Summary
Numerous developmentally regulated genes in mouse embryonic stem cells (ESCs) are marked by both active (H3K4me3)- and polycomb group (PcG)-mediated repressive (H3K27me3) histone modifications. This bivalent state is thought to be important for transcriptional poising, but the mechanisms that regulate bivalent genes and the bivalent state remain incompletely understood. Examining the contribution of microRNAs (miRNAs) to the regulation of bivalent genes, we found that the miRNA biogenesis enzyme DICER was required for the binding of the PRC2 core components EZH2 and SUZ12, and for the presence of the PRC2-mediated histone modification H3K27me3 at many bivalent genes. Genes that lost bivalency were preferentially upregulated at the mRNA and protein levels. Finally, reconstituting Dicer-deficient ESCs with ESC miRNAs restored bivalent gene repression and PRC2 binding at formerly bivalent genes. Therefore, miRNAs regulate bivalent genes and the bivalent state itself.
Highlights
-
•
MicroRNAs contribute to the regulation of bivalent genes
-
•
Dicer deletion reduces PRC2 binding to bivalent gene promoters
-
•
MicroRNA reconstitution restores Ezh2 binding
-
•
MicroRNAs regulate bivalent genes and the bivalent state itself
In this article Merkenschlager and colleagues show that microRNAs of the miR-290–295 family are required for the regulation of bivalent genes in mouse embryonic stem cells. Unexpectedly, microRNAs are also important for the binding of PRC2 core components to the promoters of many bivalent genes, and hence for the bivalent state itself.
Introduction
MicroRNAs (miRNAs) are small, non-coding RNAs that negatively regulate the stability and translation of protein-coding transcripts through partial complementarity with their 3′ UTRs (Jaskiewicz and Filipowicz, 2008). There have been reports that miRNAs affect histone modifications or DNA methylation in mammalian cells (Kanellopoulou et al., 2005, Sinkkonen et al., 2008, Benetti et al., 2008, Nesterova et al., 2008, Han et al., 2007), although the mechanisms are often indirect. For example, miRNAs post-transcriptionally control negative regulators of DNA methyltransferases in mouse ESCs (Sinkkonen et al., 2008, Benetti et al., 2008, Nesterova et al., 2008).
In mouse ESCs a set of developmental regulator genes is characterized by the simultaneous presence of activating and repressive histone modifications (Azuara et al., 2006, Bernstein et al., 2006). This bivalent state is resolved during ESC differentiation (Mikkelsen et al., 2007), suggesting that bivalency keeps genes in a poised state to enable rapid activation or stable silencing upon differentiation (Pietersen and van Lohuizen, 2008, Voigt et al., 2013). Bivalent genes are prematurely expressed in ESCs that lack the PRC2 components EED (Azuara et al., 2006, Boyer et al., 2006) or SUZ12 (Pasini et al., 2004). PRC2-deficient ESCs remain viable and can self-renew (Pasini et al., 2007, Chamberlain et al., 2008, Shen et al., 2008) but show reduced developmental potential (Pasini et al., 2007, Chamberlain et al., 2008, Shen et al., 2008), and PRC2 mutations are lethal during post-implantation in vivo (Faust et al., 1995, O'Carroll et al., 2001, Pasini et al., 2004). Hence, polycomb group (PcG) repression of bivalent genes may safeguard the pluripotent state and the ability of ESCs to differentiate.
miRNAs affect key features of ESCs, including their characteristic cell-cycle behavior (Wang et al., 2008). miRNAs are critical for ESC pluripotency and differentiation (Miyoshi et al., 2011, Anokye-Danso et al., 2011), and somatic miRNAs facilitate differentiation by terminating the expression of pluripotency factors such as Nanog (Melton et al., 2010). Although much is known about ESC miRNAs, their impact on the regulation of bivalent genes has not been systematically investigated. To address this point we have analyzed the distribution of PRC2 core components and the regulation of bivalent genes in ESCs deficient in the RNase III enzyme DICER, which is required for the biogenesis of most miRNAs (Jaskiewicz and Filipowicz, 2008). We find that miRNAs are required for the binding of EZH2 and SUZ12 at many bivalent promoters, and therefore for the maintenance of the bivalent state. These data define an unexpected role for miRNAs in maintaining the bivalent state of developmental regulator genes in mouse ESCs.
Results and Discussion
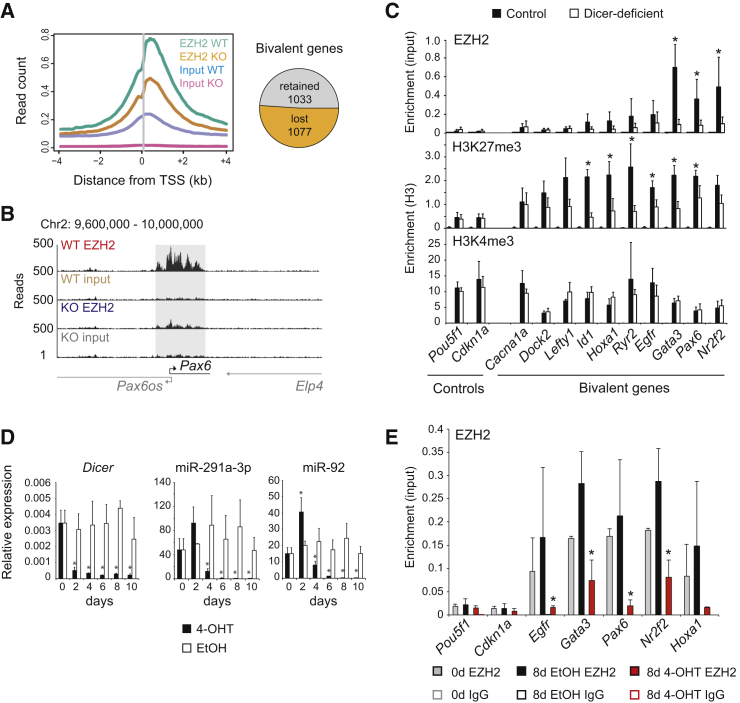
Previously characterized Dicer-deficient ESCs (Nesterova et al., 2008) were adapted to growth in feeder-free culture with inhibitors of MEK and GSK3β (see Experimental Procedures). We mapped the distribution of EZH2 by chromatin immunoprecipitation and high-throughput sequencing (ChIP-seq). Control ESCs retained EZH2 promoter binding at 2,110 bivalent genes (Ku et al., 2008) in control ESCs under 2i/LIF culture conditions. Loss of DICER resulted in reduced EZH2 occupancy of bivalent gene promoters (Figure 1A, left). The number of bivalent promoters was reduced to 1,033, reflecting the loss of EZH2 binding to 1,077 (or 51%) of formerly bivalent genes (Figure 1A, right) as illustrated for the Pax6 promoter (Figure 1B).
Figure 1.
DICER Maintains the Association of the Core PRC2 Component EZH2 with Many Bivalent Genes in ESCs
(A) EZH2 occupancy at transcriptional start sites of bivalent genes and numbers of EZH2-peaked bivalent genes that were retained (gray) or lost (orange) in Dicer-deficient ESCs. See Figure 4A for a complementary analysis of SUZ12 binding.
(B) ChIP-seq track of the Pax6 locus with highlighted EZH2 peak.
(C) ChIP-PCR of EZH2 (top), H3K27me3 (middle), and H3K4me3 (bottom) at selected genomic sites. Mean ± SD of three independent experiments. ∗p < 0.05 between control and Dicer-deficient cells. EZH2 enrichment is relative to input; H3K27me3 and H3K4me3 are relative to H3.
(D) Time-course analysis of Dicer deletion and loss of the miRNAs miR-291a-3p and miR-92 in Dicerlox/lox ERt2Cre ESCs treated with carrier (EtOH) or 4-OHT. Expression of Dicer mRNA is shown relative to Hprt; expression of miRNAs is shown relative to snoRNA-135. Mean ± SD of three independent experiments. ∗p < 0.05 (t test) between day 0 and the time points indicated.
(E) EZH2 occupancy of formerly bivalent gene promoters within 8 days of ERt2Cre activation. Mean ± SD of three independent experiments. ∗p < 0.05 (t test) between untreated cells (day 0) and cells treated with either EtOH or 4-OHT.
Loss of EZH2 was confirmed by ChIP-PCR (Figure 1C, top). At the sites we tested, the H3K27me3 chromatin mark deposited by PRC2 was reduced (Figure 1C, middle). As expected (Marks et al., 2012), the promoter mark H3K4me3 was unaffected (Figure 1C, bottom). To explore the kinetics of EZH2 loss from bivalent promoters, we induced the deletion of Dicer by treatment of ERt2Cre Dicerlox/lox ESCs with 4-hydroxytamoxifen (4-OHT). Dicer mRNA expression declined within 4 days of ERt2Cre induction, followed by the reduced expression of the miRNAs miR-291a-3p and miR-92 (Figure 1D). EZH2 occupancy of formerly bivalent promoters was reduced within 8 days (Figure 1E), indicating that EZH2 binding closely follows the reduced expression of miRNAs.
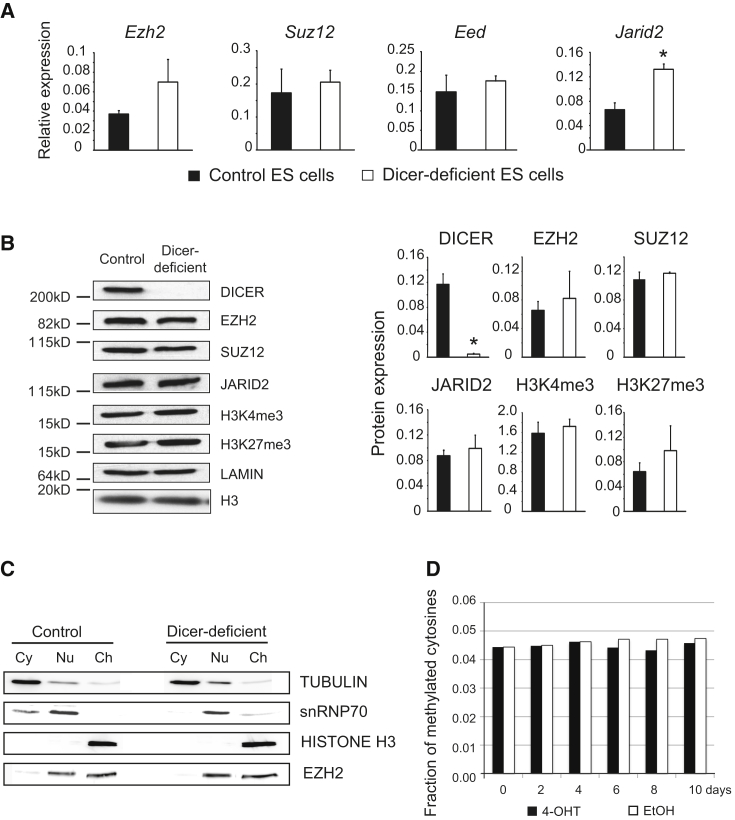
qRT-PCR showed that the expression of the PRC2 core components Ezh2, Suz12, and Eed was not reduced upon Dicer deletion (Figure 2A). Similarly immunoblotting showed no reduction of EZH2, SUZ12, or JARID2 proteins or global H3K4me3 or H3K27me3 (Figure 2B). Cellular fractionation experiments confirmed that total and chromatin-associated EZH2 protein remained unchanged (Figure 2C), suggesting selectively reduced binding of EZH2 at the promoters of bivalent genes.
Figure 2.
Dicer Deletion Does Not Diminish the Expression of PcG Proteins, the Association of EZH2 with Chromatin, or Global DNA Methylation in ESCs
(A) qRT-PCR of the PRC2 components Ezh2, Suz1, Eed, and Jarid2. Mean ± SD of three independent experiments. ∗p < 0.05 (t test) between control and Dicer-deficient cells.
(B) Immunoblots of the PRC2 components EZH2, SUZ12, EED, and JARID2 and the histone modifications H3K4me3 and H3K27me3 in control and Dicer-deficient ESCs. Left: representative blots. Right: densitometry readings of three independent experiments; ∗p < 0.05. DICER protein is a control.
(C) Immunoblots of cytoplasmic (Cy), nuclear soluble (Nu) and chromatin-associated (Ch) fractions of control and Dicer-deficient ESCs. Histone H3 identifies the chromatin fraction.
(D) ERT2Cre Dicerlox/lox ESCs were treated with either 4-OHT or carrier control (EtOH) and the fraction of methylated cytosine residues was followed by high-performance liquid chromatography.
See also Figure S1.
Exclusion of PcG from CpG-methylated regions may focus PcG binding to sites of low CpG methylation, namely CpG islands and CG-rich promoter regions, to facilitate the formation of bivalent domains (Ku et al., 2008, Lynch et al., 2012, Brinkman et al., 2012). Global DNA methylation levels are reduced in most Dicer-deficient ESC lines (Sinkkonen et al., 2008, Benetti et al., 2008, Nesterova et al., 2008, Ip et al., 2012) and DNA hypomethylation is linked to PcG redistribution in DNA methyltransferase (Dnmt)-deficient ESCs (Lynch et al., 2012, Brinkman et al., 2012, Cooper et al., 2014). PRDM14 controls Dnmt3b expression (Ma et al., 2011) and is upregulated under 2i conditions (Ying et al., 2008), but Prdm14 mRNA did not further increase in Dicer-deficient ESCs (log2 fold change −0.36, adjusted p = 0.446). Nevertheless, there were differences in Dnmt mRNA (Figure S1A) and DNMT protein (Figure S1B) expression between control and Dicer-deficient ESC under 2i/LIF culture conditions (Figure S1A). We therefore used high-performance liquid chromatography to directly monitor the fraction of methylated cytosine residues in response to Dicer deletion. Treatment of ERt2Cre Dicerlox/lox ESCs with 4-OHT did not affect the fraction of methylated cytosines (Figure 2D) even though miRNAs were efficiently depleted (Figure 1D) and EZH2 occupancy was reduced at bivalent promoters (Figures 1A–1D). Hence, the observed loss of bivalency was not due to globally reduced DNA methylation. Consistent with these data, ChIP-seq and ChIP-PCR analysis showed no substantial redistribution of EZH2 to repeat regions or to other genomic regions that had been reported to gain PRC2 binding in DNMT-deficient ESCs (Figures S1C–S1F).
We explored other potential mechanisms for the observed redistribution of PcG proteins in Dicer-deficient ESCs. First, although ERK2 is required for PRC2 occupancy and poised RNA polymerase at developmental genes (Tee et al., 2014), immunoblotting showed no significant differences in ERK1 and -2 protein expression between control and Dicer-deficient ESCs (Figure S1G). Second, NUP153 promotes PcG binding to developmentally regulated genes in ESCs (Jacinto et al., 2015), but our microarray and proteomics data show robust expression of Nup153 mRNA and NUP153 protein in both control and Dicer-deficient ESCs, without any indication of reduced expression in Dicer-deficient ESCs (GSE60161). Finally, PRC2 occupancy may reflect, rather than cause transcriptional repression (Riising et al., 2014). Eviction of PRC2 by transcription could account for the loss of EZH2 from some but not all of the formerly bivalent promoters in Dicer-deficient ESCs, since not all formerly bivalent genes showed increased expression of primary transcripts (see below).
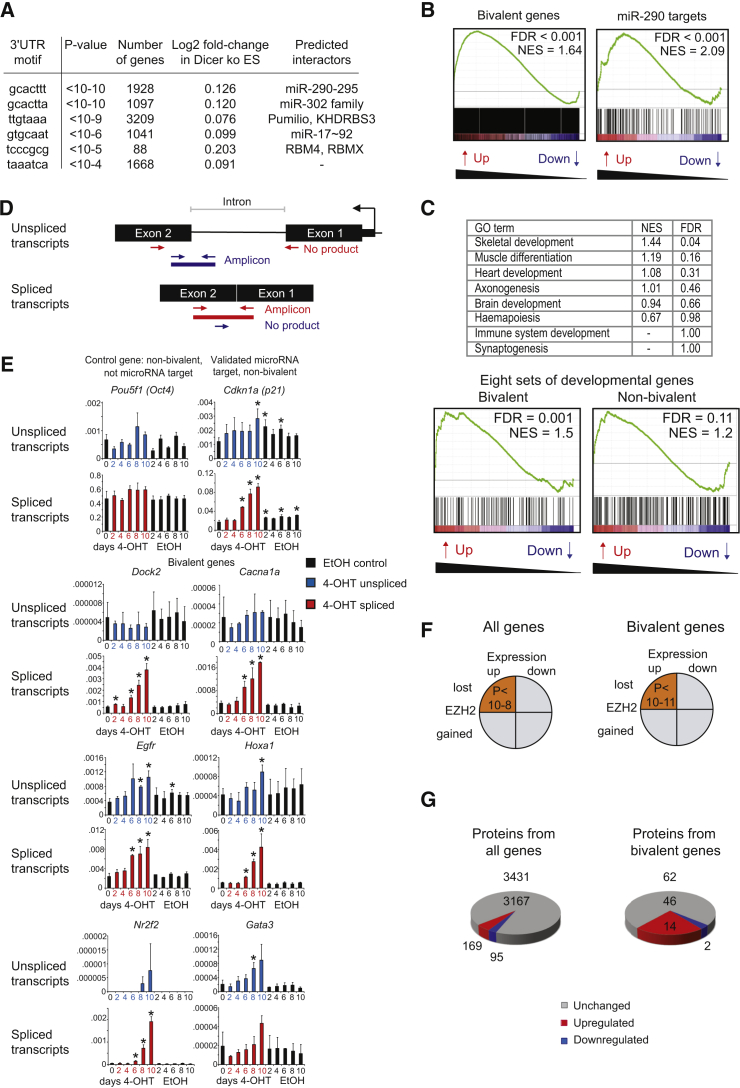
We profiled gene expression in control and Dicer-deficient ESCs and identified sequence motif enrichment in 3′ UTRs using miReduce. Highly enriched sequence motifs corresponded to binding sites for the ESC miRNAs miR-290–295 and miR-302, as well as miR-17-92 (Figure 3A). The miR-290–295 cluster is the most highly expressed miRNA population in ESCs (Zheng et al., 2011), and miR-290-3p, miR-291a-3p, miR-291b-3p, miR-292-3p, miR-294, and miR-295 contain the hexamer seed AAGUGC, which is shared by the miR-302 cluster. The miR-17-92 family contains the shifted seed AAAGUG and likely shares targets with miR-290–295 and miR-302 (Leung et al., 2011, Ciaudo et al., 2009, Babiarz et al., 2008). As expected, transcripts with 3′ UTR motifs for these abundant miRNAs were significantly upregulated in Dicer-deficient ESCs (Figure 3A). In addition, miReduce identified 3′ UTR motifs that matched binding preferences of RNA binding proteins (Figure S2A). We found the 3′ UTRs of bivalent genes enriched for miRNA motifs (p < 10−15, odds ratio = 2.91), including miR-290 (p = 1.17 × 10−6, odds ratio = 2.00) and miR-302 (p = 1.1 × 10−6, odds ratio = 1.95) (Figure S2A). Similar enrichment (p < 2.2 × 10−16, odds ratio = 2.22) was observed for an independently defined set of bivalent genes (Mikkelsen et al., 2007) and for the 2,110 bivalent genes that were bound by the EZH2 in our ESCs (p < 2.2 × 10−16, odds ratio = 3.18 for any miRNA motif; p < 2.2 × 10−16, odds ratio = 3.95 for miR-290; and p < 10−10, odds ratio = 4.32 for miR-302). The genomic basis for the increased occurrence of miRNA motifs was a greater than average 3′ UTR length, rather than a higher density of miRNA motifs in the 3′ UTRs of bivalent genes (Figure S2B).
Figure 3.
Accumulation of Spliced and Unspliced Transcripts and Proteins from Formerly Bivalent Genes in Dicer-Deficient ESCs
(A) Gene-expression profiles of Dicer-deficient ESCs were examined by miReduce for 3′ UTR sequence motif enrichment.
(B) GSEA of bivalent genes compared with miR-290 targets. FDR, false discovery rate; NES, normalized enrichment score.
(C) GSEA results for eight sets of developmentally regulated genes. Developmentally regulated genes were separated into bivalent and non-bivalent. FDR, false discovery rate; NES, normalized enrichment score.
(D) PCR strategy to distinguish primary (unspliced) from mature (spliced) transcripts.
(E) qRT-PCR analysis of primary transcripts (blue bars) and mature transcripts (red bars) in response to inducible ERt2Cre activation in Dicerlox/lox ESCs by 4-OHT. Black bars represent data for EtOH-treated control cells. Mean ± SD of three independent experiments. Expression is shown relative to Ubc, Hprt, and Rn18s. ∗p < 0.05 (t test).
(F) The relationship between changes in gene expression and EZH2 occupancy in Dicer-deficient ESCs. Orange shading indicates loss of EZH2 relative to increased global transcript levels (p < 10−8; left). This association was strongest for bivalent genes (p < 10−11; right). Gray shading indicates no significant association.
(G) SILAC quantified 3,431 protein groups, of which 169 (4.9%) were significantly upregulated and 95 (2.8%) significantly downregulated in Dicer-deficient ESCs (left). Of 62 proteins encoded by bivalent genes, 14 (23%) were upregulated and 2 (3%) downregulated (right). Proteins from bivalent genes were more likely to be deregulated than proteins from non-bivalent genes (odds ratio = 4.17). The odds for up- versus downregulation were 0.56 for all proteins and 0.14 for proteins from bivalent genes (odds ratio = 3.93).
See also Figures S2 and S3.
Gene set enrichment analysis (GSEA) of developmentally regulated genes showed that transcripts from bivalent genes were preferentially upregulated in Dicer-deficient ESCs, comparable with a set of Targetscan-predicted target genes of miR-290–295 (Figure 3B). Eight sets of developmental genes were used for comparison, and only two showed false discovery rates below 25% (Figure 3C, top). Each gene set contained a subset of bivalent genes, and separating bivalent from non-bivalent developmentally regulated genes showed significantly stronger upregulation of the bivalent subset (Figure 3C, bottom). The deregulated expression of bivalent genes was extensively validated by qRT-PCR (Figure S2C). To determine whether the deregulated expression of bivalent genes was due to the loss of miRNAs or other aspects of Dicer-deficiency, we examined gene expression in Dicer-deficient ESCs that had been reconstituted with miR-290 (Sinkkonen et al., 2008). miR-290 reconstitution significantly reduced the expression of bivalent genes (Figure S3A). qRT-PCR of Dicer-deficient ESCs reconstituted with miR-291a-3p in independent experiments confirmed this result (Figure S3B).
We employed RT-PCR primers specific for intronic sequences to assess the abundance of unspliced (primary) as well as spliced (mature) transcripts (see Figure 3D). In these experiments we followed the kinetics of transcript expression during the 4-OHT-induced deletion of Dicer in ERt2Cre Dicerlox/lox ESCs (see Figure 1D). Some bivalent genes showed selective upregulation of mature transcripts with no (e.g., Dock2) or little (e.g., Cacna1a) increase in primary transcript expression in Dicer-deficient ESCs (blue bars in Figure 3E). Others, including Nr2f2, showed significant upregulation of primary transcripts (blue bars in Figure 3E). Primary (blue bars in Figure 3E) and mature transcripts (red bars in Figure 3E) were deregulated with similar kinetics during the course of Dicer deletion (Figure 3E, data for EtOH-treated control cells are shown in black). There was no clear temporal dissociation between the deregulation of primary and mature transcripts. As miRNAs are thought to act mainly on mature, cytoplasmic transcripts in mammalian cells, the abundance of primary—predominantly nuclear—transcripts can be an indicator of transcriptional activity (Darnell, 2013). To discover whether miRNAs were directly involved in the regulation of primary transcripts, we transfected wild-type ESCs with locked nucleic acid (LNA) inhibitors of the miR-290–295 family. The LNA-mediated inhibition of miR-290–295 in wild-type ESCs recapitulated the deregulated expression of primary and mature bivalent gene transcripts (Figure S3C).
To define the relationship between EZH2 redistribution and gene expression, we integrated gene-expression and ChIP-seq data. The loss of EZH2 from promoters correlated with an increase in gene expression (p < 10−8). This correlation was particularly strong for bivalent genes (p < 10−11; Figure 3F).
SILAC-based quantitative proteomics studies were performed to address whether the increased expression of transcripts translated into altered protein expression. Control ESC extracts labeled with “heavy” amino acids were added 1:1 to either control or Dicer-deficient ESC extracts. We reliably quantified 3,431 protein groups in both control and Dicer-deficient ESCs (Figure 3G). Of these, 169 (4.9%) were significantly upregulated and 95 (2.8%) were significantly downregulated in Dicer-deficient ESCs (Figure 3G, left). Of 62 proteins encoded by bivalent genes 14 (23%) were upregulated, a much greater fraction than the 4.9% observed genome-wide (Figure 3G, right). Proteins encoded by two bivalent genes (3%) were downregulated in Dicer-deficient ESCs (Figure 3G, right). Hence, proteins encoded by bivalent genes were preferentially upregulated in Dicer-deficient ESCs (odds ratio = 3.93).
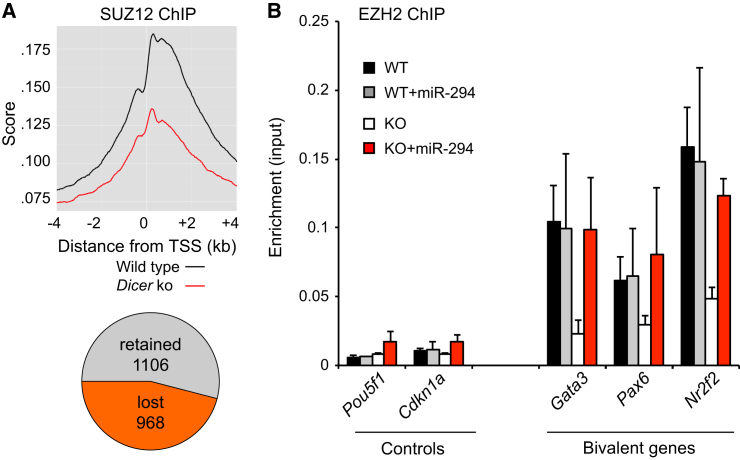
To explore whether the impact of DICER on bivalency extends to serum + LIF culture conditions, we analyzed ChIP-seq data for the PRC2 core component SUZ12 (Kanellopoulou et al., 2015). Similarly to Dicer deletion under semi-2i conditions, 968 (46.7%) of 2,074 bivalent genes showed reduced SUZ12 binding in Dicer-deficient ESCs in serum + LIF (Figure 4A). Hence, DICER is required for the maintenance of bivalency in ESCs in serum + LIF as well as in semi-2i conditions.
Figure 4.
miRNAs Rescue PRC2 Binding to Formerly Bivalent Promoters in Dicer-Deficient ESCs
(A) SUZ12 occupancy at transcriptional start sites of bivalent genes in control (black) and Dicer-deficient ESCs (red) in serum + LIF. 968 of 2,074 bivalent genes (46.7%) and 366 of 15,850 non-bivalent genes (2.3%) showed reduced SUZ12 binding in Dicer-deficient ESCs (p < 10−300).
(B) Control or Dicer-deficient ESCs were transfected with miR-294, a member of the ESC miRNA family miR-290–295. ChIP for the PRC2 core component EZH2 was done 2 days later and analyzed by real-time PCR (mean ± SE of two independent experiments).
To address whether the bivalent state requires miRNAs or other Dicer-dependent functions, we reconstituted Dicer-deficient ESCs with miR-294, a member of the miR-290–295 miRNA family. ChIP-PCR showed that EZH2 binding was restored 48 hr later at the Gata3, Pax6, and Nr2f2 promoters (Figure 4B). Thus, reconstitution of Dicer-deficient ESCs with a member of the 290–295 miRNA family was sufficient to rescue PRC2 occupancy of at least a subset of loci that lost the bivalent state in Dicer-deficient ESCs.
The expression of bivalent genes is regulated at multiple levels (Brookes et al., 2012, Margueron and Reinberg, 2011, Simon and Kingston, 2013, Jia et al., 2012). We have shown that transcripts from bivalent genes are preferentially targeted by miRNAs because their 3′ UTRs contain significantly more miRNA binding sites. The pluripotency-associated transcription factors OCT4, SOX2, and NANOG control miRNA expression in ESCs (Marson et al., 2008), linking the regulation of bivalent genes by ESC miRNAs to the ESC core regulatory network (Figure S4). An unexpected finding of our study is that miRNAs are required for the recruitment of PRC2 components to bivalent genes, and thereby the state of bivalency itself.
Experimental Procedures
Dicerlox/lox ERT2Cre mouse ESCs (Nesterova et al., 2008) were adapted to feeder-free conditions on gelatin-coated plates (0.1%) in 50% knockout DMEM, 10% fetal calf serum, and 50% neurobasal DMEM/F12, 2 mM L-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, 100 μM β-mercaptoethanol, 0.5 mM non-essential amino acids, 0.5% N2 supplement, 1% (v/v) B27 supplement, 1 μM PD032590, and 3 μM CHIR99021 (Stemgent) as described by Ying et al. (2008). For the conditional deletion of Dicer, Dicerlox/lox ERt2Cre ESCs were treated with 80 nM 4-OHT. To block miRNA function, we transfected ESCs with LNA inhibitors (500 nM; Exiqon) or negative control oligo (#199004-00, 500 nM) using Lipofectamine 2000 and assayed them 24 hr later. To mimic mRNA function, we transfected ESCs with miR-291a-3p mimic (C-310470-05, 100 nM) or control (CN-001000-01, 100 nM) oligos (Dharmacon) using DharmFECT Reagent #1 and assayed them 24 hr later. For rescue experiments, ESCs were transfected with miR-294-3p mimic (C-310474-03-0002, 100 nM, Dharmacon) using Lipofectamine 2000 and assayed 48 hr later. Details of materials and methods for gene expression, western blotting, and proteomics are given in Supplemental Experimental Procedures.
Acknowledgments
We thank Drs. James Elliott and Magda Opanowicz for cell sorting, Dr. Suzana Hadjur for cell culture, and Dr. Brian Hendrich for discussions.
Published: April 14, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, four figures, and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.03.005.
Accession Numbers
The accession number for the microarray and EZH2 ChIP-seq data reported in this paper is GEO: GSE60161, and the proteomics data are at ProteomeXchange (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=adqpgoaqpnehpwj&acc=GSE60161).
Supplemental Information
References
- Anokye-Danso F., Trivedi C.M., Juhr D., Gupta M., Cui Z., Tian Y., Zhang Y., Yang W., Gruber P.J., Epstein J.A. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuara V., Perry P., Sauer S., Spivakov M., Jørgensen H.F., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Babiarz J.E., Ruby J.G., Wang Y., Bartel D.P., Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benetti R., Gonzalo S., Jaco I., Munoz P., Gonzalez S., Schoeftner S., Murchison E., Andl T., Chen T., Klatt P. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat. Struct. Mol. Biol. 2008;15:268–279. doi: 10.1038/nsmb.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Boyer L.A., Plath K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Brinkman A.B., Gu H., Bartels S.J., Zhang Y., Matarese F., Simmer F., Marks H., Bock C., Gnirke A., Meissner A. Sequential ChIP-bisulfite sequencing enables direct genome-scale investigation of chromatin and DNA methylation cross-talk. Genome Res. 2012;12:1128–1138. doi: 10.1101/gr.133728.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes E., de Santiago I., Hebenstreit D., Morris K.J., Carroll T., Xie S.Q., Stock J.K., Heidemann M., Eick D., Nozaki N. Polycomb associates genome-wide with a specific RNA polymerase II variant, and regulates metabolic genes in ESCs. Cell Stem Cell. 2012;10:157–170. doi: 10.1016/j.stem.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain S.J., Yee D., Magnuson T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells. 2008;26:1496–1505. doi: 10.1634/stemcells.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciaudo C., Servant N., Cognat V., Sarazin A., Kieffer E., Viville S., Colot V., Barillot E., Heard E., Voinnet O. Highly dynamic and sex-specific expression of microRNAs during early ES cell differentiation. PLoS Genet. 2009;5:e1000620. doi: 10.1371/journal.pgen.1000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper S., Dienstbier M., Hassan R., Schermelleh L., Sharif J., Blackledge N.P., De Marco V., Elderkin S., Koseki H., Klose R. Targeting polycomb to pericentric heterochromatin in embryonic stem cells reveals a role for H2AK119u1 in PRC2 recruitment. Cell Rep. 2014;7:1456–1470. doi: 10.1016/j.celrep.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell J.E., Jr. Reflections on the history of pre-mRNA processing and highlights of current knowledge: a unified picture. RNA. 2013;19:443–460. doi: 10.1261/rna.038596.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust C., Schumacher A., Holdener B., Magnuson T. The Eed mutation disrupts anterior mesoderm production in mice. Development. 1995;121:273–285. doi: 10.1242/dev.121.2.273. [DOI] [PubMed] [Google Scholar]
- Han J., Kim D., Morris K.V. Promoter-associated RNA is required for RNA-directed transcriptional gene silencing in human cells. Proc. Natl. Acad. Sci. USA. 2007;104:12422–12427. doi: 10.1073/pnas.0701635104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip J., Canham P., Choo K.H., Inaba Y., Jacobs S.A., Kalitsis P., Mattiske D.M., Ng J., Saffery R., Wong N.C. DNA methylation dynamics in Dicer1-deficient mouse embryonic stem cells. PLoS Genet. 2012;8:e1002919. doi: 10.1371/journal.pgen.1002919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto F.V., Benner C., Hetzer M.W. The nucleoporin Nup153 regulates embryonic stem cell pluripotency through gene silencing. Genes Dev. 2015;29:1224–1238. doi: 10.1101/gad.260919.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskiewicz L., Filipowicz W. Role of Dicer in posttranscriptional RNA silencing. Curr. Top Microbiol. Immunol. 2008;320:77–97. doi: 10.1007/978-3-540-75157-1_4. [DOI] [PubMed] [Google Scholar]
- Jia J., Zheng X., Hu G., Cui K., Zhang J., Zhang A., Jiang H., Lu B., Yates J., 3rd, Liu C. Regulation of pluripotency and self- renewal of ESCs through epigenetic-threshold modulation and mRNA pruning. Cell. 2012;151:576–589. doi: 10.1016/j.cell.2012.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulou C., Muljo S.A., Kung A.L., Ganesan S., Drapkin R., Jenuwein T., Livingston D.M., Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulou C., Gilpatrick T., Kilaru G., Burr P., Nguyen C., Morawski A., Lenardo M. Reprogramming of polycomb-mediated gene silencing in embryonic stem cells by the miR-290 family and the methyltransferase Ash1l. Stem Cell Rep. 2015;5:971–978. doi: 10.1016/j.stemcr.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M., Koche R.P., Rheinbay E., Mendenhall E.M., Endoh M., Mikkelsen T.S., Presser A., Nusbaum C., Xie X., Chi A.S. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung A.K., Young A.G., Bhutkar A., Zheng G.X., Bosson A.D., Nielsen C.B., Sharp P.A. Genome-wide identification of Ago2 binding sites from mouse embryonic stem cells with and without mature microRNAs. Nat. Struct. Mol. Biol. 2011;18:237–244. doi: 10.1038/nsmb.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M.D., Smith A.J., De Gobbi M., Flenley M., Hughes J.R., Vernimmen D., Ayyub H., Sharpe J.A., Sloane-Stanley J.A., Sutherland L. An interspecies analysis reveals a key role for unmethylated CpG dinucleotides in vertebrate Polycomb complex recruitment. EMBO J. 2012;31:317–329. doi: 10.1038/emboj.2011.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z., Swigut T., Valouev A., Rada-Iglesias A., Wysocka J. Sequence-specific regulator Prdm14 safeguards mouse ESCs from entering extraembryonic endoderm fates. Nat. Struct. Mol. Biol. 2011;18:120–127. doi: 10.1038/nsmb.2000. [DOI] [PubMed] [Google Scholar]
- Margueron R., Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H., Kalkan T., Menafra R., Denissov S., Jones K., Hofemeister H., Nichols J., Kranz A., Stewart A.F., Smith A. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A., Levine S.S., Cole M.F., Frampton G.M., Brambrink T., Johnstone S., Guenther M.G., Johnston W.K., Wernig M., Newman J. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton C., Judson R.L., Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi N., Ishii H., Nagano H., Haraguchi N., Dewi D.L., Kano Y., Nishikawa S., Tanemura M., Mimori K., Tanaka F. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell. 2011;8:633–638. doi: 10.1016/j.stem.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Nesterova T.B., Popova B.C., Cobb B.S., Norton S., Senner C.E., Tang Y.A., Spruce T., Rodriguez T.A., Sado T., Merkenschlager M. Dicer regulates Xist promoter methylation in ES cells indirectly through transcriptional control of Dnmt3a. Epigenetics Chromatin. 2008;1:2. doi: 10.1186/1756-8935-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Carroll D., Erhardt S., Pagani M., Barton S.C., Surani M.A., Jenuwein T. The polycomb-group gene EZH2 is required for early mouse development. Mol. Cell Biol. 2001;21:4330–4336. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D., Bracken A.P., Jensen M.R., Lazzerini Denchi E., Helin K. SUZ12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D., Bracken A.P., Hansen J.B., Capillo M., Helin K. The polycomb group protein SUZ12 is required for embryonic stem cell differentiation. Mol. Cell Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietersen A.M., van Lohuizen M. Stem cell regulation by polycomb repressors: postponing commitment. Curr. Opin. Cell Biol. 2008;20:201–207. doi: 10.1016/j.ceb.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Riising E.M., Comet I., Leblanc B., Wu X., Johansen J.V., Helin K. Gene silencing triggers polycomb repressive complex 2 recruitment to CpG islands genome wide. Mol. Cell. 2014;55:347–360. doi: 10.1016/j.molcel.2014.06.005. [DOI] [PubMed] [Google Scholar]
- Shen X., Liu Y., Hsu Y.-J., Fujiwara Y., Kim J., Mao X., Yuan G.C., Orkin S.H. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol. Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J.A., Kingston R.E. Occupying chromatin: polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell. 2013;49:808–824. doi: 10.1016/j.molcel.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkkonen L., Hugenschmidt T., Berninger P., Gaidatzis D., Mohn F., Artus-Revel C.G., Zavolan M., Svoboda P., Filipowicz W. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 2008;15:259–267. doi: 10.1038/nsmb.1391. [DOI] [PubMed] [Google Scholar]
- Tee W.W., Shen S.S., Oksuz O., Narendra V., Reinberg D. Erk1/2 Activity promotes chromatin features and RNAPII phosphorylation at developmental promoters in mouse ESCs. Cell. 2014;156:678–690. doi: 10.1016/j.cell.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt P., Tee W.W., Reinberg D. A double take on bivalent promoters. Genes Dev. 2013;27:1318–1338. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Baskerville S., Shenoy A., Babiarz J.E., Baehner L., Blelloch R. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat. Genet. 2008;40:1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Q.-L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G.X., Ravi A., Calabrese J.M., Medeiros L.A., Kirak O., Dennis L.M., Jaenisch R., Burge C.B., Sharp P.A. A latent pro-survival function for the mir-290-295 cluster in mouse embryonic stem cells. PLoS Genet. 2011;7:e1002054. doi: 10.1371/journal.pgen.1002054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.