Abstract
PHD2 is a 2-oxoglutarate, non-heme Fe2+ dependent oxygenase that senses O2 levels in human cells by hydroxylating two prolyl residues in the oxygen dependent degradation domain (ODD) of HIF1α. Identifying the active site contacts that determine the rate of reaction under limiting O2 is crucial for understanding how these enzymes sense pO2, and may suggest methods for chemically altering hypoxia responses. A hydrogen bonding network extends from the Fe(II) cofactor through ordered waters to the Thr387 residue in the second coordination sphere. Here we tested the impact of the sidechain of Thr387 on the reactivity of PHD2 toward O2 through a combination of point mutagenesis, steady state kinetic experiments and {FeNO}7 EPR spectroscopy. The steady state kinetic parameters for Thr387→Asn were very similar to those of WT-PHD2, but kcat and kcat/KM(O2) for Thr387→Ala were increased by roughly 15-fold. X-band EPR spectroscopy of the {FeNO}7 centers of the (Fe+NO+2OG) enzyme forms showed the presence of a more rhombic line shape in Thr387→Ala than seen for WT-PHD2, indicating an altered conformation for bound gas in this variant. Here we show that the sidechain of residue Thr387 plays a significant role in determining the rate of turnover by PHD2 at low [O2].
Keywords: HIF, hypoxia, non-heme iron, 2-oxoglutarate, HIF-prolyl hydroxylase
Graphical abstract
Introduction
HIF prolyl-4-hydroxylase 2 (PHD2) is a non heme Fe(II), 2-oxoglutarate (2OG) dependent oxygenase that serves as the primary O2 sensor in human cells1,2. PHD2 regulates the transcriptional activity of the hypoxia-inducible factor-1α (HIF-1α) transcription factor by hydroxylating Pro402 and Pro564 within the oxygen dependent degradation domain (ODD) of HIF-1α1,3–5. As HIF-1α hydroxylation underlies angiogenesis and the balance between aerobic/anaerobic metabolism, PHD2 plays a crucial role in health conditions such as ischemia, anemia, and cancer6,7. Identifying the structural features of PHD2 that determine the reaction rate under conditions of limiting [O2] could point the way to altering cellular hypoxia sensing.
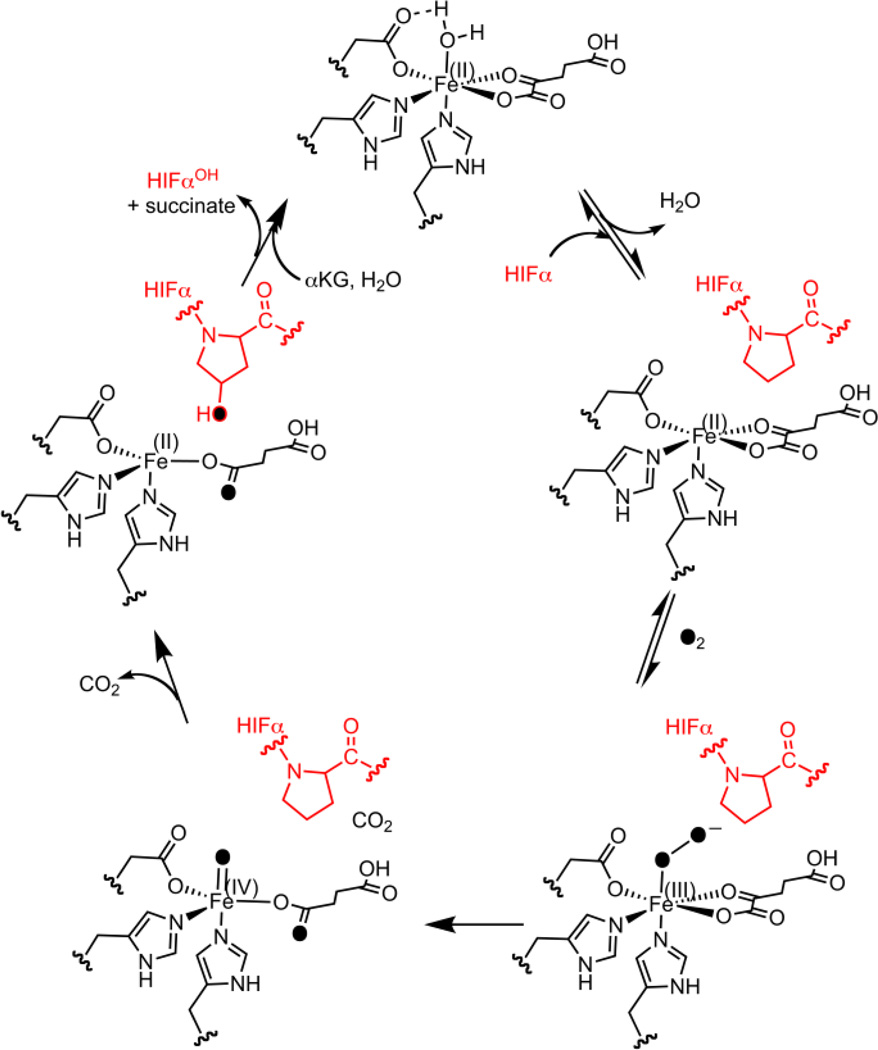
The (Fe+2OG)PHD2 form of enzyme contains an Fe(II) cofactor coordinated by a His2Asp facial triad, a bidentate 2OG ligand, and an aquo ligand. PHD2 is thought to follow the consensus chemical mechanism for 2OG oxygenases (Scheme 1).8–10 When substrate ODD binds near the cofactor, the aquo ligand is released in analogy to other 2OG oxygenases, permitting O2 to bind at the Fe for subsequent chemistry. Oxidative decarboxylation generates the ferryl (FeO)2+, an intermediate that has been observed in related enzymes,8,9,11,12 which hydroxylates the Pro target residue to complete the chemical steps of turnover (Scheme 1). Notably, PHD2 appears to be rate-limited by a step associated with O2 binding or oxidative decarboxylation, as the (FeO)2+ intermediate did not accumulate in the pre-steady state for PHD2,10 and PHD2 exhibited an inverse solvent kinetic isotope effect (KSIE).13,14 This is in contrast to other 2OG oxygenases that accumulate the (FeO)2+ in the pre-steady state, for which product release or hydroxylation appear to be rate-limiting step.8,9,11,12
Scheme 1.
Consensus chemical mechanism for PHD2.
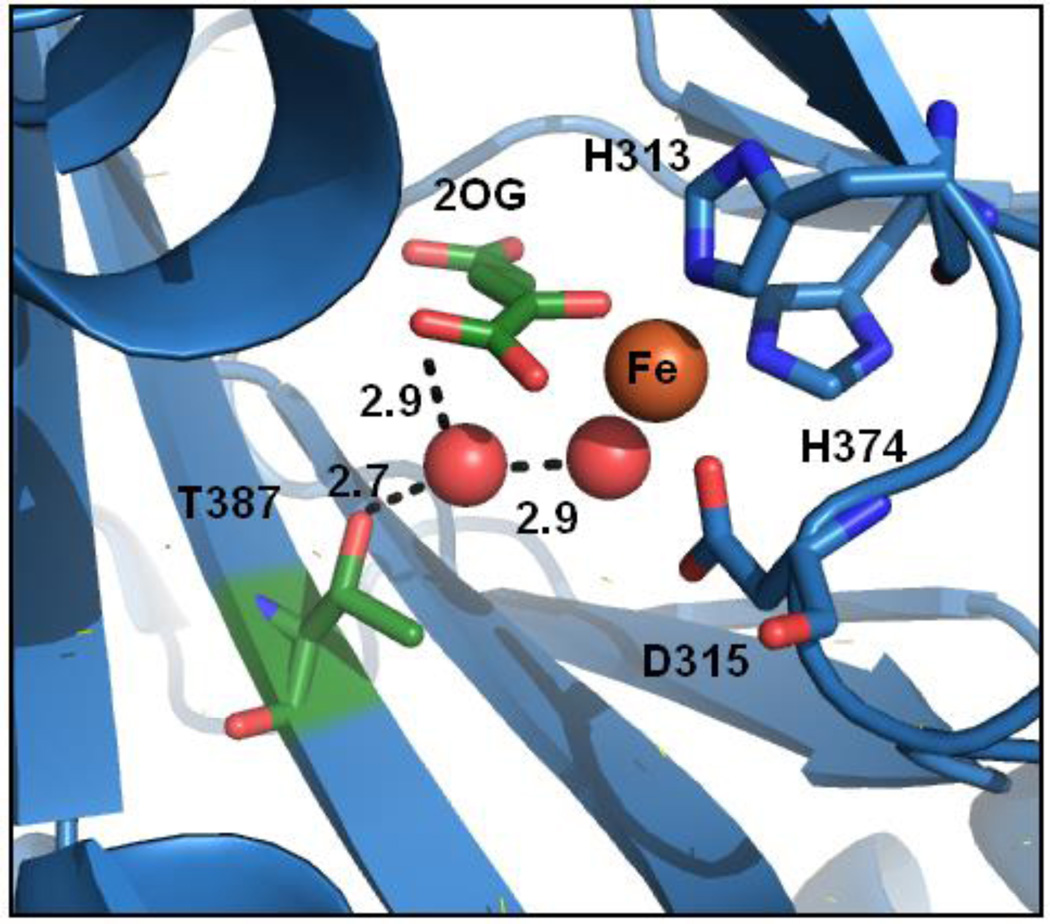
The connection between substrate binding and O2 activation is crucial to understanding the chemistry of this class of enzyme. A recent study revealed a conserved pattern of second-sphere contacts in enzymes structurally related to PHD215 which may be functionally significant. The X-ray crystal structure of (Fe+2OG)PHD216 showed a hydrogen bonding network connecting the aquo ligand and 2OG to the sidechain of Thr387 (Figure 1). Thr387 is located on the second β strand, at a position which forms a conserved H-bond in many 2OG oxygenases.15 As O2 activation is integral to the function of PHD2 and other 2OG oxygenases, the disposition of Thr387 may be crucial to gas binding and reactivity.
Figure 1.
PHD2 active site (PDB ID: 3OUJ).16 Waters (red sphere), distances (Å).
Here we tested the role of second sphere contacts from Thr387 on PHD2 hydroxylation chemistry through steady state kinetics and {FeNO}7 EPR spectroscopy. Steady-state kinetic parameters for the Thr387→Asn variant, which conserved the hydrogen bonding capacity at this position, were very similar to those of WT-PHD2. Remarkably, the macroscopic rate constants for the Thr387→Ala variant were 15 times larger than for WT-PHD2, which is attributed to faster O2 activation. Substrate inhibition was observed for the Thr387→Ala variant, which implicates product release as a slow step in this variant – a corollary is that other substrate inhibition in other 2OG oxygenases likely reflects slow succinate release17. These results suggest that the second sphere contacts from Thr387 slow turnover such that decarboxylation chemistry is rate-limiting in WT-PHD2.
Materials and Methods
Materials
All chemicals were purchased from commercial vendors, and used without purification. The sequences of the peptide substrates were derived from the native HIF-1α556–574 (CODD) sequence. The sequence of the CODD peptide was DLDLEALAP564YIPADDDFQL. The underlined residues were changed from the native sequences as follows: CODD (M561A, M568A) to avoid methionine oxidation. The CODD peptide (99 % purity) was purchased from GL Biochem LTD (Shanghai).
Protein expression and purification
Recombinant human PHD2178–426 and its variants, Thr387→Ala and Thr387→Asn were expressed as a C-terminal GST fusion protein from E. coli BL21(DE3) cells using a pGEX-4T-1 vector (Stratagene) and purified as previously described14,18. Briefly, PHD2-GST was purified using an affinity column (GSTrap, GE Bioscience), then the GST-tag was cleaved with restriction grade thrombin for 16 hours at 4 °C. The thrombin was removed with a Hitrap Benzamidine column (GE Bioscience), then PHD2 was treated with 50 mM EDTA overnight to remove metals. PHD2 was buffer exchanged with 50 mM HEPES pH 7.50 and stored at −20 °C. The mass of the WT-PHD2 and the variants were determined by a QStar-XL hybrid quadrupole-TOF mass spectrometer (Applied Biosystems). WT-PHD2 (27786.6 Da calculated, 27784.7 Da observed), Thr387→Ala (27755.2 Da calculated, 27756.2 Da observed), and Thr387→Asn (27799.6 Da calculated, 27798.2 Da observed).
Steady state kinetic assays
Steady-state kinetic constants were obtained from initial rates were measured under saturating concentrations of (NH4)2Fe(SO4)2 (10 µM) and ascorbic acid (1 mM) in 50 mM HEPES buffer pH 7.00 at 37.0 °C. MALDI-TOF was used to measure the production of of hydroxylated product CODDOH ((M+O+Na+): 2174 m/z calc., 2172 m/z obs.) from the substrate peptide CODD ((M+Na+): 2158 m/z calc., 2156 m/z obs.). Substrate inhibition constants were obtained by fitting the initial-rate data to Eq. 1, in which the initial rate (vo) is a function of the maximal rate (vmax), the Michaelis constant (KM) and the inhibition constant (Ki) for the substrate (S).
| (Equation 1) |
For assays in which [2OG] was varied, the concentration of CODD (10 µM) was fixed at saturating levels. Reactions were quenched at different time points in saturated 4-α-cyano hydroxycinnamic acid dissolved in 75 % acetonitrile and 0.2 % trifluoroacetic acid. Quenched reactions were spotted onto a MALDI target plate and analyzed using a Bruker Daltonics Omniflex MALDI-TOF mass spectrometer. For assays in which the [CODD] varied, the concentration of 2OG (100 µM) was fixed at saturating.
Assays in which O2 was the varied substrate used saturating concentrations of (NH4)2Fe(SO4)2 (10 µM), 2OG (100 µM), CODD (10 µM) and ascorbic acid (1 mM) in 50 mM HEPES pH 7.00 at 37 °C. The concentration of dissolved O2 was controlled by varying the ratio of N2 and O2 gas flowing through separate flow meters through a gas washer into an Atmosbag (Sigma-Aldrich), with the dissolved O2 concentration monitored by a YSI model 5300 biological oxygen monitor. Reactions were quenched and analyzed as for other steady state reactions.
2OG Binding
The dissociation constant (KD) of 2OG was determined by measuring the quenching of the intrinsic tryptophan fluorescence of PHD2 upon binding 2OG. Samples were excited at 295 nm and the emission measured at 330 nm using a PTI spectrofluorometer (PTI-QM-4 2005SE). A cuvette containing PHD2 (1 µM), and MnSO4 (20 µM) in 50 mM HEPES pH 7.00 was titrated with aliquots of 2OG (500 µM); the KD was obtained by fitting the data to equation 2, in which the observed fluorescence intensity (I) is normalized to the intensity in the absence of 2OG (I0) and the presence of saturating [2OG] (If).
| (Equation 2) |
X-band EPR of {FeNO}7
Anaerobic samples were prepared a glovebox ([O2] < 1 ppm) for analysis. (NH4)2FeSO4 was brought into the glovebox as a solid and dissolved using degassed H2O. The DEANO stocks were prepared in 10 mM NaOH in the glovebox and the concentration verified by its published extinction coefficient and characteristic UV absorbance at 250 nm.19,20 A 100 µL EPR sample was prepared anaerobically and contained PHD2 (0.10 mM), (NH4)2FeSO4, (0.10 mM), 2OG (0.50mM), CODD (100 µM) and DEANO (0.5 mM) in 50 mM HEPES pH 7.00 at room temperature (23°C). The sample was aged for 20 minutes to allow NO release from DEANO, then flash frozen in liquid nitrogen. EPR spectra were collected using a Bruker Elexsys E-500 EPR equipped with a DM4116 cavity and a Bruker ER 4118CF-O LHe/LN2 cryostat at 9.624 GHz frequency, 2.0 mW power, 10 G modulation amplitude, 100 GHz modulation frequency, 163 ms time constant, 4 K. As the electronic structure of the S = 3/2 ferrous-nitrosyl center is highly axial (E/D < 0.1), the observed EPR resonances (gx,y,z) were related to the rhombicity of the zero field splitting (E/D) using an effective spin Hamiltonian (Eq. 3).21
| (Eq. 3a) |
| (Eq. 3b) |
| (Eq. 3c) |
Results and Discussion
Steady state kinetics with varied [2OG]
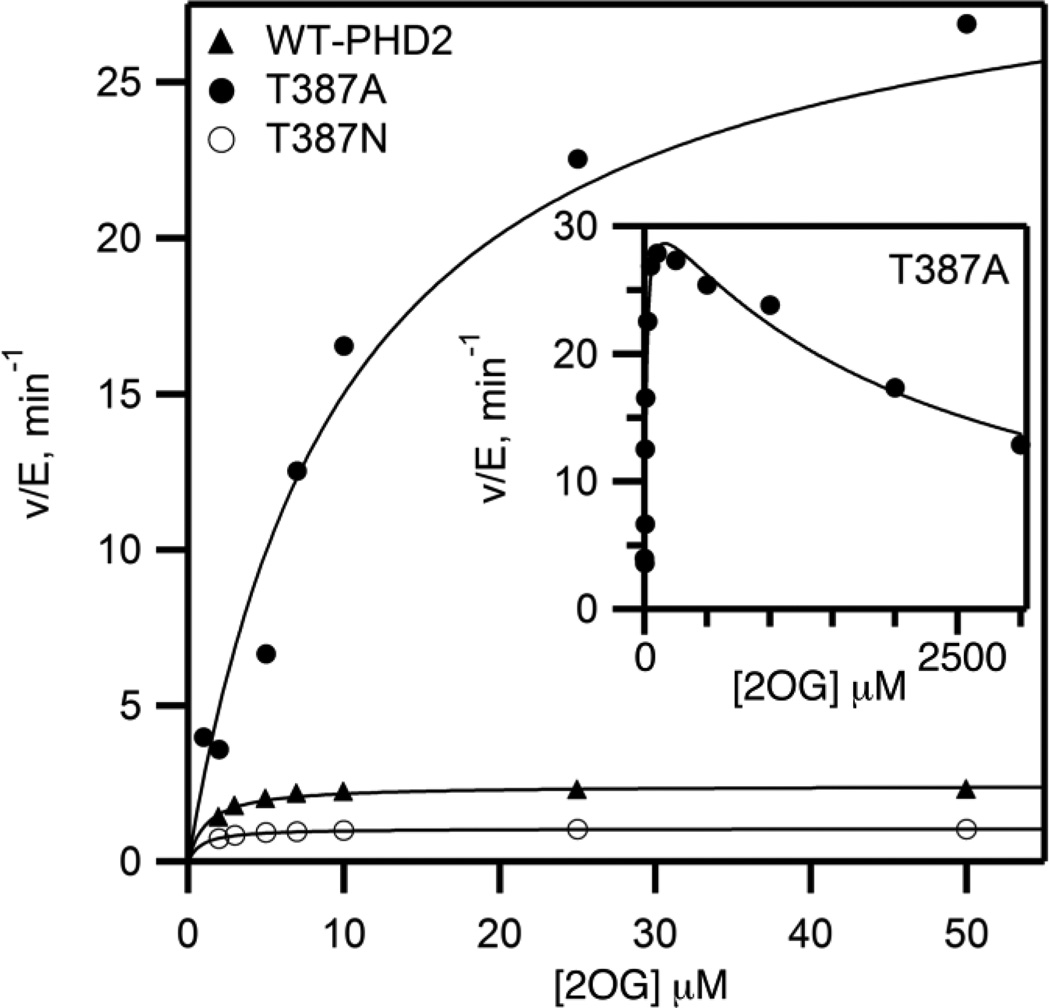
The structure of PHD2 showed a hydrogen bond network connecting 2OG, the aquo ligand, and Thr387 (Figure 1). As a hydrogen bonding residue is structurally conserved at this position in other 2OG oxygenases,13,15 we hypothesized that this played an important role during turnover. To test the effect of this contact on steps between 2OG binding and ODD binding, initial rate data were measured as a function of varied [2OG] using saturating concentrations of Fe2+ (10 µM), CODD (10 µM), and ascorbate (1 mM); [O2] was fixed at ambient levels (217 uM). WT-PHD2 exhibited simple saturation kinetics over the range of 1 – 50 µM 2OG, with values of kcat (2.3 min−1) and kcat/KM(2OG) (2.7 µM−1min−1) similar to those previously reported14,18,22–24. Similarly, the conservative Thr387→Asn variant exhibited saturation kinetics over the tested concentration range, with modestly decreased rate constants relative to WT-PHD2 (Table 1).
Table 1.
Steady state kinetics of WT-PHD2 and variants with varied [2OG].a
| Enzyme |
kcat min−1 |
kcat/KM(2OG) µM−1min−1 |
KI(2OG) µM |
KM(2OG), µM |
KD(2OG)b µM |
|---|---|---|---|---|---|
| WT-PHD2 | 2.3 ± 0.1 | 2.7 ± 0.4 | >2.6×104 | 0.9 ± 0.2 | 0.7 ± 0.2 |
| Thr387→Ala | 32.9 ± 1.4 | 2.7 ± 0.3 | 2150 ± 340 | 12.0 ± 1.6 | 1.2 ± 0.2 |
| Thr387→Asn | 1.0 ± 0.1 | 1.5 ± 0.2 | >5×104 | 0.7 ± 0.1 | 0.6 ± 0.2 |
. Reactions contained (NH4)2Fe(SO4)2 (10 µM), ascorbic acid (1 mM), 2OG (1–3000 µM), and CODD (10 µM) in 50 mM HEPES pH 7.00, 37°C, ambient [O2](217 µM).
. Fluorescence titrations: PHD2 (1.1 µM), MnSO4 (20 µM) in 50 mM HEPES pH 7.00 titrated with 2OG (500 µM).
The initial rate data for the Thr387→Ala variant was a significant contrast to that of WT-PHD2. While Thr387→Ala exhibited simple saturation kinetics for [2OG] < 250 µM, this variant exhibited ~50% activity at 3000 µM 2OG (Figure 2). Fitting the initial rate data to a simple substrate inhibition model led to parameters of kcat (32.9 min−1), kcat/KM(2OG) (2.7 µM−1min−1), and KI(2OG) (2200 µM). The high value for kcat indicated that Thr387→Ala performed chemistry more rapidly than WT-PHD2, but the observed substrate inhibition suggested that slow release of one of the products could limit turnover.
Figure 2.
Steady state kinetics for PHD2 variants with varied [2OG]. Reaction mixtures included (NH4)2Fe(SO4)2 (10 µM), ascorbic acid (1 mM), 2OG (1–3000 µM), and CODD (10 µM) in 50 mM HEPES pH 7.00, 37.0 °C, ambient [O2] (217 µM).
The steady state rate constant kcat/KM(2OG) reflects those steps between 2OG binding and the subsequent irreversible step, which is CODD binding under conditions of saturating [CODD]. As this rate constant is essentially unchanged upon mutation of Thr387, contacts from this residue do not affect these steps. In contrast, kcat reflects non-diffusional steps, and was greatly affected by mutation of Thr387. While kcat was unchanged in the conservative Thr387→Asn variant, this kinetic parameter increased ~ 15 fold for the Thr387→Ala variant, suggesting that a chemical step was affected by the Thr387 variants.
The substrate inhibition observed for Thr387→Ala prompted us to test the other variants for this effect. WT-PHD2 retained >95% activity even at extremely high levels of 2OG; fits of this data to the substrate inhibition model were unsatisfactory (KI(2OG) >26 mM) indicating that PHD2 did not undergo substrate inhibition. Thr387→Asn similarly showed minimal inhibition at extremely high 2OG concentration (KI(2OG) >50 mM). The absence of significant substrate inhibition observed for WT-PHD2 was consistent with product release being rapid, consistent with the prior reports that implicated a step early in catalysis as being rate-limiting for WT-PHD210.
Substrate inhibition could arise from many possible mechanisms in which substrate binding leads to an enzyme form of lower reactivity. Inhibition by 2OG could be due to 2OG binding to the Fe2+ cofactor in a geometry that prevented O2 from reacting, or by binding to the Fe2+ when an equivalent of succinate or 2OG was already present. Although our kinetics data and 2OG binding data cannot distinguish among these possibilities, a unified model that accounts for the observed kinetics (see below) and substrate inhibition found for Thr387→Ala is one in which chemistry is much faster than succinate release, such that the (Fe+Succ)PHD2 form of enzyme accumulates in the steady state for this variant. Inhibition by 2OG or by CODD would arise from these substrates binding to the (Fe+Succ)PHD2 form of enzyme, thereby preventing the enzyme from adopting the proper cofactor orientation to enter into catalysis.
The binding affinity for 2OG was measured for each of the variants by intrinsic tryptophan fluorescence quenching (supplemental material). Binding curves were fit to a single binding site model using Eq. 2, which indicated a similar avidity toward 2OG (KD ~ 1 µM) for each variant. Attempts to fit a second 2OG binding event were not successful, possibly due to the large difference in magnitude between KM(2OG) and KI(2OG) for each of the variants.
Steady state kinetics with varied [CODD]
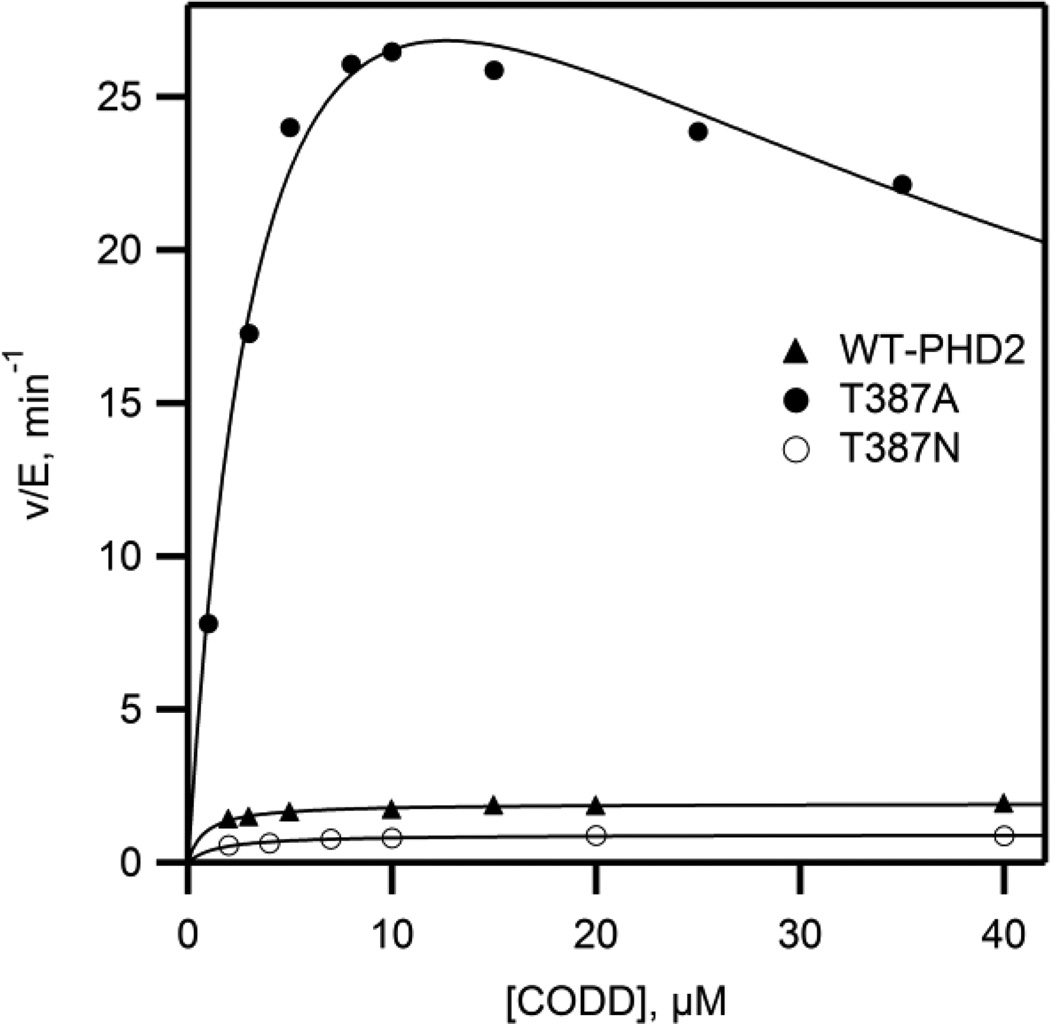
The impact of Thr387 variants on the chemical steps of turnover was tested by measuring steady state kinetics with CODD as the varied substrate. Initial rates of CODD hydroxylation were measured as a function of varied CODD concentration in 50 mM HEPES (pH 7.00, 37.0 °C), with saturating concentrations of Fe2+ (10 µM), 2OG (100 µM), and ascorbate (1 mM); the [O2] was fixed at the ambient level (217 µM). As O2 was not saturating, kcat/KM(CODD) will reflect steps spanning CODD binding up through the irreversible chemistry involved with oxidative decarboxylation. Steady state kinetic constants, kcat, and kcat/KM(CODD), were determined by fitting the data to the Michaelis-Menten equation (Figure 3). The rate constants for WT-PHD2 were in good agreement with prior reports14,22,25. While the conservative Thr387→Asn mutation led to rate constants that were modestly decreased from those of WT-PHD2, those for the Thr387→Ala variant were ~ 10 fold greater than those of WT-PHD2, indicating that removal of the hydrogen bond from Thr387 greatly impacted chemical steps. The rate constants for the Thr387→Ala also exhibited pronounced substrate inhibition at elevated CODD concentrations (Figure 3).
Figure 3.
Steady state kinetics data with varied [CODD]. Reactions included (NH4)2Fe(SO4)2 (10 µM), ascorbic acid (1 mM), 2OG (100 µM), and CODD (2–40 µM) in 50 mM HEPES pH 7.00, 37.0 °C, ambient [O2](217 µM).
The initial rate data for Thr387→Ala was fit in two ways in order to adequately treat the inhibition. The first fit used the Michaelis-Menten equation to fit the initial rate data for [CODD] < 10 µM, leading to fitted parameters of kcat = 33 min−1 and kcat/KM(CODD) = 13 µM−1min−1. These kinetic constants were remarkable because they suggested that the active site in WT-PHD2 was arranged to slow turnover.
The second fit used the substrate inhibition model and resulted in kcat = 44 min−1, kcat/KM(CODD) = 11 µM−1min−1, and KI = 38 µM (Table 2). This second fit reproduced the data well, suggesting that the peptide substrate (CODD) was a stronger inhibitor than 2OG toward the Thr387→Ala variant. As for 2OG inhibition, inhibition by the CODD substrate could arise from many possible mechanisms in which CODD binding would lead to a low-activity enzyme form. A simple model that accounts for the kinetic changes for the Thr387→Ala variant is one in which succinate release is slow, as this could lead to either 2OG or CODD binding to the unreactive (Fe+succ)PHD2 enzyme form. The substrate inhibition by 2OG and CODD implicate a step after O2 binding/activation as rate-limiting in the Thr387→Ala variant.
Table 2.
Steady state kinetic constants, kcat and kcat/KM(CODD) with varied [CODD].a
| Enzyme | kcat, min−1 | kcat/KM(CODD), µM−1min−1 | KI, µM | KM, µM |
|---|---|---|---|---|
| WT-PHD2 | 1.9 ± 0.1 | 2.4 ± 0.2 | - | 0.8 ± 0.1 |
| Thr387→Ala | 44 ± 3 | 11 ± 1 | 38 ± 6 | 4.1 ± 0.7 |
| Thr387→Asn | 0.9 ± 0.1 | 0.7 ± 0.1 | - | 1.4 ± 0.2 |
. Reactions contained (NH4)2Fe(SO4)2 (10 µM), ascorbic acid (1 mM), 2OG (100 µM), and CODD (2 – 50 µM) in 50 mM HEPES pH 7.00, 37.0 °C, ambient [O2](217 µM).
Steady state kinetics with varied [O2]
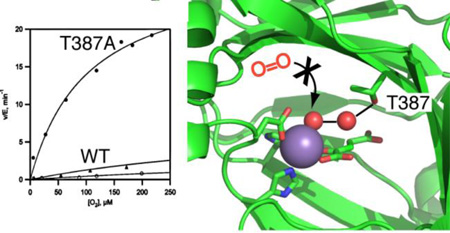
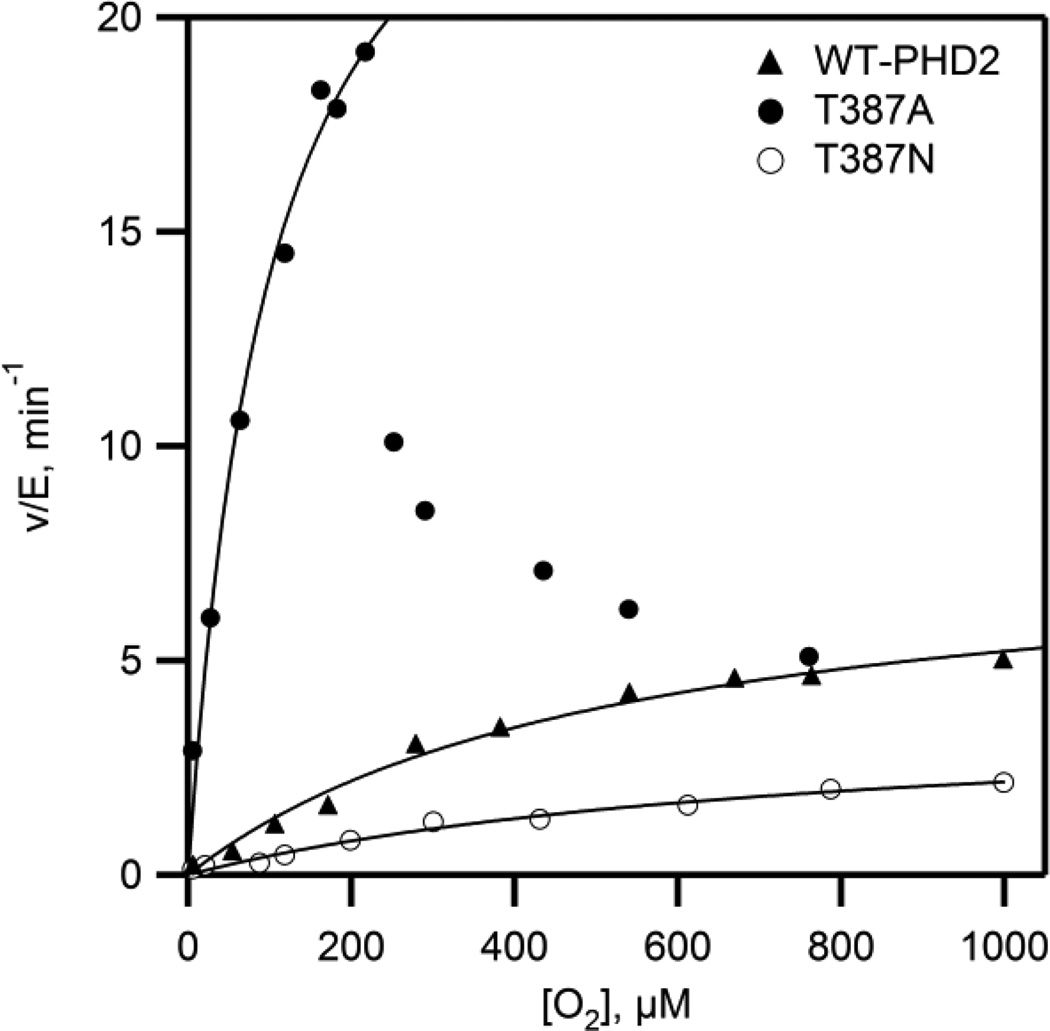
The increased rates of turnover for the Thr387→Ala variant suggested that steps involved in O2 activation were faster than for WT-PHD2. The steady-state kinetics as a function of varied O2 concentration were measured to isolate steps between O2 binding and oxidative decarboxylation. The initial rate data showed simple saturation kinetics for all but the Thr387→Ala variant, and was fit to the Michaelis-Menten equation (Figure 4). Both WT-PHD2 and the conservative Thr387→Asn variant exhibited sluggish reactivity toward O2, with the kinetic parameters for WT-PHD2 kcat/KM(O2) = 0.015 µM−1min-1 (~ 0.25 × 10−9 M−1 sec−1) and KM(O2) = 530 µM consistent with prior reports28,29. The high KM(O2) has been rationalized as being necessary for hypoxia sensing, as PHD2 activity would be proportionate to physiologically relevant levels of [O2].30 The Thr387→Ala variant is a contrast to this, as kcat/KM(O2) = 0.3 µM−1 min−1, a 20-fold increase in reaction rate over that of WT-PHD2 which indicates that step(s) involving O2 activation have markedly increased in rate for this variant. The Thr387→Ala variant also exhibited saturation kinetics up to 220 µM O2, but with an activity that was only ~25% of maximal at elevated O2 concentrations (Fig. 4). Attempts at fitting this activity curve to a variety of inhibition models led to unsatisfactory fits, suggesting that the decreased activity at high [O2] may reflect enzyme inactivation, as is seen for several other 2OG oxygenases.31–34
Figure 4.
Steady state kinetics as a function of varied [O2]. Reaction mixtures included (NH4)2Fe(SO4)2 (10 µM), 2OG (100 µM), CODD (10 µM), and ascorbic acid (1 mM) in 50 mM HEPES pH 7.00. (open circles) WT-PHD2, (closed circles) Thr387→Asn, (triangles) Thr387→Ala.
As our experimental conditions used saturating levels of 2OG and CODD, the macroscopic rate constant (kcat/KM(O2)) reports only on steps between O2 binding and the subsequent irreversible step, which in the consensus mechanism for PHD2 is oxidative decarboxylation. The increased values for kcat and kcat/KM(O2) observed for Thr387→Ala point definitively to an increased rate for oxidative decarboxylation in this variant over that observed for WT-PHD2.
The kinetic parameters for the Thr387→Ala variant are remarkable for two reasons. First, the increased kcat/KM(O2) and related mechanistic data indicate that chemistry has become faster as a result of this point mutation, underscoring the enormous influence over oxidative decarboxylation exerted by 2°-sphere contacts in PHD2. This implies that O2-activation is not just a function of the redox potential at the Fe2+ cofactor, but will include the polar environment near the 2OG and the distal O-atom of bound O2. Second, the Thr387→Ala variant has a lower KM(O2) than found for WT-PHD2, making for a more sensitive activity response to changes in [O2], particularly over the physiological range of [O2] < 50 µM. This opens the possibility of using enzyme delivery35 or engineering approaches to tailor the hypoxia response within cells.
EPR spectra of (Fe2+NO) PHD2
According to the consensus mechanism, PHD2 must bind O2 such that the distal O-atom can attack the 2-oxo position of 2OG. We propose that the Thr387 of WT-PHD2 favors a specific gas binding geometry such that the bound O2 is not positioned optimally for reaction with 2OG. As NO binds to Fe2+ in non-heme enzymes to form a spectroscopically accessible {FeNO}7 with S = 3/2, and adopts a bent geometry similar to that of bound O227,36,37,) we used EPR lineshape analysis to assess the impact of Thr387 on bound gas geometry.
The EPR lineshape of {FeNO}7 centers in 2OG oxygenases and related enzymes is dominated by the large zero-field splitting,36–38 with EPR features near geff ~ 2 and geff ~ 4 that are sensitive reporters of the rhombic zero-field splitting parameter (E/D).21 Rhombic zero-field splitting can be directly calculated by use of Eq. 3, relating the observed geff values to the E/D ratio. As the zero-field splitting arises from admixture of the low-lying excited states into the ground state, the E/D ratio ultimately reports on the geometry of the {FeNO}7 spin center. For example, the EPR lineshape of the {FeNO}7 center is very sensitive to the Fe-NO bond angle in sterically constrained models complexes39 as well as equatorial bonding in non-heme proteins21,37 due to the splitting of the geff ~ 4 features caused by rhombic zero-field splitting.
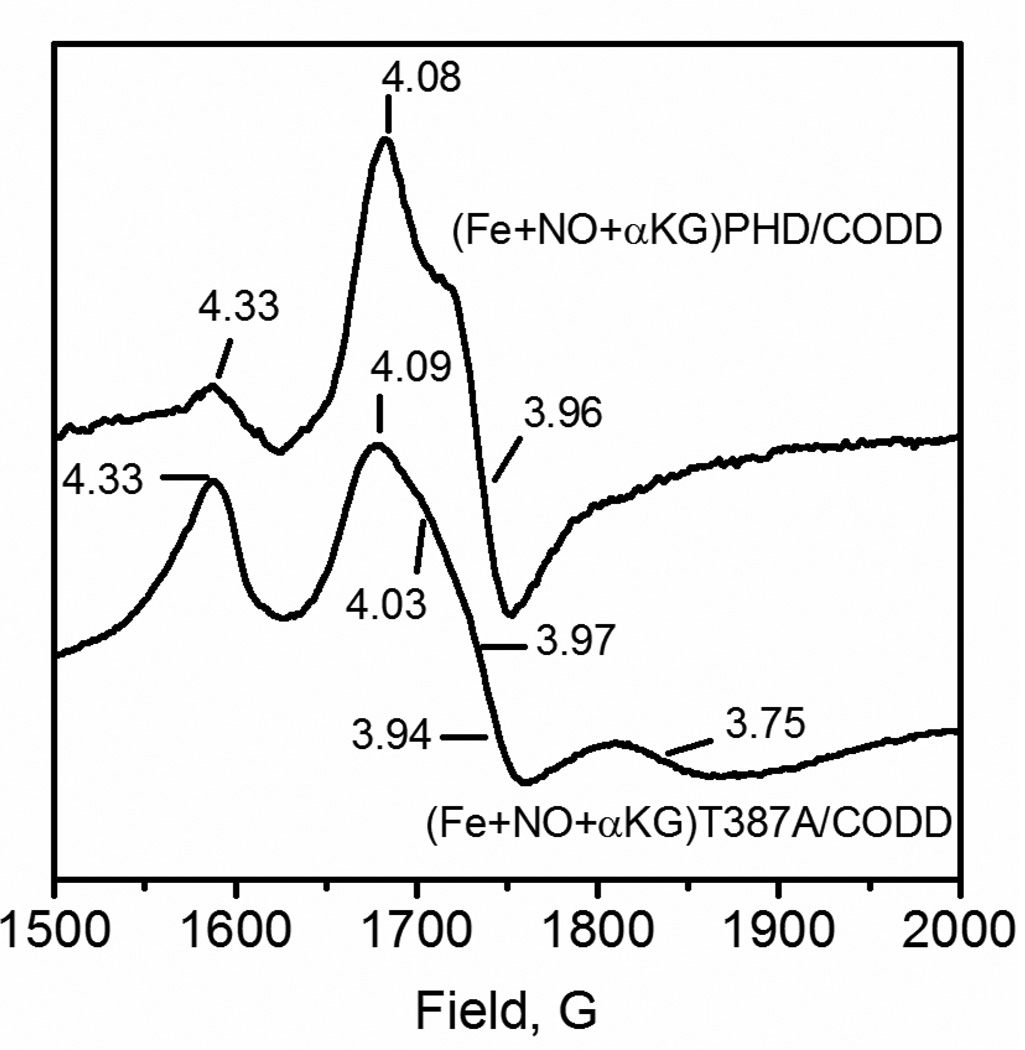
Ferrous nitrosyl complexes of WT-PHD2 and the Thr387→Ala variant were prepared by anaerobically generating NO in the presence of the (Fe+2OG)PHD2/CODD form of each enzyme. The X-band EPR spectra were collected at 4 K in a liquid helium flow cryostat, showing a complex lineshape near geff = 4 for both samples (Figure 5). The {FeNO}7 center of WT-PHD2 exhibited intense features at geff = 4.08, 3.96, which arose from an S=3/2 spin with a highly axial zero-field splitting (E/D = 0.011); as well as a minor feature at geff ~ 4.3 that likely arose from adventitious Fe3+. In contrast, the {FeNO}7 center of Thr387→Ala was more complex, with two S = 3/2 species evident. One species was very similar to that observed for WT-PHD2, with geff = 4.09, 3.94 (E/D = 0.014); the second center was much more rhombic, geff = 4.33, 3.75 (E/D = 0.48).
Figure 5.
X-band EPR of (Fe2++NO+2OG)PHD2/CODD. PHD2 (100 µM), CODD (100 µM), (NH4)2FeSO4, (100 µM), 2OG (500 µM), and DEANO (500 µM) in 50 mM HEPES pH 7.00. 9.624 GHz frequency; 2.0 mW power; 10 G, 100 GHz modulation, 4 K.
Although it is not possible to ascribe specific geometries to each of these spin centers, the Thr387→Ala mutation lead to a much more heterogeneous {FeNO}7 center than found in WT-PHD2. This likely arose from the steric interactions between the bound NO and contacts with the active site environment. Coupled with the observed increase in activity for Thr387→Ala, this suggests that the geometry of bound gas is correlated with O2 activation such that WT-PHD2 positions the bound gas in a non-optimal orientation for oxidative decarboxylation. In the consensus mechanism O2 binds as a putative (Fe3+ O2—) adduct, which then attacks the C-2 keto position of 2OG to initiate oxidative decarboxylation. Should the distal O-atom be stabilized by an electric dipole interaction with the hydroxyl group of Thr387, it could be less nucleophilic in WT-PHD2. Alternately, Thr387 could cause the distal O-atom to be oriented away from the C-2 keto position, thereby lowering the reactivity of the putative superoxide in WT-PHD2.
Implications for hypoxia sensing
Why is turnover so slow for WT-PHD2, and how could removing the hydrogen bonds from Thr387 increase the kinetic parameters for O2 activation? The answers to both of these questions are speculative at this point, but considering the structural and mechanistic data for PHD2 within the context of the physiological role of this enzyme points to the intriguing possibility that the naturally occurring residue is part of the machinery that limits maximal turnover.
As the physiological role of PHD2 is to turn off gene expression, it is very likely that this enzyme fulfills its role while turning over on the minutes timescale – this lack of evolutionary pressure to increase reaction rates may lead to an active site optimized for something other than rapid turnover. Single turnover experiments indicated that hydroxylated CODD product formed on the minutes timescale (kobs = 0.018 s−1, 5°C) without any accumulating intermediates.10 As this rate constant is virtually identical to that observed in the steady state (kcat = 0.03 s−1, 37 °C)14, it is very likely that oxidative decarboxylation is rate-limiting in WT-PHD2. This is unusual for 2OG-dependent oxygenases, but may reflect the O2-sensing from the function of PHD2.
The implication of this work are two-fold. First, contacts within the active site control the O2 activation rate in 2OG oxygenases. Second, PHD2 can be engineered into a more active form than found in nature, in particular with respect to the reactivity under low [O2] conditions. Although this was achieved by point mutagenesis in the current report, one can envision the use of small molecules to target the distal hydrogen-bonding in PHD2. Achieving increased turnover under hypoxic conditions would lead to altered cellular hypoxia sensing.
Supplementary Material
Table 3.
Steady state kinetic parameters using O2 as the varied substrate.
| Enzyme | kcat, min−1 | kcat/KM, µM−1min−1 | KM, µM |
|---|---|---|---|
| WT-PHD2 | 8 ± 0.7 | 0.015 ± 0.001 | 530 ± 90 |
| Thr387→Ala | 29 ± 3 | 0.3 ± 0.1 | 100 ± 20 |
| Thr387→Asn | 3.8 ± 0.5 | 0.005 ± 0.001 | 760 ± 170 |
[Fe2+] = 10 µM, [2OG] = 100 µM, [CODD] = 10 µM, 37.0°C, pH 7.00.
Acknowledgments
Funding This research was supported by the U.S. National Institutes of Health (1R01-GM077413 to M.J.K), the NIH Chemistry-Biology Interface Predoctoral Training Grant (T32-GM008515 to C. Y. T.) and the Turkish Ministry of National Education (S.P.).
Abbreviations
- 2OG
2-oxo-glutarate, α-ketoglutarate
- AtsK
Alkylsulfatase
- FIH
factor-inhibiting HIF
- HAT
hydrogen atom transfer
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- HIF
hypoxia inducible factor-1α
- MALDI-TOF-MS
Matrix assisted laser desorption ionization-time of flight-mass spectrometry
- PHD2
prolyl hydroxylase domain 2
- TauD
taurine dioxygenase
Footnotes
The authors declare no competing financial interest.
Experimental measurement of KD(2OG) for PHD2 variants. The material is available free of charge via the Internet at http://pubs.acs.org.
Bibliography
- 1.Berra E, Benizri E, Ginouvès A, Volmat V, Roux D, Pouysségur J. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1alpha in normoxia. EMBO J. 2003;22:4082–4090. doi: 10.1093/emboj/cdg392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Appelhoff RJ, Tian Y-M, Raval RR, Turley H, Harris AL, Pugh CW, Ratcliffe PJ, Gleadle JM. Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. J. Biol. Chem. 2004;279:38458–38465. doi: 10.1074/jbc.M406026200. [DOI] [PubMed] [Google Scholar]
- 3.Bruick RK. Oxygen sensing in the hypoxic response pathway: regulation of the hypoxia-inducible transcription factor. Genes Dev. 2003;17:2614–2623. doi: 10.1101/gad.1145503. [DOI] [PubMed] [Google Scholar]
- 4.Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, Kriegsheim Av, Hebestreit HF, Mukherji M, Schofield CJ, Maxwell PH, Pugh CW, Ratcliffe PJ. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 5.Aprelikova O, Chandramouli GVR, Wood M, Vasselli JR, Riss J, Maranchie JK, Linehan WM, Barrett JC. Regulation of HIF prolyl hydroxylases by hypoxia-inducible factors. J. Cell. Biochem. 2004;92:491–501. doi: 10.1002/jcb.20067. [DOI] [PubMed] [Google Scholar]
- 6.Hewitson KS, Schofield CJ. The HIF pathway as a therapeutic target. Drug Discov. Today. 2004;9:704–711. doi: 10.1016/S1359-6446(04)03202-7. [DOI] [PubMed] [Google Scholar]
- 7.Speer RE, Karuppagounder SS, Basso M, Sleiman SF, Kumar A, Brand D, Smirnova N, Gazaryan I, Khim SJ, Ratan RR. Hypoxia-inducible factor prolyl hydroxylases as targets for neuroprotection by “antioxidant” metal chelators: From ferroptosis to stroke. Free Radic. Biol. Med. 2013;62:26–36. doi: 10.1016/j.freeradbiomed.2013.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffart LM, Barr EW, Guyer RB, Bollinger JM, Krebs C. Direct spectroscopic detection of a C-H-cleaving high-spin Fe(IV) complex in a prolyl-4-hydroxylase. Proc. Natl. Acad. Sci. U. S. A. 2006;103:14738–14743. doi: 10.1073/pnas.0604005103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krebs C, Galonić Fujimori D, Walsh CT, Bollinger JM. Non-heme Fe(IV)-oxo intermediates. Acc. Chem. Res. 2007;40:484–492. doi: 10.1021/ar700066p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flashman E, Hoffart LM, Hamed RB, Bollinger JM, Krebs C, Schofield CJ. Evidence for the slow reaction of hypoxia-inducible factor prolyl hydroxylase 2 with oxygen. FEBS J. 2010;277:4089–4099. doi: 10.1111/j.1742-4658.2010.07804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Price JC, Barr EW, Tirupati B, Bollinger JM, Krebs C. The first direct characterization of a high-valent iron intermediate in the reaction of an alpha-ketoglutarate-dependent dioxygenase: a high-spin FeIV complex in taurine/alpha-ketoglutarate dioxygenase (TauD) from Escherichia coli. Biochemistry. 2003;42:7497–7508. doi: 10.1021/bi030011f. [DOI] [PubMed] [Google Scholar]
- 12.Grzyska PK, Ryle MJ, Monterosso GR, Liu J, Ballou DP, Hausinger RP. Steady-state and transient kinetic analyses of taurine/alpha-ketoglutarate dioxygenase: effects of oxygen concentration, alternative sulfonates, and active-site variants on the FeIV-oxo intermediate. Biochemistry. 2005;44:3845–3855. doi: 10.1021/bi048746n. [DOI] [PubMed] [Google Scholar]
- 13.Hangasky JA, Saban E, Knapp MJ. Inverse Solvent Isotope Effects Arising from Substrate Triggering in the Factor Inhibiting Hypoxia Inducible Factor. Biochemistry. 2013;52:1594–1602. doi: 10.1021/bi3015482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flagg SC, Giri N, Pektas S, Maroney MJ, Knapp MJ. Inverse solvent isotope effects demonstrate slow aquo release from hypoxia inducible factor-prolyl hydroxylase (PHD2) Biochemistry. 2012;51:6654–6666. doi: 10.1021/bi300229y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hangasky Ja, Taabazuing CY, Valliere Ma, Knapp MJ. Imposing function down a (cupin)-barrel: secondary structure and metal stereochemistry in the αKG-dependent oxygenases. Metallomics. 2013;5:287–301. doi: 10.1039/c3mt20153h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosen MD, Venkatesan H, Peltier HM, Bembenek SD, Kanelakis KC, Zhao LX, Leonard BE, Hocutt FM, Wu X, Palomino HL, Brondstetter TI, Haugh PV, Cagnon L, Yan W, Liotta La, Young A, Mirzadegan T, Shankley NP, Barrett TD, Rabinowitz MH. Benzimidazole-2-pyrazole HIF Prolyl 4-Hydroxylase Inhibitors as Oral Erythropoietin Secretagogues. ACS Med. Chem. Lett. 2010;1:526–529. doi: 10.1021/ml100198y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cascella B, Mirica LM. Kinetic analysis of iron-dependent histone demethylases: α-ketoglutarate substrate inhibition and potential relevance to the regulation of histone demethylation in cancer cells. Biochemistry. 2012;51:8699–8701. doi: 10.1021/bi3012466. [DOI] [PubMed] [Google Scholar]
- 18.Pektas S, Knapp MJ. Substrate preference of the HIF-prolyl hydroxylase-2 (PHD2) and substrate-induced conformational change. J. Inorg. Biochem. 2013;126:55–60. doi: 10.1016/j.jinorgbio.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maragos CM, Morley D, Wink DA, Dunams TM, Saavedra JE, Hoffman A, Bove AA, Isaac L, Hrabie JA, Keefer LK. Complexes of NO with Nucleophiles as Agents for the Controlled Biological Release of Nitric Oxide. Vasorelaxant Effects. J. Med. Chem. 1991;34:3242–3247. doi: 10.1021/jm00115a013. [DOI] [PubMed] [Google Scholar]
- 20.Keefer LK, Nims RW, Davies KM, Wink DA. “NONOates” (1-Substituted Diazen-1-ium-1,2-diolates) as Nitric Oxide Donors: Convenient Nitric Oxide Dosage Forms. Methods Enzymol. 1996;268:281–293. doi: 10.1016/s0076-6879(96)68030-6. [DOI] [PubMed] [Google Scholar]
- 21.Arcierog DM, Orville AM, Lipscomb JD. [l ‘ O] Water and Nitric Oxide Binding by Protocatechuate. J. Biol. Chem. 1985;260:14035–14044. [PubMed] [Google Scholar]
- 22.Flagg SC, Martin CB, Taabazuing CY, Holmes BE, Knapp MJ. Screening chelating inhibitors of HIF-prolyl hydroxylase domain 2 (PHD2) and factor inhibiting HIF (FIH) J. Inorg. Biochem. 2012;113:25–30. doi: 10.1016/j.jinorgbio.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chowdhury R, McDonough Ma, Mecinović J, Loenarz C, Flashman E, Hewitson KS, Domene C, Schofield CJ. Structural basis for binding of hypoxia-inducible factor to the oxygen-sensing prolyl hydroxylases. Structure. 2009;17:981–989. doi: 10.1016/j.str.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 24.Koivunen P, Hirsilä M, Kivirikko KI, Myllyharju J. The length of peptide substrates has a marked effect on hydroxylation by the hypoxia-inducible factor prolyl 4-hydroxylases. J. Biol. Chem. 2006;281:28712–28720. doi: 10.1074/jbc.M604628200. [DOI] [PubMed] [Google Scholar]
- 25.Flashman E, Bagg EaL, Chowdhury R, Mecinović J, Loenarz C, McDonough Ma, Hewitson KS, Schofield CJ. Kinetic rationale for selectivity toward N- and C-terminal oxygen-dependent degradation domain substrates mediated by a loop region of hypoxia-inducible factor prolyl hydroxylases. J. Biol. Chem. 2008;283:3808–3815. doi: 10.1074/jbc.M707411200. [DOI] [PubMed] [Google Scholar]
- 26.Quinn DM, Sutton LD. Theoretical Basis and Mechanistic Utility of Solvent Isotope Effects. Enzyme Mechanism from Isotope Effects. 1991:73–126. [Google Scholar]
- 27.Muthukumaran RB, Grzyska PK, Hausinger RP, McCracken J. Probing the iron-substrate orientation for taurine/alpha-ketoglutarate dioxygenase using deuterium electron spin echo envelope modulation spectroscopy. Biochemistry. 2007;46:5951–5959. doi: 10.1021/bi700562t. [DOI] [PubMed] [Google Scholar]
- 28.Ehrismann D, Flashman E, Genn DN, Mathioudakis N, Hewitson KS, Ratcliffe PJ, Schofield CJ. Studies on the activity of the hypoxia-inducible-factor hydroxylases using an oxygen consumption assay. Biochem. J. 2007;401:227–234. doi: 10.1042/BJ20061151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirsilä M, Koivunen P, Günzler V, Kivirikko KI, Myllyharju J. Characterization of the human prolyl 4-hydroxylases that modify the hypoxia-inducible factor. J. Biol. Chem. 2003;278:30772–30780. doi: 10.1074/jbc.M304982200. [DOI] [PubMed] [Google Scholar]
- 30.Aragonés J, Schneider M, Van Geyte K, Fraisl P, Dresselaers T, Mazzone M, Dirkx R, Zacchigna S, Lemieux H, Jeoung NH, Lambrechts D, Bishop T, Lafuste P, Diez-Juan A, Harten SK, Van Noten P, De Bock K, Willam C, Tjwa M, Grosfeld A, Navet R, Moons L, Vandendriessche T, Deroose C, Wijeyekoon B, Nuyts J, Jordan B, Silasi-Mansat R, Lupu F, Dewerchin M, Pugh C, Salmon P, Mortelmans L, Gallez B, Gorus F, Buyse J, Sluse F, Harris Ra, Gnaiger E, Hespel P, Van Hecke P, Schuit F, Van Veldhoven P, Ratcliffe P, Baes M, Maxwell P, Carmeliet P. Deficiency or inhibition of oxygen sensor Phd1 induces hypoxia tolerance by reprogramming basal metabolism. Nat. Genet. 2008;40:170–180. doi: 10.1038/ng.2007.62. [DOI] [PubMed] [Google Scholar]
- 31.Chen Y-H, Comeaux LM, Eyles SJ, Knapp MJ. Auto-hydroxylation of FIH-1: an Fe(ii), alpha-ketoglutarate-dependent human hypoxia sensor. Chem. Commun. (Camb) 2008:4768–4770. doi: 10.1039/b809099h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ryle MJ, Koehntop KD, Liu A, Que L, Hausinger RP. Interconversion of two oxidized forms of taurine/alpha-ketoglutarate dioxygenase, a non-heme iron hydroxylase: evidence for bicarbonate binding. Proc. Natl. Acad. Sci. U. S. A. 2003;100:3790–3795. doi: 10.1073/pnas.0636740100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mantri M, Zhang Z, McDonough Ma, Schofield CJ. Autocatalysed oxidative modifications to 2-oxoglutarate dependent oxygenases. FEBS J. 2012;279:1563–1575. doi: 10.1111/j.1742-4658.2012.08496.x. [DOI] [PubMed] [Google Scholar]
- 34.Liu a, Ho RY, Que L, Ryle MJ, Phinney BS, Hausinger RP. Alternative reactivity of an alpha-ketoglutarate-dependent iron(II) oxygenase: enzyme self-hydroxylation. J. Am. Chem. Soc. 2001;123:5126–5127. doi: 10.1021/ja005879x. [DOI] [PubMed] [Google Scholar]
- 35.Fu A, Tang R, Hardie J, Farkas ME, Rotello VM. Promises and pitfalls of intracellular delivery of proteins. Bioconjug. Chem. 2014;25:1602–1608. doi: 10.1021/bc500320j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ye S, Price JC, Barr EW, Green MT, Bollinger JM, Pennsyl V, Uni S, Park UV. Cryoreduction of the NO-Adduct of Taurine : r -Ketoglutarate Dioxygenase (TauD) Yields an Elusive {FeNO} 8 Species. J. Am. Chem. Soc. 2010;132:4739–4751. doi: 10.1021/ja909715g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han AY, Lee AQ, Abu-Omar MM. EPR and UV-vis studies of the nitric oxide adducts of bacterial phenylalanine hydroxylase: effects of cofactor and substrate on the iron environment. Inorg. Chem. 2006;45:4277–4283. doi: 10.1021/ic060478p. [DOI] [PubMed] [Google Scholar]
- 38.Hausinger RP. FeII/alpha-ketoglutarate-dependent hydroxylases and related enzymes. Crit. Rev. Biochem. Mol. Biol. 2004;39:21–68. doi: 10.1080/10409230490440541. [DOI] [PubMed] [Google Scholar]
- 39.Ray M, Golombek AP, Hendrich MP, Yap GPa, Liable-Sands LM, Rheingold AL, Borovik aS. Structure and Magnetic Properties of Trigonal Bipyramidal Iron Nitrosyl Complexes. Inorg. Chem. 1999;38:3110–3115. [Google Scholar]
- 40.Saban E, Flagg SC, Knapp MJ. Uncoupled O2-activation in the human HIF-asparaginyl hydroxylase, FIH, does not produce reactive oxygen species. J. Inorg. Biochem. 2011;105:630–636. doi: 10.1016/j.jinorgbio.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kahnert A, Kertesz MA. Characterization of a sulfur-regulated oxygenative alkylsulfatase from Pseudomonas putida S-313. J. Biol. Chem. 2000;275:31661–31667. doi: 10.1074/jbc.M005820200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.