ≤ 3.22 Å (the sum of van der Waals radii of oxygen and carbon) and the angle θ(Oi-1…Ci=Oi) lies between 99° and 119°, then the H-bond-forming proline residue is also considered to be engaged in an n → π∗ interaction. The average and standard deviation of both the distance and angle parameters for the H-bond-forming proline and all other proline residues were calculated for both data sets and are presented in Table S2 for data set II.
It is clear that 64% of H-bond-forming proline residues are in a trans conformation and are not involved in n → π∗ interactions. Only ∼30% of H-bond-forming trans proline residues fall within the Bürgi-Dunitz trajectory. In the larger data set (data set II), ∼6% of H-bond-forming proline (501 Pro residues) adopt a cis conformation and the overwhelming majority of them do not satisfy the angle and distance criteria for participating in an n → π∗ interaction. When we considered all other proline residues, we found that 42% of trans proline residues could have potential n → π∗ interactions. In data set I, the percentages of residues within the two groups of prolines (H-bond-forming prolines and the rest of the prolines) that potentially could have n → π∗ interactions were very similar (48% vs. 43%; data not shown). However, the sample size in data set I was very small compared with that in data set II.
Quantum chemical calculations on Ace-Pro-NMe
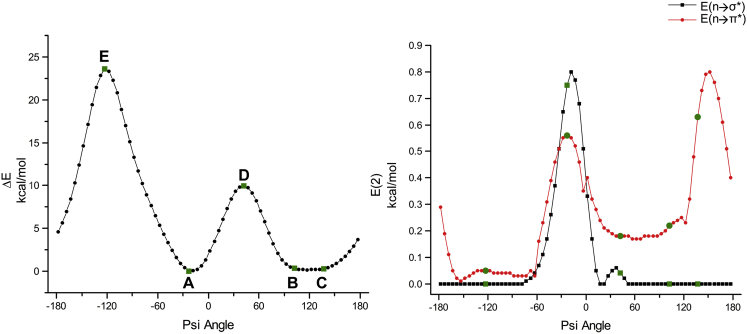
To further determine whether the close contact between the N-H group of the succeeding residue of proline and the proline backbone nitrogen can be characterized as a stabilizing interaction, we performed extensive quantum chemical calculations on the model compound Ace-Pro-NMe as described in Materials and Methods. We considered the enzyme cytochrome c peroxidase, whose structure has been determined by neutron crystallography (PDB: 4CVI). The backbone nitrogen of Pro-94 in this protein is involved in the N-H…N type of H-bond with its succeeding residue. We carried out single-point energy calculations on Ace-Pro-NMe, as described in Materials and Methods, by varying the ψ dihedral angle. All other geometric parameters were kept fixed corresponding to the experimentally determined structure (PDB: 4CVI). For each conformation, we determined the relative energy by comparing it with that of the conformation corresponding to the neutron diffraction structure. The relative energy profile thus obtained is plotted in Fig. 3.
Figure 3.
Left: potential energy profile generated from the single-point energy calculation of the model compound Ace-Pro-NMe as a function of ψ. The initial conformation corresponds to the Pro-94 residue of the experimentally determined protein structure cytochrome peroxidase (PDB: 4CVI) and the ψ angle of Pro-94 in this structure is −22.9°. Starting from this conformation, the ψ angle was varied in steps of 5° and single-point energy calculations were carried out as described in Materials and Methods on 72 conformations. Each point in the plot represents the relative energy with respect to the initial conformation. (A) This conformation corresponds to the experimentally determined initial conformation and is the energy minimum. (B and C) Conformations that are energetically closer to (A). (D and E) Two peaks in the profile. Right: the second-order perturbation energy profile was plotted for all conformations of Ace-Pro-NMe for different ψ values, and NBO calculations for each conformation were carried out as described in Materials and Methods. The energy profiles shown in filled squares and filled circles represent N-H…N H-bond and n → π∗ interactions, respectively. To see this figure in color, go online.
It is clear that the energy of the Ace-Pro-NMe molecule that assumed the conformation of experimental structure with ψ = −22.9° is the most favorable conformation among all of the conformations for which single-point energies were calculated. There are also conformations with a range of ψ values (ψ = +100 to +150°) that have favorable energy comparable to the experimental conformation. The energy difference between these conformations and the most favorable conformation is only 0.17 kcal/mol. The energy profile also shows two major energy peaks, and their ψ values are −123° and +42°. The structures corresponding to the energy minima and maxima are shown in Fig. 4.
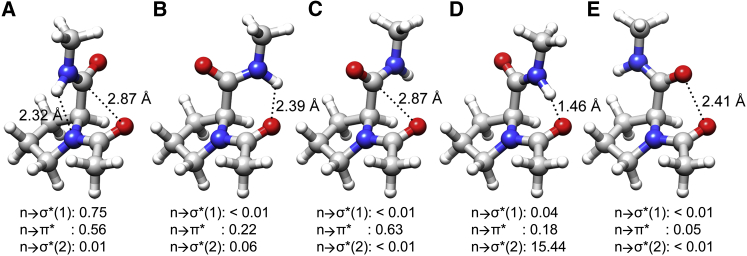
Figure 4.
Molecular plots of Ace-Pro-NMe, representing different regions of the potential energy profile and corresponding to the points labeled as (A)–(E) in Fig. 3. (A–E) The ψ values assumed for these structures are −22.9°, 102.1°, 137.1°, 42.1°, and −122.9°, respectively. To begin with, the hydrogen atoms were optimized in (A) as described in Materials and Methods. Conformation (A) is stabilized by both Ni+1-Hi+1…Ni H-bond (n → σ∗ (1)) and n → π∗ interactions, whereas (B) and (C) are stabilized by Oi-1…Hi+1-Ni+1 H-bond (n → σ∗ (2)) and n → π∗ interactions, respectively. Conformation (A) corresponds to the experimental structure and is energetically the most stable conformation, whereas the energy difference between (B) and (C) is only ∼0.17 kcal/mol. Although (D) is stabilized by the Oi-1…Hi+1-Ni+1 H-bond, the Hi+1 and Oi-1 are too close (1.46 Å, far less than the sum of van der Waals radii (2.72 Å)), and as a result there is a serious steric clash between these atoms. In (E), the two carbonyl oxygen atoms carrying partial negative charges are repulsed by the close approach of these atoms. The second-order perturbation energies of all three interactions are provided for each conformation. To see this figure in color, go online.
It can be seen from these structures that there can be three types of interactions. The first one is the H-bond between the backbone N-H of the succeeding residue with the proline backbone nitrogen (Ni+1-Hi+1…Ni). An H-bond can also occur between the backbone carbonyl group of the i-1 residue preceding proline and the backbone amino group of the i+1 succeeding residue (Ni+1-Hi+1…Oi-1). Then there is an n → π∗ interaction between the proline backbone carbonyl group and the carbonyl group of the preceding residue (Ci=Oi…Oi-1). At least one of these three favorable interactions is observed in the energetically favorable structures. The most energetically favorable structure (Fig. 4 A) has both N-H…N and n → π∗ interactions, indicating that both interactions could have contributed to the structural stability. When we analyzed the conformations in the range of ψ = +100 to +150°, the conformations corresponding to these ψ values satisfied the geometric criteria for either an Ni+1-Hi+1…Oi-1 H-bond or n → π∗ interaction (Fig. 4, B and C). The two high-energy peaks are due to the steric clash between the amino hydrogen (Ni+1-Hi+1) and Oi-1 atom in one case (Fig. 4 D) or the close approach of the two carbonyl oxygens (Oi-1…Oi) in the second case (Fig. 4 E). In the first case, when ψ = +42.1° (Fig. 4 D), the amino hydrogen of the succeeding residue comes too close to the backbone carbonyl oxygen (Oi-1) of the preceding residue, with the distance between these two atoms approaching <1.5 Å, leading to a serious steric clash and highly unfavorable energy. In the second case, when ψ = −122.9° (Fig. 4 E), the distance between the carbonyl oxygen atoms, d(Oi-1…Oi), becomes 2.41 Å, well below the sum of van der Waals radii (3.04 Å), and as a result the partial negative charges from the two oxygen atoms repel each other, giving rise to an energetically unfavorable conformation. In summary, the molecule Ace-Pro-NMe can adopt conformations that give rise to N-H…N H-bond and/or n → π∗ interactions, and both arrangements provide stability to the molecule. This further supports recent studies indicating that the proline nitrogen atom can act as an H-bond acceptor (26) and that N-H…N H-bonds involving proline nitrogen can provide stabilizing conformations, as demonstrated by spectroscopy studies and our own computational studies.
We further characterized the stereoelectronic effects of N-H…N contacts by performing NBO calculations as described in Materials and Methods. NBO calculations were performed for all 72 conformations of Ace-Pro-NMe for which single-point energy calculations were carried out (Fig. 3, left). The second-order perturbation energy profiles for N-H…N H-bond (n → σ∗) and n → π∗ interactions obtained from the NBO analyses are plotted in Fig. 3 (right). The energy profile for the Ni+1-Hi+1…Oi-1 H-bond is provided in Fig. S3. It is clear that the n → σ∗ interaction due to the N-H…N H-bond is optimally observed close to ψ = −20° with a stabilizing energy of 0.8 kcal/mol, and this conformation lies close to the experimentally observed conformation. Two conformations give rise to favorable n → π∗ interactions with ψ values of approximately −20° and −150°. The second-order perturbation energies corresponding to these two conformations are 0.6 and 0.8 kcal/mol, respectively. The conformation close to ψ = +60° gives rise to a strong Ni+1-Hi+1…Oi-1 H-bond, with the stabilization energy nearing 23 kcal/mol. However, this H-bond comes at the cost of serious steric clashes between atoms that can offset this stabilizing energy (see below).
Information about the second-order perturbation energies of n → σ∗ interactions due to Ni+1-Hi+1…Ni or Ni+1-Hi+1…Oi-1 H-bonds, or n → π∗ interactions due to the orbital overlap between the two carbonyl groups for the five different conformations, is also displayed in Fig. 4. Both n → σ∗ and n → π∗ interactions occur in the minimum energy conformation, which corresponds to the conformation of the experimentally determined structure. The n → σ∗ interaction is due to the formation of an Ni+1-Hi+1…Ni H-bond, and the strength of this interaction, 0.75 kcal/mol, is comparable to the energy values for the same type of interaction recently reported by Adhikary et al. (26). For the same conformation, there is also an n → π∗ interaction due to charge transfer between the two carbonyl groups. The other energetically stable conformations, corresponding to the locations in Fig. 4, B and C, either have only an n → π∗ interaction between the two carbonyl groups or the charge transfer interaction can be described as weak. This is despite the fact that the conformation in Fig. 4 B satisfies the geometric criteria. The charges on the atoms calculated from these calculations for the five different conformations are presented in Table S3.
When the conformation of the system deviates from the native conformation, the contribution from the N-H…N interaction diminishes to zero (Fig. 3, right). However, other contributions due to n → π∗ interactions and interactions due to H-bond formation between Ni+1-Hi+1 and Oi-1=Ci-1 groups begin to appear in some of the unfolded conformations. For example, when ψ is close to +160°, n → π∗ interactions between Ci-1=Oi-1 and Ci=Oi seem to dominate and contribute to the stability of the molecule (Figs. 3 and 4 C). When ψ is ∼+60°, the Ni+1-Hi+1…Oi-1 H-bond is observed (Figs. S3 and 4 D). However, the distance between Hi+1 and Oi-1 atoms is too close, resulting in a steric clash between the two atoms. As a result, the overall energy of the molecule becomes unfavorable (Fig. 3, left). Hence, in contrast to folded conformations, the energy of some of the unfolded conformations can be stabilized by other interactions (Ni+1-Hi+1…Ni vs. n → π∗ vs. Ni+1-Hi+1…Oi-1 H-bond), and such conformations can be as stable as the native conformation if not the most stable. In addition to intramolecular interactions, as the molecule opens up, interactions with water molecules are also expected to play a significant role.
N-H…N and/or n → π∗ interactions in proline residues
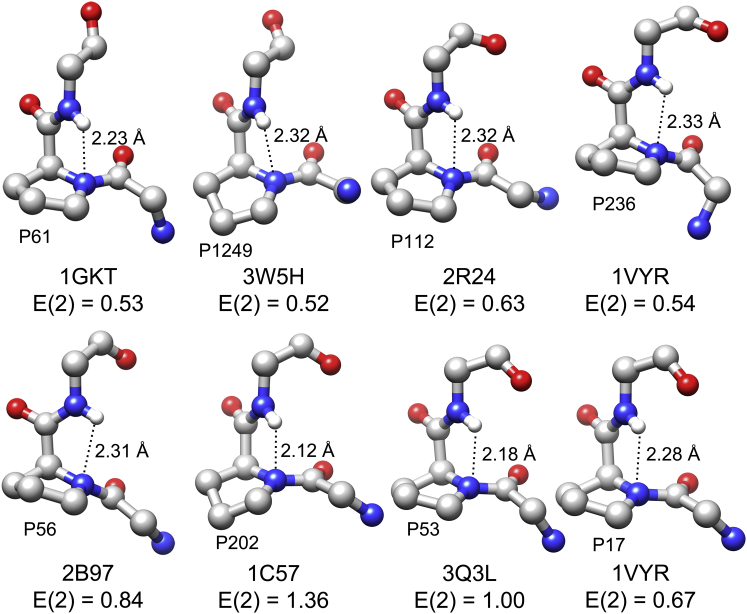
We performed NBO calculations on representative examples of proline residues from ultrahigh-resolution structures from data set I in which either both N-H…N H-bond and n → π∗ interactions are present or only one of them is present. The model compound Ace-Pro-NMe was considered for this purpose, and the conformation of each of the 18 examples from the respective PDB structures was considered for the NBO calculations. In all of the examples, the proline residue under consideration is in the trans conformation. The φ values of the proline residue vary from −53° to −104° and the corresponding ψ values range from −45° to +8°. When the geometric criteria are strictly applied, proline residues from the protein structures 4CVI (P94), 1GKT (P61), 3W5H (P1191 and P1249), and 1M40 (P107) satisfy the criteria for both N-H…N and n → π∗ interactions. The structures 2R24 (P112), 1VYR (P17, P236 and P247), 1C57 (P202), 3KKX (P83), 3Q3L (P53), and 2B97 (P56) have proline residues with parameters corresponding to N-H…N H-bond, but not n → π∗, interactions. P82, P38, and P102 from the proteins with PDB: 3ZOJ, 3QF6, and 2XU3 have only n → π∗ interactions. The proline residues from the structures with PDB: 4NSV (P213) and 4F1V (P1288) were considered as examples in which neither N-H…N H-bond nor n → π∗ interactions were observed as strong interactions. Although a structural analysis did not indicate the possibility of N-H…N H-bonds, after the hydrogen atoms were optimized, the distance between the Ni of proline and the Hi+1 of the residue succeeding proline became <2.5 Å in four structures (PDB: 3QF6, 2XU3, 4NSV, and 4F1V), thus satisfying the criteria for N-H…N H-bonds. NBO calculations on Ace-Pro-NMe, which assumed conformations of 18 proline residues from different protein structures, showed that nine of them (P94, P61, P1249, P112, P236, P56, P202, P53, and P17 from structures with PDB: 4CVI, 1GKT, 3W5H, 2R24, 1VYR, 2B97, 1C57, 3Q3L, and 1VYR, respectively) exhibit stable N-H…N H-bonds with a stabilization energy of >0.5 kcal/mol (Table S4; Figs. 4 A and 5). Six proline residues (P94, P1191, P107, P82, P38, and P102 from PDB: 4CVI, 3W5H, 1M40, 3ZOJ, 3QF6, and ZXU3, respectively) are stabilized by n → π∗ interactions, with the second-order perturbation energy ranging from 0.55 to 1.06 kcal/mol (Table S4; Fig. S4). The stabilization due to N-H…N and n → π∗ interactions is, at best, very weak in four out of 18 examples studied (P247, P213, P1288, and P83 from PDB: 1VYR, 4NSV, 4F1V, and 3KKX, respectively). Interestingly, in only one case (P94 from PDB: 4CVI), both N-H…N and n → π∗ interactions are present (Table S4; Fig. 4 A). An analysis of the (φ, ψ) values of proline residues revealed a trend for the preference to have N-H…N or n → π∗ interactions. In proline residues with N-H…N H-bonds, the magnitude of φ values appears to be larger and the ψ values seem to be smaller. In the case of proline with n → π∗ interactions, the opposite trend seems to occur (Table S4). The average (φ, ψ) values of proline with n → π∗ interactions are closer to the right-handed α-helical conformation, and this explains why Bartlett et al. (48) observed ∼95% of proline in the helical region with potential n → π∗ interactions. One must also note that in at least four cases (P1191, P107, P247, and P83 from PDB: 3W5H, 1M40, 1VYR, and 3KKX, respectively), even when the geometric criteria for N-H…N H-bond are satisfied, NBO calculations reveal that this contact is only weakly stabilizing (Table S4). The above calculations clearly demonstrate that N-H…N H-bonds involving proline backbone nitrogen are energetically stable and their strength depends on the geometrical parameters that position the donor and acceptor groups of these H-bonds.
Figure 5.
Structures of Ace-Pro-NMe, with conformations of H-bond-forming proline residues adopted from different protein structures for which NBO calculations were performed. Hydrogen atoms were optimized as described in Materials and Methods. The second-order perturbation energies and the unique PDB IDs of the proteins that possess H-bond-forming proline residues are given for each case. The distances between the Ni of proline and the Hi+1 of the succeeding residue are also provided for each proline. To see this figure in color, go online.
Intermolecular N-H…N H-bond formed by model compounds
The above calculations to characterize the N-H…N H-bond formed by Ace-Pro-NMe are also complicated by the fact that this H-bond is intramolecular in nature and it would be very difficult to separate the contribution of the N-H…N H-bond from other intramolecular interactions, such as n → π∗ and Ni+1-Hi+1…Oi-1 H-bond interactions. Hence, we considered two model compounds (NMF and NAP) and the N-H…N H-bond that formed between them. The interaction energy between the two molecules was calculated as a function of distance d between the N-H hydrogen of NMF and the nitrogen of NAP (Fig. 2 A). Our calculations show that the most favorable interaction energy of −3.08 kcal/mol occurs at a distance of 2.3 Å (Fig. 2 B). NBO calculations at this point show that the second-order perturbation energy between the interacting N-H group of NMF and the nitrogen of NAP is 1.92 kcal/mol. The natural population analysis charges for the N-H hydrogen of NMF and the acceptor nitrogen of NAP are +0.41393 and −0.49090, respectively. In the absence of any other H-bond and n → π∗ interactions, the choice of model systems and subsequent quantum chemical calculations clearly demonstrate that the N-H…N H-bond involving pyrrolidine nitrogen is favorable, and when it is linear, the strength of this H-bond seems to be stronger than the C-H…O H-bond calculated for the benzene-water system (52).
Additional interactions of N-H…N H-bond-forming residues
We examined whether the donor and acceptor atoms of N-H…N H-bonds participate in any additional interactions. In ∼50% of all H-bond-forming prolines (48.7% and 52.9% in data sets I and II, respectively), the donor amino group Ni+1-Hi+1 also forms another H-bond, and in the majority of cases the H-bond is formed with the backbone or SC functional groups of the i-1 or i-2 residue. If it is the i-2 residue, it is always the backbone C=O group, whereas the H-bond with the i-1 residue always involves the SC functional group. Bifurcated H-bonds have been analyzed in proteins and nucleic acid structures (18, 53, 54, 55, 56). However, to our knowledge, this is the first systematic study to report bifurcated H-bonds in protein structures, with one of them being the N-H…N H-bond.
Secondary-structure preference for proline with N-H…N H-bonds
We examined the occurrence of proline residues with N-H…N H-bonds in specific secondary structures in both data sets. Since the assignment of secondary structures as available in the secondary structure records for each PDB file may have used different criteria to define the secondary structures, we adopted the following procedure to first assign secondary structures using the same criteria for each protein uniformly, and then used this information to identify the preference (if any) of proline residues with N-H…N H-bonds. We used the DSSP method as implemented in Chimera software (40) to assign secondary structures to all of the proteins from each data set. This ensured that the secondary structures of all proteins in data sets I and II were defined uniformly using the same criteria. The secondary structures in which proline residues with N-H…N H-bonds were then identified (Table S5) and compared with those in which the proline residues did not participate in such contacts. Our results show that 72–75% of proline residues with N-H…N H-bonds preferred to occur in the loop region and the rest were found in the helical region. The percentage of such proline residues in the β-strand region was almost negligible (0.2–1.3%).
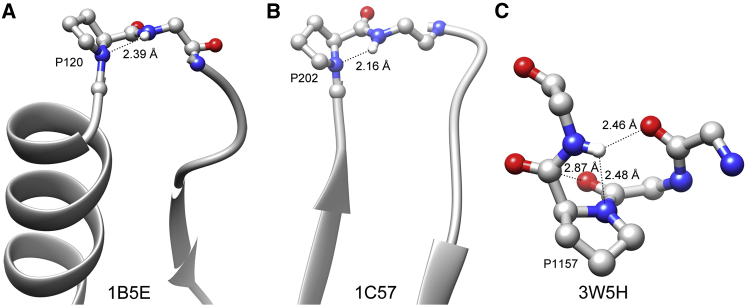
Since more than 70% of proline residues with N-H…N H-bonds occurred in the loop region, we further examined whether these proline residues showed any preference to occur in the N-cap or C-cap regions of secondary structures. The capping positions of secondary structures were defined as described by Aurora and Rose (57). We considered N″, N′, and N-cap positions in the amino terminus and C″, C′, and C-cap positions in the carboxy terminus of secondary structures. For this analysis, we only considered helices and strands that were at least six and four residues long, respectively (we used a similar strategy in a previous study (58)). At least 44–46% of all proline residues with N-H…N H-bonds were found just outside the helical or strand region in the above capping positions. Among the capping positions, the C′ position seems to be especially important for proline that can form an N-H…N H-bond with its succeeding residue. This position is the second residue to occur outside the helical or strand conformation after the secondary structure is terminated. Almost 7–9% of all H-bond-forming proline residues occurred in this position, indicating that the stability due to this H-bond is important while the secondary structures are terminated. An example of an H-bond-forming proline residue in the C′ position in a helix and strand is plotted in Fig. 6, A and B, respectively. The secondary structure preference of proline residues with bifurcated H-bonds seems to be very similar to that observed for H-bond-forming prolines (Table S5).
Figure 6.
(A and B) Proline with an Ni+1-Hi+1…Ni H-bond at the C′ capping position in a (A) helix and (B) strand. The capping positions are defined as described by Aurora and Rose (57). C′ is the second position outside of the secondary structure at the carboxyl end. (C) Example of a proline residue possessing all three types of interactions, namely, Ni+1-Hi+1…Ni H-bond, n → π∗ interaction between the proline C=O group and carbonyl oxygen of the preceding residue, and H-bond between the amino group of the i+1 residue and the carbonyl oxygen of the i-2 residue. The unique four-letter PDB IDs of the proteins are given in the figure for each example. To see this figure in color, go online.
Proline residues are known to occur in the middle of transmembrane helices (59) and they are accommodated with a bend or kink (60, 61). Their presence in helical membrane proteins has been suggested to be functionally important (62, 63). Hence, we were curious to find out whether proline residues with N-H…N H-bonds have any specific preference for transmembrane helical proteins. In the larger data set, data set II, we identified only 17 transmembrane proteins with 33 proline residues having Ni+1-Hi+1…Ni H-bonds. Among them, a majority of 76% occurred in the loop regions connecting two transmembrane segments. Only a small fraction occurred inside the transmembrane helical segments.
Proteins with multiple H-bond-forming proline residues
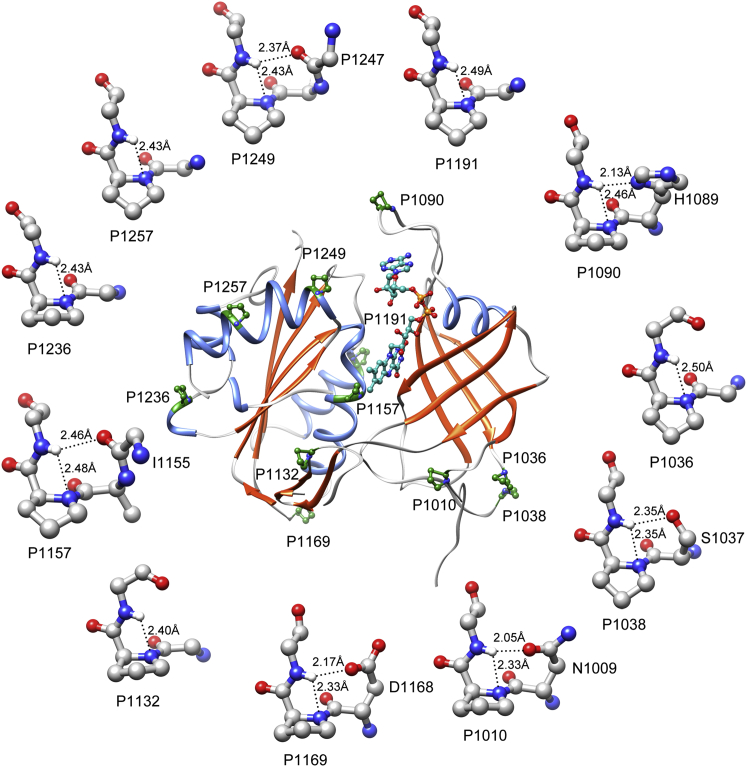
Quantum chemical calculations show that the N-H…N H-bonds involving the proline backbone hydrogen are indeed energetically favorable. We examined the protein structures from both data sets in which at least one such proline residue was identified and determined how many such proline residues were found in each protein structure. Data sets I and II contained 34 and 3424 proteins, respectively, with at least one H-bond-forming proline residue. Our results show that at least two or more H-bond-forming proline residues were found in ∼36–42% of these proteins (Fig. S5). The proteins with the maximum number of proline residues that participated in N-H…N H-bonds were NADH-cytochrome b5 reductase (PDB: 3W5H) for data set I and an oligopeptide-binding protein involved in the biosynthetic pathway of clavulanic acid (PDB: 2WOK) for data set II, and they contained 11 and 18 H-bond-forming proline residues, respectively. The structure of the oxidized form of NADH-cytochrome b5 reductase was determined at an ultrahigh resolution of 0.78 Å (64), and it is shown with its 11 H-bond-forming proline residues in Fig. 7. As anticipated, the proline residues with N-H…N H-bonds were present only in loops and α-helices, and were completely absent in β-sheets in this protein.
Figure 7.
The structure of the oxidized form of NADH-cytochrome b5 reductase (PDB: 3W5H) is shown as an example of a protein with multiple H-bond-forming proline residues. Helices and strands are shown in ribbon representation. All of the 11 H-bond-forming proline residues and cofactors are depicted in ball-and-stick representation. Additionally, each H-bond-forming proline residue is portrayed independently around the protein, and the distance between Hi+1 and Ni atoms is shown for each proline residue. When there is an additional H-bond with the i-1/i-2 residue, it is also illustrated along with the residue name. To see this figure in color, go online.
Discussion
In a small data set of 70 high-resolution protein structures, Karplus (65) analyzed a conformational strain of residues from the allowed region of a Ramachandran map and observed the close approach of the amide proton (Ni+1-Hi+1) and peptide nitrogen (Ni). The (φ, ψ) angles that gave rise to such contacts were mainly from the bridge region of the Ramachandran map, and it was suggested that such a close approach could give rise to an unconventional H-bond with the π electron of the peptide bond. Luisi et al. (66) described a handful of examples from a crystal structure analysis in which they observed hydrogen-amino close contacts in compounds containing nucleobases. DNA double-helical structures are stabilized by Watson-Crick basepairs, and the H-bonds between GC and AT basepairs include the N-H…N H-bond. Although it is known that three-dimensional protein structures are stabilized by an abundant number of N-H…O H-bonds formed by backbone functional groups, the possible existence of N-H…N H-bonds and their roles in the structural stability and function of a protein have not been explored. Recently, two reports have emerged in which H-bonds involving nitrogen as the acceptor atom were identified by spectroscopy studies. Preimesberger et al. (18) used NMR studies to demonstrate that the TXXH motif in ankryin repeat proteins is involved in a bifurcated H-bond in which the OH group of the Thr SC and its backbone NH group are involved in a bifurcated H-bond with the His imidazole nitrogen as acceptor. Adhikary et al. (26) used infrared spectroscopy to demonstrate the existence of Ni+1-Hi+1…Ni H-bonds involving proline residues in an SH3 domain protein. Up to now, however, no systematic analysis of protein structures had been carried out to identify and characterize N-H…N H-bonds.
In this study, we created two data sets containing ultrahigh- and very-high-resolution protein structures. In the ultrahigh-resolution structures, the hydrogen atom positions have been determined experimentally. To determine the N-H…N close contacts, we first defined a cutoff distance of 2.5 Å, which is well below the sum of van der Waals radii of the nitrogen and hydrogen atoms (2.75 Å) (67). We examined all possible N-H…N contacts in which the distance between an amino hydrogen and a nitrogen atom was less than or equal to the cutoff distance. Our search yielded 114 examples in data set I, and 75 of these were of the Ni+1-Hi+1…Ni type, with Ni being the proline backbone nitrogen. In the larger data set (data set II), we found 14,347 examples of N-H…N H-bonds, 8106 of which were in the Ni+1-Hi+1…Ni category with the proline nitrogen as the acceptor atom. We further characterized H-bond-forming proline residues geometrically and compared them with all other proline residues. The (φ, ψ) angles of proline residues involved in the Ni+1-Hi+1…Ni type of H-bonds fall in the α-helical/bridge region of the Ramachandran map, and the involvement of Ni+1-Hi+1 in H-bond formation explains the observed preference for the proline to occur in the helix/bridge region of a Ramachandran map (68). To understand the nature of Ni+1-Hi+1…Ni contacts, we also performed quantum chemical calculations on the model compound Ace-Pro-NMe. The quantum chemical calculations, along with NBO calculations, showed that Ni+1-Hi+1…Ni contacts are indeed energetically stable. Further calculations using the model compounds NMF and NAP with the N-H…N H-bond provided an estimate of the strength of this interaction.
Our analysis showed that 30–48% of H-bond-forming residues are also involved in n → π∗ interactions in which the lone pair of backbone carbonyl oxygen from the residue preceding proline overlaps with the π∗ antibonding orbital of the proline carbonyl group. Further analysis also showed that 48–53% of H-bond-forming proline residues also participated in a bifurcated H-bond with the functional groups of i-1 or i-2 residues. We found that 12–21% of the proline residues had all three types of interactions (Ni+1-Hi+1…Ni, n → π∗, and H-bond interaction with the i-1/i-2 residue (Fig. 6 C)). We also examined whether any of the H-bond-forming proline residues had a preference for any of the secondary structures. The majority of the H-bond-forming proline residues occurred in the loop region, and a closer examination revealed that they occurred in the capping regions of secondary structures. Although a small number of H-bond-forming proline residues were found in the middle of α-helices, these residues seemed to completely avoid the β-strand regions in both data sets I and II. We also analyzed the protein structures to determine how often H-bond-forming proline residues occurred in individual proteins. Many proteins were found to have multiple occurrences of H-bond-forming proline residues. Taken together, these results clearly demonstrate that the atypical and unconventional N-H…N H-bonds involving proline backbone nitrogen as an acceptor are found in significant numbers in proteins, and they are indeed energetically stabilizing interactions. Proline nitrogen atoms are more basic than other amides in a polypeptide chain, and since they cannot participate as H-bond donors, they are not constrained and are free to act as H-bond acceptors. If we assume that the strength of a conventional N-H…O H-bond is ∼5 kcal/mol, the strength of the N-H…N H-bond calculated as intermolecular interactions using the model compounds NMF and NAP is less than half of that observed for the conventional H-bond (1.92 kcal/mol as obtained by NBO analysis). When such H-bonds occur in the loop or capping regions of secondary structure, they can provide structural stability to the region. When several such H-bond-forming prolines occur in a protein, the stability due to multiple instances of these H-bonds will be very significant for the protein. When all three types of interactions (N-H…N, n → π∗, and H-bond with the i-1/i-2 residue) are present simultaneously in a proline, the cumulative effect of these stabilizing interactions cannot be ignored. Mutation of such residues is likely to have a significant effect on the structural stability of the protein. To achieve a full understanding of the structural stability of proteins, we must have complete knowledge about all such interactions, even if some of them seem to be counterintuitive or unusual (69, 70, 71). Traditionally, proline backbone nitrogen was never considered as a candidate for H-bond participation. The significant number of examples of H-bond-forming proline residues reported in this study has changed this picture. The heretofore unexplored N-H…N H-bond in proteins involving proline nitrogen should be included in the force field and, along with conventional H-bonds, could be important for protein structure and function.
Author Contributions
R.S. designed the research and wrote the manuscript. R.N.V.K.D. performed the research. R.S. and R.N.V.K.D. analyzed the data. Both authors approved the final version of the manuscript.
Acknowledgments
We thank all of our lab members, especially Dr. Ravi Kumar Verma, for useful discussions.
We thank IIT-Kanpur for financial support. This work was supported by the High Performance Computing Facility at IIT-Kanpur supported by the Department of Science and Technology, and the Ministry of Human Resource Development, Government of India. R.N.V.K.D. received a Senior Research Fellowship from the Council of Scientific and Industrial Research.
Editor: H. Jane Dyson.
Footnotes
Five figures and five tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30117-5.
Supporting Material
References
- 1.Jeffrey G.A., Saenger W. Springer-Verlag; Berlin: 1991. Hydrogen Bonding in Biological Structures. [Google Scholar]
- 2.Liu Z., Wang G., Wang R. Geometrical preferences of the hydrogen bonds on protein-ligand binding interface derived from statistical surveys and quantum mechanics calculations. J. Chem. Theory Comput. 2008;4:1959–1973. doi: 10.1021/ct800267x. [DOI] [PubMed] [Google Scholar]
- 3.Rozas I. On the nature of hydrogen bonds: an overview on computational studies and a word about patterns. Phys. Chem. Chem. Phys. 2007;9:2782–2790. doi: 10.1039/b618225a. [DOI] [PubMed] [Google Scholar]
- 4.Feldblum E.S., Arkin I.T. Strength of a bifurcated H bond. Proc. Natl. Acad. Sci. USA. 2014;111:4085–4090. doi: 10.1073/pnas.1319827111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harris T.K., Mildvan A.S. High-precision measurement of hydrogen bond lengths in proteins by nuclear magnetic resonance methods. Proteins. 1999;35:275–282. doi: 10.1002/(sici)1097-0134(19990515)35:3<275::aid-prot1>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 6.Derewenda Z.S., Lee L., Derewenda U. The occurrence of C-H…O hydrogen bonds in proteins. J. Mol. Biol. 1995;252:248–262. doi: 10.1006/jmbi.1995.0492. [DOI] [PubMed] [Google Scholar]
- 7.Eswar N., Ramakrishnan C. Deterministic features of side-chain main-chain hydrogen bonds in globular protein structures. Protein Eng. 2000;13:227–238. doi: 10.1093/protein/13.4.227. [DOI] [PubMed] [Google Scholar]
- 8.Steiner T., Koellner G. Hydrogen bonds with π-acceptors in proteins: frequencies and role in stabilizing local 3D structures. J. Mol. Biol. 2001;305:535–557. doi: 10.1006/jmbi.2000.4301. [DOI] [PubMed] [Google Scholar]
- 9.Vargas R., Garza J., Hay B.P. How strong is the Cα-H…O=C hydrogen bond? J. Am. Chem. Soc. 2000;122:4750–4755. [Google Scholar]
- 10.Scheiner S., Kar T., Gu Y. Strength of the Calpha H.O hydrogen bond of amino acid residues. J. Biol. Chem. 2001;276:9832–9837. doi: 10.1074/jbc.M010770200. [DOI] [PubMed] [Google Scholar]
- 11.Jain A., Sankararamakrishnan R. Dynamics of noncovalent interactions in all-α and all-β class proteins: implications for the stability of amyloid aggregates. J. Chem. Inf. Model. 2011;51:3208–3216. doi: 10.1021/ci200302q. [DOI] [PubMed] [Google Scholar]
- 12.Singh S.K., Kumar S., Das A. Competition between n → π(Ar)∗ and conventional hydrogen bonding (N-H···N) interactions: an ab initio study of the complexes of 7-azaindole and fluorosubstituted pyridines. Phys. Chem. Chem. Phys. 2014;16:8819–8827. doi: 10.1039/c3cp54169j. [DOI] [PubMed] [Google Scholar]
- 13.Hayashi K., Matubayasi N., Kawabata T. Insights into the origins of configurational stability of axially chiral biaryl amines with an intramolecular N-H-N hydrogen bond. J. Org. Chem. 2010;75:5031–5036. doi: 10.1021/jo100586b. [DOI] [PubMed] [Google Scholar]
- 14.Kawabata T., Jiang C., Tokitoh N. Axially chiral binaphthyl surrogates with an inner N-H-N hydrogen bond. J. Am. Chem. Soc. 2009;131:54–55. doi: 10.1021/ja808213r. [DOI] [PubMed] [Google Scholar]
- 15.Perrin C.L., Ohta B.K. Symmetry of O-H-O and N-H-N hydrogen bonds in 6-hydroxy-2-formylfulvene and 6-aminofulvene-2-aldimines. Bioorg. Chem. 2002;30:3–15. doi: 10.1006/bioo.2001.1222. [DOI] [PubMed] [Google Scholar]
- 16.Toh J.S.-S., Jordon M.J.T., Del Bene J.E. Can proton-shared or ion-pair N-H-N hydrogen bonds be produced in uncharged complexes? A systematic ab initio study of the structures and selected NMR and IR properties of complexes with N-H-N hydrogen bonds. J. Phys. Chem. A. 2001;105:10906–10914. [Google Scholar]
- 17.Perrin C.L., Ohta B.K. Symmetry of N-H-N hydrogen bonds in 1,8-bis(dimethylamino)naphthalene.H+ and 2,7-dimethoxy-1,8-bis(dimethylamino)naphthalene.H+ J. Am. Chem. Soc. 2001;123:6520–6526. doi: 10.1021/ja0036965. [DOI] [PubMed] [Google Scholar]
- 18.Preimesberger M.R., Majumdar A., Lecomte J.T.J. Direct NMR detection of bifurcated hydrogen bonding in the α-helix N-caps of ankyrin repeat proteins. J. Am. Chem. Soc. 2015;137:1008–1011. doi: 10.1021/ja510784g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Panigrahi S.K., Desiraju G.R. Strong and weak hydrogen bonds in the protein-ligand interface. Proteins. 2007;67:128–141. doi: 10.1002/prot.21253. [DOI] [PubMed] [Google Scholar]
- 20.Sarkhel S., Desiraju G.R. N-H…O, O-H…O, and C-H…O hydrogen bonds in protein-ligand complexes: strong and weak interactions in molecular recognition. Proteins. 2004;54:247–259. doi: 10.1002/prot.10567. [DOI] [PubMed] [Google Scholar]
- 21.Takeuchi T., Oishi S., Fujii N. Kinesin spindle protein inhibitors with diaryl amine scaffolds: crystal packing analysis for improved aqueous solubility. ACS Med. Chem. Lett. 2014;5:566–571. doi: 10.1021/ml500016j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohno I., Tomizawa M., Kagabu S. Neonicotinoid substituents forming a water bridge at the nicotinic acetylcholine receptor. J. Agric. Food Chem. 2009;57:2436–2440. doi: 10.1021/jf803985r. [DOI] [PubMed] [Google Scholar]
- 23.Yilmaz S., Altinkanat-Gelmez G., Yalcin I. Pharmacophore generation of 2-substituted benzothiazoles as AdeABC efflux pump inhibitors in A. baumannii. SAR QSAR Environ. Res. 2014;25:551–563. doi: 10.1080/1062936X.2014.919357. [DOI] [PubMed] [Google Scholar]
- 24.Font M., González Á., Sanmartín C. New insights into the structural requirements for pro-apoptotic agents based on 2,4-diaminoquinazoline, 2,4-diaminopyrido[2,3-d]pyrimidine and 2,4-diaminopyrimidine derivatives. Eur. J. Med. Chem. 2011;46:3887–3899. doi: 10.1016/j.ejmech.2011.05.060. [DOI] [PubMed] [Google Scholar]
- 25.Perrin L., André F., Delaforge M. Intramolecular hydrogen bonding as a determinant of the inhibitory potency of N-unsubstituted imidazole derivatives towards mammalian hemoproteins. Metallomics. 2009;1:148–156. doi: 10.1039/b817743k. [DOI] [PubMed] [Google Scholar]
- 26.Adhikary R., Zimmermann J., Romesberg F.E. Evidence of an unusual N-H···N hydrogen bond in proteins. J. Am. Chem. Soc. 2014;136:13474–13477. doi: 10.1021/ja503107h. [DOI] [PubMed] [Google Scholar]
- 27.Berman H.M., Westbrook J., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Word J.M., Lovell S.C., Richardson D.C. Asparagine and glutamine: using hydrogen atom contacts in the choice of side-chain amide orientation. J. Mol. Biol. 1999;285:1735–1747. doi: 10.1006/jmbi.1998.2401. [DOI] [PubMed] [Google Scholar]
- 29.Fu L., Niu B., Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li W., Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 31.Weiss M.S., Brandl M., Hilgenfeld R. More hydrogen bonds for the (structural) biologist. Trends Biochem. Sci. 2001;26:521–523. doi: 10.1016/s0968-0004(01)01935-1. [DOI] [PubMed] [Google Scholar]
- 32.Baker E.N., Hubbard R.E. Hydrogen bonding in globular proteins. Prog. Biophys. Mol. Biol. 1984;44:97–179. doi: 10.1016/0079-6107(84)90007-5. [DOI] [PubMed] [Google Scholar]
- 33.Casadei C.M., Gumiero A., Moody P.C.E. Heme enzymes. Neutron cryo-crystallography captures the protonation state of ferryl heme in a peroxidase. Science. 2014;345:193–197. doi: 10.1126/science.1254398. [DOI] [PubMed] [Google Scholar]
- 34.Schaftenaar G., Noordik J.H. Molden: a pre- and post-processing program for molecular and electronic structures. J. Comput. Aided Mol. Des. 2000;14:123–134. doi: 10.1023/a:1008193805436. [DOI] [PubMed] [Google Scholar]
- 35.Becke A.D. Density-functional exchange-energy approximation with correct asymptotic behavior. Phys. Rev. A Gen. Phys. 1988;38:3098–3100. doi: 10.1103/physreva.38.3098. [DOI] [PubMed] [Google Scholar]
- 36.Perdew J.P. Density-functional approximation for the correlation energy of the inhomogeneous electron gas. Phys. Rev. B Condens. Matter. 1986;33:8822–8824. doi: 10.1103/physrevb.33.8822. [DOI] [PubMed] [Google Scholar]
- 37.Weigend F., Ahlrichs R. Balanced basis sets of split valence, triple zeta valence and quadruple zeta valence quality for H to Rn: design and assessment of accuracy. Phys. Chem. Chem. Phys. 2005;7:3297–3305. doi: 10.1039/b508541a. [DOI] [PubMed] [Google Scholar]
- 38.Weigend F. Accurate Coulomb-fitting basis sets for H to Rn. Phys. Chem. Chem. Phys. 2006;8:1057–1065. doi: 10.1039/b515623h. [DOI] [PubMed] [Google Scholar]
- 39.Neese F. The ORCA program system. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2012;2:73–78. [Google Scholar]
- 40.Pettersen E.F., Goddard T.D., Ferrin T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 41.Zhao Y., Truhlar D.G. The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: two new functionals and systematic testing of four M06-class functionals and 12 other functionals. Theor. Chem. Acc. 2008;120:215–241. [Google Scholar]
- 42.Woon D.E., Dunning T.H., Jr. Gaussian basis sets for use in correlated molecular calculations. III. The atoms aluminium through argon. J. Chem. Phys. 1993;98:1358–1371. [Google Scholar]
- 43.Frisch M.J., Trucks G.W., Fox D.J. Gaussian Inc.; Wallingford, CT: 2009. Gaussian 09, revision E.01. [Google Scholar]
- 44.Reed A.E., Curtiss L.A., Weinhold F. Intermolecular interactions from a natural bond orbital, donor-acceptor viewpoint. Chem. Rev. 1988;88:899–926. [Google Scholar]
- 45.Glendening E.D., Landis C.R., Weinhold F. Natural bond orbital methods. WIREs Comput. Mol. Sci. 2012;2:1–42. [Google Scholar]
- 46.Glendening E.D., Reed A.E., Weinhold F. University of Wisconsin; Madison, WI: 1998. NBO version 3.1. Theoretical Chemistry Institute. [Google Scholar]
- 47.Boys S.F., Bernardi F. Calculation of small molecular interactions by differences of separate total energies—some procedures with reduced errors. Mol. Phys. 1970;19:553–566. [Google Scholar]
- 48.Bartlett G.J., Choudhary A., Woolfson D.N. n-->π∗ interactions in proteins. Nat. Chem. Biol. 2010;6:615–620. doi: 10.1038/nchembio.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Newberry R.W., Bartlett G.J., Raines R.T. Signatures of n→π∗ interactions in proteins. Protein Sci. 2014;23:284–288. doi: 10.1002/pro.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bartlett G.J., Newberry R.W., Woolfson D.N. Interplay of hydrogen bonds and n→π∗ interactions in proteins. J. Am. Chem. Soc. 2013;135:18682–18688. doi: 10.1021/ja4106122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Newberry R.W., VanVeller B., Raines R.T. n→π∗ interactions of amides and thioamides: implications for protein stability. J. Am. Chem. Soc. 2013;135:7843–7846. doi: 10.1021/ja4033583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Veljkovic D.Z., Janjic G.V., Zaric S.D. Are C-H…O interactions linear? The case of aromatic CH donors. CrystEngComm. 2011;13:5005–5010. [Google Scholar]
- 53.Liu A., Lu Z., Yan H. NMR detection of bifurcated hydrogen bonds in large proteins. J. Am. Chem. Soc. 2008;130:2428–2429. doi: 10.1021/ja710114r. [DOI] [PubMed] [Google Scholar]
- 54.Torshin I.Y., Weber I.T., Harrison R.W. Geometric criteria of hydrogen bonds in proteins and identification of “bifurcated” hydrogen bonds. Protein Eng. 2002;15:359–363. doi: 10.1093/protein/15.5.359. [DOI] [PubMed] [Google Scholar]
- 55.Auffinger P., Westhof E. Singly and bifurcated hydrogen-bonded base-pairs in tRNA anticodon hairpins and ribozymes. J. Mol. Biol. 1999;292:467–483. doi: 10.1006/jmbi.1999.3080. [DOI] [PubMed] [Google Scholar]
- 56.Sponer J., Hobza P. Bifurcated hydrogen bonds in DNA crystal structures—an ab-initio quantum chemical study. J. Am. Chem. Soc. 1994;116:709–714. [Google Scholar]
- 57.Aurora R., Rose G.D. Helix capping. Protein Sci. 1998;7:21–38. doi: 10.1002/pro.5560070103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pal T.K., Sankararamakrishnan R. Self-contacts in Asx and Glx residues of high-resolution protein structures: role of local environment and tertiary interactions. J. Mol. Graph. Model. 2008;27:20–33. doi: 10.1016/j.jmgm.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 59.Cordes F.S., Bright J.N., Sansom M.S.P. Proline-induced distortions of transmembrane helices. J. Mol. Biol. 2002;323:951–960. doi: 10.1016/s0022-2836(02)01006-9. [DOI] [PubMed] [Google Scholar]
- 60.von Heijne G. Proline kinks in transmembrane α-helices. J. Mol. Biol. 1991;218:499–503. doi: 10.1016/0022-2836(91)90695-3. [DOI] [PubMed] [Google Scholar]
- 61.Sankararamakrishnan R., Vishveshwara S. Geometry of proline-containing alpha-helices in proteins. Int. J. Pept. Protein Res. 1992;39:356–363. doi: 10.1111/j.1399-3011.1992.tb01595.x. [DOI] [PubMed] [Google Scholar]
- 62.Van Arnam E.B., Lester H.A., Dougherty D.A. Dissecting the functions of conserved prolines within transmembrane helices of the D2 dopamine receptor. ACS Chem. Biol. 2011;6:1063–1068. doi: 10.1021/cb200153g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Purohit P., Chakraborty S., Auerbach A. Function of the M1 π-helix in endplate receptor activation and desensitization. J. Physiol. 2015;593:2851–2866. doi: 10.1113/JP270223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yamada M., Tamada T., Miki K. Elucidations of the catalytic cycle of NADH-cytochrome b5 reductase by X-ray crystallography: new insights into regulation of efficient electron transfer. J. Mol. Biol. 2013;425:4295–4306. doi: 10.1016/j.jmb.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 65.Karplus P.A. Experimentally observed conformation-dependent geometry and hidden strain in proteins. Protein Sci. 1996;5:1406–1420. doi: 10.1002/pro.5560050719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Luisi B., Orozco M., Shakked Z. On the potential role of the amino nitrogen atom as a hydrogen bond acceptor in macromolecules. J. Mol. Biol. 1998;279:1123–1136. doi: 10.1006/jmbi.1998.1833. [DOI] [PubMed] [Google Scholar]
- 67.Bondi A. van der Waals volumes and radii. J. Phys. Chem. 1964;68:441–451. [Google Scholar]
- 68.Porter L.L., Rose G.D. Redrawing the Ramachandran plot after inclusion of hydrogen-bonding constraints. Proc. Natl. Acad. Sci. USA. 2011;108:109–113. doi: 10.1073/pnas.1014674107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jain A., Purohit C.S., Sankararamakrishnan R. Close contacts between carbonyl oxygen atoms and aromatic centers in protein structures: π…π or lone-pair…π interactions? J. Phys. Chem. B. 2007;111:8680–8683. doi: 10.1021/jp072742l. [DOI] [PubMed] [Google Scholar]
- 70.Pal T.K., Sankararamakrishnan R. Quantum chemical investigations on intraresidue carbonyl-carbonyl contacts in aspartates of high-resolution protein structures. J. Phys. Chem. B. 2010;114:1038–1049. doi: 10.1021/jp909339r. [DOI] [PubMed] [Google Scholar]
- 71.Jain A., Ramanathan V., Sankararamakrishnan R. Lone pair … π interactions between water oxygens and aromatic residues: quantum chemical studies based on high-resolution protein structures and model compounds. Protein Sci. 2009;18:595–605. doi: 10.1002/pro.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.