Abstract
The key steps in cellular signaling and regulatory pathways rely on reversible noncovalent protein-ligand binding, yet the equilibrium parameters for such events remain challenging to characterize and quantify in solution. Here, we demonstrate a microfluidic platform for the detection of protein-ligand interactions with an assay time on the second timescale and without the requirement for immobilization or the presence of a highly viscous matrix. Using this approach, we obtain absolute values for the electrophoretic mobilities characterizing solvated proteins and demonstrate quantitative comparison of results obtained under different solution conditions. We apply this strategy to characterize the interaction between calmodulin and creatine kinase, which we identify as a novel calmodulin target. Moreover, we explore the differential calcium ion dependence of calmodulin ligand-binding affinities, a system at the focal point of calcium-mediated cellular signaling pathways. We further explore the effect of calmodulin on creatine kinase activity and show that it is increased by the interaction between the two proteins. These findings demonstrate the potential of quantitative microfluidic techniques to characterize binding equilibria between biomolecules under native solution conditions.
Introduction
Weak and transient protein-ligand interactions are essential for cellular signaling and regulatory pathways. A quantitative understanding of this complex network of noncovalent interactions is key to providing fundamental insights into the molecular processes underpinning biological function. A range of biophysical methods have been developed to probe noncovalent protein interactions and the formation of protein complexes (1, 2, 3, 4, 5, 6). However, studies of these processes may be limited by requirements with respect to the quantity of the sample, molecular size, persistence of the labile complexes during the measurement, and resistance of dynamic species to dissociation during sample preparation.
Surface-based techniques are widely used to detect and characterize protein-protein interactions. Such methods, including surface plasmon resonance (SPR), are convenient, as they can be automated, but they require the attachment of one of the binding partners to a functionalized surface, a process which can affect the binding equilibrium (4, 7). Gas phase methods avoid the use of surfaces, and elegant studies of protein complex formation have been made using this approach (2, 6), but it can be complex and costly to implement. The development of complementary solution-based techniques to investigate proteins and their interactions is therefore desirable.
Traditional assays in bulk solution impose limitations as to the concentrations and amounts of sample required, as well as the timescale of the measurement, factors that limit the range of affinities and rate constants that can be discovered by each technique. Furthermore, to achieve a broad understanding of the biophysical basis of protein-ligand binding, the determination of absolute values for the parameters defining these interactions is necessary. Optical microscale assays can readily be applied to a wider size range of sample molecules and typically require considerably less sample material than bulk-solution techniques (8, 9). Thermophoretic measurements of differential analyte migration in a temperature gradient are examples of such miniaturization (10). In this study, we address these challenges and present a general method for the quantitative study of noncovalent protein interactions under native solution conditions. To achieve this objective, we used a microfluidic platform to measure the absolute value of a key parameter characterizing the solvated proteins, the electrophoretic mobility, μe. By measuring an absolute value for a physicochemical parameter characterizing the analyte we were able to compare results directly between solution conditions. Furthermore, microfluidic assays allow characterization of sample molecules of a wide range of dimensions and concentrations. These are advantageous features for the study of noncovalent complex formation by proteins.
The electrophoretic mobility reports on the size and charge state of the migrating species, and can therefore be exploited to differentiate between isolated protein molecules and protein complexes (11). Free-flow electrophoresis, where an electric field is applied perpendicular to the direction of flow, and capillary electrophoresis, where the field is parallel to the direction of flow, have both been adapted to a microfluidic format (12, 13). Unlike native gel electrophoresis or size-exclusion chromatography, these methods do not require sample molecules to pass through a matrix, which could introduce additional variables to the measurement, but achieve spatial separation of sample molecules in free solution.
The electrophoretic mobility of molecular species can be measured by both free-flow electrophoresis and capillary electrophoresis, with the latter requiring the use of a mobility standard (13, 14, 15, 16). Whereas protein-ligand interactions have been probed by capillary electrophoresis (14), many microfluidic free-flow electrophoresis studies have focused on the qualitative separation of sample molecules (17). Gradient microfluidic free-flow electrophoresis has been applied to probe DNA-protein binding through near-continuous observation of the equilibrium distribution at a large number of target concentrations. In this case, the formation of bubbles within the microfluidic device resulted in a variable electric field and thereby in fluctuations in the observed sample deflection (18). In this article, we demonstrate the use of a microfluidic free-flow electrophoresis assay, where the electric field is quantified, thereby enabling the sample electrophoretic mobility to be quantified and compared between solution conditions (15, 16).
We apply the microfluidic platform shown in Fig. 1, A and B, to shed light on the ligand-binding equilibria of calmodulin. Many diverse and vital signaling pathways, including the control of heart function, cell death by apoptosis, memory, and learning are activated in response to variation in the intracellular concentration of Ca2+, and the protein calmodulin is at the focal point of calcium-mediated signaling (19, 20). We therefore chose to elucidate the ion-dependent ligand binding of this protein as a representative model of protein-protein interactions in signal transduction.
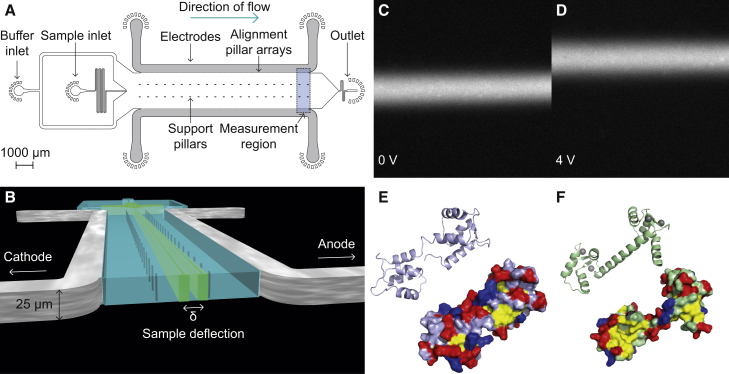
Figure 1.
Experimental design. (A) The design of the microfluidic free-flow electrophoresis device used in this study. Integrated metal electrodes are shown in gray. Inlets for the sample and buffer are indicated. Support pillars for the channel ceiling and the pillar array utilized for the electrode integration and alignment are also shown. The sample deflection is measured at the end of the electrophoresis channel (blue shaded area). (B) A pseudo-three-dimensional representation of the device showing a cross section of the main electrophoresis channel. Upon application of an electric potential, the sample is deflected from its path at the center of the channel. The scale bar refers to the vertical dimension. (C) Fluorescence image of 1 μM calmodulin in the buffer containing EDTA at 0 V. (D) Fluorescence image of 1 μM calmodulin in the buffer containing EDTA at an applied potential of 4 V. The negatively charged protein is deflected toward the anode. (E) The structure of calmodulin in the absence of Ca2+ shown as a ribbon diagram (top) and as a surface plot with hydrophobic residues (isoleucine, leucine, phenylalanine, and methionine) in yellow, acidic residues in red, and basic residues in blue (PDB: 1CFD) (22). (F) The structure of calmodulin with four Ca2+ (gray spheres), chelated by the four EF-hands, can be seen in the ribbon diagram (top), whereas the surface representation (bottom) reveals the exposure of two hydrophobic patches upon Ca2+ binding (PDB: 3CLN) (23).
Calmodulin is a small, 16.7 kDa, ubiquitously expressed signaling protein with >300 reported interaction partners (see (21) and references therein). It contains four calcium-binding helix-loop-helix motifs, called EF-hands (Fig. 1, E and F) (22, 23, 24). These are present as EF-hand pairs forming two globular domains connected by a flexible linker. Binding of Ca2+ to the calmodulin EF-hands occurs with pairwise cooperativity, leading to conformational changes, as seen in Fig. 1, E and F (22, 23, 25). These conformational changes result in the exposure of hydrophobic residues, in many cases increasing the binding affinity of calmodulin for target proteins (Fig. 1 F) (4, 20, 26). The flexible central linker of calmodulin allows the protein to wrap around the target segment of a binding partner (27, 28, 29, 30, 31). The prevalence of flexible methionine side chains at the hydrophobic patches of Ca2+ -bound calmodulin and the adaptable fold underlie its promiscuity in target recognition (32, 33, 34).
The microfluidic free-flow electrophoresis method presented in this article can be applied as a general method for the detection and characterization of protein-ligand interactions. This technique does not rely on specific properties of the sample molecule and is thus ideally suited for the investigation of newly discovered protein-ligand interactions. We used the electrophoretic binding assay to validate and explore the calcium dependence of calmodulin binding to creatine kinase (35, 36), which we have identified in this study as a ligand in a surface-based protein array screen. In a negative control, we did not observe binding to calbindin D9k (37), whereas we did observe binding to a known binding partner, phosphorylase kinase (26, 31, 38, 39). The results of these studies highlight the need for fast nondisruptive assays for the quantitative characterization of protein interactions in solution.
Materials and Methods
Microfluidic free-flow electrophoresis measurements
Microfluidic devices were prepared using standard soft lithography methods (40). Electrodes were integrated as described in the Supporting Material and previously reported (15). Fluorescence images for the electrophoresis experiments were recorded using a charge-coupled-device camera (Photometrics Evolve 512, Photometrics, Tucson, AZ) through inverted optics (Observer D1, Zeiss, Oberkochen, Germany). Electrophoresis measurements for each target protein were performed under two sets of buffer conditions, 5 mM Tris-HCl, pH 8.0, with either 0.1 mM CaCl2 or 0.1 mM EDTA to chelate any trace calcium ions. Unless otherwise stated, buffer solutions were prepared using chemicals of analytical grade purchased from Sigma (St. Louis, MO). Measurements were performed for a range of binding partner/calmodulin ratios depending on the observation of a plateau in the measured electrophoretic mobility.
To track calmodulin within the microfluidic device, the protein was labeled with Alexa488, as described in the Results and the Supporting Material. The sample was introduced at the center of the fluid flow within the electrophoresis channel. The sample stream would span the vertical direction of the channel, as illustrated in Fig. 1 B. Sample molecules were transported past the electrodes by the flow through the channel (Fig. 1 A). Upon the application of an electric field across the electrophoresis channel, the sample molecules migrated perpendicular to the direction of flow according to their electrophoretic mobility.
Fluorescence image acquisition, the application of an electric potential, and current measurements by a multimeter (Agilent Technologies, Santa Clara, CA) were triggered through a lock-in amplifier (Stanford Research Systems, Sunnyvale, CA). Experiments were performed at room temperature, 23°C. The flow rate through the device was set to 500 μL h−1 and controlled by withdrawal through the outlet using a glass syringe (Hamilton, Bonaduz, Switzerland) and a syringe pump (neMESYS, Cetoni, Korbussen, Germany) (Fig. 1 A). The lower flow velocities at the edges of the microfluidic channel were taken into consideration, and the average flow rate in the region of the channel explored by the sample was evaluated to be 510.9 μL h−1 (41). This value was used in calculations of the residence time of 3.4 s and .
Four repeats of a voltage ramp of 0–4 V at 0.5 V intervals were applied for each sample. After each change in the applied potential, there was a pause between image acquisitions, so that the imaged sample had only been exposed to one voltage while traveling through the electrophoresis channel. The applied voltage range was adjusted so as to achieve significant sample deflection (Fig. 1, C and D), and electrolysis products were removed by the flow through the channel and thus prevented from accumulating beyond their solubility limit. The mean displacement of the sample signal was measured at the end of the separation channel (see Fig. 3 C). The electrophoresis channel was 10,000 μm long, with a height of 25 μm and a width of 2100 μm, and an array of support pillars were placed in the wide channel (Fig. 1, A and B). Each electrophoretic mobility measurement consumed 1.6 μL of sample. For a 1 μM solution of calmodulin, this volume corresponds to 1.6 pmol of protein. To fit three free parameters, at least nine electrophoretic mobility measurements would be required.
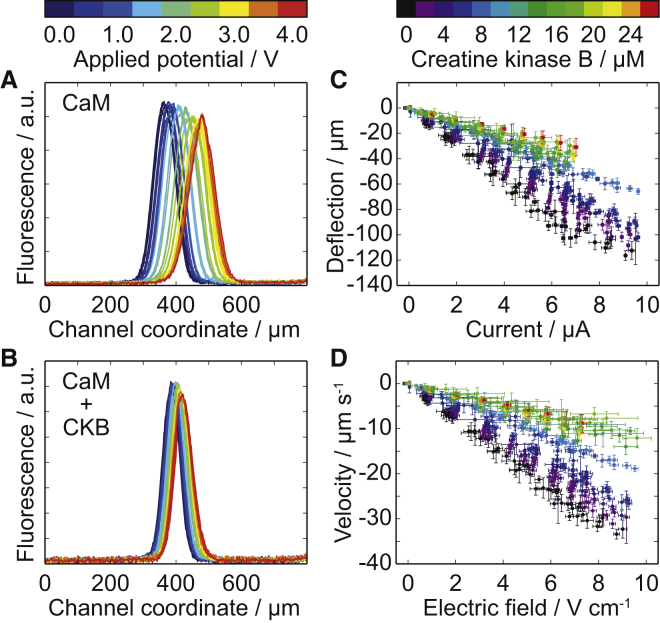
Figure 3.
Electrophoresis experiments in buffer containing EDTA. (A) Sample deflection is monitored via fluorescence. Fluorescence intensity profiles for the isolated protein: 1 μM calmodulin (CaM) for four repeats of a voltage ramp. The color bar above (A) shows the applied voltage for each profile and applies to (A) and (B). The profiles are recorded for a cross section of the separation channel. (B) Fluorescence intensity profiles for 1 μM labeled calmodulin in the presence of 20 μM unlabeled creatine kinase B (CaM + CKB). (C) The measured sample deflection after passing between the electrodes against the measured current for 1 μM calmodulin and 0–27 μM creatine kinase B. Throughout the figure, data points are colored according to the creatine kinase concentration, going from low (black) to high (red). The color bar applies to (C) and (D). (D) Sample deflection velocity against the electric field across the solution for calmodulin and creatine kinase in the absence of Ca2+.
Calibration of the cell constants and buffer conductivities were also performed using a lock-in amplifier (15). Detailed descriptions of the calibration procedure and the subsequent determination of the electric field can be found in (15). Using Ohm’s law, the effective voltage drop, Veffective, across the solution was determined from the buffer conductance and the measured current. The electric field corresponding to each current measurement was then found by dividing Veffective by the distance between the electrodes (see Fig. 3 D). The electrophoretic mobilities of the solvated proteins were determined by a linear fit to against E (see Fig. 4). Electrophoresis data analysis was performed using software written in Python.
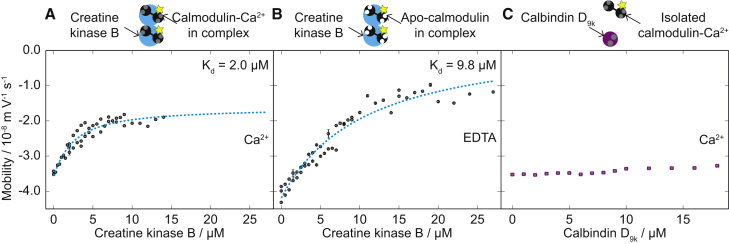
Figure 4.
Creatine kinase interaction with calmodulin monitored by free-flow electrophoresis. (A) The electrophoretic mobility of 1 μM calmodulin shown against an increasing creatine kinase B concentration in the presence of 0.1 mM Ca2+. The dashed line shows a least-squares fit of Eq. 2 to the data. The diagram at the top shows calmodulin (black), calcium ions as gray spheres, the label as a star, and creatine kinase (lighter color). (B) Electrophoretic mobility of 1 μM calmodulin against increasing creatine kinase B concentration in the presence of 0.1 mM EDTA. (C) The electrophoretic mobility of 1 μM calmodulin against increasing calbindin concentration in buffer containing 0.1 mM Ca2+. The diagram at the top shows calcium-bound calbindin and calmodulin. To see this figure in color, go online.
Cloning, expression, and purification of creatine kinase
The creatine kinase B gene was purchased from the Arizona State University clone collection and cloned into the PetSac vector using standard polymerase chain reaction and cloning techniques. The protein was expressed in Escherichia coli strain BL21 Des3 pLysS star in lysogeny broth. The protein was purified from inclusion bodies. These were isolated by probe sonication of the cell pellet from 2 L of culture in 40 mL 10 mM Tris-HCl and 1 mM EDTA, pH 7.5, for 1.5 min (1/2 horn, maximum output, duty cycle 50%) in a beaker surrounded by ice-water slurry, followed by centrifugation for 10 min at 18,000 × g at 4°C. The pellet was solubilized in 8 M urea, 10 mM Tris-HCl, and 1 mM EDTA, pH 7.5. The protein was then purified by anion exchange chromatography on a 3.4 × 10 cm DEAE cellulose column equilibrated in 10 mM Tris-HCl and 1 mM EDTA, pH 7.5, and eluted by a linear salt gradient from 0 to 300 mM NaCl in 10 mM Tris-HCl and 1 mM EDTA, pH 7.5, to a total gradient volume of 400 mL. Five-milliliter fractions were collected and examined by ultraviolet absorbance at 280 nm and by sodium dodecyl sulfate polyacrylamide gel electrophoresis. Fractions containing creatine kinase were combined, concentrated, and further purified by size-exclusion chromatography on a Superdex 200 column equilibrated and eluted in 10 mM Tris-HCl, 1 mM EDTA, and 50 mM NaCl, pH 7.5. Purity was confirmed by agarose gel electrophoresis in EDTA and by sodium dodecyl sulfate polyacrylamide gel electrophoresis.
Calmodulin-binding protein profiling on ProtoArrays
For the protein array screen, ProtoArray slides (Invitrogen, Carlsbad, CA) were blocked through incubation in 1% w/v skimmed milk powder dissolved in phosphate-buffered saline containing 1 mM dithiothreitol, 0.1% v/v Tween20, and 50% v/v glycerol, pH 7.5 for 1 h and then washed five times in Tris-buffered saline and 0.1% v/v Tween20, pH 7.5. The ProtoArray was then incubated with 1 μM Alexa546-labeled calmodulin in 50 mM Tris-HCl, 150 mM NaCl, 0.1% v/v Tween20, pH 7.5, and either 1 mM CaCl2 or 1 mM EDTA for 90 min, followed by five 5 min washes with the same buffer without calmodulin. Before imaging, the slides were rinsed in deionized water. Imaging was performed using a Genepix 4000B scanner (Axon Instruments, Sunnyvale, CA) with excitation at 532 nm and 635 nm, the latter for detection of red-labeled control proteins spotted on the array to guide the analysis.
Creatine kinase activity by NMR spectroscopy
All experiments were performed at 37°C in 10 mM Tris-HCl, 150 mM KCl, 100 μM MgCl2, 100 μM CaCl2, and 10% v/v D2O, pH 7.5, at a static magnetic field strength of 14.1 T. Series of one-dimensional 1H NMR spectra were recorded over time at several concentrations (1–37 mM) of creatine phosphate and ADP in the presence of 0.5 μM creatine kinase with and without 0.5 μM Ca2+ -loaded calmodulin (see Fig. 5). Creatine phosphate and ADP concentrations were altered in parallel. Decay of creatine phosphate (resonances at 3.94 and 3.04 ppm) and buildup of creatine (3.90 and 3.01 ppm) were monitored over time and converted into initial rate constants. Neither ADP nor ATP displayed any signals that were isolated enough for quantification. Doubling of the calmodulin concentration resulted in virtually the same kinetics. The auto reaction was studied without the enzyme present and found to be insignificant during the experimental time. Spectra were processed and analyzed with NMRPipe (42). Michaelis-Menten kinetics were fitted to models without (in the presence of calmodulin) and with a Hill parameter (without calmodulin) to account for allosteric effects.
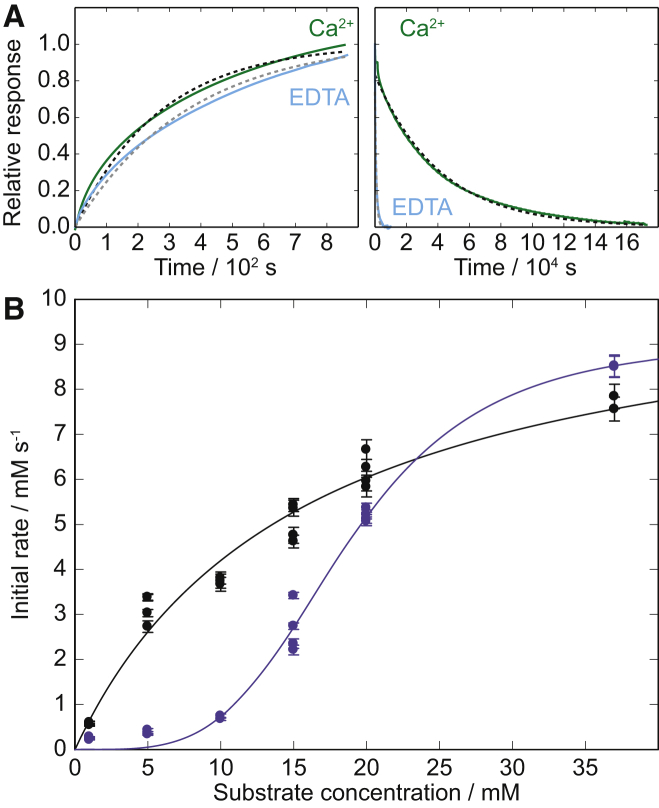
Figure 5.
Further investigation of the calmodulin-creatine kinase interaction. (A) SPR data for 5 μM creatine kinase in buffer containing Ca2+ (dark) or EDTA (light). The association data are shown at left, and the disassociation data are shown at right. The dashed lines represent the fits to determine kon and koff assuming a 1:1 binding event. (B) Michaelis-Menten kinetics of 0.5 μM creatine kinase at 37°C in 10 mM Tris-HCl, 150 mM KCl, 100 μM MgCl2, and 100 μM CaCl2 with (black) and without (light) 0.5 μM Ca2+-loaded calmodulin. The concentrations of creatine phosphate and ADP were altered in parallel, and the initial rate is plotted versus either substrate concentration. The black curve shows the best fits of the Michaelis-Menten model to the data in the presence of calmodulin, whereas the lighter colored curve shows a fit that includes a Hill parameter to account for allosteric coupling between subunits of the enzyme. To see this figure in color, go online.
Results
We combined microfluidic and electrical components to generate a microfluidic platform for protein ligand-binding assays. To establish a well-defined electric field, we integrated self-aligning three-dimensional (3D) electrodes into microfluidic devices (Fig. 1 A) (15, 16, 43). Using these microfabricated electrodes, we were able to perform quantitative free-flow zone electrophoresis in aqueous solutions (Fig. 1, A and B) (15, 16). Upon application of an electrical potential, the sample molecules migrated perpendicular to the direction of the fluid flow according to their electrophoretic mobility, as shown in Fig. 1, B–D.
An S17C variant of calmodulin was generated using site-directed mutagenesis. Residue S17 has been identified at the surface of calmodulin-ligand complexes in previous studies (44). Fluorophore-labeled S17C variants have also been used successfully in FRET studies of calmodulin conformational changes (44) and protein array screens (21). To visualize calmodulin in the experiments, the protein was labeled at position 17 with Alexa488 for the electrophoresis measurements or Alexa546 for the protein array screen. Crucially, the target proteins were unlabeled, which enabled us to quantify the extent of ligand binding through the observed mean electrophoretic mobility of calmodulin. With free-flow electrophoresis, analyte sizes ranging from Ångströms to micrometers can be investigated simultaneously. This method is therefore well suited for the study of protein-ligand complex formation.
Surface-based screen for potential calmodulin targets
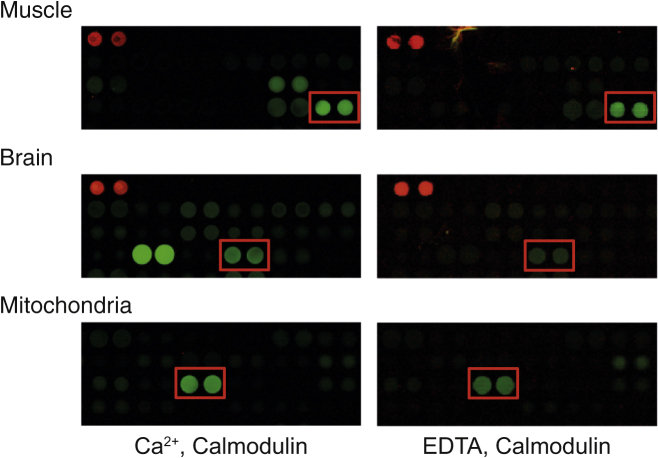
We sought to explore the potential of microfluidic free flow electrophoresis in the discovery and characterization of new binding targets. To this effect, we performed a protein array screen for calmodulin binding partners (Fig. 2). The protein spots on the array exhibiting the highest signal in the presence of Ca2+ are listed in Fig. S1, and those with the highest signal in the absence of Ca2+ are listed in Fig. S2. Calmodulin bound to a large number of known interaction partners present in this screen, among which were a number of kinases. Interestingly, the screen also highlighted a number of novel putative targets. Among the previously unknown targets were several creatine kinase isoforms, including the B (brain), M (muscle), and U (mitochondrial) isoforms, observed both in the absence and presence of Ca2+. We chose to further investigate creatine kinase B, as the signal intensity of calmodulin bound to this protein showed the largest apparent calcium dependence in the protein array screen.
Figure 2.
Protein array screen for calmodulin-binding proteins. Fluorescence image of the areas of the protein array containing the immobilized brain isoform for experiments performed in the presence of 1 mM Ca2+ (left) or 1 mM EDTA (right). Every protein is present in duplicate dots. The creatine kinase spots have been highlighted with a red bounding box. The very bright spots to the left of the brain isoform in the presence of Ca2+ are calcium/calmodulin-dependent protein kinase type 1 (Q14012), a known and highly Ca2+-sensitive binding target of calmodulin.
Creatine kinase catalyzes the reversible addition of a phosphate ion to creatine (35, 36). Abnormalities in creatine kinase metabolism have been associated with an increased risk of heart failure (45). To the best of our knowledge, calmodulin has not previously been reported to interact with this protein. Here, we studied calmodulin binding to creatine kinase B using electrophoresis. In these studies, we evaluated the effect of Ca2+ on the equilibrium dissociation constant, Kd, of the interaction.
Binding to creatine kinase B is validated in solution by free-flow electrophoresis
The results of the surface-based protein array screen pointed to a potential interaction between calmodulin and creatine kinase B. However, experiments in free solution were required to ascertain that the observed binding to the immobilized substrate was not a nonspecific effect or the result of a change in the conformation of creatine kinase upon surface immobilization. To verify the interaction between calmodulin and creatine kinase in free solution, we performed free-flow electrophoresis using the device shown in Fig. 1 A. To minimize the background signal in the fluorescence measurements, we cast the microfluidic devices in black polydimethylsiloxane. A range of electric potential differences were applied across the separation channel, causing the sample molecules to migrate perpendicularly to the direction of flow according to their electrophoretic mobility. The mean deflection, δ, of the labeled calmodulin was recorded (Fig. 3).
In addition to δ, we simultaneously recorded the current, I (Fig. 3 C). From measurements of the frequency-dependent admittance, we determined the cell constant for each individual device and the conductivities of the buffers used (15). By combining these measurements with the residence time of the proteins between the electrodes we found the transverse electrophoretic drift velocity, , and the electric field across the separation channel, E (Fig. 3 D). The electrophoretic mobility, μe, of the sample was given by the slope of against E, (Fig. 3 D). This parameter reports on the relation between the diffusion coefficient, D, and the charge component, q, defining the migrating molecules:
| (1) |
where kB and T represent the Boltzmann constant and the absolute temperature, respectively. The electrophoretic mobility can therefore be exploited to monitor changes in the size and charge of the analyte, for instance, through interaction with other molecules in solution.
The results displayed in Figs. 3 and 4 show a change in the electrophoretic mobility of the fluorescently labeled calmodulin in response to an increasing concentration of unlabeled creatine kinase. This change in μe arises from changes in the size and charge of the migrating species, indicating complex formation between calmodulin and creatine kinase (compare Fig. 3, A and B). At intermediate target protein concentrations, broadening of the fluorescence peak was observed as the sample migrated in the electric field. This observation indicated the coexistence of more than one species with different electrophoretic mobilities, consistent with the presence of isolated calmodulin and calmodulin-target complex. The width of the sample inlet is 50 μm. At the end of the electrophoresis channel, the width of the sample distribution is therefore comparable to δ. Thus, although variation in δ is readily detected, the fluorescence peak does not divide into two peaks. In this study, we combine the accurate determination of the mean sample position with quantification of the electric field, yielding a sensitive analytical assay for the measurement of electrophoretic mobilities without the need for separation of the sample components.
Determination of calcium-dependent equilibrium dissociation constants
In this study, we monitored the electrophoretic mobility of Alexa488-labeled calmodulin as a function of the concentration of unlabeled target protein. The fractional contributions from isolated calmodulin, with an electrophoretic mobility of μc, and calmodulin in complex with the target protein, μcp, result in an observed mean electrophoretic mobility, μobs, of
| (2) |
The concentration of free target protein, [P], and the concentration of complex, [CP], can be expressed in terms of the two known total concentrations of calmodulin, Ct, and target protein, Pt, and isolated calmodulin, [C], in the equation for the equilibrium dissociation constant, Kd. This allows the resulting quadratic equation to be solved for [C], as described in the Supporting Material, yielding an expression for [C] in terms of Pt, Ct, and Kd.
We investigated the calcium dependence of calmodulin binding to creatine kinase by performing electrophoresis measurements in 5 mM Tris-HCl containing either 0.1 mM CaCl2 or 0.1 mM EDTA to chelate any residual Ca2+ (Fig. 4). Plots of the observed mobility of 1 μM calmodulin as a function of the total concentration of target protein, Pt, are shown in Fig. 4. Inserting the solution for [C] found in the Supporting Material into Eq. 2 allowed us to fit the data with μc, μcp, and Kd as the free parameters.
In agreement with the protein array data, we observed calmodulin binding to creatine kinase B. In the presence of Ca2+ in low-ionic-strength buffers, we found the dissociation constant to be 2.0 μM (Fig. 4 A). Binding of Ca2+ by the four EF-hand motifs of calmodulin leads to conformational changes in calmodulin (Fig. 1, E and F) (22, 23, 32) involving exposure of hydrophobic side chains, which results in an increase in the binding affinity for most target proteins (Fig. 1 F) (22, 23). Indeed, in the absence of free calcium ions, we observed a reduction in the binding affinity, Kd = 9.8 μM (Fig. 4 B).
In a negative control experiment, we investigated whether the change in μobs of calmodulin could be the result of nonspecific effects associated with the presence of another protein in solution, for instance, through electrostatic screening by the added protein (46). We chose calbindin D9k as a control system, because although the function of this protein relies on binding of calcium ions, calbindin has not been known to interact with calmodulin. Calbindin D9k contains a pair of EF-hands that enable it to bind and transport two calcium ions (37). In the presence of Ca2+ and calbindin at concentrations of 0–18 μM, we did not observe a change in the μobs of calmodulin (Fig. 4 C). In positive control experiments, we detected calmodulin binding to phosphorylase kinase, a well-documented ligand (31, 39), with a Kd of 2.2 μM (see Fig. S4).
Direct observation of the fluorescently labeled calmodulin during the electrophoresis experiments furthermore enabled us to confirm the absence of any sample signal with the mobility of the isolated protein, once the electrophoretic mobility had reached a plateau against the concentration of the target protein (Fig. 3 B). Calmodulin was found to have a negative μe: −3.5 ± 0.1 × 10−8 m V−1 s−1 for Ca2+-calmodulin and −4.1 ± 0.2 × 10−8 m V−1 s−1 for apocalmodulin. These observations are in agreement with the overall negative charge expected from the amino acid sequence of calmodulin (47). Comparison of the interaction between calmodulin and creatine kinase B in the presence of Ca2+ and EDTA reveals that the magnitude of change in μe upon binding to creatine kinase B is larger for apocalmodulin than for Ca2+-bound calmodulin. The mobility of the complex approaches comparable values under both sets of conditions; this finding may arise from charge-dependent modulation of pKa values in the complexes (48).
After the protein array screen and free-flow electrophoresis measurements, we probed the interaction between calmodulin and the target proteins creatine kinase and phosphorylase kinase by SPR (see Fig. 5 A and Fig. S3). Calmodulin containing the S17C mutation was site-specifically coupled to the dextrane matrix of the CM5 sensor chips via the thiol side chain of Cys-17, to allow the two domains of calmodulin to move freely relative to one another and cooperate in target binding. A series of creatine kinase B solutions with different concentrations were flowed over CM5 sensor chips with immobilized calmodulin. The SPR experiments were performed both in buffers containing Ca2+ and in buffer with EDTA, as shown in Fig. 5 A. We obtained values for the rate constants through fits to the association and dissociation data, as detailed in the Supporting Material, yielding kon = 7.3 × 102 M−1 s−1 and koff = 2.6 × 10−5 s−1 for creatine kinase B in the presence of calcium and kon = 3.5 × 102 M−1 s−1 and koff = 1.1 × 10−3 s−1 in the absence of free Ca2+. Relating these rate constants to the equilibrium dissociation constant provided us with values for the apparent equilibrium dissociation constant of Kd = 36 nM for creatine kinase B binding to Ca2+-loaded calmodulin and Kd = 3.1 μM for creatine kinase B binding to apocalmodulin on the sensor-chip surface at physiological salt concentration, 150 mM NaCl.
We investigated whether the observed difference in the binding affinity of calcium-bound calmodulin for creatine kinase B between the surface-based experiments and the measurements in solution was due to the changes in ionic strength. To probe the effect of low versus high salt concentrations, we investigated calmodulin binding to creatine kinase B through thermophoresis (see the Supporting Material). The thermophoresis measurements were performed in 0.1 mM CaCl2, 5 mM Tris-HCl, pH 8.0, with either no or 150 mM added KCl. Fitting the data from the thermophoresis measurements in the low-ionic-strength buffer resulted in Kd = 1.4 ± 1.8 μM. In the high-salt buffer, we found the Kd to be 1.7 ± 0.9 μM (see Fig. S5). Thus, when the binding affinity between Ca2+ -calmodulin and creatine kinase B was evaluated in free solution, we found the Kd to be the same within error for both low and physiological levels of salt.
Calmodulin binding has an effect on creatine kinase B activity
After the discovery and validation of the interaction between calmodulin and creatine kinase B in solution and on a surface, we addressed the question of whether the binding of calmodulin had an effect on the activity of creatine kinase B. To explore the potential role of calmodulin in modulating the function of creatine kinase B, we monitored the effect of calmodulin on creatine kinase B activity using 1H NMR. The conversion of ADP and creatine phosphate to ATP and creatine by creatine kinase was monitored via the decay of creatine phosphate signals and buildup of creatine signals. A clear difference in the reaction rate was observed between the presence and absence of Ca2+ -loaded calmodulin (Fig. 5 B). Over the range of investigated substrate concentrations (ADP and creatine phosphate were altered in parallel between 1 and 37 mM), calmodulin was found to activate creatine kinase and increase the rate of substrate conversion by up to sixfold. Moreover, the influence of calmodulin was more prominent at low substrate concentrations, and the initial rate versus substrate concentration changes from sigmoidal in the absence of calmodulin to hyperbolic in the presence of calmodulin. The latter case could be fitted by standard Michaelis-Menten kinetics ( = 11 ± 1 mM s−1, KM = 15 ± 2 mM), but without calmodulin, an additional Hill-coefficient of 3.9 ± 0.4 was needed to obtain a reasonable fit (apparent = 9 ± 1 mM s−1, apparent KM = 19 ± 5 mM). The effects on the apparent values for and KM are in the range of errors, but the difference in allosteric behavior is striking. Typical cellular concentrations at rest are 4 mM ATP, 0.013 mM ADP, 13 mM creatine, and 25 mM creatine phosphate (49). Thus, the activation by calmodulin occurs within a physiologically relevant range of substrate concentrations.
Discussion
To achieve a fundamental understanding of protein function, it is necessary not only to obtain a qualitative knowledge of the identities of interacting proteins, but also to achieve a quantitative insight into the affinity of the interactions. Microfluidic free-flow electrophoresis provides us with a general tool for the study of protein-ligand binding. Here, we have explored the ligand binding of calmodulin. Through the negative control of calbindin D9k, we were able to demonstrate the specificity of the binding assay. This assay is excellently suited to probe weak interactions between biomolecules, such as the binding of apocalmodulin to creatine kinase, an interaction that was below the cutoff used in the protein array screen. In addition, this solution-based technique is not liable to the artifacts associated with surface-based assays, and the sample can be readily recovered after passing through the microfluidic device.
The interaction between calmodulin and creatine kinase, discovered in this work and quantified using free-flow electrophoresis, suggests a number of implications for human energy metabolism. Indeed, creatine kinase is an important enzyme in tissues that rapidly consume ATP. Creatine phosphate, which has a higher phosphate-transfer potential than ATP, serves as an energy reservoir for the fast generation of ATP in such tissues. The activation of creatine kinase by Ca2+-bound calmodulin may provide a coupling to cellular Ca2+ oscillations and signaling. The discovery of a relatively moderate affinity means that these signals can be regulated by calcium oscillations in myocardial and neuronal cells.
In the free-flow electrophoresis study, we determined the Kd values for the equimolar interaction between calmodulin and individual subunits of its binding partners. Creatine kinase B is a homodimer (36), whereas phosphorylase kinase has four γ-subunits (26, 31). The measured Kd values for calmodulin binding to creatine kinase B and phosphorylase kinase in the presence of Ca2+ in solution are higher than those determined by SPR (see Figs. 2, 4, and S4). However, in the SPR experiments it was not possible to differentiate between a multisubunit protein bound to one or to multiple immobilized calmodulin molecules. Avidity of binding could thus lead to an overestimate of the binding affinity by surface-based assays. Simultaneous dissociation from all the calmodulin molecules would be required for the multimeric binding partner to leave the surface, leading to a reduction in the apparent dissociation rate.
A systematic comparison of protein binding affinity measurements in free solution, in the gas phase, and on a surface, has previously revealed that SPR measurements are liable to report a considerably lower Kd compared with alternative methods (7). Indeed, this report agrees with the findings from our study. Here, we found the same value for the Kd within error through two solution-based methods, whereas we arrived at a higher affinity between Ca2+-calmodulin and creatine kinase B by SPR. Another factor that might contribute to this effect is the common use of a dextran matrix for immobilization, which may lead to enhanced complex formation due to so-called crowding effects or additional avidity effects due to low-affinity binding of the target protein to the dextran (50).
In the case of apocalmodulin, the values for Kd were comparable: 9.8 μM by electrophoresis and 3.1 μM by SPR, whereas, in the presence of calcium, the difference is marked between the surface-based assay, Kd = 36 nM, and the experiments in solution, where the measured values for Kd were 2.0, 1.4, and 1.7 μM. These findings suggest that calcium-bound and apocalmodulin interact with creatine kinase through different binding modes (51).
The SPR measurements were performed at physiological salt concentration to minimize unspecific surface effects, whereas microfluidic free-flow electrophoresis can access low-ionic-strength solution conditions. The voltage drop across the electrophoresis channel is proportional to the current and inversely proportional to the conductance of the solution. To achieve readily measurable sample deflections, we have therefore chosen to use low-ionic-strength buffers in these experiments. Even for the low electric potentials applied above, we recorded sample deflections of tens to hundreds of micrometres. The specifications of the optical detection methods influence the resolution of the deflection measurements. In this case, the sample position at a given time can be determined with micrometer or submicrometer precision. The microfluidic device designs can be optimized for use with higher-ionic-strength solutions. For the setup discussed in this work, experiments with salt concentrations on the order of tens of millimolar would be practicable.
Fluorescence images were acquired under steady-state conditions. Lower sample concentrations than those studied above can therefore be accessed by increasing the exposure time, without the risk of photobleaching. The measured current at each step of a voltage ramp would typically be stable to 0.1 μA. The main source of variation in electrophoretic mobility measurements between microfluidic devices originated in the cell constant and buffer conductivity measurements.
Conclusions
In conclusion, we have developed a general and quantitative approach to probe protein binding equilibria in free solution. The microfluidic free-flow electrophoresis approach presented in this article offers many advantages to the validation and quantification of protein-ligand interactions, including low sample consumption and short analysis time. The method is generally applicable to probe weak and transient interactions between solvated macromolecules. This strategy does not rely on specific solution conditions or exploit particular properties of the sample molecules. We have shown that this electrophoresis method can be applied to detect and characterize specific binding events in solution in a rapid manner. Furthermore, we have quantitatively confirmed the protein microarray identification of a calmodulin creatine kinase interaction. We found that the binding affinity of calmodulin for creatine kinase B was increased in the presence of calcium ions, and we showed that the catalytic activity of creatine kinase was increased in the presence of calmodulin. The quantitative approach applied here has allowed us to determine μe, and from this Kd directly, without the need for complex fitting or reference measurements. This approach therefore has the potential to be applied in a large number of areas of basic and applied research, for example, to measure antibody-antigen binding in drug discovery and for rapid diagnostics. The results presented here therefore open up the possibility of fast and quantitative measurements of protein-protein interactions in free solution under native conditions.
Author Contributions
T.W.H., D.J.O., U.W., T.P.J.K., and S.L. designed the research. T.W.H., D.J.O., M.C.B., J.P., U.W., and S.L. performed the research. T.W.H., D.J.O., U.W., and S.L. contributed analytical tools. T.W.H., D.J.O., U.W., T.P.J.K., and S.L. analyzed the data, T.W.H., T.P.J.K., and S.L. wrote the manuscript. All authors commented on the manuscript.
Acknowledgments
We acknowledge financial support from the Biotechnology and Biological Sciences Research Council (T.W.H. and T.P.J.K.), the Newman Foundation, and the European Research Council (T.P.J.K. and S.L.), the Swedish Research Council (S.L., J.P., and M.C.B.), and the Science Foundation Ireland Investigator Programme (D.J.O.).
Editor: Rohit Pappu.
Footnotes
Supporting Materials and Methods and five figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30151-5.
Contributor Information
Tuomas P.J. Knowles, Email: tpjk2@cam.ac.uk.
Sara Linse, Email: sara.linse@biochemistry.lu.se.
Supporting Citations
References (52, 53) appear in the Supporting Material.
Supporting Material
References
- 1.Lomakin A., Teplow D.B., Benedek G.B. Quasielastic light scattering for protein assembly studies. Methods Mol. Biol. 2005;299:153–174. doi: 10.1385/1-59259-874-9:153. [DOI] [PubMed] [Google Scholar]
- 2.Benesch J.L.P., Aquilina J.A., Robinson C.V. Tandem mass spectrometry reveals the quaternary organization of macromolecular assemblies. Chem. Biol. 2006;13:597–605. doi: 10.1016/j.chembiol.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 3.Berggård T., Linse S., James P. Methods for the detection and analysis of protein-protein interactions. Proteomics. 2007;7:2833–2842. doi: 10.1002/pmic.200700131. [DOI] [PubMed] [Google Scholar]
- 4.Behnen P., Davis E., Linse S. Calcium-dependent interaction of calmodulin with human 80S ribosomes and polyribosomes. Biochemistry. 2012;51:6718–6727. doi: 10.1021/bi3005939. [DOI] [PubMed] [Google Scholar]
- 5.Guilliams T., El-Turk F., De Genst E. Nanobodies raised against monomeric α-synuclein distinguish between fibrils at different maturation stages. J. Mol. Biol. 2013;425:2397–2411. doi: 10.1016/j.jmb.2013.01.040. [DOI] [PubMed] [Google Scholar]
- 6.Paslawski W., Mysling S., Otzen D.E. Co-existence of two different α-synuclein oligomers with different core structures determined by hydrogen/deuterium exchange mass spectrometry. Angew. Chem. Int. Ed. Engl. 2014;53:7560–7563. doi: 10.1002/anie.201400491. [DOI] [PubMed] [Google Scholar]
- 7.Jecklin M.C., Schauer S., Zenobi R. Label-free determination of protein-ligand binding constants using mass spectrometry and validation using surface plasmon resonance and isothermal titration calorimetry. J. Mol. Recognit. 2009;22:319–329. doi: 10.1002/jmr.951. [DOI] [PubMed] [Google Scholar]
- 8.Abate A.R., Hung T., Weitz D.A. High-throughput injection with microfluidics using picoinjectors. Proc. Natl. Acad. Sci. USA. 2010;107:19163–19166. doi: 10.1073/pnas.1006888107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song Y., Liu Z., Shum H.C. Manipulation of viscous all-aqueous jets by electrical charging. Chem. Commun. (Camb.) 2013;49:1726–1728. doi: 10.1039/c3cc38094g. [DOI] [PubMed] [Google Scholar]
- 10.Wienken C.J., Baaske P., Duhr S. Protein-binding assays in biological liquids using microscale thermophoresis. Nat. Commun. 2010;1:100. doi: 10.1038/ncomms1093. [DOI] [PubMed] [Google Scholar]
- 11.Hannig K. Die trägerfreie kontinuierliche Elektrophorese und ihre Anwendung. Anal. Chem. 1961;181:244–254. [Google Scholar]
- 12.Harrison D.J., Manz A., Widmer H.M. Capillary electrophoresis and sample injection systems integrated on a planar glass chip. Anal. Chem. 1992;64:1926–1932. [Google Scholar]
- 13.Raymond D.E., Manz A., Widmer H.M. Continuous sample pretreatment using a free-flow electrophoresis device integrated onto a silicon chip. Anal. Chem. 1994;66:2858–2865. [Google Scholar]
- 14.Carbeck J.D., Colton I.J., Whitesides G.M. Protein charge ladders, capillary electrophoresis, and the role of electrostatics in biomolecular recognition. Acc. Chem. Res. 1998;31:343–350. [Google Scholar]
- 15.Herling T.W., Müller T., Knowles T.P.J. Integration and characterization of solid wall electrodes in microfluidic devices fabricated in a single photolithography step. Appl. Phys. Lett. 2013;102:184102. [Google Scholar]
- 16.Herling T.W., Arosio P., Knowles T.P.J. A microfluidic platform for quantitative measurements of effective protein charges and single ion binding in solution. Phys. Chem. Chem. Phys. 2015;17:12161–12167. doi: 10.1039/c5cp00746a. [DOI] [PubMed] [Google Scholar]
- 17.Köhler S., Weilbeer C., Belder D. PDMS free-flow electrophoresis chips with integrated partitioning bars for bubble segregation. Lab Chip. 2011;11:309–314. doi: 10.1039/c0lc00347f. [DOI] [PubMed] [Google Scholar]
- 18.Turgeon R.T., Fonslow B.R., Bowser M.T. Measuring aptamer equilibria using gradient micro free flow electrophoresis. Anal. Chem. 2010;82:3636–3641. doi: 10.1021/ac902877v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berridge M.J., Lipp P., Bootman M.D. The versatility and universality of calcium signalling. Nat. Rev. Mol. Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- 20.Clapham D.E. Calcium signaling. Cell. 2007;131:1047–1058. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 21.O’Connell D.J., Bauer M.C., Cahill D.J. Integrated protein array screening and high throughput validation of 70 novel neural calmodulin-binding proteins. Mol. Cell. Proteomics. 2010;9:1118–1132. doi: 10.1074/mcp.M900324-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuboniwa H., Tjandra N., Bax A. Solution structure of calcium-free calmodulin. Nat. Struct. Biol. 1995;2:768–776. doi: 10.1038/nsb0995-768. [DOI] [PubMed] [Google Scholar]
- 23.Babu Y.S., Bugg C.E., Cook W.J. Structure of calmodulin refined at 2.2 A resolution. J. Mol. Biol. 1988;204:191–204. doi: 10.1016/0022-2836(88)90608-0. [DOI] [PubMed] [Google Scholar]
- 24.Kretsinger R.H., Nockolds C.E. Carp muscle calcium-binding protein. II. Structure determination and general description. J. Biol. Chem. 1973;248:3313–3326. [PubMed] [Google Scholar]
- 25.Linse S., Johansson C., Forsén S. Electrostatic contributions to the binding of Ca2+ in calbindin D9k. Biochemistry. 1991;30:154–162. doi: 10.1021/bi00215a023. [DOI] [PubMed] [Google Scholar]
- 26.Priddy T.S., Middaugh C.R., Carlson G.M. Electrostatic changes in phosphorylase kinase induced by its obligatory allosteric activator Ca2+ Protein Sci. 2007;16:517–527. doi: 10.1110/ps.062577507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meador W.E., Means A.R., Quiocho F.A. Target enzyme recognition by calmodulin: 2.4 A structure of a calmodulin-peptide complex. Science. 1992;257:1251–1255. doi: 10.1126/science.1519061. [DOI] [PubMed] [Google Scholar]
- 28.Ikura M., Clore G.M., Bax A. Solution structure of a calmodulin-target peptide complex by multidimensional NMR. Science. 1992;256:632–638. doi: 10.1126/science.1585175. [DOI] [PubMed] [Google Scholar]
- 29.Clapperton J.A., Martin S.R., Bayley P.M. Structure of the complex of calmodulin with the target sequence of calmodulin-dependent protein kinase I: studies of the kinase activation mechanism. Biochemistry. 2002;41:14669–14679. doi: 10.1021/bi026660t. [DOI] [PubMed] [Google Scholar]
- 30.Aoyagi M., Arvai A.S., Getzoff E.D. Structural basis for endothelial nitric oxide synthase binding to calmodulin. EMBO J. 2003;22:766–775. doi: 10.1093/emboj/cdg078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cook A.G., Johnson L.N., McDonnell J.M. Structural characterization of Ca2+/CaM in complex with the phosphorylase kinase PhK5 peptide. FEBS J. 2005;272:1511–1522. doi: 10.1111/j.1742-4658.2005.04591.x. [DOI] [PubMed] [Google Scholar]
- 32.Yamniuk A.P., Vogel H.J. Calmodulin’s flexibility allows for promiscuity in its interactions with target proteins and peptides. Mol. Biotechnol. 2004;27:33–57. doi: 10.1385/MB:27:1:33. [DOI] [PubMed] [Google Scholar]
- 33.Junker J.P., Ziegler F., Rief M. Ligand-dependent equilibrium fluctuations of single calmodulin molecules. Science. 2009;323:633–637. doi: 10.1126/science.1166191. [DOI] [PubMed] [Google Scholar]
- 34.Stigler J., Ziegler F., Rief M. The complex folding network of single calmodulin molecules. Science. 2011;334:512–516. doi: 10.1126/science.1207598. [DOI] [PubMed] [Google Scholar]
- 35.Wallimann T., Wyss M., Eppenberger H.M. Intracellular compartmentation, structure and function of creatine kinase isoenzymes in tissues with high and fluctuating energy demands: the “phosphocreatine circuit” for cellular energy homeostasis. Biochem. J. 1992;281:21–40. doi: 10.1042/bj2810021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bong S.M., Moon J.H., Hwang K.Y. Structural studies of human brain-type creatine kinase complexed with the ADP-Mg2+-NO3−-creatine transition-state analogue complex. FEBS Lett. 2008;582:3959–3965. doi: 10.1016/j.febslet.2008.10.039. [DOI] [PubMed] [Google Scholar]
- 37.Christakos S. Mechanism of action of 1,25-dihydroxyvitamin D3 on intestinal calcium absorption. Rev. Endocr. Metab. Disord. 2012;13:39–44. doi: 10.1007/s11154-011-9197-x. [DOI] [PubMed] [Google Scholar]
- 38.Brostrom C.O., Hunkeler F.L., Krebs E.G. The regulation of skeletal muscle phosphorylase kinase by Ca2+ J. Biol. Chem. 1971;246:1961–1967. [PubMed] [Google Scholar]
- 39.Cohen P., Burchell A., Nairn C. Identification of the Ca2+-dependent modulator protein as the fourth subunit of rabbit skeletal muscle phosphorylase kinase. FEBS Lett. 1978;92:287–293. doi: 10.1016/0014-5793(78)80772-8. [DOI] [PubMed] [Google Scholar]
- 40.McDonald J.C., Whitesides G.M. Poly(dimethylsiloxane) as a material for fabricating microfluidic devices. Acc. Chem. Res. 2002;35:491–499. doi: 10.1021/ar010110q. [DOI] [PubMed] [Google Scholar]
- 41.Spiga M., Morino G.L. A symmetric solution for velocity profile in laminar flow through rectangular ducts. Int. Commun. Heat Mass Transfer. 1994;21:469–475. [Google Scholar]
- 42.Delaglio F., Grzesiek S., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 43.So J.-H., Dickey M.D. Inherently aligned microfluidic electrodes composed of liquid metal. Lab Chip. 2011;11:905–911. doi: 10.1039/c0lc00501k. [DOI] [PubMed] [Google Scholar]
- 44.Hellstrand E., Kukora S., Åkerfeldt K.S. Förster resonance energy transfer studies of calmodulin produced by native protein ligation reveal inter-domain electrostatic repulsion. FEBS J. 2013;280:2675–2687. doi: 10.1111/febs.12269. [DOI] [PubMed] [Google Scholar]
- 45.Bottomley P.A., Panjrath G.S., Weiss R.G. Metabolic rates of ATP transfer through creatine kinase (CK Flux) predict clinical heart failure events and death. Sci. Transl. Med. 2013;5:215re3. doi: 10.1126/scitranslmed.3007328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Linse S., Jönsson B., Chazin W.J. The effect of protein concentration on ion binding. Proc. Natl. Acad. Sci. USA. 1995;92:4748–4752. doi: 10.1073/pnas.92.11.4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watterson D.M., Sharief F., Vanaman T.C. The complete amino acid sequence of the Ca2+-dependent modulator protein (calmodulin) of bovine brain. J. Biol. Chem. 1980;255:962–975. [PubMed] [Google Scholar]
- 48.Lund M., Jönsson B. Charge regulation in biomolecular solution. Q. Rev. Biophys. 2013;46:265–281. doi: 10.1017/S003358351300005X. [DOI] [PubMed] [Google Scholar]
- 49.Berg J.M., Tymoczko J.L., Stryer L. 7th ed. W. H. Freeman; New York: 2010. Biochemistry. [Google Scholar]
- 50.Jiao M., Li H.T., Liang Y. Attractive protein-polymer interactions markedly alter the effect of macromolecular crowding on protein association equilibria. Biophys. J. 2010;99:914–923. doi: 10.1016/j.bpj.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yuan T., Walsh M.P., Vogel H.J. Calcium-dependent and -independent interactions of the calmodulin-binding domain of cyclic nucleotide phosphodiesterase with calmodulin. Biochemistry. 1999;38:1446–1455. doi: 10.1021/bi9816453. [DOI] [PubMed] [Google Scholar]
- 52.Waltersson Y., Linse S., Grundström T. Mutational effects on the cooperativity of Ca2+ binding in calmodulin. Biochemistry. 1993;32:7866–7871. doi: 10.1021/bi00082a005. [DOI] [PubMed] [Google Scholar]
- 53.Meyer W.L., Fischer E.H., Krebs E.G. Activation of skeletal muscle phosphorylase b kinase by Ca2+∗. Biochemistry. 1964;3:1033–1039. doi: 10.1021/bi00896a004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.