Abstract
Antibody fragments (scFvs) fused to luciferase reporter proteins have been used as highly sensitive optical imaging probes. Gaussia princeps luciferase (GLuc) is an attractive choice for a reporter protein because it is small and bright and does not require ATP to stimulate bioluminescence-producing reactions. Both GLuc and scFv proteins contain multiple disulfide bonds, and consequently the production of active and properly folded GLuc–scFv fusions is challenging. We therefore produced both proteins individually in active form, followed by covalent coupling to produce the intended conjugate.
We used an Escherichia coli-based cell-free protein synthesis (CFPS) platform to produce GLuc and scFv proteins containing non-natural amino acids (nnAAs) for subsequent conjugation by azide–alkyne click chemistry. GLuc mutants with exposed alkyne reactive groups were produced by global replacement of methionine residues in CFPS. Antibody fragment scFvs contained a single exposed azide group using a scheme for site-specific incorporation of tyrosine analogs. Incorporation of tyrosine analogs at specific sites in proteins was performed using an engineered orthogonal tRNA–tRNA synthetase pair from an archaebacterium. The unique azide and alkyne side chains in GLuc and the antibody fragment scFv facilitated conjugation by click chemistry. GLuc–scFv conjugates were shown to differentiate between cells expressing a surface target of the scFv and cells that did not carry this marker.
Keywords: Cell-free protein synthesis (CFPS), Non-natural amino acids, Gaussia princeps luciferase, Bioconjugate, Bioluminescence, Cell-surface binding assay
Introduction
Antibody fragments (single chain antibody Fv fragments, or scFvs) specific for unique cell surface markers can be used to differentiate cells that bear this marker (e.g. tumor cells) from other cells. Fusion of a luciferase reporter protein to antibody fragments allows highly sensitive detection of certain tumors in vivo and ex vivo [1,2]. Firefly luciferase (FLuc) from Photinus pyralis and Renilla luciferase (RLuc) from Renilla reniformis are two extensively studied luciferases [3,4]. The Gaussia princeps luciferase (GLuc) is a more attractive choice for a reporter protein because it is small, bright, and ATP-independent [2,5]. GLuc and RLuc catalyze the oxidation of coelenterazine to coelenteramide accompanied by the emission of light. FLuc, RLuc and GLuc have been used in numerous in vitro and in vivo applications as reporter proteins [6–8], and luciferase and antibody fragment fusion proteins have been produced as reagents for detection of specific antigens for in vivo and in vitro imaging applications [1,2,9].
However, production of complex disulfide bonded proteins such as scFvs as well as GLuc using recombinant expression systems is challenging and production of properly folded and active bi-functional GLuc–scFv fusions is even harder. It is therefore desirable to produce both proteins separately in active form, followed by covalent coupling to produce the desired conjugate. Incorporation of non-natural amino acids (nnAAs) in proteins followed by direct linkage using azide–alkyne click chemistry [10,11] is an attractive option since these reactions are efficient and can be performed under physiological conditions.
Our lab has developed a cell-free protein synthesis (CFPS) platform that facilitates incorporation of azide- and alkyne-containing nnAAs in proteins by adopting two different schemes; site-specific incorporation of tyrosine analogs [12,13] as well as global replacement of methionine analogs [14]. Site-specific incorporation offers greater control and flexibility since a non-natural amino acid can be introduced at any desired site in a protein. The open cell-free system facilitates addition of optimal amounts of the orthogonal components, the tRNA and synthetase pair, which are required for site-specific incorporation of nnAAs in proteins [15]. Alternatively, the global replacement strategy mentioned earlier can provide higher yields of proteins since no orthogonal components are required and it appears that the methionine analogs are incorporated more efficiently [14]. However, the use of this method is limited to proteins where mutation of all methionine residues is not deleterious to protein folding or function. CFPS is also well suited for producing proteins containing methionine analogs since the absence of the cell wall barriers allows greater control over the concentrations of both methionine and the nnAA.
Cell-free protein synthesis has been successfully used to produce GLuc, scFv fragments, and other disulfide bonded proteins in soluble and active form with high yields [16–20]. We previously reported the cell-free production of GLuc mutants containing the methionine analogs azidohomoalanine (AHA) and homopropargylglycine (HPG) (Fig. 1A) [21]. The GLuc (HPG) mutant exhibited prolonged bioluminescence with an approximately 3-fold longer luminescence half-life as compared to the wild-type enzyme while retaining two-thirds of the wild-type specific activity. Further examination led to the identification of GLuc mutants containing methionine-to-leucine mutations at two critical positions, resulting in even higher luminescence half-lives and specific activities similar to wild-type. We also attached 5 kDa azide–PEG (polyethylene glycol) to each of the four HPG residues, suggesting that all four methionines in the native GLuc sequence are surface exposed and accessible for conjugation.
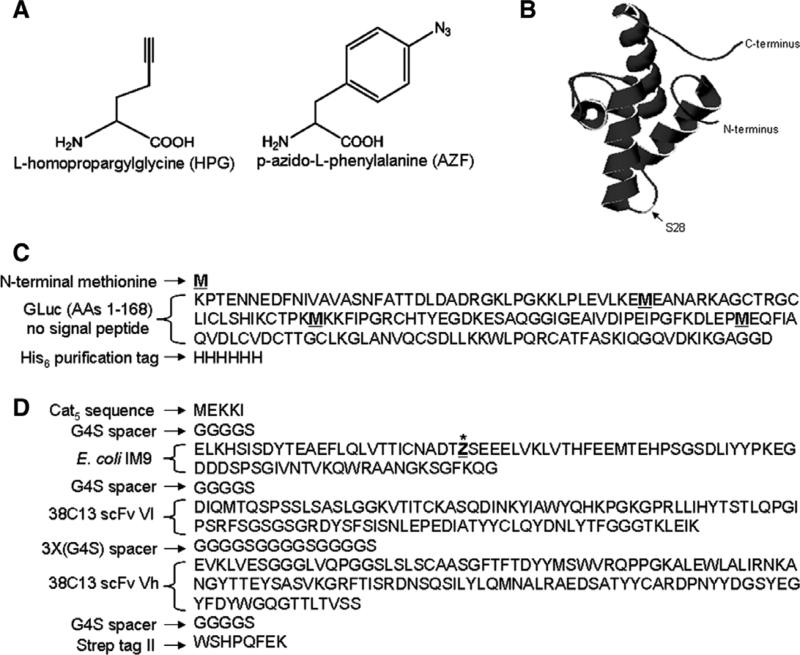
Fig. 1.
(A) Chemical structures of l-homopropargylglycine (HPG) and p-azido-l-phenylalanine (AZF). (B) Crystal structure of E. coli IM9 (PDB ID: 1bxi) showing the surface-exposed serine residue at position 28 (S28). AZF is incorporated in place of S28 in the IM9scFv fusion protein. (C) Amino acid sequence of GLuc. Methionine residues are underlined and indicated in bold. (D) Amino acid sequence of IM9scFv fusion with the site for incorporation of AZF indicated as an underlined “Z” with an asterisk.
Here we demonstrate the ability of Gaussia luciferase – antibody fragment bioconjugates to detect cells bearing a unique surface marker; specifically, an interaction between a mouse B cell lymphoma tumor idiotype scFv and an anti-idiotype antibody [22] expressed as a cell-surface immunoglobulin (Fig. 2). The tumor idiotype scFv is produced as a fusion with the Escherichia coli IM9 protein, which has been shown to improve cell-free production of soluble scFv fusion proteins [18,23]. The IM9 domain is designed to contain a site for the incorporation of tyrosine analogs at position 28, which is in a surface-exposed loop region (Fig. 1B). We produce GLuc–IM9scFv conjugates by first incorporating p-azido-l- phenylalanine (AZF, Fig. 1A) in the IM9scFv fusion protein and by replacing methionine residues in GLuc with HPG, followed by conjugation using Cu(I) catalyzed click chemistry. We constructed a mouse cell line that stably expresses surface anti-idiotype antibody, which is known to bind the tumor idiotype scFv [22]. The GLuc–IM9scFv conjugates were successfully used in an in vitro assay to differentiate between populations of cells expressing the anti-Id from cells that did not. Thus, we demonstrate the feasibility of using CFPS for the production of GLuc and scFv species that can be directly converted into bioconjugates capable of detecting cells that bear unique surface markers.
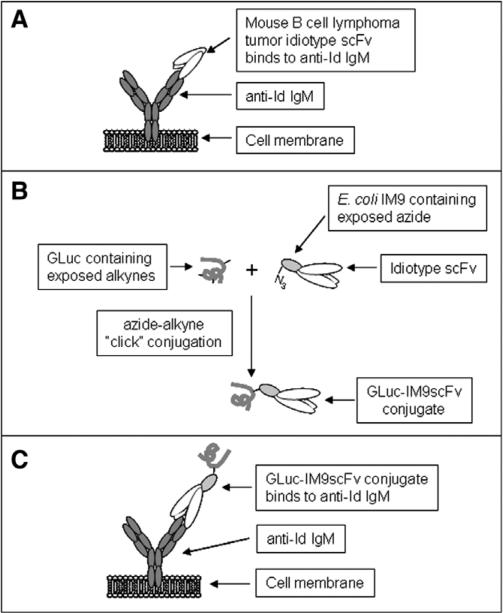
Fig. 2.
(A) B cell lymphoma tumor (38C13) idiotype (Id) scFv binds anti-idiotype (S1C5) IgM expressed on the surface of engineered A20 cells. (B) E. coli IM9–Id scFv fusion protein and GLuc, both containing non-natural amino acids are conjugated using click chemistry. (C) GLuc–IM9scFv conjugate binds surface anti-Id IgM on engineered A20 cells.
Experimental procedures
Plasmids for production of GLuc and IM9scFv
Plasmid pET24–AG1–GLuc–6H, described in [17] was used for cell-free production of GLuc containing HPG. It encodes amino acids 1–168 of the natural secreted protein sequence with the 17 amino acid signal sequence omitted and has been extended to encode an N-terminal methionine and a His6 sequence at the C-terminus (Fig. 1C).
Plasmid pY71-IM9(28TAG)-(38C13)scFv was constructed for expression of the IM9scFv fusion protein (Fig. 1D) which contains:
(1) the first five amino acids of the chloramphenicol acetyl transferase (cat) gene for efficient translation,
(2) the IM9 domain containing an amber stop codon in place of a codon for serine at position 28,
(3) the 38C13 tumor idiotype variable regions in the Vl–Vh orientation, separated by a spacer consisting of three consecutive Gly-Gly-Gly-Gly-Ser (G4S) sequences, and
(4) a Strep-Tag II affinity purification tag sequence.
The DNA sequence was PCR amplified from overlapping oligo-nucleotides that were designed using DNAworks [24]. The PCR product was digested with restriction endonucleases NdeI and XhoI and cloned into a pY71 expression vector [25] digested with NdeI and SalI.
Plasmids were transformed into DH5a cells (One Shot MAXX Efficiency DH5α-T1R Competent Cells, Invitrogen, Carlsbad, CA) and purified with Plasmid Maxi Kits (Qiagen, Valencia, CA).
Production and purification of GLuc and the IM9scFv fusion protein
GLuc containing HPG was produced and purified as described previously [21]. Extracts for production of IM9scFv containing AZF were prepared from E. coli KGK10 cells [26] harboring a plasmid for the constitutive expression of the orthogonal tRNA from Methanococcus jannaschii (pDule-tRNA, described in [15]). Cell extracts were prepared using high density fermentation and extract preparation protocols described previously [27,28]. CFPS was conducted using the PANOx-SP (PEP, amino acids, nicotinamide adenine dinucleotide (NAD), oxalic acid, spermidine, and putrescine) cell-free system [29] with component concentrations specified below. All reagents were obtained from Sigma–Aldrich (St. Louis, MO) unless otherwise noted. Reaction mixtures include: 1.2 mM ATP, 0.85 mM each of GMP, UMP and CMP, 33 mM phosphoenol pyruvate (Roche Molecular Biochemicals, Indianapolis, IN), 170 mM potassium glutamate, 10 mM ammonium glutamate, 16 mM magnesium glutamate, 1.5 mM spermidine, 1.0 mM putrescine, 34 μg/mL folinic acid, 171 μg/mL E. coli tRNA mixture (Roche Molecular Biochemicals), 13.3 μg/mL plasmid, 300 μg/mL T7 RNA polymerase (prepared by the procedure described in [29]), 2 mM of each of the 20 unlabeled amino acids, 0.33 mM NAD, 0.26 mM Coenzyme A (CoA), 2.7 mM sodium oxalate, 0.28 volumes of E. coli S30 extract, and with or without 21 μM l-[U-14C]-leucine (Amersham Pharmacia, Uppsala, Sweden). To encourage formation of disulfide bonds [19,20], the cell extract was first pretreated at room temperature for 30 min with 50 μM iodoacetamide. A glutathione buffer (4 mM oxidized glutathione (GSSG) and 1 mM reduced glutathione (GSH)) was then added to the reaction to stabilize the thiol/disulfide redox potential and finally DsbC, a periplasmic disulfide bond isomerase (prepared as described [16]) was added to a final concentration of 100 μg/mL.
For AZF incorporation, 4 mM AZF (Chem-Impex International, Wood Dale, IL) and 500 μg/mL tRNA synthetase were added to cell-free reactions. Purified tRNA synthetase (AzRS-His), formulated in 10 mM potassium phosphate buffer, pH 8 with 20% sucrose, was prepared as described [15]. Multiple 0.5 ml batch reactions were incubated at 30 °C for 10 h in the dark in six-well polystyrene tissue-culture plates (Falcon Cat. No. 353046). Total and soluble protein concentrations were measured by liquid scintillation counting of l-[U-14C]-leucine labeled proteins as described [28].
Ten parallel 0.5 ml CFPS reactions for producing IM9scFv containing AZF were pooled after the 10 h incubation and dialyzed against 100 volumes of Strep Load Buffer (SLB: 150 mM NaCl, 100 mM Tris–HCl, 1 mM EDTA, pH 8) for 12 h before loading onto a 1 ml Strep-Tactin Sepharose (IBA Gmbh, Göttingen, Germany) column which was equilibrated with 10 ml of SLB. Columns were washed three times with 5 ml SLB and eluted with 2.5 ml of Strep Elution Buffer (SEB: 150 mM NaCl, 100 mM Tris–HCl, 1 mM EDTA, 2.5 mM desthiobiotin, pH 8). The last 2 ml of the elution contained purified protein and was dialyzed against 2 L of 10 mM potassium phosphate buffer, pH 8 for 12 h with three buffer exchanges, the last one containing 20% sucrose. Protein purification was conducted with minimal exposure of IM9scFv (AZF) samples to laboratory light. Purified proteins were stored at −80 °C. Purified protein samples were analyzed for purity by SDS–PAGE using NuPAGE 10% Bis–Tris gels in MES buffer (Invitrogen) under reducing conditions.
Cu(I) catalyzed conjugation reactions
Azide–alkyne click reactions were conducted in 1.5 mL microcentrifuge tubes in an anaerobic glove box (Coy Laboratories, Grass Lake, MI) in the absence of oxygen to preserve the reduced state of the tetrakis(acetonitrile)copper(I)hexafluorophosphate ([(CH3CN)4Cu]PF6) (Sigma–Aldrich), hereafter referred to as Cu(I) catalyst. 0.5 mM tris(triazolylmethyl) amine Cu ligand (TTMA) was added to improve the rate of the click reactions [30]. Click reaction components were mixed and deoxygenated by incubation in the dark in 10–20 μL volumes for 45–90 min in the anaerobic chamber before the addition of the reduced copper catalyst. The 10–20 μL reactions contained 3 μM GLuc protein containing HPG, 2 mM Cu(I) catalyst, 0.5 mM TTMA and 8-fold molar excess of the IM9scFv fusion protein containing AZF. Click reactions were incubated under anaerobic conditions at ambient temperature in the dark for 2 h after the addition of Cu(I) catalyst.
Samples from click reaction products were loaded onto NuPAGE 10% Bis-Tris gels (Invitrogen) to assess attachment of IM9scFv (AZF) to the HPG residues in GLuc. Samples were run in MES buffer (Invitrogen) with 50 mM DTT. Gels were stained, dried, and exposed to a storage phosphor screen (Molecular Dynamics, Sunnyvale, CA) which was subsequently scanned using a Typhoon Scanner (GE Healthcare, Waukesha, WI). The relative abundance of individual bands on the autoradiogram was estimated by densitometry analysis using ImageJ software (ImageJ 1.41o, National Institutes of Health, USA).
Construction of an expression vector producing a membrane-bound anti-38C13 idiotype IgM
RNA was extracted from a hybridoma producing an anti-38C13 idiotype monoclonal antibody (clone S1C5, murine IgG2a/κ) [22] using the RNeasy mini kit (Qiagen). 5′-RACE-ready first-strand cDNA was synthesized, and the heavy and light chain variable region nucleotide sequences (Vh and Vl) isolated using the SMART RACE cDNA Amplification kit (Clontech, Mountain View, CA) and primers specific to the murine IgG2a heavy chain constant region 1 (5′-gaccgatggggctgttgttttggc-3′) and murine kappa light chain constant region (5′-ctgatcagtccaactgttcaggacgcc-3′). A 5′ SalI and a 3′ NheI flanking restriction site were added to the Vh sequence by PCR using primers 5′-gtcgacatggaatggagctggatcttt-3′ and 5′-gctagctgaggagactgtgagagtggtg-3′. A 5′ BglII and a 3′ BsiW1 flanking restriction site were added to the Vl sequence by PCR using primers 5′-agatctatggattttcaggtgcagatttt-3′ and 5′-cgtacgtttcagctccagcttggtc-3′.
In addition, we modified a human IgG expression vector (pUC19-based Tandem Chimeric Antibody Expression vector) [31] by replacing the constant IgG1 region with that from the human membrane IgM. RNA was extracted from the surface IgM expressing human B cell lymphoma line OCI-LY8 [32] using the RNeasy mini kit. Using the SMART RACE cDNA Amplification kit, we synthesized 5′-RACE-ready first-strand cDNA and isolated the membrane-bound mu constant region sequence after two rounds of 5′-RACE PCR with two pairs of nested upstream and downstream primers (5′-ctaatacgactcactatagggc-3′ and 5′-gtggggtagagcccacctc gtggcctgc-3′, followed by 5′-aagcagtggtatcaacgcagagt-3′ and 5′-c cagacccggaggctgcgcccctttga-3′). 5′ NheI and 3′ BamHI restriction sites were added using primers 5′-gtctcctcagctagcgcatccgccccaacccttttc-3′ and 5′-ctgggatcctcatttcaccttgaacaaggtgacggtgg-3′. The original vector was digested with NheI and BamHI to remove the IgG1 constant region, which was then replaced by the IgM constant region. The Vh and Vl segments of this vector were replaced in a similar fashion with those of the anti-38C13 idiotype.
Cell culture and transfection conditions
A20, a BALB/c B cell lymphoma line, was obtained from ATCC (Manassas, VA). Cells were cultured in culture medium (CM) which consisted of RPMI1640 medium (Invitrogen) supplemented with 10% heat-inactivated fetal calf serum (FCS; HyClone Laboratories, Logan, UT), 100 U/mL penicillin and 100 μg/mL streptomycin (both from Invitrogen), and 50 μM 2-mercaptoethanol (Sigma–Aldrich). The A20 cell line expressing an anti-38C13 idiotype surface IgM was created by stable transfection with the expression vector described above using the Cell Line Nucleofector Kit V and a Nucleofector II electroporator (AMAXA Biosystems, Cologne, Germany). The transfected cells were cultured in the same media as the parental cell line with the addition of 800 μg/mL of Geneticin (Invitrogen). Cells were grown in suspension culture at 37 °C in 5% CO2.
Cell assay with GLuc–IM9scFv bioconjugates
Approximately 1.3 million cells were incubated for 1 h at 4 °C in 1.5 ml microcentrifuge tubes with 200 μL culture medium (CM) containing 0.3 μM GLuc or GLuc–IM9scFv. After the 1 h incubation, cells were pelleted by centrifugation at 300 rcf for 5 min and supernatants containing unbound proteins were discarded. Cells were washed three times with 1 ml of CM. After washing, cells were resuspended in PBS (1% BSA).
Luminescence measurement
Samples containing GLuc were diluted in PBS, pH 7.4 (Invitrogen) with 1% w/v bovine serum albumin (BSA). Dilutions were made so that initial readings were within the linear range of the luminometer. Activity was measured in triplicate by adding 10 μL of diluted sample to 89 μL of PBS, pH 7.4 with 0.01% Tween 20. One microliter of 0.5 μg/μL coelenterazine (Nanolight, Pinetop, AZ) in propylene glycol was then added and the reaction was mixed for ~5 s. Luminescence was read immediately following coelenterazine addition and mixing, for a period of 150 s with a Berthold Mithras LB 940 luminometer (Bert-hold Technologies, Oak Ridge, TN). The luminometer was calibrated to absolute units (photons/s) as described previously [33].
Results and discussion
Production and purification of proteins for conjugation
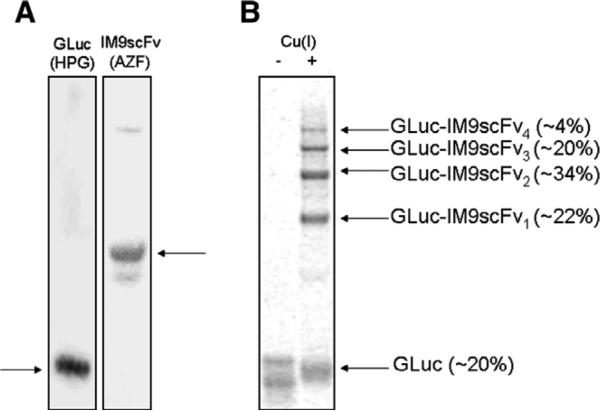
We previously reported that incorporation of the methionine analog HPG in GLuc using CFPS provided mutants with prolonged bioluminescence [21]. Incorporation of HPG only reduced GLuc specific activity by about a third while increasing the luminescence half-life from 1.3 min to 4 min. GLuc (HPG) was produced by CFPS with high yields (~300–400 μg/mL) and purified using Ni–NTA chromatography. IM9scFv was produced using CFPS with the engineered orthogonal tRNA and tRNA synthetase pair from M. jannaschii for incorporation of AZF. IM9scFv (AZF) was produced at yields of ~80–120 μg/mL and purified using a Strep-Tactin Sepharose column. Purified proteins were dialyzed into 10 mM potassium phosphate buffer (pH 8) and analyzed for purity by SDS–PAGE (Fig. 3A).
Fig. 3.
(A) Purified GLuc (HPG) and IM9scFv (AZF) proteins, analyzed by SDS–PAGE and coomassie staining. GLuc containing HPG was purified by Ni–NTA chromatography. The IM9scFv fusion protein containing AZF was purified using a Strep-Tactin Sepharose column. Proteins of interest are indicated by horizontal arrows. (B) Autoradiogram of GLuc–IM9scFv conjugates generated by azide–alkyne click chemistry. 3 μM GLuc (HPG) and 24 μM IM9scFv (AZF) samples were incubated with (+) and without (−) Cu(I). The GLuc (HPG) protein was labeled with l-[U-14C]-leucine for visualization by autoradiography. In the presence of Cu(I), the appearance of four bands above the unreacted GLuc suggests that all four HPG residues are accessible for attachment of IM9scFv (AZF). The relative abundance of individual bands is indicated in parentheses.
Conjugation of GLuc and IM9scFv
GLuc and IM9scFv proteins were conjugated using Cu(I) catalyzed click chemistry. Products from conjugation reactions were analyzed by SDS–PAGE and autoradiography (Fig. 3B). The appearance of four bands above the unreacted GLuc protein band suggests that all four HPG residues in GLuc are accessible for conjugation to IM9scFv with AZF at position 28 in the IM9 domain. Densitometry analysis of individual bands on autoradiogram (Fig. 3B) indicates that the predominant reaction product is GLuc with IM9scFv molecules attached at two sites (GLuc–IM9scFv2, ~34%). Also, only ~20% free GLuc remains after the click reactions when an 8-fold molar excess of the IM9scFv protein is used.
Detection of cells with surface markers using GLuc–IM9scFv conjugates
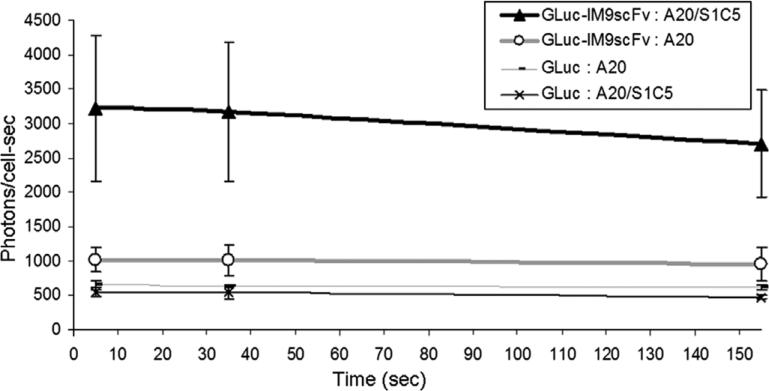
Next, we verified the binding and reporter functions of GLuc–IM9scFv conjugates as detection reagents. We used mouse B cell lymphoma (A20) cells that were engineered to express a surface anti-Id immunoglobulin (S1C5), which binds to the scFv immunoglobulin (38C13) [22]. GLuc–IM9scFv conjugates were incubated with engineered and with parental A20 cells. A ~3-fold higher signal is observed for cells expressing the anti-Id IgM as compared to parental cells that do not express this surface antibody (Fig. 4). This demonstrates the feasibility of using GLuc–IM9scFv conjugates for the detection of tumor cells bearing a particular surface marker. We are now extending this assay to the detection of individual cells bearing unique surface makers among background cells for applications including ex vivo detection of rare circulating cancer cells in patients. This might require more efficient conjugation such that every GLuc has four scFvs conjugated to it. Multiple binding sites on each GLuc–IM9scFv conjugate is expected to increase the avidity and help generate a stronger luminescence signal.
Fig. 4.
GLuc luminescence (in photons/cells) over 150 s from GLuc–IM9scFv conjugates bound to cell surface anti-Id immunoglobulin expressed on A20 cells (A20/S1C5) or to parental A20 cells (A20). The average luminescence values (in photons/cell s) measured at 5, 35 and 155 s after the addition of coelenterazine are shown (data from three samples ± 1 standard deviation). The 38C13 scFvs conjugated to GLuc (GLuc–IM9scFv) bind the engineered A20 cells (A20/S1C5) to provide a ~3-fold higher signal compared to parental cells (A20). Luminescence from free GLuc protein (GLuc) incubated with parental (A20) and with engineered (A20/S1C5) A20 cells is also shown. The GLuc (HPG) and GLuc–IM9scFv conjugates provide stable luminescence signals for several minutes, consistent with our previously reported observations [21].
Conclusions
The cell-free protein synthesis platform is well suited for the production of complex disulfide bonded proteins like GLuc and IM9scFv fusion proteins. The open cell-free system also allows for efficient production of these proteins with azide- and alkyne-containing nnAAs for conjugation. Interestingly, the use of a global replacement strategy to incorporate HPG residues in place of methionine in GLuc provided a mutant with prolonged bioluminescence as compared to the wild-type enzyme, making it an attractive choice for a reporter protein. In addition, global replacement of methionine residues with HPG in GLuc provided four reactive groups for conjugation to azide-functionalized molecules using click chemistry. We also site-specifically incorporated a single AZF residue in the IM9scFv fusion protein for conjugation to GLuc. Incorporation of AZF at a specific location in the IM9 domain enabled conjugation to GLuc with a defined orientation so that the binding site of the scFv domain projects outwards from the resulting conjugate. We showed that GLuc–IM9scFv conjugates bind to a specific cell surface marker expressed on A20 cells to produce 3-fold more intense bioluminescence compared to parental cells that do not express this surface receptor. This demonstrates the feasibility both of producing Gaussia luciferase – antibody fragment bioconjugates by direct conjugation of CFPS products and also of using these bioconjugates for the detection of cells bearing a unique surface marker.
Acknowledgments
This work was supported in part by a grant from the Leukemia and Lymphoma Society.
References
- 1.Venisnik KM, Olafsen T, Loening AM, Iyer M, Gambhir SS, Wu AM. Bifunctional antibody–Renilla luciferase fusion protein for in vivo optical detection of tumors. Protein Eng. Des. Sel. 2006;19:453–460. doi: 10.1093/protein/gzl030. [DOI] [PubMed] [Google Scholar]
- 2.Venisnik KM, Olafsen T, Gambhir SS, Wu AM. Fusion of Gaussia luciferase to an engineered anti-carcinoembryonic antigen (CEA) antibody for in vivo optical imaging. Mol. Imaging Biol. 2007;9:267–277. doi: 10.1007/s11307-007-0101-8. [DOI] [PubMed] [Google Scholar]
- 3.Fraga H. Firefly luminescence: a historical perspective and recent developments. Photochem. Photobiol. Sci. 2008;7:146–158. doi: 10.1039/b719181b. [DOI] [PubMed] [Google Scholar]
- 4.Lorenz WW, McCann RO, Longiaru M, Cormier MJ. Isolation and expression of a cDNA encoding Renilla reniformis luciferase. Proc. Natl. Acad. Sci. USA. 1991;88:4438–4442. doi: 10.1073/pnas.88.10.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tannous BA, Kim DE, Fernandez JL, Weissleder R, Breakefield XO. Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol. Ther. 2005;11:435–443. doi: 10.1016/j.ymthe.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 6.Contag CH, Ross BD. It's not just about anatomy: in vivo bioluminescence imaging as an eyepiece into biology. J. Magn. Reson. Imaging. 2002;16:378–387. doi: 10.1002/jmri.10178. [DOI] [PubMed] [Google Scholar]
- 7.Luker KE, Luker GD. Applications of bioluminescence imaging to antiviral research and therapy: multiple luciferase enzymes and quantitation. Antiviral Res. 2008;78:179–187. doi: 10.1016/j.antiviral.2008.01.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Villalobos V, Naik S, Piwnica-Worms D. Current state of imaging protein– protein interactions in vivo with genetically encoded reporters. Annu. Rev. Biomed. Eng. 2007;9:321–349. doi: 10.1146/annurev.bioeng.9.060906.152044. [DOI] [PubMed] [Google Scholar]
- 9.Billiald P, Mousli M, Goyffon M, Vaux D. Engineering of a bioluminescent antigen-binding protein. Biotechnol. Lett. 1997;19:1037–1041. [Google Scholar]
- 10.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew. Chem. Int. Ed. Engl. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 11.Tornoe CW, Christensen C, Meldal M. Peptidotriazoles on solid phase: [1,2,3]-triazoles by regiospecific copper(i)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. J. Org. Chem. 2002;67:3057–3064. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- 12.Chin JW, Santoro SW, Martin AB, King DS, Wang L, Schultz PG. Addition of p-azido-L-phenylalanine to the genetic code of Escherichia coli. J. Am. Chem. Soc. 2002;124:9026–9027. doi: 10.1021/ja027007w. [DOI] [PubMed] [Google Scholar]
- 13.Deiters A, Schultz PG. In vivo incorporation of an alkyne into proteins in Escherichia coli. Bioorg. Med. Chem. Lett. 2005;15:1521–1524. doi: 10.1016/j.bmcl.2004.12.065. [DOI] [PubMed] [Google Scholar]
- 14.Strable E, Prasuhn DE, Jr., Udit AK, Brown S, Link AJ, Ngo JT, Lander G, Quispe J, Potter CS, Carragher B, Tirrell DA, Finn MG. Unnatural amino acid incorporation into virus-like particles. Bioconjug. Chem. 2008;19:866–875. doi: 10.1021/bc700390r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goerke AR, Swartz JR. High-level cell-free synthesis yields of proteins containing site-specific non-natural amino acids. Biotechnol. Bioeng. 2009;102:400–416. doi: 10.1002/bit.22070. [DOI] [PubMed] [Google Scholar]
- 16.Goerke AR, Swartz JR. Development of cell-free protein synthesis platforms for disulfide bonded proteins. Biotechnol. Bioeng. 2008;99:351–367. doi: 10.1002/bit.21567. [DOI] [PubMed] [Google Scholar]
- 17.Goerke AR, Loening AM, Gambhir SS, Swartz JR. Cell-free metabolic engineering promotes high-level production of bioactive Gaussia princeps luciferase. Metab. Eng. 2008;10:187–200. doi: 10.1016/j.ymben.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 18.Yang J, Kanter G, Voloshin A, Michel-Reydellet N, Velkeen H, Levy R, Swartz JR. Rapid expression of vaccine proteins for B-cell lymphoma in a cell-free system. Biotechnol. Bioeng. 2005;89:503–511. doi: 10.1002/bit.20283. [DOI] [PubMed] [Google Scholar]
- 19.Yin G, Swartz JR. Enhancing multiple disulfide bonded protein folding in a cell-free system. Biotechnol. Bioeng. 2004;86:188–195. doi: 10.1002/bit.10827. [DOI] [PubMed] [Google Scholar]
- 20.Kim DM, Swartz JR. Efficient production of a bioactive, multiple disulfide-bonded protein using modified extracts of Escherichia coli. Biotechnol. Bioeng. 2004;85:122–129. doi: 10.1002/bit.10865. [DOI] [PubMed] [Google Scholar]
- 21.Welsh JP, Patel KG, Manthiram K, Swartz JR. Multiply mutated Gaussia luciferases provide prolonged and intense bioluminescence. Biochem. Biophys. Res. Commun. 2009;389:563–568. doi: 10.1016/j.bbrc.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 22.Maloney DG, Kaminski MS, Burowski D, Haimovich J, Levy R. Monoclonal anti-idiotype antibodies against the murine B cell lymphoma 38C13: characterization and use as probes for the biology of the tumor in vivo and in vitro. Hybridoma. 1985;4:191–209. doi: 10.1089/hyb.1985.4.191. [DOI] [PubMed] [Google Scholar]
- 23.Kanter G, Yang J, Voloshin A, Levy S, Swartz JR, Levy R. Cell-free production of scFv fusion proteins: an efficient approach for personalized lymphoma vaccines. Blood. 2007;109:3393–3399. doi: 10.1182/blood-2006-07-030593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoover DM, Lubkowski J. DNAWorks: an automated method for designing oligonucleotides for PCR-based gene synthesis. Nucleic Acids Res. 2002;30:e43. doi: 10.1093/nar/30.10.e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuchenreuther JM, Stapleton JA, Swartz JR. Tyrosine, cysteine, and S-adenosyl methionine stimulate in vitro [FeFe] hydrogenase activation. PLoS, ONE. 2009;4:e7565. doi: 10.1371/journal.pone.0007565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knapp KG, Goerke AR, Swartz JR. Cell-free synthesis of proteins that require disulfide bonds using glucose as an energy source. Biotechnol. Bioeng. 2007;97:901–908. doi: 10.1002/bit.21296. [DOI] [PubMed] [Google Scholar]
- 27.Zawada J, Swartz J. Maintaining rapid growth in moderate-density Escherichia coli fermentations. Biotechnol. Bioeng. 2005;89:407–415. doi: 10.1002/bit.20369. [DOI] [PubMed] [Google Scholar]
- 28.Yang WC, Patel KG, Lee J, Ghebremariam YT, Wong HE, Cooke JP, Swartz JR. Cell-free production of transducible transcription factors for nuclear reprogramming. Biotechnol. Bioeng. 2009;104:1047–1058. doi: 10.1002/bit.22517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jewett MC, Swartz JR. Mimicking the Escherichia coli cytoplasmic environment activates long-lived and efficient cell-free protein synthesis. Biotechnol. Bioeng. 2004;86:19–26. doi: 10.1002/bit.20026. [DOI] [PubMed] [Google Scholar]
- 30.Zhou Z, Fahrni CJ. A fluorogenic probe for the copper(I)-catalyzed azide–alkyne ligation reaction: modulation of the fluorescence emission via 3(n,pi)– 1(pi,pi) inversion. J. Am. Chem. Soc. 2004;126:8862–8863. doi: 10.1021/ja049684r. [DOI] [PubMed] [Google Scholar]
- 31.Reff ME, Carner K, Chambers KS, Chinn PC, Leonard JE, Raab R, Newman RA, Hanna N, Anderson DR. Depletion of B cells in vivo by a chimeric mouse human monoclonal antibody to CD20. Blood. 1994;83:435–445. [PubMed] [Google Scholar]
- 32.Oren R, Takahashi S, Doss C, Levy R, Levy S. TAPA-1, the target of an antiproliferative antibody, defines a new family of transmembrane proteins. Mol. Cell. Biol. 1990;10:4007–4015. doi: 10.1128/mcb.10.8.4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Kane DJ, Lee J. Absolute calibration of luminometers with low-level light standards. Methods Enzymol. 2000;305:87–96. doi: 10.1016/s0076-6879(00)05479-3. [DOI] [PubMed] [Google Scholar]