Summary
After DNA replication, chromosomal processes including DNA repair and transcription take place in the context of sister chromatids. While cell cycle regulation can guide these processes globally, mechanisms to distinguish pre- and post-replicative states locally remain unknown. Here, we reveal that new histones incorporated during DNA replication provide a signature of post-replicative chromatin, read by the TONSL–MMS22L1–4 homologous recombination (HR) complex. We identify the TONSL Ankyrin Repeat Domain (ARD) as a reader of histone H4 tails unmethylated at K20 (H4K20me0), which are specific to new histones incorporated during DNA replication and mark post-replicative chromatin until G2/M. Accordingly, TONSL–MMS22L binds new histones H3–H4 both prior to and after incorporation into nucleosomes, remaining on replicated chromatin until late G2/M. H4K20me0 recognition is required for TONSL–MMS22L binding to chromatin and accumulation at challenged replication forks and DNA lesions. Consequently, TONSL ARD mutants are toxic, compromising genome stability, cell viability and resistance to replication stress. Together, this reveals a histone reader based mechanism to recognize the post-replicative state, offering a new approach and opportunity to understand DNA repair with potential for targeted cancer therapy.
The TONSL–MMS22L complex is an obligate heterodimer required for replication fork stability and repair of replication-associated DNA damage by aiding Rad51 loading1–4. TONSL–MMS22L associates with soluble non-nucleosomal histones H3–H41,5, the histone chaperone ASF11–4 and MCM2/4/6/71–5 in a manner that depends on the TONSL ankyrin repeat domain (ARD)1. We have found that histones H3–H4 bridge the interactions between TONSL–MMS22L and ASF11, between ASF1 and MCM26,7, and between TONSL–MMS22L and MCM2 (Extended Data Fig. 1a), suggesting simultaneous binding of these proteins to histones H3–H4 in a large pre-deposition complex. In addition, TONSL–MMS22L interacts with nucleosomal histones in chromatin (Extended Data Fig. 1b). This suggests that TONSL–MMS22L functions as an H3–H4 histone chaperone whilst also acting as a histone reader in chromatin. Consistent with the first, TONSL was recently shown to have histone chaperone activity in vitro5. We therefore set out to explore the mechanism of action of TONSL–MMS22L by a structure-function approach.
Full length TONSL and the ARD alone bound directly to histones H3–H4 but not H2A–H2B in vitro (Extended Data Fig. 1c-f). As our attempts to crystallize the ARD with H3–H4 were not successful, we linked the ARD to the MCM2 histone-binding domain (HBD), because a similar design previously enabled us to solve the structure of an H3–H4 dimer in complex with MCM2 and ASF17. We obtained crystals of covalently linked MCM2 HBD–G4–TONSL ARD in complex with H3 (57–135) and H4 that diffracted to 2.43 Å resolution, and solved the structure by molecular replacement based on our structure of MCM2 HBD in complex with an H3–H4 tetramer7 (Fig. 1a, b; X-ray statistics in Extended Data Table 1). The structure shows a pair of MCM2 HBDs wrapped around the lateral surface of the H3–H4 tetramer similar to the MCM2 HBD–H3–H4 complex alone7,8, while two TONSL ARDs interact with each of the H4 tails (Fig. 1a, b). The G4-linker along with flanking residues formed a 19-residues long disordered segment that could reach a distance of up to 70 Å. The distance from the observed C-terminus of MCM2 HBD to the observed N-terminus of TONSL ARD is only 10 Å, arguing that the covalent linkage within the MCM2 HBD–G4–TONSL ARD cassette does not impact on the structural integrity of the complex. TONSL ARD forms no intermolecular interactions with the MCM2 HBD, consistent with H3–H4 bridging the interaction of TONSL and MCM2 in cells (Extended Data Fig. 1a), and it shows only minimal contacts with the core of the H3–H4 tetramer (Fig. 1a, b). However, the TONSL ARD forms extensive contacts with a segment of the H4 tail (Fig. 1b, c, Extended Data Fig. 1g) and, consistently, it binds the GST-H4 tail, but not the GST-H3 tail, in vitro (Extended Data Fig. 2a). In addition to defining TONSL binding to soluble histone H3–H4 in complex with MCM2 (Fig. 1a, b), this binding-mode is also compatible with TONSL binding histone H3–H4 dimers in a co-chaperone complex with MCM2 and ASF17 as well as recognizing H4 tails in a nucleosome (see models in Extended Data Fig. 2b, c).
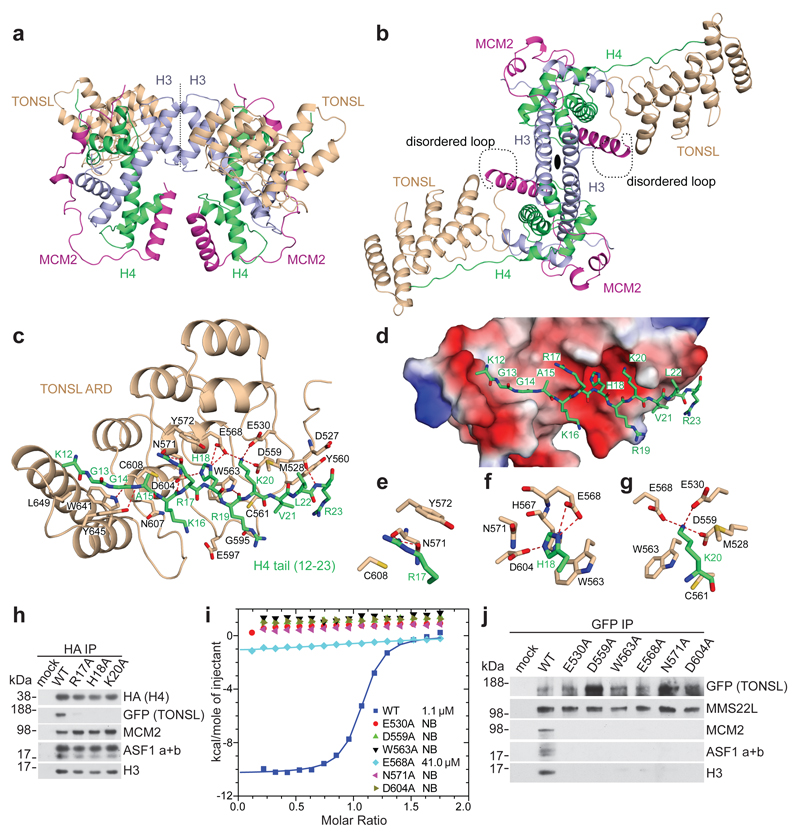
Figure 1. TONSL ARD interacts with the histone H4 tail.
a, b, Two different representative views of the overall structure of the TONSL ARD–MCM2 HBD–H3–H4 tetramer complex. c, Intermolecular interactions between TONSL ARD and the H4 tail. d, The electrostatic potential surface of ARD showing the acidic concave surface binding site for the H4 tail. e-g, Highlights of the inter-molecular interactions of H4 Arg17, His18 and Lys20 with ARD. h, Immunoprecipitation of soluble HA-SNAP-H4 wild type (WT) or mutant transfected into GFP-TONSL U-2-OS cells. i, ITC of TONSL ARD WT and mutants with H4 tail peptide. j, Immunoprecipitation of soluble GFP-TONSL WT or mutant. Data are representative of 3 independent experiments (h, j). Protein inputs, Extended Data Fig. 9b, c. Gel source data, Supplementary Fig. 1.
The TONSL ARD consists of four ankyrin repeats, three of which adopt the canonical fold (ANK1-3), while the remaining one is an atypical and capping repeat (ANK4) (Extended Data Fig. 1g). The TONSL ARD uses its elongated concave surface to form extensive contacts with the H4 tail in an extended β-strand like conformation (Extended Data Fig. 1g, Fig. 1c). Notably, 15 out of 18 residues that constitute the H4 tail-binding surface of TONSL ARD are highly conserved (Extended Data Fig. 2d). The TONSL ARD targets the H4 tail spanning residues Lys12 to Arg23 (Fig. 1c-g, Extended Data Fig. 3a, b) with three consecutive binding channels accommodating Arg17, His18 and Lys20 (Fig. 1d). These H4 residues are part of a basic region, which can interact with the acidic patch on neighbor nucleosomes9 in compact chromatin. H4 Arg17 forms two hydrogen bonds with ARD Asn571 and stacks with Tyr572 and Cys608 (Fig. 1c, e), while H4 His18 penetrates into a pocket lined by four strictly conserved residues (Trp563, Glu568, Asn571 and Asp604) (Fig. 1c, f). Substitution of H4 His18 with the larger Trp residue (mutant H18W) disrupts binding with ARD (Fig. 2a), underscoring the importance of fitting His18 in the pocket. The H4 Lys20 residue is bound within an acidic surface channel on ARD (Fig. 1c, d). The side-chain of H4 Lys20 interacts with Met528 and contacts the edge of Trp563 of ARD, while the main-chain atoms of H4 Lys20 packs against Cys561 of ARD (Fig. 1g). The Nζ atom of H4 Lys20 forms three strong hydrogen bonds (distance < 3 Å) with the side-chains of strictly conserved residues Glu530, Asp559 and Glu568 of ARD, which engage H4 Lys20 in a triangular arrangement (Fig. 1g). Consistent with the structural data, histone H4 mutations R17A, H18A, and K20A disrupted binding to TONSL in cells (Fig. 1h). Likewise, mutation of 6 conserved TONSL residues lining the H4 Arg17, His18 and Lys20 binding channels disrupted binding to H4 peptides and recombinant histone H3–H4 (Fig. 1i, Extended Data Fig. 3c). In vivo, these mutants abrogated binding to soluble histone H3–H4 and, consequentially, also association with ASF1 (a and b) and MCM2 without affecting MMS22L binding to TONSL1,2 (Fig. 1j; Extended Data Fig. 3d). These mutations did not affect ARD structure, as indicated by Circular Dichroism (Extended Data Fig. 3e). Together, this defines TONSL ARD as a recognition module for histone H4 tails, distinct from the GLP/G9A ARDs that binds histone H3 tails mono or dimethylated at K910 (Extended Data Fig. 4a, b).
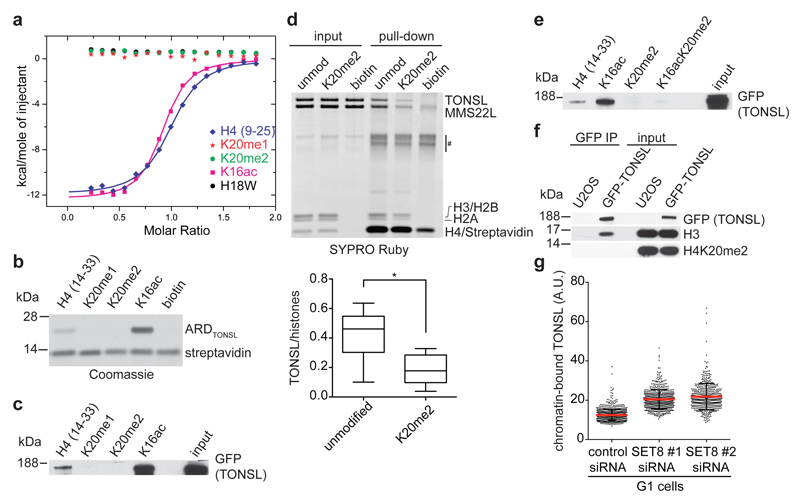
Figure 2. TONSL ARD recognizes unmodified H4K20.
a, ITC of TONSL ARD binding to H4 tail peptides. b, c, Pull-down of recombinant TONSL ARD (b) or GFP-TONSL from extracts (c) with biotinylated H4 tail peptides. d, (top) Pull-down of recombinant TONSL–MMS22L with biotinylated recombinant mononucleosomes. #, unspecific band. (bottom) TONSL binding quantified relative to histones. Unpaired t-test: *, P<0.05; mean n=6; whiskers, outliers. e, as c. f, Immunoprecipitation of GFP-TONSL from solubilised chromatin. g, TONSL chromatin-binding in pre-extracted PR-SET7/SET8 depleted G1 cells. Error bars, SD; n=747 (siCtrl), 579 (siSET8#1) and 485 (siSET8#2). Data are representative of 4 (b), 3 (c), and 2 (e-g) independent experiments. Peptide inputs, Extended Data Fig. 9d.
The structure predicts that methylation on H4K20 should break critical hydrogen bonds with the TONSL ARD. Isothermal titration calorimetry (ITC) and H4 tail peptide pull-downs confirmed that H4K20me1/2 is incompatible with TONSL binding (Fig. 2a-c). Further, H4K20me2 significantly reduced binding of full-length recombinant TONSL–MMS22L to reconstituted mono-nucleosomes (Fig. 2d). Recently, TONSL ARD with its neighboring acidic stretch was proposed to bind H3K9me15, but we were unable to detect an interaction between TONSL ARD (with or without the acidic stretch) and H3K9me1 peptides (Extended Data Fig. 4c, d). Together, our data show that TONSL binds to both free histones and nucleosomes via ARD recognition of H4 tails unmodified at K20 (Fig. 1a-j, Fig. 2a-e, Extended Data Fig. 1b, 2a-c, 3a-d). In line with this, H4K20me2 was not detected on TONSL-bound nucleosomal histones (Fig. 2f), while H4K16ac was present (Extended Data Fig. 5a). H4K16ac stimulated TONSL binding in peptide pull-downs (Fig. 2b, c, Extended Data Fig. 5b) and slightly enhanced ARD binding in ITC (Fig. 2a), but it did not overturn the inhibitory effect of H4K20me2 (Fig. 2e). However, H4K16ac is not essential for TONSL binding in vivo, as soluble histone H4 does not carry H4K16ac11 and depletion of MOF, the major H4K16 acetyltransferase12, did not significantly affect TONSL binding to chromatin (Extended Data Fig. 5c, d). In contrast, depletion of the H4K20 methyltransferase PR-SET7/SET8 significantly enhanced TONSL binding to chromatin in G1 cells where H4K20me2 peaks13–16 (Fig. 2g, Extended Data Fig. 5e, f). We conclude that TONSL ARD is a histone reader domain specific for H4 tails unmethylated at K20.
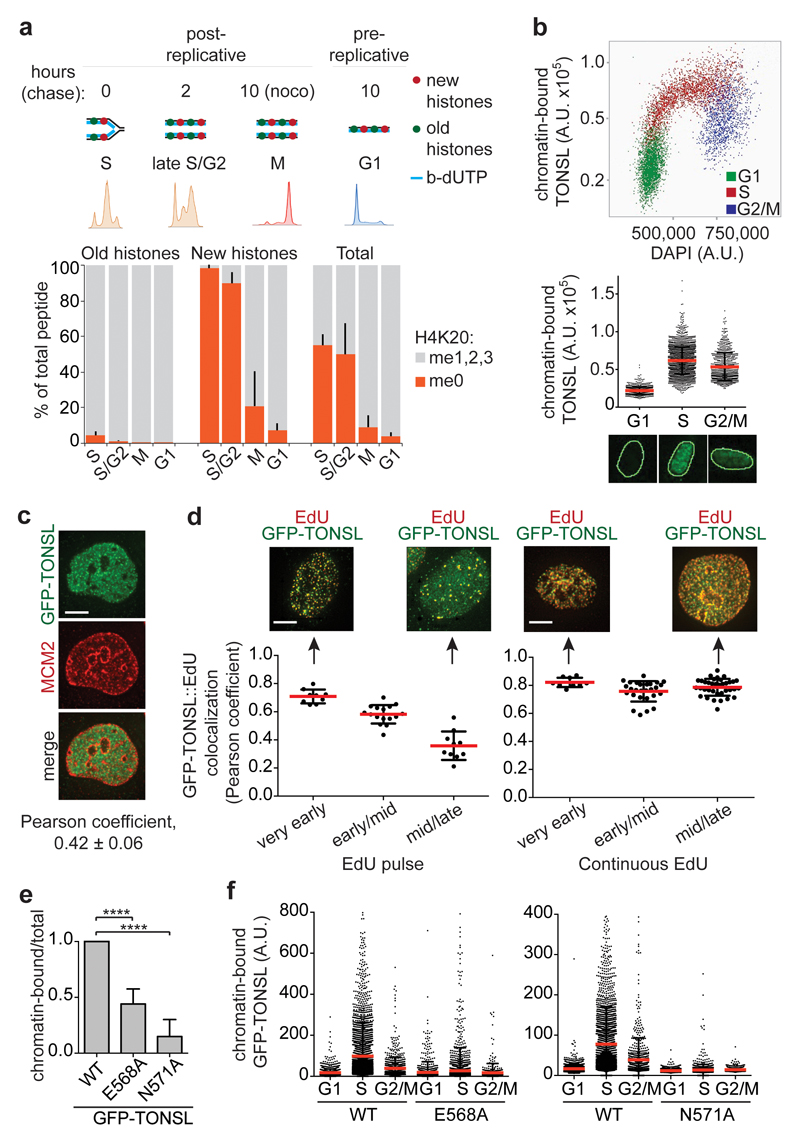
Given that TONSL–MMS22L binds new histones (devoid of H4K20me11,17) in a pre-deposition complex with ASF1 and MCM21 (Fig. 1j, Extended Data Fig. 1a), TONSL–MMS22L could be loaded onto replicating DNA together with new histones. To test how long after deposition new histones remain unmethylated at H4K20 with the potential to bind TONSL, we extracted H4K20 data from our recent large-scale proteomic study18 tracking modifications on new and old recycled histones by Nascent Chromatin Capture (NCC)19 (Fig. 3a, Extended Data Fig. 6a, b). In nascent chromatin, new histones were exclusively unmethylated at H4K20 (98% H4K20me0), while old recycled histones were almost fully methylated at H4K20 (me1, 7%; me2, 88%; me3, 2%). Consistent with previous work13–16, our analysis of primary cells (Extended Data Fig. 6c) and degradation of PR-SET7/SET8 in S phase15,16, new histones became methylated in late G2/M, rendering G1 chromatin devoid of H4K20me0 (Fig. 3a). This identifies H4K20me0 on new histones as a signature of post-replicative chromatin, implying that TONSL–MMS22L can bind H4 tails on new histones at replication forks and sister chromatids until late G2/M. Confirming this prediction, TONSL accumulated on chromatin in S phase, remained chromatin-bound in a population of G2 cells and was excluded from chromatin in G1 (Fig. 3b, Extended Data Fig. 6d-f). To discriminate pre- and post-replicative chromatin, we labeled replicating DNA with EdU (pulse to mark ongoing replication, continuous labeling to identify post-replicative chromatin) and stained pre-replicative chromatin with MCM220,21, and analyzed co-localization with TONSL. TONSL staining was mutually exclusive with MCM2 (Fig. 3c, Extended Data Fig. 7a), but co-localized with EdU pulse labeling in very early S phase and with replicated DNA (continuous EdU labeling) throughout S phase (Fig. 3d, Extended Data Fig. 7b, c). TONSL was present at sites of ongoing DNA replication throughout S phase, but the degree of co-localization declined in mid/late S (Fig. 3d, left panel), consistent with TONSL binding to post-replicative chromatin also after fork passage (Fig. 3d, right panel). Mutation of the TONSL ARD abrogated recruitment of TONSL to chromatin, including DNA replication sites (Fig. 3e, f, Extended Data Fig. 7d-g). Together, this demonstrates that TONSL is recruited to replication forks and post-replicative chromatin via ARD recognition of H4K20me0 on new histones.
Figure 3. H4K20me0 is a signature of post-replicative chromatin, read by TONSL ARD.
a, H4K20me on new and old histones analysed by SILAC-based mass spectrometry of chromatin pulse-labelled with b-dUTP and isolated by NCC (from18). Error bars, SD; n=9 (S), 3 (S/G2, M), 5 (G1); M (old histones) shows the mean of n=2; see also Extended Data Fig. 6a,b. b, TONSL chromatin-binding in pre-extracted TIG3 fibroblasts shown as a function of DAPI intensity or cell cycle stage. Error bars, SD; n=886 (G1), 2194 (S) and 756 (G2); representative cells are shown. c, d, Co-localization analysis of chromatin-bound GFP-TONSL with MCM2 (c) and EdU (d). (d) Cells were pulsed with EdU (left) or released into S phase in presence of EdU (right) and analysed by deconvolution microscopy. Error bar, SD; c, n=13; d from left, n=9, 16, 10, 9, 27, 36; representative cells are shown. Scale bar, 5 μm. e, Chromatin-binding of GFP-TONSL analysed by cellular fractionation, quantified relative to total GFP-TONSL and normalized to WT. Error bar, SD; n=5 (WT/N571A); n=3 (E568A). Unpaired t-test: ****, P<0.0001. f, Chromatin-binding of GFP-TONSL analysed as in b. Error bar, SD; from left, n=1302, 3567, 750, 1311, 3850, 838, 1495, 3221, 832, 1695, 2729, 877. Data are representative of 2 (b-d, f) independent experiments.
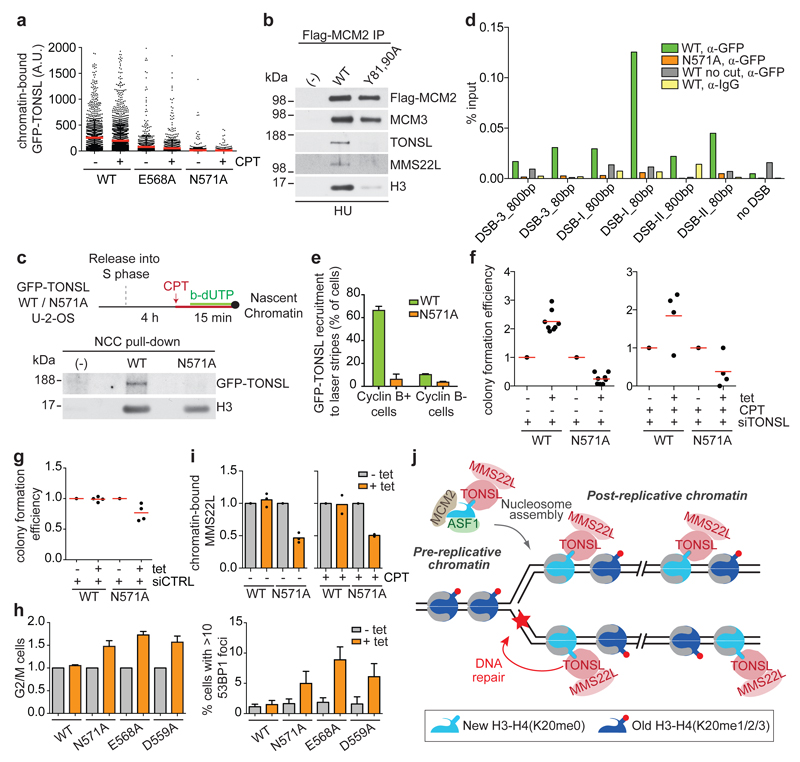
Mutation of TONSL ARD also abrogated chromatin binding and recruitment to replication forks in the presence of replication poisons like camptothecin (CPT) and hydroxyurea (HU) (Fig. 4a-c). Furthermore, ARD mutation prevented accumulation of TONSL at site-specific DSBs (Fig. 4d, Extended Data Fig. 8a) and microlaser-generated DNA damage (Fig. 4e, Extended Data Fig. 8b, c). Co-staining with cell cycle markers confirmed that TONSL is recruited to DNA repair sites only in S and G2 cells as expected2 (Fig. 4e, Extended Data Fig. 8d, e). We conclude that H4K20me0 binding is required for TONSL accumulation at damaged forks and DNA lesions in post-replicative chromatin. However, this was not due to increased H4K20me0 (Extended Data Fig. 8f), suggesting that unmasking of H4 tails upon chromatin decompaction9,22 and/or interaction with repair factors contribute to TONSL–MMS22L accumulation at repair sites. Consistent with the latter, MMS22L interaction with Rad51 can stabilize the complex at challenged forks (P. Cejka and M. Peter, personal communication). Our data argue that this is subsequent to H4K20me0 binding (Fig. 4a-e), and we thus next addressed the contribution of H4K20me0 recognition to TONSL–MMS22L function. In complementation analysis, TONSL WT partially rescued viability of TONSL depleted cells in the presence and absence of CPT (Fig. 4f, Extended Data Fig. 8g, h), whereas TONSL ARD mutants were highly toxic (Fig. 4f, Extended Data Fig. 8g, h). In control cells, TONSL ARD mutants also reduced viability, causing G2/M arrest accompanied by replication-associated DNA damage (Fig. 4g, h). Further, TONSL ARD mutants titrated MMS22L away from chromatin (Fig. 4i, Extended Data Fig. 8i), explaining the dominant negative phenotype that mimics TONSL–MMS22L depletion1–4. Collectively, this indicates that recognition of H4K20me0 is central to TONSL-MMS22L function in safeguarding genome stability.
Figure 4. H4K20me0 recognition is required for TONSL accumulation at DNA repair sites and genome stability.
a, Chromatin-binding of GFP-TONSL in CPT treated S-phase cells. Error bar, SD; from left, n=1461, 2631, 1245, 1764, 2116, 3178. b, Co-immunoprecipitation of TONSL-MMS22L with Flag-HA-MCM2 WT or histone binding mutant (Y81A, Y90A)7 from chromatin after HU treatment. c, NCC analysis of GFP-TONSL recruitment to replication forks in CPT treated cells. (-), no biotin-dUTP. d, ChIP-qPCR analysis of GFP-TONSL recruitment to site-specific DSBs induced by AsiSI29. See Extended data Fig. 8a for additional controls. e, GFP-TONSL recruitment to laser-induced DNA lesions (error bars, SD; n=3; total cells counted, 210 (WT) and 252 (N571)). f, g, Colony formation upon GFP-TONSL induction (+tet) in siRNA and CPT treated cells. h, Cell cycle and 53BP1 foci analysed by microscopy. (left) % G2/M cells shown relative to non-induced. Error bars, SD, n= 4 (left), 5 (right). i, Chromatin-bound MMS22L analysed as in Fig. 3e. Mean with individual data points are shown (n=3 (untreated), 2 (CPT)), see Extended Data Fig. 8i for western blots. j, TONSL-MMS22L identifies post-replicative chromatin by binding H4K20me0 on new histones, directing TONSL-MMS22L genome surveillance function to DNA having a sister chromatid. Data are representative of 3 (a), 2 (b-d, f-right, g), and 4 (f-left) independent experiments. Protein inputs, Extended Data Fig. 9e, f.
This study reveals that post-replicative chromatin has a distinct histone modification signature, read by the TONSL–MMS22L effector protein (Fig. 4j). This opens a new avenue to understand how DNA repair and other chromosomal transactions can be directly linked to the replication state of a genomic locus. Intriguingly, it is the new histones that make post-replicative chromatin distinct, and in this way H4K20me0 resembles the behavior of H3K56ac23 in yeast. Our data argues that TONSL–MMS22L is delivered to nascent chromatin with new histones via the pre-deposition complex with MCM2 and ASF1 (Fig. 4j). We favor the idea that TONSL has a dual function as a histone chaperone5 and histone reader. Our structural work proposes that TONSL acts in a histone chaperone-like capacity by sequestering the H4 tail to prevent spurious contacts with DNA during H3-H4 deposition. Further, TONSL ARD may counteract chromatin compaction by preventing association of the H4 tail with the H2A–H2B acidic patch on neighboring nucleosomes. Thus, TONSL changes our perception of a histone chaperone by binding both soluble and nucleosomal histones. In its function as a histone reader, TONSL localizes MMS22L to post-replicative chromatin via H4K20me0 and allows TONSL–MMS22L to accumulate at damaged forks and DNA lesions. We envision that H4K20me0 works as an affinity trap, making TONSL–MMS22L readily available to support Rad51 loading during HR. This provides a new approach and opportunity to understand the role of H4K20 in DNA repair, complementing the well-described role of H4K20me1/2 in recruiting 53BP124 to promote NHEJ in competition with BRCA1-BARD125. In post-replicative chromatin, H4K20me1/2 on old histones will support 53BP1 recruitment. Whether H4K20me0 on new histones also influences DNA repair pathway choice will be of interest to future investigations. It is notable that the structure of the TONSL ARD, including the histone-binding surface, is highly similar to the ARD of BARD126 (Extended Data Fig. 9a), required for BRCA1 tumor suppressor function and HR27. Multiple mutations in the TONSL ARD are reported in cancer (C608G, COSM4879909; P557S, COSM4565032; E597K, COSM3382163) and the N571 residue, key to histone H4 binding, corresponds to the BARD1 N470S cancer mutation26,28. This underscores that the tumor suppressor function of H4K20me0 recognition and the possibilities it brings for targeted cancer therapy should be explored in the future.
Materials and Methods
Protein expression and purification
All proteins used in this study, unless otherwise indicated, were expressed in BL21(DE3)-RIL cell strain (Stratagene). The human TONSL Ankyrin Repeat Domain (ARD, residues 512–692) and MCM2 Histone-binding Domain (HBD, fragments 61–130) were covalently linked through a four-Glycine linker (G4 linker) into one expression cassette. The MCM2 HBD–G4–TONSL ARD expression cassette was cloned into a modified RSFDuet-1 vector (Novagen), with an N-terminal His6-SUMO tag. The resulting plasmid was coexpressed with plasmid harbouring histone genes H3.3(Δ56) and H4. The expressed protein complex was first purified on Ni-NTA affinity column. After removing the His6-SUMO tag by using Ulp1 (SUMO protease), the protein complex was further purified on HiLoad 16/600 Superdex 200 column (GE Healthcare).
The GST-tagged TONSL ARD and its mutants including E530A, D559A, W563A, E568A, N571A and D604A were cloned into pGEX-6P-1 vector (GE Healthcare). The expressed proteins were first purified using Glutathione Sepharose 4B, then further purified by gel-filtration step. In some case, the GST-tag was removed with 3C protease before gel-filtration step. For purification of GST-H3 tail and GST-H4 tail proteins, the human histones H3 fragment 1–59 and H4 fragment 1–31 were cloned into pGEX-6P-1 vector respectively. The proteins were expressed and purified in the same way.
For production of recombinant full-length TONSL-MMS22L heterodimer, the sequence coding for full-length MMS22L was fused with a MBP tag at the 5’ end and 10×His tag at the 3’ end. The sequence coding for full-length TONSL was fused with GST tag at the 5’ end. Both MMS22L and TONSL constructs were cloned into a pFastBac1 vector. The complex was expressed in Sf9 cells by co-infection with both recombinant baculoviruses according to manufacturer’s recommendation (Invitrogen). The proteins were extracted from Sf9 cells and purified similarly as described previously for Sgs130. Briefly, the complex was purified on amylose resin, and MBP and GST tags were subsequently cleaved with PreScission protease. The heterodimer was then further purified using a Ni-NTA affinity resin. Washes were performed with 300 mM NaCl buffer.
Crystallization
At first, we tried to crystallize TONSL ARD in complex with a H4 tail or H3–H4 tetramer, but failed even with extensive screening. Additional binding protein may help to stabilize the whole complex and help for crystallization. Then we tried to crystallize TONSL ARD in complex with the MCM2 HBD and H3–H4 tetramer. We just got very tiny crystals for this complex, but failed to get big and well diffracted crystals. We realized that the whole complex of TONSL ARD with MCM2 HBD and H3–H4 tetramer might be destabilized by the harsh crystallization conditions and form subcomplex, thus hindered the optimization of the crystals. Then we tried to covalently link TONSL ARD and MCM2 HBD into one cassette through different length of Glycine linker (Gx linker). The G12, G11, G10, G9, G8, G7, G6, G5 and G4 linkers had been tried and all these cassettes could be crystallized. One of the constructs with a G4 linker gave well-diffracted crystals.
The G4 linker complex, MCM2 HBD–G4–TONSL ARD cassette–H3.3(Δ56) –H4 complex (herein denoted as TONSL ARD–MCM2 HBD–H3–H4 tetramer complex) in a concentration of 23 mg ml-1 was crystallized in 0.1M MES pH 5.6, 7% isopropanol using sitting-drop vapour-diffusion method at 20 °C. All the crystals were soaked in a cryoprotectant made from mother liquor supplemented with 25% glycerol before flash freezing in liquid nitrogen.
Structure determination
The data sets for the TONSL ARD–MCM2 HBD–H3–H4 tetramer complex were collected at 0.979 Å on 24-ID-C/E NE-CAT (Advanced Photo Source, Argonne National Laboratory). All the data sets were processed by using the HKL 2000 program. The initial structure for the TONSL ARD–MCM2 HBD–H3–H4 tetramer complex was solved by molecular replacement in PHASER31 with our previous structure of the MCM2 HBD–H3–H4 tetramer complex7 as a search model and manually refined and built using Coot32. The final structure of this complex was refined to 2.43 Å resolution using PHENIX33. The Ramachandran plot showed 95.9% favored and 4.1% allowed. Extended Data Table 1 summarizes the statistics for data collection and structural refinement.
Preparation of recombinant modified mono-nucleosomes
Recombinant human histone proteins were expressed in E. coli BL21(DE3)-RIL cells from pET21b(+) (Novagen) vectors and purified by denaturing gel filtration and ion exchange chromatography essentially as described34. All histone proteins were dialysed into water containing 1mM DTT, lyophilised and stored dry at -80°C. Modified H4 proteins were generated by native chemical ligation essentially as described for H335. Briefly, tail-less H4 Δ1-28 I29C protein was expressed in E. coli BL21(DE3)-RIL cells from pET24b(+) (Novagen) and purified by denaturing gel filtration and reversed phase chromatography using a ResourceRPC column (GE Healthcare). Purified H4 Δ1-28 I29C was then ligated to N-terminally acetylated H4 1-28 thioester peptides (Almac) and full length ligated H4 was separated from unligated H4 Δ1-28 I29C by reversed phase chromatography via a C18 column (Aquapore RP-300/Perkin Elmer) using a gradient from 35% B to 45% B over 20 column volumes (A: 0.1%TFA in water, B: 90% acetonitrile; 0.1% TFA). Ligated H4 was directly lyophilised and stored dry at -80°C. Ligated H4 was refolded into octamers together with purified histones H2A, H2B and H3.1 and then assembled into nucleosomes with biotinylated 601-DNA as described34,35.
GST pull-downs
For pull-downs of GST-ARD and its mutants including E530A, D559A, W563A, E568A, N571A and D604A with H3–H4, first 25 μL of Glutathione Sepharose 4B beads were suspended with 200 μL of binding buffer (20 mM Tris pH 7.5 and 0.5 M NaCl), and 1 nmol of GST-ARD proteins were added and incubated at 23 °C for 10 minutes (min); then 0.5 nmol of pre-purified H3/H4 tetramers were added and incubated for another 1 hour; then the beads were washed quickly with five times 1 mL of washing buffer (binding buffer, 1% Triton X-100) before adding 50 µL of sample loading buffer. An aliquot of 20 μL of each sample was analysed with SDS-PAGE. The GST pull-downs of histone tails of GST-H31-59 and GST-H41-31 with TONSL ARD were performed similarly.
Circular Dichroism
Circular dichroism spectra were acquired using a Jasco J-815 Circular Dichroism (CD) Spectropolarimeter with a 1 mm quartz cuvette. Spectra were recorded for wild type and mutant TONSL ARD (512-692, 6.25 µM) between 260 nm and 195 nm in KH2PO4/K2HPO4 buffer (25 mM, pH 7.8) with a data pitch of 0.5 nm, bandwidth of 1 nm and with three accumulations at a scanning speed of 50 nm/min.
In vitro translation (IVT) and pull-downs with H3–H4 sepharose beads
NHS-activated sepharose 4 Fast-Flow beads (GE Healthcare) were washed with 0.1M HCl and incubated O/N with 1 μM recombinant histone H3.1–H4 tetramers (New England Biolabs, catalog # M2509S) or 1 μM recombinant histone H2A–H2B dimers (New England Biolabs, catalog # M2508S) in Coupling Buffer (0.2M NaHCO3, 0.2M NaCl). 1 μg of pSC-B-TONSL, pEXPR-IBA-105-ASF1A WT and pEXPR-IBA-105-ASF1A V94R plasmids was incubated with TnT® Quick Coupled Transcription/Translation System (Promega) and 35S-methionine according to the manufacturer’s instructions. 10 μl of IVT mixture were added to the H3.1–H4, or H2A–H2B, sepharose beads and incubated for 2 hours. Beads were washed with 200 mM NaCl, 0.2% NP40 buffer. Beads were boiled in 1× LSB and loaded on a 4–12% Bis-Tris NuPage gel (LifeTechnologies). Proteins were transferred to a 0.2 μm nitrocellulose membrane by O/N wet transfer at 20V and the membrane was incubated in an autoradiography cassette for 24 hours before detection by Phosphor Imager (PerkinElmer).
Isothermal Titration Calorimetry (ITC) experiments
All the ITC titrations were performed on a Microcal ITC 200 calorimeter at 25 °C or 20 °C. The peptides of H4 (residues 9–25) and its modified peptides K16ac (with acetylation on Lys16), H18W (with His18 mutated to Trp18), H4K20me1 (mono-methylation on Lys20) and H4K20me2 (di-methylation on Lys20), and peptide of H3(1-21)K9me1 (mono-methylation on Lys9) were all synthesized at Tufts University Core Facility The exothermic heat of the reaction was measured by 17 sequential 2.2 μl injections of the peptides (1.41 mM in buffer 20 mM Tris pH 7.5 and 0.5 M NaCl) into 200 μl of the TONSL ARD solution (145 μM in the same buffer), spaced at intervals of 150 s or 180s. The data were processed with Microcal Origin software and the curves were fit to a single site binding model.
Peptide pull-downs assays
Purified recombinant TONSL ARD (residues 512-692) was stored at 400 µM in 1 M NaCl, 20 mM Tris HCl pH 7.5 at -80 °C. For each pull-down, 400 pmol of the ARD stock (1 μl, 400 μM) was diluted with 99 μl of binding buffer (150 mM NaCl, 50 mM Tris HCl pH 7.5, 5 % Glycerol, 0.25 % NP-40, 0.2 mM EDTA, 0.5 mM DTT, 0.2 mM PMSF, 1 mM Leupeptin, 1 mM Pepstatin). ARD input material was scaled to the number of pull-downs performed. For each pull-down, a H4 peptide (JPT Peptide Technologies GmbH) spanning residues 14-33 (2.5 μl, 250 μM) with a C-terminal biotinoyl-lysine residue or, as control, biotin (2.5 μl, 400 μM) was added to 1.1 ml of binding buffer in addition to 100 μl of the ARD input material and the mixture incubated overnight rotating at 4 °C. The next day 25 μl of MyOne Streptavidin C1 beads (Life Technologies) was washed in binding buffer (3 × 500 µl) for each pull-down removing the final wash from the beads. The ARD/peptide or ARD/biotin mixture was added to an aliquot of pre-washed MyOne Streptavidin C1 beads and incubated with rotation at 4 °C for 3 hours. Finally the beads were washed (2 × 300 μl and 1 × 200 μl of 300 mM NaCl, 50 mM Tris HCl pH 7.5, 5 % Glycerol, 0.25 % NP-40, 0.2 mM EDTA, 0.5 mM DTT, 0.2 mM PMSF, 1 mM Leupeptin, 1 mM Pepstatin) and pulldown material visualized by coomassie staining after SDS PAGE separation of proteins on a NuPAGE 4–12 % gel.
For pull-downs from cell extracts, MyOne T1 beads were incubated O/N with 1 μg of biotinylated peptides in High Salt (HS; 300mM NaCl, 0.5% NP40, Tris HCl, EDTA, 5% glycerol) buffer and subsequently washed 2 times with PBS. 1 mg of NP40/NaCl extract from HeLa S3 or GFP-TONSL U-2-OS cells was added to the beads and incubated for 2 hours rotating at 4°C. The beads were then washed 5 times with HS buffer, 2 min rotating at 4°C. After washing, the beads were resuspended in 1 × LSB and boiled for 10 min. The eluted proteins were loaded on a 4–12% Bis-Tris NuPage gel (LifeTechnologies). Proteins were then transferred to a 0.2 μm nitrocellulose membrane by O/N wet transfer at 20V and detected by western blotting.
Nucleosome pull-down assay
Modified nucleosomes of H4K20me0 or H4K20me2 were prepared by peptide ligation and stored at 0.1 μg/μl (by histone octamer) in 100 mM NaCl, 50 mM Tris HCl pH 7.5 at 4 °C. Full-length TONSL-MMS22L complex was stored at 746 nM in 100 mM NaCl, 50 mM Tris HCl pH 7.5, 5 mM β-mercaptoethanol, 10% Glycerol, 0.5 mM PMSF at -80 °C. Nucleosome pull-downs were performed across two sets of conditions (n=3 for each condition) in the presence of Herring Sperm competitor DNA. Condition #1: Nucleosomes (1 μg by histone octamer) or Biotin (0.5 μg) were mixed with TONSL-MMS22L (1.9 pmoles) and made up to 30 μl with binding buffer (500 mM NaCl, 50 mM Tris HCl pH 7.5, 20 % Glycerol, 0.1 % NP-40, 1 mM DTT, protease inhibitors and 10 μg/ml Herring Sperm DNA (Sigma)). Inputs of 10 μl were taken before diluting each sample with binding buffer to a final volume of 300 μl and incubating overnight at 4 °C. Pull-downs were performed by adding 20 μl of MyOne Streptavidin C1 beads prewashed and resuspended in 100 μl of binding buffer to each pull-down reaction, incubating at 4 °C for 2 hours, washing with 5 × 500 μl binding buffer 2 min at room temperature. Condition #2: Nucleosomes (0.5 μg by histone octamer) or Biotin (0.5 μg) were mixed with TONSL-MMS22L (1.3 pmoles) and made up to 30 μl with binding buffer (500 mM NaCl, 50 mM Tris HCl pH 7.5, 5 % Glycerol, 0.5 % NP-40, 0.2 mM EDTA, 1 mM DTT, protease inhibitors and 10 μg/ml Herring Sperm DNA (Sigma)). Inputs of 15 μl were taken before diluting each sample with binding buffer to a final volume of 500 μl and incubating overnight at 4 °C. Pull-downs were performed by adding 10 μl of MyOne Streptavidin T1 beads prewashed and resuspended in 100 μl of binding buffer to each pull-down reaction, incubating at 4 °C for 4 hours, washing with 5 × 500 μl binding buffer 2 min at room temperature. Pull-downs were visualized by SYPRO Ruby staining after SDS PAGE separation of proteins on a NuPAGE 4–12 % gel using an ImageQuant LAS 4000 (GE Healthcare). The intensity of stained bands were quantified using ImageJ, TONSL intensity was normalized to the combined intensity of H3, H2A and H2B. Statistical analysis was performed using data from the 6 independent experiments using the unpaired t-test with equal standard deviations in Prism 6.
Cell culture, transfection and drug treatment
U-2-OS (kind gift of Dr. Bartek, the Danish Cancer Society, Denmark), HeLa S3 (kind gift of Dr. Nakatami, Dana-Faber Cancer Institute, US) and TIG-3 (kind gift of Dr. Hansen, BRIC, University of Copenhagen, Denmark) cells were grown in DMEM (Gibco) containing 10% FBS (Hyclone) and 1% penicillin/streptomycin and drugs for selection. The construct for siRNA resistant GFP-TONSL was described2 and ARD mutation were introduced in this construct by site-directed mutagenesis. The construct for pBABE-SNAP-HA-H4 plasmid was described36 and H4 tail mutations were introduced in this construct by site-directed mutagenesis. Cells inducible for GFP-TONSL WT and ARD mutants were generated in Flp-In T-Rex U-2-OS cells (Invitrogen) by transfection of pcDNA5/FRT/TO-GFP-TONSL plasmids with Lipofectamine 2000, according to the manufacturer’s protocol, and selection with hygromycin (200 µg/ml). Previously described inducible GFP-TONSL U-2-OS cells1 were used for Fig. 1h and Fig. 2c,e. U-2-OS Flag-HA-MCM2 WT and Y81A, Y90A were previously described7. pBABE-AsiSI–ER–HA29 was introduced into inducible GFP-TONSL cell lines by lentiviral infection and puromycin selection. All cell lines were authenticated by western blotting and/or immunofluorescence. All cell lines used in this study tested negative for mycoplasma contamination. Expression of GFP-TONSL was induced by addition of 1 μg/ml of tetracycline for 24 hours. U-2-OS and TIG3 cells were synchronized by a single thymidine block (2mM) and released into S phase in the presence of 24 μM dCTP. For transient expression of GFP-TONSL or SNAP-HA-H4 (Fig. 1h, j), expression plasmids were introduced by transfection with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol and cells harvested 24 hours after transfection. siRNA transfection was performed with RNAiMax reagent (Invitrogen) according to the manufacturer’s protocol. All siRNAs were used to a final concentration of 50 nM.
siRNA sequences (Sigma): siSET8#1: 5’-GUACGGAGCGCCAUGAAGU-3’; siSET8#2: 5’-ACUUCAUGGCGCUCCGUACUU-3’37; siMOF#1: 5’-GUGAUCCAGUCUCGAGUGA-3’12; siMOF#2: 5’-GAGAUCAACCAUGUGCAGA-3’; siTONSL: 5’-GAGCUGGACUUAAGCAUGA-3’2.
Drug treatment. CPT: cells were either treated with 1 μM CPT for 3 hours (Fig. 4a, i; Extended Data Fig. 8f) or 20 minutes (Fig. 4c), or with 50 nM CPT for 24 hours (Fig. 4f). HU: Cells were treated with 3mM HU for 2 (Fig. 4b) or 3 (Extended Data Fig. 8f) hours.
Cell extracts and chromatin solubilisation
For detergent/salt soluble extracts, HeLa S3 and U-2-OS cells were washed with cold PBS, scraped and incubated for 15 mins on ice in HS buffer supplemented with trichostatin A (TSA) and protease and phosphatase inhibitors (5 mM sodium fluoride, 10 mM β-glycerolphosphate, 0.2 mM sodium vanadate, 10 μg/ml leupeptin, 10 μg/ml pepstatin, 0.1 mM PMSF, Sigma). After centrifugation at 16.000 g for 15 min at 4°C, the supernatant was collected. To analyse chromatin-bound complexes, cells were washed twice in cold PBS, scraped and centrifuged at 1.500 g for 10 min at 4°C. The pellet was incubated on ice for 10 mins in CSK buffer (10 mM PIPES pH 7, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2)/0.5% Triton X-100, supplemented with TSA and protease and phosphatase inhibitors (5 mM sodium fluoride, 10 mM β-glycerolphosphate, 0.2 mM sodium vanadate, 10 μg/ml leupeptin, 10 μg/ml pepstatin, 0.1 mM PMSF, Sigma) and subsequently centrifuged at 1.500 g for 10 min to collect soluble proteins. For DNaseI or benzonase release of chromatin material, the remaining pellet was resuspended in CSK/0.1% Triton X-100 containing DNase I (1000 U/mL, Roche), or Benzonase (2500 U/mL, Millipore), and incubated at 30°C for 30 mins. Solubilized chromatin was then collected by centrifugation at 16.000 g for 10 mins.
Immunoprecipitation from cell extracts
Immunoprecipitation was performed with agarose magnetic GFP-Trap beads (Chromotek), anti-FLAG magnetic beads (Sigma) and anti-HA magnetic beads (Life Technologies). Cell extracts were incubated with beads for 2 hours at 4°C rotating. The beads were subsequently washed 5 times with HS buffer and resuspended in 1 × LSB prior to boiling and SDS PAGE separation of proteins on a NuPAGE 4–12 % gel.
Western blotting and antibodies
The following antibodies were used: TONSL (Abcam ab101898), TONSL (Sigma, HPA024679; validated in Extended Data Fig. 6d), MMS22L1, H3 (Abcam ab1791, Abcam ab10799), GFP (Santa Cruz sc-8334, Abcam ab290), biotin (Abcam ab53494), MCM2 (BD Biosciences 610701), H2B (Abcam ab1790), H4K16ac (Millipore 07-329), H4K20me1 (Abcam ab9051), H4K20me2 (Cell Signalling 9759), 53BP1 (Santa Cruz sc-22760; Novus Biologicals NB100-904), γ-H2AX (Millipore 05-636), Cyclin B (BD Biosciences 610220), RPA70 (Abcam ab79398), SET8 (Millipore, 06-1304), MCM3 (Abcam ab 4460). Secondary antibodies conjugated with HRP were from Jackson ImmunoResearch Labs. Signals were revealed by chemiluminescence substrate from Pierce (SuperSignal West Pico or SuperSignal West Femto).
Fluorescence Activated Cell Sorting (FACS) and analysis
For analysis of cell cycle progression, cells were fixed in 70% ethanol and stained with propidum iodide/RNase for 30 minutes in the dark, before analysis on a FACS Calibur machine. FACS profiles were analyzed by FlowJo 10.0.8 software.
Mass spectrometry
Histones from TIG3 fibroblasts were extracted from chromatin as previously described18. Protein was resuspended in 50 μl of 100 mM triethylammonium bicarbonate (TEAB, Sigma), pH adjusted with 2 µl of 1.5 M Tris pH 8 and digested for 16 hours at 37°C with 3 µl of 20 ng/μl Asp-N (Wako) in 100 mM TEAB. After 15 min centrifugation at 10.000 g at 25°C, the SN was placed in a new tube and digestion was repeated for the pellet during 4 hours at the conditions described above. The digested peptides of both digestions were merged, acidified with 10 µl of 1 % TFA and purified using sequential Stagetip C18 and Carbon Toptip (Glygen). Purified peptides were evaporated, resuspended in 15 µl of 0.1 % TFA. Injected material was normalized to analyse by LC-MS the histones corresponding to 9.0 × 105 cells. The LC method was used as described elsewhere18. The MS was performed in an Orbitrap Classic with similar settings as described before18 but with survey scan range at 550–690 m/z and MS2 set in scheduled and targeted data independent mode for the four-time charged ions of the 4 different methylation states (unmodified, mono-, di- and trimethylated H4K20) for the 5 different acetylated forms of S1–R23 peptide of histone 4 (N-term, K5, K8, K12 and K16). Peptides were quantified using the peak area from the corresponding extracted ion chromatograms (±10 ppm).
Immunofluorescence, microscopy and laser microirradiation
U-2-OS cells conditional for GFP-TONSL were grown on glass coverslips or 96-well plates and either directly fixed in 4% paraformaldehyde (PFA) for 10 mins or washed in CSK, pre-extracted 5 min with cold CSK/0.5% Triton X-100 and rinsed with CSK and PBS before fixation in 4% PFA for 10 mins. Coverslips were mounted on glass slides with Mowiol mounting medium (Sigma Aldrich) containing DAPI. Fluorescence images were collected on a DeltaVision system with a 40X or 60X oil immersion objective. For co-localization analysis by deconvolution microscopy, z-stacks were acquired (step of 0.2 μm), deconvolved and analysed by SoftWoRX 5.0.0. Pearson coefficient correlation analysis was performed on single cells using SoftWoRX 5.0.0. Brightness and contrast were adjusted using Adobe Photoshop CS6. For high content quantitative analysis, fluorescence images were acquired using an Olympus ScanR high-content microscope and processed on the ScanR analysis software. More than 5000 cells per sample were analysed. Cell cycle phases were gated on DAPI and EdU intensity. Graphs were generated with TIBCO Spotfire software. For microirradiation experiments, cells grown on glass coverslips were fixed in 4% formaldehyde for 15 min, permeabilised with PBS containing 0.2% Triton X-100 for 5 min and incubated with primary antibodies diluted in DMEM for 1 hour at room temperature. Following staining with secondary antibodies (Alexa Fluor 488, 568 and 647; Life Technologies) for 30 min, coverslips were mounted on glass slides in Vectashield mounting medium (Vector Laboratories) containing the nuclear stain DAPI. For detection of nucleotide incorporation during DNA replication, an EdU-Plus labeling kit (Life Technologies) was used according to the manufacturer’s instructions. Confocal images were acquired on an LSM-780 (Carl Zeiss) mounted on a Zeiss-AxioObserver Z1 equipped with a Plan-Neofluar 40X/1.3 oil immersion objective. Image acquisition and analysis was carried out with LSM-ZEN software. Laser microirradiation of cells was performed essentially as described38.
ChIP
GFP-TONSL WT and N571A U-2-OS harbouring the inducible ER-HA-AsiSI endonuclease29 were treated with 4-OHT and 10μM DNA-PK inhibitor NU7026 (Millipore) for 4 hours to increase HR efficiency39. Cells were cross-linked for 10 min in 1% formaldehyde and chromatin was fragmented by sonication using Bioruptor Sonicator (Diagenode). Chromatin immunoprecipitation was performed as previously described40 with the following modifications: 30 μg of chromatin was immunoprecipitated with 5 μg of anti-GFP (Abcam, ab290) and rabbit-IgG. Immunoprecipitated DNA was analysed in duplicate by RT-qPCR. In all cases, γH2A.X induction was verified by immunofluorescence and a sample without 4-OHT was included as a ‘no cut’ control. Primer pairs for the analysis of DSB-3, DSB-I and DSB-II are described41. Primer sequences used for the amplification of a genomic region devoid of DSBs were as follows: noDSB-for: 5’-TGACAAGGACAGGGTCTTCC; noDSB-rev: 5’-CACCGTCCGTTGTATGTCTG. ChIP efficiency was calculated as percentage of input DNA immunoprecipitated.
NCC
The NCC protocol19 was adjusted for adherent U-2-OS cells. CPT (1μM) was added 5 minutes prior to b-dUTP labelling and was included in all steps until fixation. Cells were incubated for 5 minutes in a hypotonic buffer (50 mM KCl, 10 mM Hepes) containing biotin-dUTP and resuspended into fresh cell culture medium for an additional 15 minutes. Cells were fixed 15 minutes in 1% formaldehyde, rinsed twice in PBS and collected by scraping in cold room. Nuclei were mechanically isolated in sucrose buffer (0.3 M sucrose, 10 mM HEPES-NaOH at pH 7.9, 1% Triton X-100 and 2mM MgOAc). Chromatin was solubilized by 28 cycles 30 sec ON, 90 sec OFF in sonication buffer (10 mM HEPES-NaOH at pH 7.9, 100 mM NaCl, 2 mM EDTA at pH 8, 1 mM EGTA at pH 8, 0.2% SDS, 0.1% sodium sarkosyl and 1 mM phenylmethylsulphonylfluoride) using a Bioruptor at 4° C. Solubilized chromatin was pre-cleared using streptavidin-coated magnetic beads (MyC1 Streptavidin beads) pre-incubated with biotin. b-dUTP labelled chromatin was next purified over night at 4° C using streptavidin-coated magnetic beads. Beads were washed 5 times for 2 minutes in wash buffer (10 mM HEPES-NaOH pH 7.9; 200 mM NaCl; 2 mM EDTA pH 8; 1 mM EGTA pH 8; 0.1% SDS; 1 mM PMSF). Total chromatin (input) and isolated nascent chromatin were boiled 40 min on beads in LSB 1 X (50 mM Tris-HCl pH 6.8, 100 mM DTT, 2% SDS, 8% Glycerol, Bromophenol blue) and separated by SDS-PAGE for western blotting. Pulse-SILAC-NCC (Fig. 3a) was performed as described18.
Clonogenic assay
U-2-OS inducible for GFP-TONSL ARD WT and mutant were transfected with siRNA, trypsinised 24 hours later and seeded in technical triplicates of 1000 or 3000 cells in the presence or absence of tetracycline. After 24 hours, cells were washed to remove tetracycline and CPT was added for 24 hours as indicated. Cells were then cultured in fresh medium for 12-15 days before fixation and staining with MeOH/Crystal Violet. Colony formation efficiency was determined by manual colony counting or quantification of Crystal Violet staining by ImageJ software and normalized to non-induced control. Each data point represents a technical triplicate of 1000 or 3000 seeded cells within each biological replicate.
Extended Data
Extended Data Figure 1. TONSL binding to histones in vivo and in vitro.
a, Histones bridge the interaction between TONSL-MMS22L and MCM2 in cell extracts as shown by co-immunoprecipitation of Flag-HA-MCM2 WT or histone binding mutant (Y81A, Y90A)7. U-2-OS cell inducible for Flag-HA-MCM2 WT or Y81A, Y90A7 were induced for 24 hours prior to immunoprecipitation with Flag antibodies (one representative experiment out of two is shown). b, Immunoprecipitation of GFP-TONSL from solubilized chromatin of HeLa cells transiently transfected with GFP-TONSL plasmid, showing that TONSL associates with nucleosomal histones H3 and H2B (one representative experiment out of two is shown). c, Domain structure of TONSL. TPR, tetratricopeptide repeats. ARD, ankyrin repeat domain. UBL, ubiquitin-like domain. LRR, leucine-rich repeats1–4. d, Pull-down of GST-ARD with recombinant histones H3–H4 tetramers. e, f, Pull-down of in vitro-translated full length TONSL with recombinant histones H3–H4 tetramers (e) or H2A–H2B dimers (f) coupled to NHS-activated sepharose beads (one representative experiment out of three (e) and two (f) is shown). ASF1a WT and histone-binding mutant (V94R)1 were included as controls. g, TONSL ARD consists of four ankyrin repeats and uses its elongated concave surface to target the H4 tail spanning residues 12 to 23.
Extended Data Figure 2. Models and sequence alignment of TONSL ARD.
a, Pull-down assay of recombinant ARD with GST-H3 tail (a.a. 1–59) and GST-H4 tail (a.a. 1–31). b, Modelling of TONSL ARD on the co-chaperone structure of MCM2 HBD and ASF1 in complex with an H3–H4 dimer. When comparing the structure of the TONSL ARD–MCM2 HBD–H3–H4 tetramer complex with our previous structure of the MCM2 HBD–H3–H4 dimer–ASF1 complex (PDB 5BNX, Huang H. et al. NSMB 2015), the common parts of both structures superimposed well with a small r.m.s.deviation of 0.44 Å. A model of the quinary complex composed of one molecule of each protein TONSL ARD, MCM2 HBD, ASF1, H3 and H4, was made after superposition. This model shows that TONSL ARD, MCM2 HBD and ASF1 could simultaneously bind an H3–H4 dimer without steric clash. c, Model of TONSL ARD on the structure of the nucleosome. The model was generated by a direct superposition of the H3–H4 tetramer in the structure of the TONSL ARD–MCM2 HBD–H3–H4 tetramer complex onto the H3–H4 tetramer in the nucleosome structure (PBD 3AV2). There was no adjustment in the conformation of the model and no steric clash in the model. The MCM2 HBD molecules were omitted from the model for clarity. d, Alignment of TONSL ARD (512–692) sequences from H. sapiens, M. musculus, X. laevis and D. rerio. The secondary structures of human TONSL ARD are showed on top of the sequence alignment. Stars (*) indicate the highly conserved residues that constitute the H4 tail-binding surface of TONSL ARD and the three strictly conserved acidic residues forming hydrogen bonds with the key residue H4 Lys20 are highlighted with red stars ‘*’.
Extended Data Figure 3. Interaction details of TONSL ARD and GST pull-downs.
a, b, Molecular details of the interactions of TONSL ARD with H4 tail regions residues 12-15 (a) and residues 21-23 (b). The Lys12-Gly13-Gly14-Ala15 segment of H4 is positioned within a narrow surface channel of the TONSL ARD scaffold. The intermolecular contacts spanning the Lys12-Gly13-Gly14-Ala15 segment of H4 include hydrophobic interactions between residues Gly13, Gly14 and Ala15 of H4 and residues Asn507, Cys508, Trp641, Tyr645 and Leu649 of ARD, as well as hydrogen bonds between the main-chain O of H4 Gly14 and Nε1 of ARD Trp641, and between the main-chain N of H4 Ala15 and Oη of ARD Tyr645 (a; Fig. 1c). The main-chain O of H4 Lys16 hydrogen bonds with the Nδ2 of ARD Asn571, while the side-chain of H4 Lys16 forms contacts with ARD Asn607 and electrostatic interactions with the side-chain of ARD Glu597 (Fig. 1c). The side-chain of H4 Arg17 stacks over the side-chains of ARD Tyr572 and Cys608, while its Nη1 atom forms two hydrogen bonds with main-chain O and Oδ1 of ARD Asn571 (Fig. 1c,e). The side-chain of H4 H18 penetrates into a pocket lined by four strictly conserved residues (Trp563, Glu568, Asn571 and Asp604) and is positioned over His567 of ARD (Fig. 1c,f). The side chain of H4 His18 is stacked between Trp563 and Asn571 and forms hydrogen bonds to Glu568 and Asp604 of ARD (Fig. 1f). The main-chain O of H4 Arg19 forms a hydrogen bond with Nε1 of Trp563 and its side-chain forms contacts with Cys561 and Gly595 of ARD (Fig. 1c). Interactions with the key residue H4 Lys20 are described in the main text (Fig. 1g). The intermolecular contacts spanning the Val21-Leu22-Arg23 segment of H4 include contacts between side-chains of H4 Val21 with Tyr560 and Cys561 of ARD (b), while H4 Leu22 interacts with Asp527 and Met528 of ARD. The main-chain N of H4 Arg23 forms a hydrogen bond with the main-chain O of Asp527 of ARD, while the side-chain packs against the side-chain of Tyr560 of ARD (b). c, Pull-down of recombinant histones H3–H4 with GST-TONSL ARD WT or indicated mutants. d, Pull-down of pre-purified MCM2 HBD–H3–H4 tetramer complex with GST-TONSL ARD WT or indicated mutants. e, Circular dichroism analysis of TONSL ARD WT and the indicated ARD mutants.
Extended Data Figure 4. Structural comparison of the ARDs of TONSL and GLP.
a, b, Representative view of the TONSL ARD with histone H4 tail (a, this work), and crystal structure of the GLP ARD in complex with histone H3 tail dimethylated at Lys9 (Collins R.E. et al. NSMB 2008) (b). Both TONSL ARD and GLP ARD use the concave surface to bind their cognate target H4 tail and H3 tail, respectively. TONSL ARD recognizes H4K20me0 mainly through 3 strong hydrogen bonds with acidic residues Glu530, Asp559 and Glu568, while GLP ARD recognizes H3K9me2 mainly through an aromatic cage forming by residues Trp839, Trp844, Glu847 and Trp877. c, ITC analysis of TONSL ARD binding to H3K9me1 peptide. d, ITC analysis of TONSL acidic stretch and ARD (a.a. 450-692) with H3K9me1 (a.a. 1-21) and H4 (a.a. 9-25) peptides.
Extended Data Figure 5. Effect of SET8 and MOF depletion on TONSL chromatin binding.
a, Immunoprecipitation of GFP-TONSL from solubilized chromatin of GFP-TONSL U-2-OS cells (one representative experiment out of two is shown). Same exposures are shown for input and IP western blots of H3 and H4K16ac. b, TONSL ARD preference for H4K16ac could be mediated by I599 through hydrophobic association with the K16 acetyl group as I599E ARD mutation preferentially reduces binding to H4K16ac peptides as compared to the unmodified H4 tail. (Left panel) Pull-down of GFP-TONSL from cell extracts with biotinylated H4 tail peptides. (Right panel) Quantification of the western blot, GFP-TONSL binding to the H4K16ac peptide is shown relative to the unmodified peptide. Mean with individual data points are shown (n=2). c, High-content quantitative imaging of TONSL in pre-extracted U-2-OS cells. Plots show total chromatin-bound TONSL and DAPI intensities in cells treated with control or TONSL siRNA, confirming the specificity of TONSL antibody staining. Each dot represents one nucleus. d-f, Analysis of TONSL chromatin-binding in MOF-depleted (d), SET8-depleted (e) and IR-treated cells (f). Chromatin-bound TONSL was quantified by high content imaging of pre-extracted U-2-OS cells stained for endogenous TONSL. Mean TONSL intensity is shown. A.U., arbitrary units. (d, e) knock-down efficiency and expected effect on histone modification were confirmed by western blotting (representative of 2 experiments). (e, f) G1 cells were defined by gating on DAPI and EdU intensity. (f) Cells were irradiated (1.5 Gy) and analysed 1.5 hours later (representative of 2 experiments). (d, f) Error bars, SD; d from left, n=4920, 2341, 3608, 2917; f, n= 382 (-IR), 523(+IR).
Extended Data Figure 6. TONSL binding to chromatin during the cell cycle.
a, b, H4K20 methylation levels on new and old histones analysed by NCC-pulse-SILAC (data are extracted from Alabert C. et al. G&D 2015). Cells grown in light SILAC medium were released into S phase in heavy medium and pulsed with biotin-dUTP. Chromatin was fixed, sonicated and b-dUTP labelled fragments isolated on streptavidine beads by NCC. Histones were isolated and analysed by mass spectrometry for modifications on new (heavy) and old (light) histones. For clarity a 24 hrs (G1-S) chase time-point is included. Error bars, SD; n=9 (S), 3 (S/G2, M), 5 (G1), 3 (G1/S). Data for M (old histones) is shown as the mean of n=2, as light peptides were not detected in one of the 3 biological replicas. c, H4K20 methylation levels measured by mass spectrometry in synchronized TIG3 fibroblasts. d, Plot of mean EdU and total DAPI intensities from TIG3 fibroblasts as in Fig. 3b, with the intensity of chromatin-bound TONSL shown in 3rd dimension as a colour-gradient. A.U., arbitrary units. Each dot represents one nucleus. Note that a population of G2 cells (EdU negative) retain TONSL on chromatin. e, High-content quantitative imaging of pre-extracted U-2-OS cells stained for EdU and TONSL analysed as in Fig. 3b. f, Analysis of TONSL chromatin-binding by cellular fractionation. U-2-OS cells released from a nocodazole block were followed by FACS analysis of DNA content and analysed by western blotting of soluble (CSK-Triton extracted) and chromatin (pellet) fractions (representative of 2 experiments).
Extended Data Figure 7. Analysis of GFP-TONSL localization.
a, Co-localization analysis of chromatin-bound GFP-TONSL with MCM2 analysed by deconvolution microscopy and measurement of Pearson coefficient in single cells. Error bars, SD, n=13 from two independent experiments. Representative image, Fig. 3c. b, c, Representative images for the analysis shown in Fig. 3d. Cells were either pulsed with EdU (40 μM) for 15 minutes (b) or synchronized in G1/S and released into S phase in continuous presence of EdU (5 μM) (c). Images are representative of b: n=9 (very early), 16 (early/mid), 10 (mid/late); c: 9 (very early), 27 (early/mid), 36 (mid/late). Scale bar, 5 μm. (b) EdU and MCM2 staining was used to determine the cell cycle state of asynchronous cells. (c) Progression through S phase was followed by FACS analysis of DNA content. d, Chromatin-binding of GFP-TONSL analysed by cellular fractionation in inducible U-2-OS cells as quantified in Fig. 3e. S, soluble. C, chromatin. e, f, Chromatin-binding analysis as in Fig. 3f. U-2-OS cells conditional for GFP-TONSL ARD WT and mutant were directly fixed or pre-extracted to remove soluble proteins. Data are representative of 3 (e) and 2 (f) experiments, fields of cells in (e) are representative of (from left) n=16, 18, 17 and 17 images. Scale bar, 20 μm. g, Asynchronous U-2-OS cells conditional for GFP-TONSL were pulsed with 40 μM EdU for 15 minutes and soluble proteins were extracted. Representative images of EdU positive cells are shown (n=30 for WT and N571A), for the specific patterns of TONSL WT see Fig. 3d. Scale bar, 5 μm.
Extended Data Figure 8. TONSL-MMS22L recruitment to damaged DNA.
a (left) ChIP-qPCR analysis of GFP-TONSL recruitment to site-specific DSBs induced by AsiSI, as shown in Fig. 4d but with additional controls. Note that the colours have been changed for clarity. Mean of technical duplicates is shown. (right) Dot plot illustrating the relative enrichment of GFP-TONSL WT and N571A obtained in four independent ChIPs performed on two biologically independent chromatin preparations. Each experiment was normalized to GFP-TONSL WT enrichment at DSB-I_80bp. Mean is shown with two-sided Mann-Whitney test; ***, P < 0.001; n.s., P > 0.05; n=24. Two-sided Mann-Whitney analysis of individual experiments gave similar results. b, U-2-OS cells conditional for GFP-TONSL were laser-microirradiated. 53BP1 and Cyclin B staining was used as markers of DNA damage and cells in S/G2 phase, respectively. Representative of 3 experiments as quantified Fig. 4e. Full arrowheads, GFP-TONSL recruitment; empty arrowheads, no recruitment. Scale bars, 10 μm c, U-2-OS cells transiently transfected with GFP-TONSL WT or the indicated mutants were laser-microirradiated and processed for γH2A.X immunofluorescence. Representative cells are shown (n=200 cells per condition from two independent experiments). d, U-2-OS cells conditional for GFP-TONSL were laser-microirradiated. γH2A.X and RPA staining was used as markers of DNA damage and cells undergoing resection in S/G2 phase, respectively. The % of GFP-TONSL cells with recruitment to RPA positive (+) and RPA negative (-) laser tracks is indicated. Data are representative of 2 independent experiments, a total of 118 cells were counted. e, (top) U-2-OS cells conditional for GFP-TONSL WT and N571A were laser-microirradiated. γH2A.X and EdU staining was used as markers of DNA damage and S phase cells, respectively. (bottom) Quantification of GFP-TONSL cells with recruitment to laser tracks. Mean with individual data points are shown (n=2, a total of 138 (WT) and 174 (N571A) cells were counted). f, H4K20 methylation levels measured by mass spectrometry in synchronized TIG3 cells as in Extended Data Fig. 6c. Cell were released into S phase for 3 hours and treated with HU (3 mM) or CPT (1μM) for 3 hours or left untreated (6 hours). Mean with individual data points are shown (n=2). g, Colony formation in cells treated with control or TONSL siRNA and induced to express GFP-TONSL. As shown in Fig. 4f, but including additional mutants. Two cell concentrations in technical triplicate from two (E568A, D559A) or four (WT, N571A) biological replicates are shown. h, Representation of the complementation analysis from Fig. 4f in a single panel including both CPT treated and untreated cells. This illustrates that the toxicity of the TONSL ARD mutant is comparable to CPT treatment of cells expressing WT TONSL. i, Analysis of GFP-TONSL and MMS22L by cellular fractionation in cells inducible for GFP-TONSL ARD WT and mutant. Representative experiment of the quantification shown in Fig. 4i.
Extended Data Fig. 9. Similarity of the ARDs in TONSL and BARD1, and protein inputs.
a, Superposition of the structures of TONSL ARD and BARD1 ARD (PDB 3C5R, Fox D. 3rd et al. JBC 2008). The main residues involved in TONSL ARD interactions with the H4 tail are compared to the corresponding residues of BARD1 ARD. The two ARDs show highly similar topology and conservation of the histone-binding surface. b, Input material of the experiment in Fig. 1h. c, Input material of the experiment in Fig. 1j. d, Spot assay with biotinylated H4 tail (aa. 14-33) peptides confirming equal input into pull down reactions. e, Input material of the experiment in Fig. 4b. f, Input material of the NCC experiment in Fig. 4c. Note that because ARD mutation disrupts chromatin binding in the presence and absence of CPT (Fig. 3e-f, 4a), GFP-TONSL N517 levels are low in the input chromatin. The NCC experiment in Fig. 4c supports our microscopy-based data (Fig. 4a) and further shows that there is no local accumulation of the GFP-TONSL ARD mutant at damaged forks that could have been missed in our microscopy-based quantification of total TONSL on chromatin.
Extended Data Table 1. Data collection and refinement statistics.
Supplementary Material
a Highest resolution shell is shown in parenthesis. One Crystal was used for the data.
Acknowledgements
We thank the beam staff at the synchrotrons at the Argonne National Laboratory (NE-CAT) for technical assistance. We thank John Rouse, Dan Durocher, Gaelle Legube and Claus Storgaard Sørensen for reagents, Guillermo Montoya for assistance with Circular Dichroism, Caroline B. Strømme, Anne Strandsby, Kyosuke Nakamura, Sung-bau Lee, and Martina Hödl for help with experiments, and Yasuko Antoku for assistance with microscopy. We thank Jiri Lukas for comments on the manuscript and Zuzana Jasencakova for illustrations. GS was supported by European Commission Marie Curie ITN FP7 ‘aDDRess’. DJP was supported in part by grants from the Leukemia and Lymphoma Society and the STARR foundation. AG is an EMBO Young Investigator and her research is supported by the European Research Council (ERC StG, no. 281765), the Danish National Research Foundation to the Center for Epigenetics (DNRF82), the Danish Cancer Society, the Danish Medical Research Council, the Novo Nordisk Foundation and the Lundbeck Foundation. AI is supported by the European Commission FP7 Network of Excellence EpiGeneSys (Project 257082), the DFG Excellence Clusters CIPSM and SyNergy, as well as the DFG Collaborative Research Center 1064 (projects A3 and Z3). TB is supported by the Medical Research Council and the European Research Council (ERC StG, no. 309952).
Footnotes
Author contribution
G.S. and A.G. conceived and led the functional studies. H.H conceived and led the generation of cassette to crystallize the complex, and H.H. solved the structure under the supervision of D.J.P. C.H. performed peptide pull-downs with ARD and recombinant nucleosomes. C.A. performed SET8 experiments and NCC. S.B-J. and N.M. analysed recruitment to laser induced DNA damage. N.R-G. prepared histones for mass spectrometry and performed ChIP analysis, I.F. analysed histone modifications by mass spectrometry under the supervision of A.I. B.M.F and T.B. prepared modified recombinant nucleosomes. L.M. and P.C. prepared recombinant TONSL-MMS22L. G.S. H.H, D.J.P. and A.G. wrote the manuscript and all authors commented on the manuscript.
Accession Codes: Coordinate and structure factors have been deposited with the following accession code: 5JA4
Competing financial interests
GS, HH, CMH, DJP and AG are inventors on a filed patent application covering the discoveries presented in this manuscript.
References
- 1.Duro E, et al. Identification of the MMS22L-TONSL complex that promotes homologous recombination. Mol Cell. 2010;40:632–644. doi: 10.1016/j.molcel.2010.10.023. [DOI] [PubMed] [Google Scholar]
- 2.O'Donnell L, et al. The MMS22L-TONSL complex mediates recovery from replication stress and homologous recombination. Mol Cell. 2010;40:619–631. doi: 10.1016/j.molcel.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O'Connell BC, et al. A genome-wide camptothecin sensitivity screen identifies a mammalian MMS22L-NFKBIL2 complex required for genomic stability. Mol Cell. 2010;40:645–657. doi: 10.1016/j.molcel.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Piwko W, et al. RNAi-based screening identifies the Mms22L-Nfkbil2 complex as a novel regulator of DNA replication in human cells. EMBO J. 2010;29:4210–4222. doi: 10.1038/emboj.2010.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campos EI, et al. Analysis of the Histone H3.1 Interactome: A Suitable Chaperone for the Right Event. Mol Cell. 2015;60:697–709. doi: 10.1016/j.molcel.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Groth A, et al. Regulation of replication fork progression through histone supply and demand. Science. 2007;318:1928–1931. doi: 10.1126/science.1148992. [DOI] [PubMed] [Google Scholar]
- 7.Huang H, et al. A unique binding mode enables MCM2 to chaperone histones H3-H4 at replication forks. Nat Struct Mol Biol. 2015;22:618–626. doi: 10.1038/nsmb.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richet N, et al. Structural insight into how the human helicase subunit MCM2 may act as a histone chaperone together with ASF1 at the replication fork. Nucleic Acids Res. 2015;43:1905–1917. doi: 10.1093/nar/gkv021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalashnikova AA, Porter-Goff ME, Muthurajan UM, Luger K, Hansen JC. The role of the nucleosome acidic patch in modulating higher order chromatin structure. J R Soc Interface. 2013;10:20121022. doi: 10.1098/rsif.2012.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins RE, et al. The ankyrin repeats of G9a and GLP histone methyltransferases are mono- and dimethyllysine binding modules. Nat Struct Mol Biol. 2008;15:245–250. doi: 10.1038/nsmb.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jasencakova Z, et al. Replication stress interferes with histone recycling and predeposition marking of new histones. Mol Cell. 2010;37:736–743. doi: 10.1016/j.molcel.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 12.Taipale M, et al. hMOF histone acetyltransferase is required for histone H4 lysine 16 acetylation in mammalian cells. Mol Cell Biol. 2005;25:6798–6810. doi: 10.1128/MCB.25.15.6798-6810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rice JC, et al. Mitotic-specific methylation of histone H4 Lys 20 follows increased PR-Set7 expression and its localization to mitotic chromosomes. Genes Dev. 2002;16:2225–2230. doi: 10.1101/gad.1014902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pesavento JJ, Yang H, Kelleher NL, Mizzen CA. Certain and progressive methylation of histone H4 at lysine 20 during the cell cycle. Mol Cell Biol. 2008;28:468–486. doi: 10.1128/MCB.01517-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beck DB, Oda H, Shen SS, Reinberg D. PR-Set7 and H4K20me1: at the crossroads of genome integrity, cell cycle, chromosome condensation, and transcription. Genes Dev. 2012;26:325–337. doi: 10.1101/gad.177444.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jorgensen S, Schotta G, Sorensen CS. Histone H4 lysine 20 methylation: key player in epigenetic regulation of genomic integrity. Nucleic Acids Res. 2013;41:2797–2806. doi: 10.1093/nar/gkt012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loyola A, Bonaldi T, Roche D, Imhof A, Almouzni G. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol Cell. 2006;24:309–316. doi: 10.1016/j.molcel.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 18.Alabert C, et al. Two distinct modes for propagation of histone PTMs across the cell cycle. Genes Dev. 2015;29:585–590. doi: 10.1101/gad.256354.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alabert C, et al. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat Cell Biol. 2014;16:281–293. doi: 10.1038/ncb2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prasanth SG, Mendez J, Prasanth KV, Stillman B. Dynamics of pre-replication complex proteins during the cell division cycle. Phil Trans R Soc B. 2004;359:7–16. doi: 10.1098/rstb.2003.1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takahashi TS, Wigley DB, Walter JC. Pumps, paradoxes and ploughshares: mechanism of the MCM2-7 DNA helicase. Trends Biochem Sci. 2005;30:437–444. doi: 10.1016/j.tibs.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 22.Seeber A, Hauer M, Gasser SM. Nucleosome remodelers in double-strand break repair. Curr Opin Genetics Dev. 2013;23:174–184. doi: 10.1016/j.gde.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 23.Burgess RJ, Zhang Z. Histone chaperones in nucleosome assembly and human disease. Nat Struct Mol Biol. 2013;20:14–22. doi: 10.1038/nsmb.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Botuyan MV, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Panier S, Boulton SJ. Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol. 2014;15:7–18. doi: 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- 26.Fox D, 3rd, et al. Crystal structure of the BARD1 ankyrin repeat domain and its functional consequences. J Biol Chem. 2008;283:21179–21186. doi: 10.1074/jbc.M802333200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laufer M, et al. Structural requirements for the BARD1 tumor suppressor in chromosomal stability and homology-directed DNA repair. J Biol Chem. 2007;282:34325–34333. doi: 10.1074/jbc.M705198200. [DOI] [PubMed] [Google Scholar]
- 28.Ishitobi M, et al. Mutational analysis of BARD1 in familial breast cancer patients in Japan. Cancer letters. 2003;200:1–7. doi: 10.1016/s0304-3835(03)00387-2. [DOI] [PubMed] [Google Scholar]
- 29.Iacovoni JS, et al. High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. EMBO J. 2010;29:1446–1457. doi: 10.1038/emboj.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cejka P, Kowalczykowski SC. The full-length Saccharomyces cerevisiae Sgs1 protein is a vigorous DNA helicase that preferentially unwinds holliday junctions. J Biol Chem. 2010;285:8290–8301. doi: 10.1074/jbc.M109.083196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr, Sect D: Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 33.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr, Sect D: Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 34.Dyer PN, et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- 35.Bartke T, et al. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell. 2010;143:470–484. doi: 10.1016/j.cell.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bodor DL, Valente LP, Mata JF, Black BE, Jansen LE. Assembly in G1 phase and long-term stability are unique intrinsic features of CENP-A nucleosomes. Mol Biol Cell. 2013;24:923–932. doi: 10.1091/mbc.E13-01-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jorgensen S, et al. The histone methyltransferase SET8 is required for S-phase progression. J Cell Biol. 2007;179:1337–1345. doi: 10.1083/jcb.200706150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mosbech A, Lukas C, Bekker-Jensen S, Mailand N. The deubiquitylating enzyme USP44 counteracts the DNA double-strand break response mediated by the RNF8 and RNF168 ubiquitin ligases. J Biol Chem. 2013;288:16579–16587. doi: 10.1074/jbc.M113.459917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakamura K, et al. Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol Cell. 2011;41:515–528. doi: 10.1016/j.molcel.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 40.Jakobsen JS, et al. Temporal mapping of CEBPA and CEBPB binding during liver regeneration reveals dynamic occupancy and specific regulatory codes for homeostatic and cell cycle gene batteries. Genome Res. 2013;23:592–603. doi: 10.1101/gr.146399.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aymard F, et al. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat Struct Mol Biol. 2014;21:366–374. doi: 10.1038/nsmb.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
a Highest resolution shell is shown in parenthesis. One Crystal was used for the data.