Abstract
Background
The H19/IGF2 imprinted loci have attracted recent attention due to their role in cellular differentiation and proliferation, heritable gene regulation, and in utero or early postnatal growth and development. Expression from the imprinted H19/IGF2 locus involves a complex interplay of three means of epigenetic regulation: proper establishment of DNA methylation, promoter occupancy of CTCF and expression of microRNA-675 (miR675). We have previously demonstrated in a multigenerational rat model of intrauterine growth restriction the epigenetic heritability of adult metabolic syndrome in a F2 generation. We have further demonstrated abrogation of the F2 adult metabolic syndrome phenotype with essential nutrient supplementation of intermediates along the one-carbon pathway, and shown that alterations in the metabolome precede the adult onset of metabolic syndrome. However, the upstream molecular and epigenomic mediators underlying these observations have yet to be fully elucidated.
Objective
In the current study, we sought to characterize the impact of the intrauterine growth restricted lineage and essential nutrient supplementation on both levels and molecular mediators of H19 and IGF2 gene expression in the F2 generation.
Study Design
F2 intrauterine growth restricted and sham lineages were obtained by exposing P1 (grandmaternal) pregnant dams to bilateral uterine artery ligation or sham surgery at gestational day 19.5. F1 pups were allocated to the essential nutrient supplemented or control diet at postnatal day 21, and bred at 6–7 weeks of age. Hepatic tissues from the resultant F2 offspring at birth and at weaning (day 21) were obtained. Bisulfite modification and sequencing was employed for methylation analysis. H19 and IGF2 expression was measured by QPCR. Promoter occupancy was quantified using chromatin immunoprecipitation, or ChIP, against CTCF insulator proteins.
Results
Growth-restricted F2 on control diet demonstrated significant down-regulation in H19 expression as compared to sham lineage (0.7831 vs 1.287; p< 0.05); however, essential nutrient supplementation diet abrogates this difference (4.995 vs 5.100; p>0.05). Conversely, Igf2 was up regulated by essential nutrient supplemented diet on the sham lineage (2.0 fold, p=0.01), an effect that was not observed in the growth restricted offspring. A significant differential methylation was observed in the promoter region of region H19 among the intrauterine growth restricted lineage (18% vs 25%; p<0.05) on a control diet, while the essential nutrient supplemented diet was alternately associated with hypermethylation in both lineages (sham: 50%; IUGR: 84%, p<0.05). Consistent with essential nutrient supplementation impacting the epigenome, a decrease of CTCF promoter occupancy was observed in CTCF4 of the growth restricted lineage (2.45% vs 0.56%; p<0.05) on the control diet, an effect that was repressed with essential nutrient supplementaion.
Conclusions
Heritable growth restriction is associated with changes in H19 gene expression; these changes are reversible with diet supplementation to favorably impact adult metabolic syndrome.
Keywords: IUGR, epigenomics, imprinting, histone modifications, CTCF, H19, insulin-like growth factor 2, miR-675
Condensation
Heritable growth restriction is associated with changes in H19 gene expression; these changes are reversible with diet supplementation to impact the adult metabolic syndrome favorably.
INTRODUCTION
The in utero environment is known to play a major role in the long term health of the offspring. According to the Developmental Origins of Health and Disease (DOHaD) hypothesis, an adverse in utero environment is associated with fetal programming, making the individual susceptible later in life to the onset of metabolic syndrome (MetS). This fetal programming is associated with epigenomic alterations1–5 and has been demonstrated to occur across multiple generations.12–17 Therefore, the identification of effective interventions during gestation to stop the cycle of the adult onset of disease is essential to ameliorate not only the health of the individual, but that of future generations.
IUGR resulting from uteroplacental insufficiency is an example of such an adverse in utero environment, where the fetus us subjected to hypoxia, acidosis and substrate deprivation.6, 7. We and others have shown that these individuals are at an increased risk of MetS in adulthood.8–11 Using our established model of uteroplacental insufficiency-induced fetal growth restriction,10, 11 we have shown that the growth restricted phenotype is multigenerational. In this model, late-gestation bilateral uterine artery ligation (or a control “sham” surgery) is performed on grandparental (P1) pregnant Sprague Dawley rats at embryonic day 19 (e19), and the F1 pups are delivered at e21.10, 11 The F1 generation exposed to the uterine artery ligation are born growth restricted compared to the offspring from the sham surgery. The F1 generation was allocated onto either a control diet, or an essential nutrient supplemented (ENS) diet at weaning (postnatal day 21, (D21)). The ENS diet is enriched with components of the one carbon metabolic pathway. These F1 pups bred spontaneously to yield the F2 generation. Of note, the IUGR lineage-F2 generation, born to mothers on the control diet, were growth restricted, even though no surgical intervention was performed on the F1 animals. In this model we have previously found that only at postnatal day 160 (D160) a gender specific MetS phenotype was apparent, with the males exhibiting obesity, increased central fat mass accumulation, glucose intolerance, insulin resistance, and increased triglyceride, VLDL, and fatty acids.10 No sex-specific differences were observed early in the F2 offspring early in life, at either birth (D0) or D21. This phenotype was only observed in the IUGR lineage animals with no ENS diet intervention. We have also found distinct serum metabolomes between the F2 D160 males exposed to either a control or ENS diet in utero.11
Such a multigenerational phenotype of IUGR begs the question: Are there epigenetic changes involved in the propagation of this phenotype? We and others have shown that IUGR is associated with epigenetic alterations in many tissues including liver.12–15, 5, 16, 17 Methylation levels are sensitive to the availability of the nutrients in the one carbon metabolism pathway including methionine, folic acid and choline.18 We therefore hypothesize that supplementation of the maternal diet with components of the one carbon metabolism pathway during gestation would be associated with changes in gene-specific DNA methylation levels in the offspring.
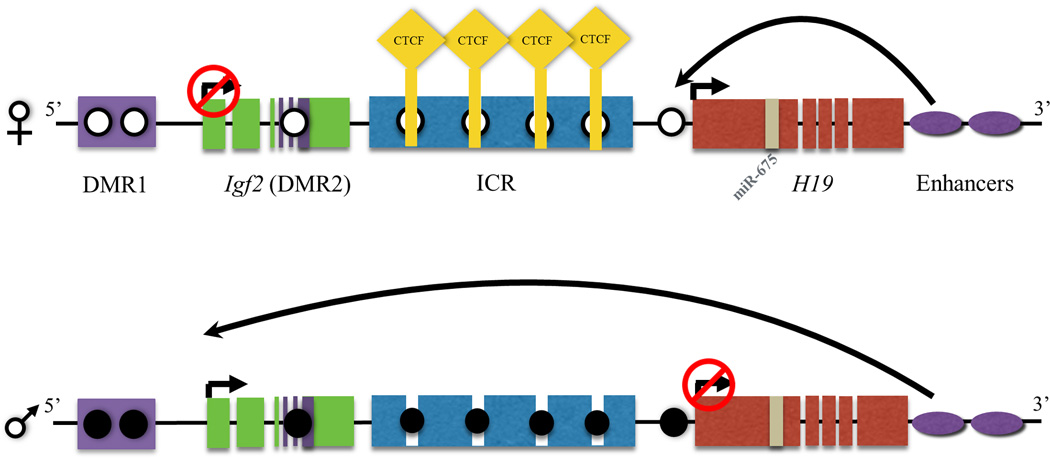
Expression from the imprinted H19/IGF2 locus involves a complex interplay of three means of epigenetic regulation: proper establishment of DNA methylation, promoter occupancy of CTCF and expression of microRNA-675 (miR675)19–26. DNA methylation is necessary for the establishment of genomic imprinting at this locus, an epigenetic mechanism leading to parent-of-origin monoallelic expression.27 Of note, the expression of imprinted genes is dictated by the parent of origin and not the sex of the offspring. Specific DNA methylation patterns throughout the Imprinting Control Region (ICR) of these imprinted genes are necessary for proper transcriptional regulation. Both H19 and IGF2 (insulin-like growth factor 2) are examples of imprinted genes integral to fetal growth and development. The IGF2 gene is expressed from the paternal allele throughout development,28 promoting fetal and placental growth. Alterations in Igf2 have also been implicated in postnatal growth control and the susceptibility to obesity.29–31. H19 is a long noncoding RNA (lncRNA) expressed in fetal life from the maternal allele, and thereafter repressed in early neonatal life.32 Within the first exon of H19 lies microRNA-675 (miR-675), which is expressed in the placenta and is involved in regulating placental growth.33 Within the H19 promoter lies a differentially methylated region (DMR) whose deletion in a murine model has been shown to completely disrupt H19 and IGF2 expression from this locus.34 This promoter region also contains multiple binding elements for the CTCF transcription factor.35–37 CTCF is a highly conserved transcription factor which can act as either a transcriptional activator or repressor35–37 (Figure 1). The function of CTCF varies by cell type and is regulated through an epigenetic mechanism.38,35
Figure 1. Insulator protein and epigenetic regulation on Igf2/H19 gene cluster.
In the mammalian genome the Igf2 and H19 are co-localized and undergo interdependent expression regulated by imprinting. Igf2 gene is transcribed from the paternal allele, while the non-coding H19 RNA is transcribed from the maternal allele. The regulation of expression from this locus involves a complex interplay of three specific epigenomic regulators, namely through DNA methylation, promoter occupancy of CTCF and expression of miR675. Epigenetic regulation is observed in this process where a demethylated (open circles) imprinted control region (ICR) binds CTCF (yellow diamond) to block the interaction of the downstream enhancers (purple ovals) with the promoter region of Igf2, thereby repressing the transcription of this gene, while the enhancers instead induce the transcription of H19. On the paternal allele, methylation (filled circle) on the ICR blocks the binding of CTCF allowing the interaction of the enhancers with the promoter of Igf2 facilitating gene transcription, while H19 is repressed due to methylation of its promoter region. miR675 is a microRNA expressed from exon one in the H19 gene.
We hypothesized that an adverse in utero environment would be associated with changes in transcription from the H19/IGF2 locus and that these changes could be ameliorated with supplementation of the maternal diet with components of the one carbon metabolic pathway. We utilized our transgenerational model of IUGR to determine the effects of growth restriction and ENS supplementation on this imprinted locus, and to specifically investigate the three epigenetic regulators at this locus: DNA methylation, CTCF occupancy, and miR675 expression.
METHODS
Heritable Multigenerational Model of IUGR
Our multigenerational model of fetal growth restriction in Sprague-Dawley rats has been previously described.11 Briefly, pregnant rats were randomly allocated assigned on e19 to receive either a sham surgery or bilateral uterine artery ligation with resultant F1 IUGR and control offspring born by cesarean on e21. Pups remained with their mothers after birth and throughout lactation. On D21, weaned F1 were allocated to control (Harlan Teklad®8640; Harlan Laboratories, Indianapolis, IN, USA) or to ENS (Harlan Teklad®8640 + folic acid, choline, B12, betaine, L-methionine, L-arginine, zinc) diet. Mating pairs were established with F1 generation on D80. The resultant offspring (F2) were maintained on the allocated diet of their parent. All experiments were approved by The University of Utah and Baylor College of Medicine Animal Care Committee(s).
cDNA analysis
The cDNA expression level of H19 and IGF2 in rat hepatic tissue was analyzed. Total RNA was extracted from D0, D21, and D160 of F2 generation. Approximately 30 mg of hepatic tissue were lysed using a chaotropic buffer and RNA was extracted using the Machery-Nagel NucleoSpin kit (Bethlehem, PA) and reverse-transcribed using the Applied Biosystems High Capacity cDNA Reverse Transcription Kit (Foster City, CA), as described before.39 RT-PCR analyses were then performed using 10µl cDNA and 2µM final concentration of forward and reverse primers in a fixed 30 cycle PCR reaction. The primers used were: H19, 5’-GAACATTTCCAGGGGAGTCA-3’ (forward) and 5’-CAGACATGAGCTGGGTAGCA-3’ (reverse); and IGF2, 5’-GGAAGTCGATGTTGGTGCTT-3’ (forward) and 5’-TTCACTGATGGTTGCTGGAC-3’ (reverse). Products were analyzed by agarose gel electrophoresis.
Real-Time RT-PCR
RNA was extracted from rat liver using TRIzol protocol (Invitrogen, Carlsbad, USA). H19, IGF2 and miR-675 mRNA abundance was analyzed via real-time RT-PCR using Icycler Thermal cycle (Bio-Rad, Hercules, CA). Real time quantitative RT-PCR analyses were performed as described above using 2µl cDNA and 2µM final concentration of forward and reverse primers and TaqMan probes in a total reaction volume of 5µL. We used the iQ5 Real-Time PCR Detection System from BioRad. Relative quantification of each gene was calculated after normalization to GAPDH by using the comparative CT method.40
Bisulfite Sequencing and Analysis
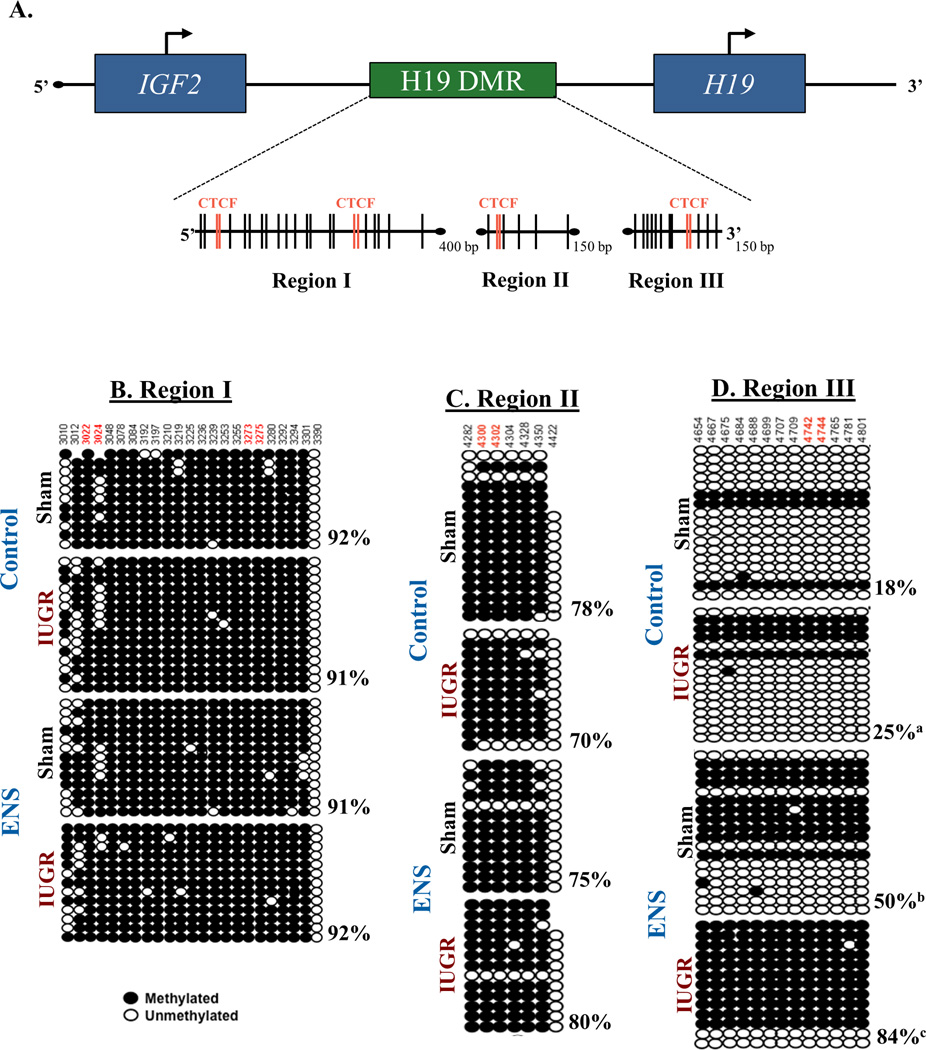
DNA was isolated from hepatic tissue using the DNeasy Blood and Tissue kit from Qiagen (Valencia, VA, USA). The purified DNA was subjected to bisulfite modification with the EZ DNA methylation kit (Zymo Research, Orange, CA). Bisulfite treated DNA was PCR amplified utilizing primers specific for three distinct regions of the H19 proximal imprinting control region (primers used are listed in Supplemental Table 1). The amplified PCR products were subsequently cloned using Stratagenes Strataclone system (San Diego, CA) and a minimum of eleven samples from each reaction were sequenced by Lone Star Labs (Houston, TX) and aligned using CLUSTAL W (Kyoto University). A representation of each individual CpG site, was created using ABI Methyl Prime software (Figure 4A).
Figure 4. Altered DNA methylation of the H19 differentially methylated regions (DMR) occurs among IUGR lineage rats.
(A) Three areas of the H19 DMR imprinting control region analyzed by are depicted. Vertical lines indicate CpG dinucleotides while red lines indicate the CpG falls within a CTCF finding site. (B–C) Methylation status was determined by sequencing cloned DNA following bisulfite treatment. The various regions of and their methylation status is depicted. While no changes were found in regions I and II, region III showed significant methylation changes at every site analyzed (p<0.02, Chi squared). Overall, IUGR induced a 40% increase in methylation, while IUGR fed ENS diet was increased 460%. (a, p<0.05; b, p<0.01; c, p<0.001).
Chromatin Immunoprecipitation (ChIP)
Chromatin immunoprecipitation assay was performed as previously described5 with the following modifications. Approximately 100mg of frozen D21 rat hepatic tissue from each animal was fixed at room temperature in 1% formaldehyde for 15 minutes. The reaction was quenched with 125mM glycine. Cells were lysed using a dounce homogenizer. Chromatin was sonicated using the Bioruptor sonicator at 4°C into 200–500 base pair fragments.
Approximately 100µg of chromatin was used for each reaction. Ten percent of the reaction was set aside before the addition of antibody. This served as the input. A mock IP was performed for each sample using IgG as a control. Five microliters of CTCF (Millipore, Billerica, MA) antibody were used per IP and incubated overnight with rotation at 4°C. Chromatin was purified by addition of Dynal Magnetic Beads conjugated to Protein A (Illumina, San Diego, CA). Beads were pulled down with a magnet, washed repeatedly and resuspended in elution buffer. Crosslinks were reversed by incubation at 65°C for five hours. DNA was purified using Qiaquick columns (Qiagen, Hilden, Germany).
Enrichment of promoter occupancy by virtue of histone modifications was determined using qPCR (primer sequences for the non-specifc site and the 4 CTCF binding sites can be found in Supplementary Table 1). PCR was performed using SYBR Green (Applied Biosystems) in the iCycler (BioRad). To determine percent IP, samples were first normalized to the input for each sample (ΔCt). Percent IP was calculated as 2−ΔCt.
Statistical Analysis
Univariate comparisons were performed using Chi-square or Fisher’s exact test for discrete variables and the Wilcoxon test for the continuous variables. Real Time PCR was analyzed using the 2−ΔΔCT method as previously described.40 Results are displayed as fold change of growth-restricted lineage on ENS versus control diet compared with sham lineage. An independent-sample t test was performed for each gene. An independent-sample 1-tailed t test was performed to analyze the differential methylation between groups. An independent-sample 2-tailed t test was performed for the ChIP analysis, comparing ENS with control diet. SPSS (SPSS, INC) was used for analyses and a nominal p-value <0.05 was considered significant.
RESULTS
H19 Expression is Restricted to Fetal and Early Neonatal Life
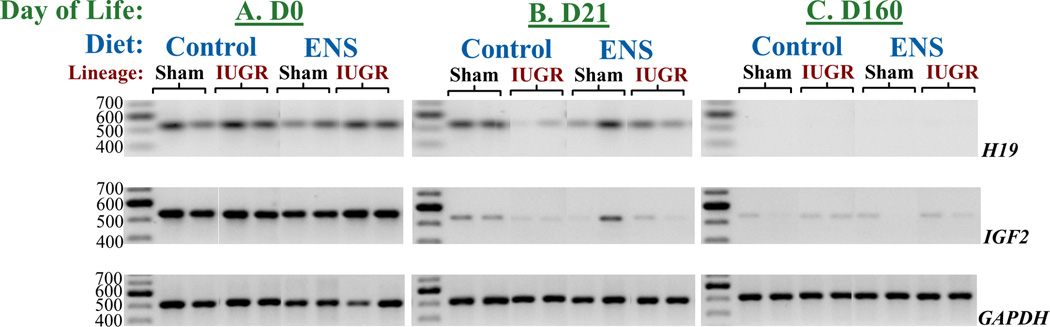
To analyze the developmental patterns of H19 expression, full-length transcripts were analyzed by RT-PCR. At birth (D0), H19 is ubiquitously expressed in both sham and IUGR lineage F2 offspring regardless of gestational diet (demonstrated by equal band intensity across all groups; Figure 2A). However, at the time of weaning (D21), we observed an unanticipated silencing of H19 gene expression in IUGR lineage rats on control diet (absence of bands in far right lanes; Figure 2B). Of interest, H19 expression persisted in IUGR lineage F2 animals born to dams on ENS diet. By adulthood (D160), H19 is ubiquitously silenced seen as absence of bands in all groups (Figure 2C). Thus, fetal growth restriction is associated with premature loss of H19 expression among IUGR lineage F2 offspring on control diet while expression appropriately persists in offspring exposed to an ENS diet. These same ENS-exposed IUGR lineages will be not develop adult MetS later in life.
Figure 2. Qualitative transcriptional changes of H19 and IGF2 by IUGR lineage and ENS supplemented diet.
H19 and IGF2 cDNA expression from both sham surgery and IUGR lineages on day of life 0, 21, and 160 in animals fed either a control or ENS diet. Both H19 and IGF2 expression is strongest during development. Expression decreasing significantly by weaning (D21) and nearly absent in adulthood (D160). At day 21, animals who are destined to develop metabolic syndrome have low H19 expression, while ENS rescued this diminished expression.
Such differential developmental silencing is uniquely restricted to H19, as IGF2 expression persists into adult life regardless of growth restricted lineage or diet, as evidenced by persistence of bands in D21 and D160 offspring (Figure 2B). As a loading control, GAPDH is equivalently amplified in all samples (Figure 2C).
Effect of ENS diet on Quantitative H19 and IGF2 Expression on F2 IUGR offspring
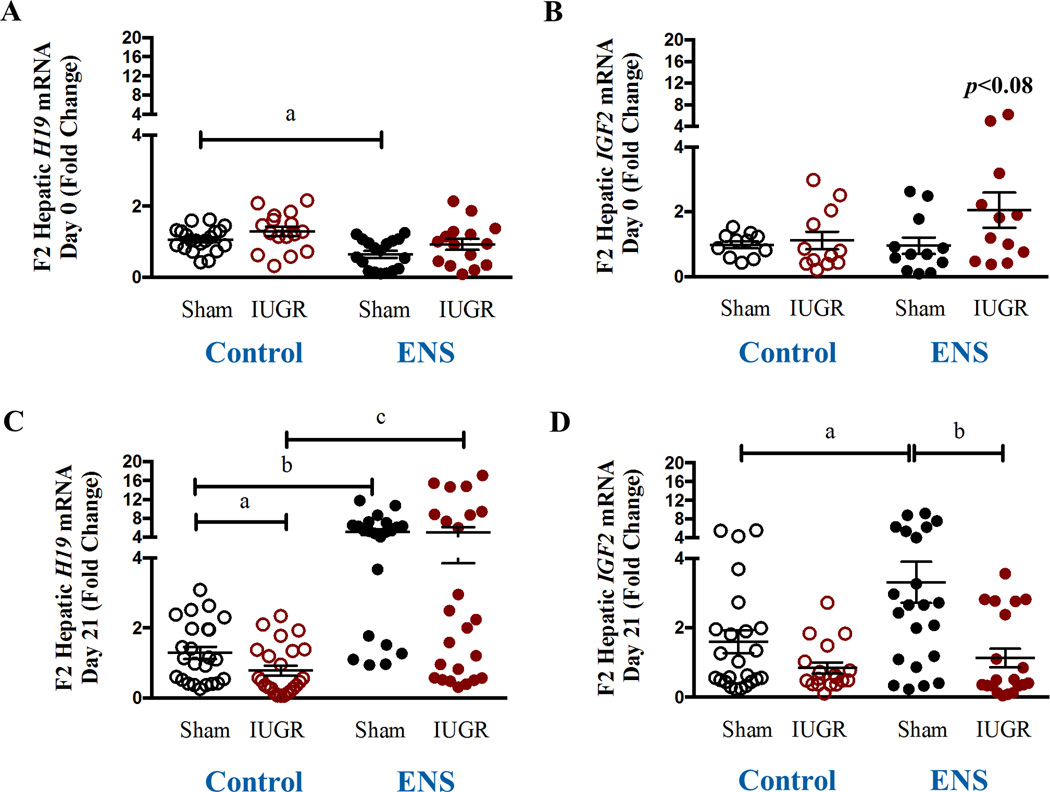
Hepatic expression of H19 and IGF2 were quantified using real time RT-PCR. While expression of H19 and IGF2 do not show a significant alteration in the IUGR lineage in the D0 offspring, expression of H19 is significantly decreased by virtue of the ENS diet in the Sham lineage (0.8 fold, p<0.002, Figure 3A). No effect was observed on the expression of IGF2 by virtue of lineage or diet at D0 (Figure 3B). At D21, H19 expression is significantly decreased in the IUGR lineage on the control diet (0.4 fold, p<0.05, Figure 3C) and significantly upregulated in both the Sham (4.0-fold, p<0.001) and IUGR lineage (6.6-fold, p<0.001) animals on the ENS diet (Figure 3C). At D21, IGF2 expression was up-regulated in the sham lineage on the ENS diet (2.0 fold, p=0.01, Figure 3D), an effect that was not observed in the growth restricted offspring. Therefore, maintaining the F1 generation on a diet rich in methyl donors is associated with relief of IUGR lineage H19 silencing in D21 F2 offspring and an up-regulation of IGF2 on sham lineage.
Figure 3. Quantiative H19 and IGF2 gene expression in association with IUGR lineage and ENS supplemented diet.
Expression of imprinted genes H19 and IGF2 from fetal rat liver was determined via real time PCR for (A) H19 D0 (a: 1.06±0.07 vs 0.8±0.1; p< 0.002, N=18–20) (B) IGF2 D0, (C) H19 D21 (a: 1.2±0.2 vs 0.7±0.1; p< 0.05, N=24; b: 1.2±0.2 vs 5.1±0.7; p < 0.0001, N22–24; c: 0.7±0.1 vs 5.1±0.7; p< 0.001, N=24) and (D) IGF2 D21 (a: 1.6 ± 0.3 vs 3.3 ± 0.6, N=22–24, p=0.01; b: P3.3 ± 0.6 vs 1.1 ± 0.3, N=20–22, p=0.002) in both sham and IUGR lineages on control versus ENS diet.
Hypermethylation of the Imprinting Control Region
Given the changes in H19 gene expression, we sought to investigate the mechanism underlying this differential regulation by characterizing the methylation pattern of the ICR. We analyzed three regions (Regions I-III) which contain 4 distinct CTCF binding elements (Figure 4A). In total, 43 CpG dinucleotide sites were analyzed within the designated regions: Region I (−3001 to −3396), Region II (−4278 to −4427) and Region III (−4622 to −4894) (Figure 4A).
Overall percent methylation was calculated for each group (Supplemental Table 2). There was no significant difference in methylation by virtue of lineage or diet in Region I of the ICR (Figure 4B). Similarly, Region II, no significant difference in CpG site-specific methylation was observed (Figure 4C). In contrast, we observed significant differential methylation in region III of the H19 ICR region by virtue of ENS supplementation. When compared to sham lineage on control diet, we observed a significant hypermethylation (18% vs 25% meCpG, p=0.019). F2 offspring given ENS diet, irrespective of lineage, also demonstrated significant hypermethylation in this region when compared to sham lineage on control diet (18% vs 50% meCpG for sham ENS p=0.002, 18% vs 84% meCpG IUGR ENS p<0.001; Figure 4D).
Ideally, studies which aim to examine parent of origin effects will utilize allele specific methylation marks to ascribe imprinting as either maternal or paternal. However, this is not possible among in-bred rat strains such as those used in our current study. As an alternative, we elected to examine for parent of origin effects by delineating the IUGR phenotype early in life as arising from either the maternal or paternal lineage. Fetal weight was measured at day 21 as a phenotypic measure of inherited changes following an IUGR lineage derived from either the maternal or paternal parent of origin (Supplemental Figure S1). For control fed male rats of IUGR linage there was a significantly lower weight for those with inherited IUGR through the paternal line (p<0.05) (Supplemental Figure S1). While for female rats the effect at day 21 was more pronounced with significantly decreased expression from either maternal or paternal inheritance (p<0.05 and p<0.001, respectively) (Supplemental Figure S1). However, ENS fed rats appeared to be lower than non ENS fed rats at day 21 regardless of treatment. The epigenetic inheritance of the IUGR phenotype though either maternal or paternal lineage in male and female rats in our model echoes our previous findings10, and is consistent with extensive metabolomic changes discovered in F2 generation IUGR lineage rats in serum at day 21, regardless of F2 gender11.
CTCF binding within the H19 ICR
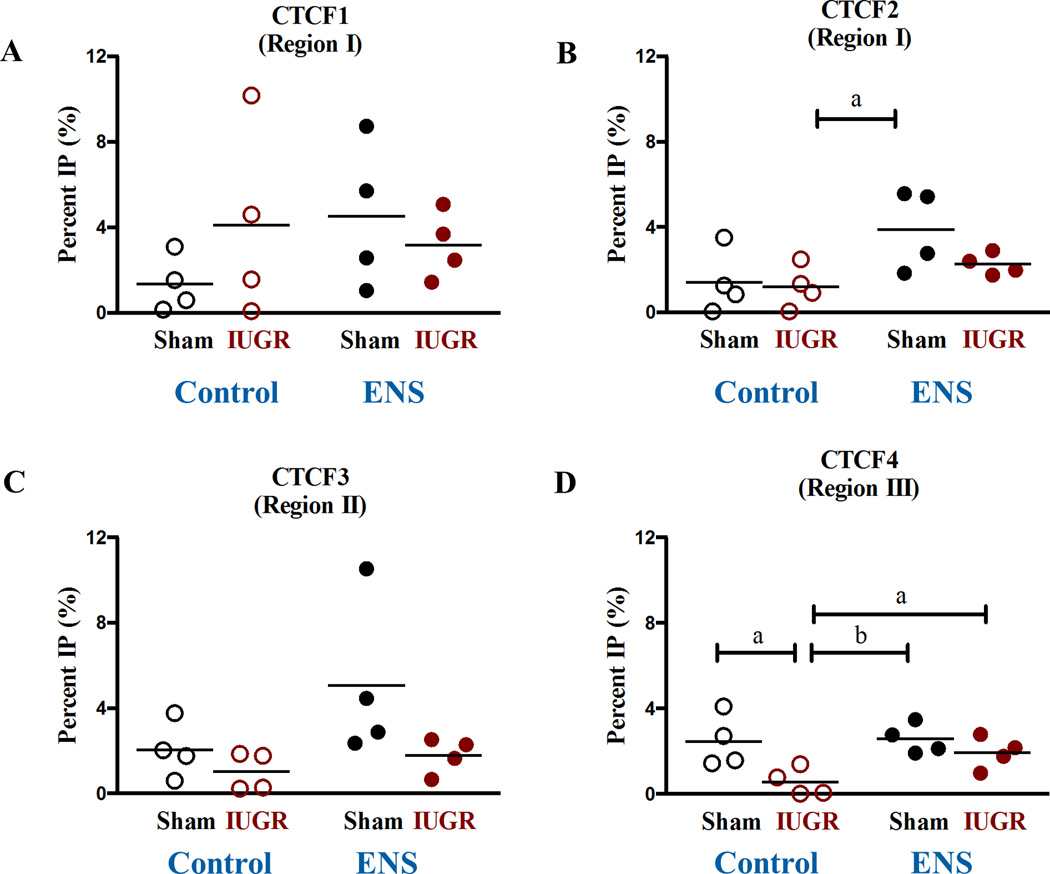
Given the above findings and the known mechanisms of epigenomic inheritance at this locus, we sought to characterize other means of epigenomic regulation in the ICR, namely through CTCF occupancy within the H19 promoter. CTCF occupancy was quantified utilizing chromatin immunoprecipitation (ChIP) with an antibody against the CTCF protein. qPCR was used to determine CTCF enrichment within the 4 known binding sites within the H19 ICR. At CTCF binding site 2, occupancy is increased between the Control diet IUGR animals and the ENS Sham lineage (Figure 5B). We observed a 77% decreased occupancy of CTCF at Site 4 in growth-restricted offspring on control diet compared to sham (2.4±0.6 vs 0.6±0.3, p<0.01, Figure 5D) as was expected due to the hypermethylation observed in region III (Figure 4D) while the supplemented diet increased the CTCF binding, 3.43 fold (Figure 5D), on the growth-restricted lineage as compared to control diet. No significant changes were observed in CTCF binding sites 1 and 3 (Figures 5A and C).
Figure 5. CTCF promoter occupancy altered in IUGR lineage rats.
Chromatin immunoprecipitation (ChIP) compared to a mock IgG was carried out for the various CTCF binding sites. A 77% decreased occupancy of CTCF at insulator site 4 (a: 2.4±0.6 vs 0.6±0.3, N=4, p<0.01) was induced by IUGR linage in D21 rats.
Down-regulation of miR-675 in F2 D21 offspring
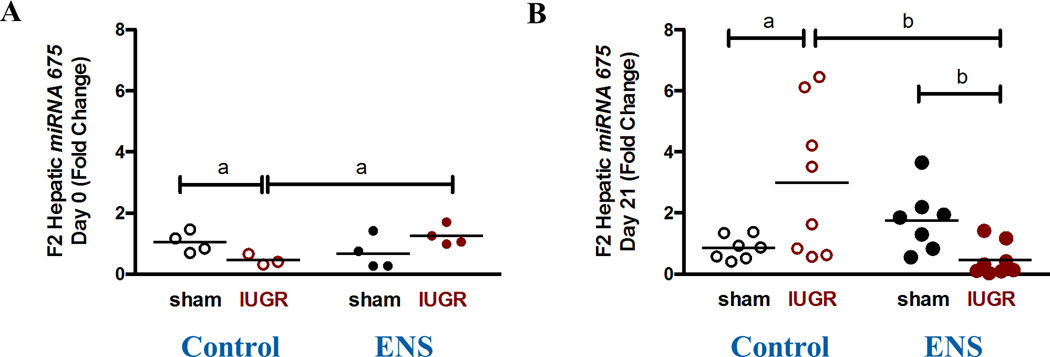
To quantify the expression of miR-675, real time RT-PCR was employed. We did not observe altered miR-675 expression of the F2 D0 offspring by virtue of lineage or diet (Figure 6A). However, at D21, we observed a significant down-regulation of mi-R675 expression (0.26-fold, p<0.05) among the IUGR lineage compared to the sham lineage on the ENS diet (Figure 6B).
Figure 6. Expression of miRNA-675.
The expression of miR-675 was determine by real time PCR for (A) D0 and (B) D21 (a: 1.7±0.4 vs 0.4±0.2, N=7–8, p=0.008). Its repression was decreased in IUGR animals fed an ENS diet at D21.
COMMENT
Principal findings
The H19/IGF2 imprinted loci have attracted recent attention due to their role in cellular differentiation and proliferation, heritable gene regulation, and in utero or early postnatal growth and development. Expression from the imprinted H19/IGF2 locus is a hotbed of epigenomic modulations, and involves a complex interplay of the three currently known means of epigenetic regulation: proper establishment of DNA methylation, promoter occupancy of CTCF and expression of microRNA-675 (miR675).
As a first step, we sought to examine gene transcription from the H19/IGF2 locus. Because the IUGR lineage is destined for MetS in the absence of the ENS diet supplementation, we hypothesized that these animals would have a reduced expression from the H19/IGF2 locus. Indeed, we found that compared to the sham lineage animals on the control diet, the IUGR lineage F2 animals from the control diet had a significantly reduced H19 expression (Figure 2 and 3C). We also observed an effect of the ENS diet, as both sham and IUGR lineage animals had a significantly increased H19 expression (Figure 3C).
Since we observed both a lineage and diet effect on H19 expression, we sought to determine if changes within the promoter could account for the differences in gene expression. To this end we interrogated the hepatic DNA methylation profile of 3 regions within the H19 promoter in the D21 F2 animals. We analyzed distinct regions known to include CTCF binding sites to determine if methylation within this region precludes CTCF binding (Figure 1). Although there were no changes in Regions I and II, DNA methylation was significantly altered in Region III (Figure 4). Similar to what we observe with H19 expression, DNA methylation is altered by virtue of lineage and diet. While we and others have reported that DNA methylation is sensitive to methyl donors in the diet, this finding was surprising as the alteration in methylation has occurred post-conception and within an imprinted region. Imprinting is established in fetal life and is a highly regulated process within the germline. However, our observations reveal evident postnatal plasticity of the methylation profile of an imprinted gene, resulting in differential gene expression and our previously reported significant alterations in the F2 metabolome and subsequent development of adult MetS.10, 11 This is an important discovery as it reveals a potential mechanism behind the ENS intervention in the prevention of the cycle of obesity in future generations in the growth restricted lineage.
CTCF occupancy within the ICR of H19 was further interrogated using chromatin immunoprecipitation. While we did not observe increased differential DNA methylation within region III excluded CTCF binding, we did find a decreased binding of this transcription factor in the IUGR lineage control diet fed animals. As previously noted, these animals are destined for MetS in adulthood. It is possible that the combination of alterations in DNA methylation and CTCF occupancy contribute to the decreased H19 expression which is associated with growth restriction and susceptibility to adult disease. We also observed that although we have increased methylation in the IUGR lineage ENS diet animals, an increased occupancy of CTCF (Figure 5D) may account for the increase in expression of H19 in these animals compared to the Sham lineage ENS diet animals. We did not find that alterations in expression of miR-675 account for the differences we observe in H19 expression, as miR-675 expression is only altered between the sham and IUGR lineage animals on the ENS diet (Figure 6).
In sum, we have determined that heritable growth restriction is associated with changes in H19 gene expression, and that these changes are also altered by virtue of diet supplementation. These changes in gene expression are associated with altered CTCF occupancy and differential methylation within the promoter region, and cannot be attributed to later transcriptional silencing with known miRNA effectors (e.g., miR-675).
This current study’sobservations has notable significance in relation to our prior work. Specifically, our previous studies revealed that F2 offspring from the IUGR lineage were destined to develop MetS in adulthood.10, 11 Similarly, we have previously observed that the ENS diet ameliorated the MetS phenotype such that the IUGR lineage offspring exposed to the ENS diet were no different from the sham lineage.10, 11 The present study significantly extends these findings at a molecular mechanistic level, characterizing the differential epigenetic regulation of the H19 imprinted locus among second generation IUGR rats. What is particularly novel about our current findings is the observation that although the H19 locus is classically described as imprinted, it is notably observed to be amenable to further alterations in both differential methylation and promoter occupancy.
When considered with our prior work10,11, there are several notable implications to our collective findings. First, our cumulative observations reveal a multigenerational growth restricted phenotype which is associated with the susceptibility to MetS in adulthood,10, 11 as well as a postnatal plasticity where an essential nutrient supplementation can rescue this phenotype. The current mechanistic studies reported herein yield insight into potential epigenetic mechanisms behind both the susceptibility to MetS as well as how ENS rescues the phenotype. Specifically, while we and others have shown that offspring of obese mothers are more likely to suffer from MetS in adulthood, an understanding of how this MetS phenotype could be inherited throughout multiple generations has been lacking. A major strength of this work is the ability to interrogate the epigenetic changes at a molecular level in the F2 generation and gain mechanistic insight into the propagation of this multigenerational phenotype as well as how the ENS diet reverses this phenotype. Furthermore, we have interrogated three epigenetic regulators involved in driving expression from this locus: changes in DNA methylation, CTCF occupancy and miR675 expression.
A potential limitation of this study is the inability to specifically follow the disruption of methylation in a parent of origin manner (e.g., allele specific maternal versus paternal imprinting). Previous studies in genetic mouse models over two decades have demonstrated that the H19/IGF2 locus is imprinted, and have carefully delineated the imprinting control region. Akin studies are not possible in inbred rat strains such as ours, since the maternal and paternal alleles are not distinct. In point of fact, that is a strength of our study, since our previous published findings demonstrated that the growth restricted phenotype, as observed by birth weight, can be transmitted following a P1 (grandmaternal) uterine artery ligation, through either the maternal or the paternal F1 lineages to the F2 offspring.10 Indeed, we have shown that ENS supplementation rescues the maternal IUGR lineage, consistent with parent of origin effect.10 We therefore hypothesize that disruption in methylation is likely transmitted through the paternal allele. These data are further supported by our findings at both a metabolite level11 and in the current study at D21 (Supplemental Figure S1), where we similarly observe that a reduction in weight is seen from both the maternal and paternal IUGR lineage in the control diet animals and a parent of origin difference is found with ENS supplementation. The changes in DNA methylation we have found warrant further study, as this reveals a postnatal plasticity in the regulation of an imprinted gene, and may provide mechanistic insight into how the ENS diet rescues the MetS phenotype of the IUGR lineage animals. Future studies aimed at illuminating the complex interplay of genomic-epigenomic-environmental interactions may help dissect multifactorial etiologies and identify at-risk populations for the common adverse pregnancy outcomes.
Supplementary Material
Acknowledgments
Funding. The effort and resources for this study was partially funded by the NIH (grant numbers K12HD00849, R01NR014792, DP2OD001500, R01DK089201A1 to K.M.A; K99HD075858-02 to M.A.S.) and the March of Dimes Prematurity Research Initiative (K.M.A).
Footnotes
Disclosure statement. The author(s) report(s) no conflict of interest.
SMFM Fast Track submission. Oral presentation (abstract #107) to be presented at 36th Annual Scientific Meeting of the Society of Maternal Fetal Medicine.
References
- 1.Gong L, Pan YX, Chen H. Gestational low protein diet in the rat mediates Igf2 gene expression in male offspring via altered hepatic DNA methylation. Epigenetics. 2010;5(7):619–626. doi: 10.4161/epi.5.7.12882. [DOI] [PubMed] [Google Scholar]
- 2.Sohi G, Marchand K, Revesz A, Arany E, Hardy DB. Maternal protein restriction elevates cholesterol in adult rat offspring due to repressive changes in histone modifications at the cholesterol 7alpha-hydroxylase promoter. Mol Endocrinol. 2011;25(5):785–798. doi: 10.1210/me.2010-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suter M, Bocock P, Showalter L, Hu M, Shope C, McKnight R, Grove K, Lane R, Aagaard-Tillery K. Epigenomics: maternal high-fat diet exposure in utero disrupts peripheral circadian gene expression in nonhuman primates. FASEB J. 2011;25(2):714–726. doi: 10.1096/fj.10-172080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng S, Rollet M, Pan YX. Maternal protein restriction during pregnancy induces CCAAT/enhancer-binding protein (C/EBPbeta) expression through the regulation of histone modification at its promoter region in female offspring rat skeletal muscle. Epigenetics. 2011;6(2):161–170. doi: 10.4161/epi.6.2.13472. [DOI] [PubMed] [Google Scholar]
- 5.Suter MA, Takahashi D, Grove KL, Aagaard KM. Postweaning exposure to a high-fat diet is associated with alterations to the hepatic histone code in Japanese macaques. Pediatr Res. 2013;74(3):252–258. doi: 10.1038/pr.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dimauro S, Garone C. Metabolic disorders of fetal life: glycogenoses and mitochondrial defects of the mitochondrial respiratory chain. Semin Fetal Neonatal Med. 2011;16(4):181–189. doi: 10.1016/j.siny.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 7.Parks WT. Placental hypoxia: the lesions of maternal malperfusion. Semin Perinatol. 2015;39(1):9–19. doi: 10.1053/j.semperi.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 8.Salam RA, Das JK, Bhutta ZA. Impact of intrauterine growth restriction on long-term health. Curr Opin Clin Nutr Metab Care. 2014;17(3):249–254. doi: 10.1097/MCO.0000000000000051. [DOI] [PubMed] [Google Scholar]
- 9.Dessi A, Pravettoni C, Cesare Marincola F, Schirru A, Fanos V. The biomarkers of fetal growth in intrauterine growth retardation and large for gestational age cases: from adipocytokines to a metabolomic all-in-one tool. Expert Rev Proteomics. 2015;12(3):309–316. doi: 10.1586/14789450.2015.1034694. [DOI] [PubMed] [Google Scholar]
- 10.Goodspeed D, Seferovic MD, Holland W, McKnight RA, Summers SA, Branch DW, Lane RH, Aagaard KM. Essential nutrient supplementation prevents heritable metabolic disease in multigenerational intrauterine growth-restricted rats. FASEB J. 2015;29(3):807–819. doi: 10.1096/fj.14-259614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seferovic MD, Goodspeed DM, Chu DM, Krannich LA, Gonzalez-Rodriguez PJ, Cox JE, Aagaard KM. Heritable IUGR and adult metabolic syndrome are reversible and associated with alterations in the metabolome following dietary supplementation of 1-carbon intermediates. FASEB J. 2015;29(6):2640–2652. doi: 10.1096/fj.14-266387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tosh DN, Fu Q, Callaway CW, McKnight RA, McMillen IC, Ross MG, Lane RH, Desai M. Epigenetics of programmed obesity: alteration in IUGR rat hepatic IGF1 mRNA expression and histone structure in rapid vs. delayed postnatal catch-up growth. Am J Physiol Gastrointest Liver Physiol. 2010;299(5):G1023–G1029. doi: 10.1152/ajpgi.00052.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joss-Moore LA, Wang Y, Ogata EM, Sainz AJ, Yu X, Callaway CW, McKnight RA, Albertine KH, Lane RH. IUGR differentially alters MeCP2 expression and H3K9Me3 of the PPARgamma gene in male and female rat lungs during alveolarization. Birth Defects Res A Clin Mol Teratol. 2011;91(8):672–681. doi: 10.1002/bdra.20783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suter MA, Chen A, Burdine MS, Choudhury M, Harris RA, Lane RH, Friedman JE, Grove KL, Tackett AJ, Aagaard KM. A maternal high-fat diet modulates fetal SIRT1 histone and protein deacetylase activity in nonhuman primates. FASEB J. 2012;26(12):5106–5114. doi: 10.1096/fj.12-212878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suter MA, Sangi-Haghpeykar H, Showalter L, Shope C, Hu M, Brown K, Williams S, Harris RA, Grove KL, Lane RH, Aagaard KM. Maternal high-fat diet modulates the fetal thyroid axis and thyroid gene expression in a nonhuman primate model. Mol Endocrinol. 2012;26(12):2071–2080. doi: 10.1210/me.2012-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suter MA, Ma J, Vuguin PM, Hartil K, Fiallo A, Harris RA, Charron MJ, Aagaard KM. In utero exposure to a maternal high-fat diet alters the epigenetic histone code in a murine model. Am J Obstet Gynecol. 2014;210(5):463, e461–e463, e411. doi: 10.1016/j.ajog.2014.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ke X, McKnight RA, Gracey Maniar LE, Sun Y, Callaway CW, Majnik A, Lane RH, Cohen SS. IUGR increases chromatin-remodeling factor Brg1 expression and binding to GR exon 1.7 promoter in newborn male rat hippocampus. Am J Physiol Regul Integr Comp Physiol. 2015;309(2):R119–R127. doi: 10.1152/ajpregu.00495.2014. [DOI] [PubMed] [Google Scholar]
- 18.Mandaviya PR, Stolk L, Heil SG. Homocysteine and DNA methylation: a review of animal and human literature. Mol Genet Metab. 2014;113(4):243–252. doi: 10.1016/j.ymgme.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 19.Ideraabdullah FY, Vigneau S, Bartolomei MS. Genomic imprinting mechanisms in mammals. Mutat Res. 2008;647(1–2):77–85. doi: 10.1016/j.mrfmmm.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tabano S, Colapietro P, Cetin I, Grati FR, Zanutto S, Mando C, Antonazzo P, Pileri P, Rossella F, Larizza L, Sirchia SM, Miozzo M. Epigenetic modulation of the IGF2/H19 imprinted domain in human embryonic and extra-embryonic compartments and its possible role in fetal growth restriction. Epigenetics. 2010;5(4):313–324. doi: 10.4161/epi.5.4.11637. [DOI] [PubMed] [Google Scholar]
- 21.Gillen AE, Harris A. The role of CTCF in coordinating the expression of single gene loci. Biochem Cell Biol. 2011;89(5):489–494. doi: 10.1139/o11-040. [DOI] [PubMed] [Google Scholar]
- 22.Ideraabdullah FY, Abramowitz LK, Thorvaldsen JL, Krapp C, Wen SC, Engel N, Bartolomei MS. Novel cis-regulatory function in ICR-mediated imprinted repression of H19. Dev Biol. 2011;355(2):349–357. doi: 10.1016/j.ydbio.2011.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weth O, Renkawitz R. CTCF function is modulated by neighboring DNA binding factors. Biochem Cell Biol. 2011;89(5):459–468. doi: 10.1139/o11-033. [DOI] [PubMed] [Google Scholar]
- 24.Matouk I, Raveh E, Ohana P, Lail RA, Gershtain E, Gilon M, De Groot N, Czerniak A, Hochberg A. The increasing complexity of the oncofetal h19 gene locus: functional dissection and therapeutic intervention. Int J Mol Sci. 2013;14(2):4298–4316. doi: 10.3390/ijms14024298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loke YJ, Galati JC, Saffery R, Craig JM. Association of in vitro fertilization with global and IGF2/H19 methylation variation in newborn twins. J Dev Orig Health Dis. 2015;6(2):115–124. doi: 10.1017/S2040174415000161. [DOI] [PubMed] [Google Scholar]
- 26.Leeuwerke M, Eilander MS, Pruis MG, Lendvai A, Erwich JJ, Scherjon SA, Plosch T, Eijsink JJ. DNA Methylation and Expression Patterns of Selected Genes in First Trimester Placental Tissue from Pregnancies with Small for Gestational Age Infants at Birth. Biol Reprod. 2016 doi: 10.1095/biolreprod.115.131698. [DOI] [PubMed] [Google Scholar]
- 27.Gabory A, Ripoche MA, Le Digarcher A, Watrin F, Ziyyat A, Forne T, Jammes H, Ainscough JF, Surani MA, Journot L, Dandolo L. H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development. 2009;136(20):3413–3421. doi: 10.1242/dev.036061. [DOI] [PubMed] [Google Scholar]
- 28.Dechiara TM, Robertson EJ, Efstratiadis A. Parental imprinting of the mouse insulin-like growth factor II gene. Cell. 1991;64(4):849–859. doi: 10.1016/0092-8674(91)90513-x. [DOI] [PubMed] [Google Scholar]
- 29.Deodati A, Inzaghi E, Liguori A, Puglianiello A, Germani D, Brufani C, Fintini D, Cappa M, Barbetti F, Cianfarani S. IGF2 methylation is associated with lipid profile in obese children. Horm Res Paediatr. 2013;79(6):361–367. doi: 10.1159/000351707. [DOI] [PubMed] [Google Scholar]
- 30.Hernandez-Valero MA, Rother J, Gorlov I, Frazier M, Gorlova O. Interplay between polymorphisms and methylation in the H19/IGF2 gene region may contribute to obesity in Mexican-American children. J Dev Orig Health Dis. 2013;4(6):499–506. doi: 10.1017/S204017441300041X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morita S, Horii T, Kimura M, Arai Y, Kamei Y, Ogawa Y, Hatada I. Paternal allele influences high fat diet-induced obesity. PLoS One. 2014;9(1):e85477. doi: 10.1371/journal.pone.0085477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bartolomei MS, Zemel S, Tilghman SM. Parental imprinting of the mouse H19 gene. Nature. 1991;351(6322):153–155. doi: 10.1038/351153a0. [DOI] [PubMed] [Google Scholar]
- 33.Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, Reik W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol. 2012;14(7):659–665. doi: 10.1038/ncb2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thorvaldsen JL, Duran KL, Bartolomei MS. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 1998;12(23):3693–3702. doi: 10.1101/gad.12.23.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanduri C, Pant V, Loukinov D, Pugacheva E, Qi CF, Wolffe A, Ohlsson R, Lobanenkov VV. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr Biol. 2000;10(14):853–856. doi: 10.1016/s0960-9822(00)00597-2. [DOI] [PubMed] [Google Scholar]
- 36.Dunn KL, Davie JR. The many roles of the transcriptional regulator CTCF. Biochem Cell Biol. 2003;81(3):161–167. doi: 10.1139/o03-052. [DOI] [PubMed] [Google Scholar]
- 37.Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128(6):1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ideraabdullah FY, Thorvaldsen JL, Myers JA, Bartolomei MS. Tissue-specific insulator function at H19/Igf2 revealed by deletions at the imprinting control region. Hum Mol Genet. 2014;23(23):6246–6259. doi: 10.1093/hmg/ddu344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suter M, Abramovici A, Showalter L, Hu M, Shope CD, Varner M, Aagaard-Tillery K. In utero tobacco exposure epigenetically modifies placental CYP1A1 expression. Metabolism. 2010;59(10):1481–1490. doi: 10.1016/j.metabol.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.