ABSTRACT
Alcohol dependence is a severe disorder contributing substantially to the global burden of disease. Despite the detrimental consequences of chronic alcohol abuse and dependence, effective prevention strategies as well as treatment options are largely missing to date. Accumulating evidence suggests that gene-environment interactions, including epigenetic mechanisms, play a role in the etiology of alcohol dependence. A recent epigenome-wide study reported widespread alterations of DNA methylation patterns in alcohol dependent patients compared to control individuals. In the present study, we validate and replicate one of the top findings from this previous investigation in an independent cohort: the hypomethylation of GDAP1 in patients. To our knowledge, this is the first independent replication of an epigenome-wide finding in alcohol dependence. Furthermore, the AUDIT as well as the GSI score were negatively associated with GDAP1 methylation and we found a trend toward a negative association between GDAP1 methylation and the years of alcohol dependency, pointing toward a potential role of GDAP1 hypomethylation as biomarker for disease severity. In addition, we show that the hypomethylation of GDAP1 in patients reverses during a short-term alcohol treatment program, suggesting that GDAP1 DNA methylation could also serve as a potential biomarker for treatment outcome. Our data add to the growing body of knowledge on epigenetic effects in alcohol dependence and support GDAP1 as a novel candidate gene implicated in this disorder. As the role of GDAP1 in alcohol dependence is unknown, this novel candidate gene should be followed up in future studies.
KEYWORDS: Alcohol dependence, biomarker, disease severity, DNA methylation, epigenetics, GDAP1, replication, treatment outcome, validation
Introduction
Causing approximately 3.3 million deaths every year (or 5.9% of all deaths world-wide) and attributing to 5.1% of the global burden of disease, harmful use of alcohol plays a decisive role for health (WHO, 2014). Despite the detrimental consequences of chronic alcohol abuse and dependence, effective preventive strategies and treatment options are still less than optimal.
Genetic and environmental factors modulate susceptibility to chronic alcohol abuse and alcohol dependence. Whereas heritability estimates for alcohol dependence range between 40 and 60%, environmental and stochastic effects account for the remainder of this variability.1-3 Accumulating evidence suggests that genetic and environmental factors not only act independently of each other but that also their interactions are implicated in the etiology of alcohol dependence.4-6 Among others, the interaction between genes and environment is mediated by epigenetic mechanisms.7 The major epigenetic mechanisms involve covalent modifications: DNA methylation and posttranslational histone modifications.8,9 Both mechanisms are important regulators of gene expression.10 DNA is methylated at position 5 of the cytosine pyrimidine ring, a reaction catalyzed by DNA methyltransferases (DNMTs). DNA methylation mainly occurs at the cytosine of a CpG dinucleotide. These CpG sites are not evenly distributed throughout the genome but are enriched in regions called CpG islands. CpG islands overlap with the promoter regions of 50 – 60% of human genes and are typically less methylated than CpG sites outside of CpG islands.11 Methylation of CpG sites is usually correlated with a decrease in gene expression.12,13
Initially, DNA methylation was believed to be established during early embryonic development and to remain stable afterwards. However, more recent research hints toward a more complex pattern of transcriptional regulation through DNA methylation and it is now known that DNA methylation is a dynamic mechanism.14 DNA methylation patterns vary over the lifetime of an organism and allow it to adapt to environmental changes.15 Various diseases are associated with altered epigenetic regulation and epigenetic mechanisms also play an important role in many neuropsychiatric disorders, 16 such as depression,17 schizophrenia 18 and addictions 19 including alcohol dependence.
Increased levels of homocysteine have been described in alcohol dependent patients.20-22 Homocysteine is of importance for DNA methylation as it is metabolized to methionine, which is then transformed into S-adenosyl methionine (SAM), the most important methyl group donor in vertebrates.23 Consequently, elevated homocysteine levels were associated with DNA hypermethylation in alcohol dependent patients.24 In contrast to those findings, other studies have reported that alcohol dependent patients lack the regulation of methionine adenosyl transferase resulting in global DNA hypomethylation.25,26
Several previous candidate-gene driven studies investigated the interplay between alcohol consumption and DNA methylation. An impact of alcohol intake on the methylation state of various genes, including monoamine oxidase A,27 dopamine transporter,28 serotonin transporter,29 nerve growth factor 30 and, most recently, leptin 31 have been described.
To date, there are only few studies investigating the influence of alcohol consumption on epigenetic mechanisms at an epigenome-wide level. In these studies, a number of genes were found to be significantly differentially methylated epigenome-wide between alcohol dependent patients and control individuals. The epigenetically differentially regulated regions included hyper- as well as hypo-methylated genes in patients.32-34 The most recent study by Clark et al. identified CNTN4 as a risk factor for alcohol use by examining the methylation status of approximately 27 million autosomal CpG sites and comparing them to GWAS data.35 Earlier candidate-gene based studies investigating the influence of therapeutic interventions on DNA methylation reported decreasing homocysteine levels in alcohol dependent patients during alcohol treatment,20,21,36,37 leading to the hypothesis that DNA methylation levels also decrease during alcohol treatment. However, candidate-gene driven DNA methylation studies conducted thus far have resulted in conflicting findings.28,30,38
To date, only one study has investigated the effects of an alcohol treatment on the epigenome using a systematic approach.32 No gene was epigenome-wide significantly differentially regulated when comparing the patients' methylome at the beginning of the alcohol treatment and after 4 weeks of treatment. However, when comparing patients entering the program and healthy control individuals, 56 genes reached epigenome-wide significance after Bonferroni correction, among them, GDAP1. This gene caught our attention, as it was the most significant finding within a promoter region of a characterized gene product. GDAP1 was significantly hypomethylated in alcohol dependent patients compared to the control group. GDAP1 is a member of the ganglioside-induced differentiation-associated protein family. Mutations in GDAP1 have been linked to Charcot-Marie-Tooth disease, a peripheral nerve disorder involving loss of muscle tissue.39,40 So far, no associations of GDAP1 with alcohol dependence or other addictions have been reported.
To clarify whether GDAP1 is indeed a novel epigenetic biomarker for alcohol dependence, we aimed to replicate the DNA methylation status of GDAP1 in a cohort of 49 alcohol dependent patients entering an alcohol treatment program and 37 healthy control individuals. In addition, we studied GDAP1 DNA methylation after 3 weeks of participating in an inpatient alcohol treatment program to elucidate whether GDAP1 DNA methylation could also serve as an epigenetic biomarker of treatment response.
Results
Lower GDAP1 DNA methylation in patients at the beginning of the alcohol treatment (T1) compared to control individuals
The demographic characteristics as well as nicotine and alcohol consumption of our cohort is provided in Table 1.
Table 1.
Characterization of patients and control individuals. Errors are given as standard deviation (SD). Amount of drinks is the standardized unit originating from the AUDIT questionnaire.
| Control individuals | Patients | P-value | |
|---|---|---|---|
| Total N | 37 | 49 | |
| Age (years) | 47 ± 12.32 | 49 ± 10.47 | 0.30 |
| Smokers (% of total) | 29 (78%) | 38 (79%) | 0.93 |
| Cigarettes smoked daily | 16 ± 10.99 | 20 ± 10.93 | 0.18 |
| Days since last drink | 2.9 ± 6.9 | ||
| Amount of drinks consumed daily one week before hospital admission | 17 ± 13.1 | ||
| Years of alcohol dependency | 12.3 ± 9.9 |
Control individuals and patients did not differ significantly in age (patients: 49 ± 10.47 years, control individuals: 47 ± 12.32 years; P = 0.3) or smoking behavior (control individuals: 16 ± 10.99 cigarettes per day, patients: 20 ± 10.93; P = 0.18). AUDIT scores differed significantly between control individuals (4.9 ± 3.7; P = 5.1E-15) and patients (25.1 ± 6.1) as well as the GSI scores (0.16 ± 0.13 for control individuals, 0.78 ± 0.54 for patients; P = 1.7E-10).
For all 3 sites analyzed, DNA methylation levels between control individuals and patients at T1 differed significantly (Table 2 and Fig. 1). For cg23779890 / site 1, the CpG site identified by Philibert et al.,32 DNA methylation levels were as follows: 7.8 ± 0.2 in control individuals, 6.6 ± 0.3 in patients, P = 0.001. For site 2, DNA methylation levels were 4.0 ± 0.1 in control individuals and 3.6 ± 0.1 in patients, P = 0.015. For site 3, DNA methylation levels were 2.1 ± 0.1 in control individuals and 1.8 ± 0.1 in patients, P = 0.012.
Table 2.
DNA methylation levels, AUDIT and GSI scores for control individuals vs. patients at T1. DNA Methylation level errors are given as standard error of the mean (SE), questionnaire score errors are given as standard deviation (SD).
| Control individuals | Patients at T1 | P-value | |
|---|---|---|---|
| Site 1 (cg23779890) | 7.8 ± 0.2 | 6.6 ± 0.3 | 0.001 |
| Site 2 | 4.0 ± 0.1 | 3.6 ± 0.1 | 0.015 |
| Site 3 | 2.1 ± 0.1 | 1.9 ± 0.1 | 0.012 |
| Mean DNA methylation (sites 1 – 3) | 4.6 ± 0.1 | 4.0 ± 0.2 | 0.001 |
| AUDIT score | 4.9 ± 3.7 | 25.1 ± 6.1 | 5.1E-15 |
| GSI score | 0.16 ± 0.13 | 0.78 ± 0.54 | 1.7E-10 |
Figure 1.
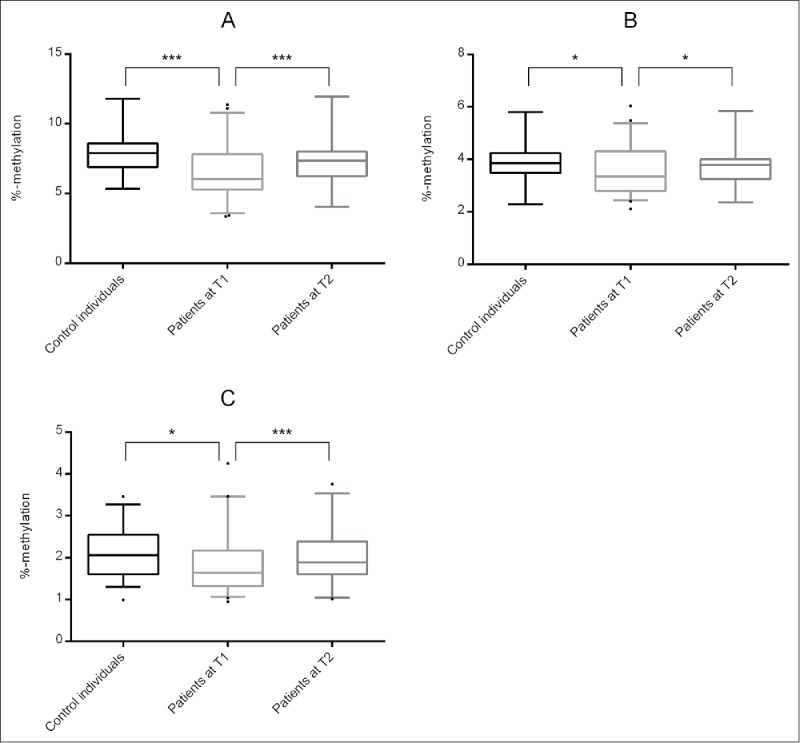
DNA methylation levels at (A) site 1 / cg23779890, (B) site 2 and (C) site 3 for control individuals, patients at T1 and patients at T2. Significant differences are indicated with * (P ≤ 0.05) and *** (P ≤ 0.001).
Mean DNA methylation across all 3 sites differed significantly between control individuals and patients (control individuals: 4.6 ± 0.1, patients: 4.0 ± 0.2; P = 0.001).
Since the DNA methylation levels of each site were highly correlated with the mean DNA methylation level across all sites (site 1: rs = 0.979, P = 2.0E-82; site 2: rs = 0.938, P = 1.4E-55; site 3: rs = 0.892, P = 4.0E-42), we decided to use the mean DNA methylation levels for further analyses.
First, comparing control individuals and patients at T1, the mean DNA methylation level was significantly negatively associated with the GSI score (rs = −0.2066, P = 0.016), and AUDIT Score (rs = −0.2041, P = 0.009). Furthermore, a trend toward a negative association between the mean DNA methylation level and the years of dependency (rs = 0.266, P = 0.08) was observed. We did not find any association between the DNA methylation levels and the amount of drinks consumed daily in the week before hospital admission (rs = −0.1038, P = 0.35).
Alcohol treatment significantly influences GDAP1 DNA methylation levels in alcohol dependent patients
After three weeks of alcohol treatment (T2), DNA methylation levels at all 3 sites were increased (Fig. 1, Table 3): 7.3 ± 0.3 (site 1, P = 0.001), 3.8 ± 0.1 (site 2, P = 0.033) and 2.0 ± 0.1 (site 3, P = 0.001). Again, the mean DNA methylation across these sites differed significantly (4.4 ± 0.2, P = 0.001) from the values of patients at T1 (4.0 ± 0.2) and were highly correlated with each other (site 1: rs = 0.972, P = 4.6E-21; site 2: rs = 0.901, P = 9.1E-13; site 3: rs = 0.877, P = 2.2E-11). In addition, the GSI score decreased significantly (0.78 ± 0.54 vs. 0.48 ± 0.49, P = 0.008, N = 30), as well as the OCDS score (3.93 ± 1.32 vs. 2.71 ± 0.96, P = 2.1E-5, N = 33).
Table 3.
DNA methylation levels, OCDS and GSI scores for patients at T1 vs. patients at T2. DNA Methylation level errors are given as standard error of the mean (SE), questionnaire score errors are given as standard deviation (SD).
| Patients at T1 | Patients at T2 | P-value | |
|---|---|---|---|
| Site 1 (cg23779890) | 6.6 ± 0.3 | 7.3 ± 0.3 | 0.001 |
| Site 2 | 3.6 ± 0.1 | 3.8 ± 0.1 | 0.033 |
| Site 3 | 1.9 ± 0.1 | 2.0 ± 0.1 | 0.001 |
| Mean DNA methylation(sites 1 – 3) | 4.0 ± 0.2 | 4.4 ± 0.2 | 0.001 |
| OCDS score | 3.93 ± 1.32 | 2.71 ± 0.96 | 2.1E-5 |
| GSI score | 0.78 ± 0.54 | 0.48 ± 0.49 | 0.008 |
Comparing the mean DNA methylation across all 3 sites between the control individuals and patients at T2, none of the DNA methylation levels differed significantly (site 1: P = 0.098; site 2: P = 0.244; site 3: 0.377; mean: P = 0.167).
The exclusion of 8 patients who had been abstinent for more than 3 d before hospital admission led to a diminishment of the days since the last drink from 2.9 ± 6.9 d to only 1.3 ± 0.8 d. Furthermore, it enhanced the observed effect of differential GDAP1 methylation between control individuals and patients at T1. At T2, this only had a moderately positive effect (see Table S1).
Discussion
By conducting pyrosequencing of 3 adjacent CpG sites in GDAP1, including cg23779890, we were able to replicate the finding of significant differences in DNA methylation between alcohol dependent patients and matched control individuals previously reported by Philibert et al.32 In addition, we identified significant differences between GDAP1 DNA methylation levels in patients at the day of hospital admission (T1) and after 3 weeks of attending an inpatient alcohol treatment program (T2). Furthermore, the AUDIT score as well as the GSI score at T1 were negatively associated with the DNA methylation levels and we found a trend toward a negative association between the DNA methylation levels and the years of alcohol dependency, but not with the amount of drinks consumed in the week before hospital admission. Our study thus provides additional evidence supporting the hypothesis that GDAP1 DNA methylation could serve as new biomarker for the severity of alcohol dependence.
In contrast to the hypothesis of increased DNA methylation levels in alcohol dependent patients due to higher levels of homocysteine,24 our results, as well as the previous results from Philibert et al. 32 show a hypomethylation of the GDAP1 gene in patients compared to control individuals. This was surprising, but as we did not measure homocysteine levels in our study samples, we can neither support nor contradict a potential correlation between homocysteine levels and DNA methylation of the GDAP1 gene promoter. Nevertheless, other studies did not find a correlation between homocysteine and global DNA methylation, and some did find a hypothesis-opposing outcome: with higher homocysteine levels in their samples, global DNA methylation was decreased.41 Other studies speculate that the missing regulation of the methionine adenosyl transferase in alcohol dependent patients results in global DNA hypomethylation.25,26 A recent study specifically investigating the role of homocysteine in altered DNA methylation in 363 alcohol dependent patients also found no correlation between homocysteine and global DNA methylation.42 Further studies are therefore necessary to clarify the relationship between homocysteine levels and GDAP1 DNA methylation.
Furthermore, we observe that increased severity of alcohol dependence in patients, assessed by the AUDIT score, as well as the GSI score, is associated with lower GDAP1 DNA methylation. However, we did not find a correlation between the amount of alcohol consumed one week before admission to the hospital and GDAP1 DNA methylation. As the exact amount of alcohol consumed one week before hospital admission does not affect GDAP1 DNA methylation, but rather the intensity and time span of alcohol dependence, GDAP1 DNA methylation could serve as an indicator of long-term and severe alcohol dependence rather than for short-term alcohol exposure.
Whereas Philibert et al. did not identify significant differences in GDAP1 DNA methylation in patients between T1 and T2, our results show an increase in DNA methylation levels in patients at T2, which no longer differed from the levels in control individuals. Our finding therefore supports the hypothesis of DNA methylation as a reversible process and suggests that DNA methylation levels return to their previous state, if the environmental condition underlying the epigenetic alteration—in this case alcohol dependence—is amended. However, to prove this hypothesis, it would be necessary to perform a longitudinal study and compare GDAP1 DNA methylation in patients before and after the onset of the disease. In our study, we only included patients after disease onset (mean years of dependency: 12.3 ± 9.9 years). After three weeks of attending an inpatient alcohol treatment program, the GSI score as well as the OCDS score, a measure of craving severity, decreased significantly in our patient cohort, suggesting a positive therapy outcome. Questionnaires are the most common means to assess these traits but are not an objective measure as they can be subjectively biased. The reversion of GDAP1 DNA methylation levels during abstinence could therefore serve as a biological, more objective indicator of a positive therapy outcome.
Although DNA methylation percentages in our study did deviate from the ones reported by Philibert and colleagues,32 we are able to replicate and validate the effect of alcohol dependence on GDAP1 DNA methylation. Philibert et al. found the mean DNA methylation level of cg23779890 to be 19.4% in patients and 24.3% in control individuals. We measured DNA methylation levels of 6.6% and 7.8%, respectively. These differences could be explained as follows.
One major difference between these studies is the source of material. Philibert and colleagues used mononuclear cells, whereas we used DNA prepared from whole blood. Whole blood is a heterogeneous mixture of different cell-types and blood composition varies from individual to individual and is depending on numerous factors such as age, sex, and individual health status. This is of importance as DNA methylation patterns are cell-type specific and could therefore explain the differences in DNA methylation levels between our study and the study by Philibert et al.32
The use of whole blood could be seen as a limitation of our study. However, we have explicitly chosen to investigate GDAP1 DNA methylation in whole blood to serve as an epigenetic biomarker for alcohol dependence and as a potential gauge of the therapy efficacy in a clinical setting. To be suitable as a biomarker, the study material needs to be easy and cost effective to obtain. The preparation and use of mononuclear cells for clinical diagnostics is impossible, as it is very time-consuming and labor-intensive in addition to being more expensive than the usage of whole blood.
In addition, as both the Illumina's 450K Chip as well as the pyrosequencing approach have systematic biases the differences could also be explained by the different methods used. As the overall congruency between Illumina's 450K Chip and pyrosequencing data is good, there are however specific sites where a direct translation from β-values originating from the Chip analysis to DNA methylation levels measured by pyrosequencing is difficult.43 These include, but are not limited to non-specific and cross-hybridizing probes, which represent a combination of multiple loci and therefore can elevate readings of low methylation or diminish readings of high methylation, biasing the results.44 Another well-known limitation of pyrosequencing is amplification bias. To account for this and to prevent batch effects, the samples were run at least in duplicates and they were assigned to different positions on different plates.
However, the overall effect in DNA methylation changes between patients at T1 and control individuals in both studies is very similar despite being obtained in 2 distinct cohorts using different methods (450K Chip analysis vs. pyrosequencing) as well as different sources of DNA: Philibert et al. found a 4.9% higher DNA methylation in control individuals compared to patients; our data show a 1.2% higher DNA methylation. These data indicate that indeed GDAP1 DNA methylation levels obtained from whole blood are usable as potential epigenetic biomarkers of alcohol dependence severity. Although the differences in DNA methylation are quite small, the fact that they can be found in different populations, different tissue as well as with different analytical methods suggests that GDAP1 DNA methylation could serve as biological predictor of alcohol dependence, especially in combination with epigenetic data of other genes of known influence. Unfortunately, we were not able to collect a second sample after 3 weeks from those patients, who did not complete the alcohol treatment. Without having obtained the DNA methylation levels for this group, we can only speculate that GDAP1 DNA methylation could also serve as a biomarker for treatment outcome. Measuring DNA methylation levels at a second time point from patients, who do not complete the alcohol treatment, should be taken into consideration in future studies.
In contrast to Philibert et al., we used a slightly different matching strategy: The cohort used for this study only consists of Caucasian men, and patients and control individuals were matched for age and smoking behavior. The cohort investigated by Philibert et al. is more heterogeneous, consisting of both sexes and different ethnicities. Furthermore, 27 patients were daily smokers, whereas only one control individual was a daily smoker. This is problematic, because smoking has a major influence on DNA methylation patterns.45 The authors take this limitation of their study into consideration by comparing the overlap of their 10000 most significant probes to the 910 epigenome-wide significant genes found by Dogan et al., who evaluated the effect of smoking on DNA methylation.45 Only 22 significant hits were overlapping between the 2 studies, leading Philibert et al. to the conclusion that the effects they are reporting are indeed due to alcohol consumption, and are not biased by differences in smoking behavior. However, this approach is based on the assumption that Dogan et al. were able to identify all genes epigenetically altered by smoking, which is highly unlikely. Furthermore, this strategy does not take into account potential overlapping effects of both smoking and alcohol consumption, which displays a high comorbidity and would therefore have to be further evaluated.
As already mentioned, no associations between the outer mitochondrial membrane protein GDAP1 and alcohol dependence have been reported thus far. Mutations in GDAP1 cause Charcot-Marie-Tooth (CMT) disease, a hereditary motor, and sensory neuropathy.46 The 2 major causes leading to CMT disease are mutations in PMP22 and MFN2, which directly affect the myelin sheath and the axon.47,48 Mutations in GDAP1 are associated with decreased mitochondrial fission activity (recessively inherited) or an impairment of mitochondrial fusion (dominantly inherited).39,40, 49 The expression of dominantly inherited mutated forms of GDAP1 lead to increased production of reactive oxygen species (ROS).50 Furthermore, wild type GDAP1 has been reported to protect against oxidative stress.51 As the production of ROS also is a direct effect of alcohol intake,52 this could be a potential link explaining GDAP1 hypomethylation in alcohol dependent patients: DNA hypomethylation should lead to increased expression and consequently increased protein production in alcohol dependent patients. Therefore, GDAP1 overexpression could counteract and compensate for the increased oxidative stress in alcohol dependence. This hypothesis is supported by the fact that DNA methylation levels rise after 3 weeks of alcohol treatment. In this time period, oxidative stress in alcohol dependent patients should be dramatically reduced. However, as we neither measured GDAP1 expression, nor GDAP1 protein levels or the levels of ROS, this hypothesis should be followed up in subsequent studies. Further links between CMT and alcohol dependence are provided by recent studies, showing that a triple-therapy with a combination of naltrexone, baclofen, and sorbitol (PXT3003) can improve health of patients suffering from CMT disease.53,54 While PXT3003 was shown to downregulate PMP22 mRNA expression and improve myelination as well as axonal regeneration,53 both naltrexone and baclofen are also used (partly off-label) to treat alcohol dependence.55 Acting as an opioid antagonist (naltrexone) and a GABA-B-receptor agonist (baclofen), respectively, these drugs reduce the rewarding effects of alcohol and inhibit dopaminergic neurotransmission.56 Whether GDAP1 is also influenced by naltrexone and/or baclofen requires investigation.
The GDAP1 gene is regulated by the transcription factor YY1.57 Up to date, there is no evidence linking YY1 to alcohol dependence. However, other putative transcription factor binding sites include the binding sites of EGR1 and ZNF143, among others, as analyzed with JASPAR.58 For both transcription factors, a potential link to alcohol dependence, such as alcoholic fatty liver disease 59 or the regulation of aldehyde reductase 60 has been previously reported. The lack of functional data is a limitation of our study. Therefore, future studies are needed to better understand the regulation of GDAP1 as well as its function in the context of alcohol dependence and to investigate the impact of altered DNA methylation on gene expression.
In conclusion, in the present study we were able to validate and replicate the finding of GDAP1 being significantly hypomethylated in alcohol dependent patients compared to healthy control individuals, which was previously discovered in an epigenome-wide association study.32 Furthermore, we show that these differences in DNA methylation diminish after 3 weeks of abstinence, leading us to the conclusion that GDAP1 DNA methylation could serve as a possible epigenetic biomarker for severity of alcohol dependence and potentially for treatment outcome. Our data add to the growing body of knowledge on epigenetic effects in alcohol dependence and support GDAP1 as a novel candidate gene implicated in alcohol dependence. However, future studies are needed to replicate our finding of epigenetic changes in GDAP1 during alcohol treatment in independent cohorts, as well as to clarify potential mechanisms of action.
Subjects & methods
This sample was comprised of 49 male patients (mean age 49.14 ± 10.47 years) with a diagnosis of alcohol dependence according to the fourth edition of the Diagnostic and Statistical Manual of Mental Disorders (DSM-IV) participating in a 3- or 6-week alcohol treatment program at the Clinic for Psychiatry and Psychotherapy, Tuebingen. Subjects with any other addiction except nicotine have been excluded, as well as subjects with any other psychiatric comorbidity necessitating psychiatric medication. Except for 8 patients, the last exposure to alcohol before entering the study had not exceeded 72 h. For the first days of detoxification, clomethiazole was administered if necessary. Population-based male control individuals (n = 37, mean age 47.41 ± 12.32 years) were recruited from the city of Tuebingen, Germany and the surrounding area. Control individuals were matched for age and smoking behavior. Phenotypic information about patients and control individuals was obtained by self-administered questionnaires. The following questionnaires were used in patients: Alcohol consumption was assessed using the AUDIT,61 alcohol craving using the OCDS 62 and the global distress level (GSI) using the SCL-90-R.63 The SCL-90-R and OCDS were repeated after 21 d of detoxification (T2). Control individuals were screened for problematic alcohol intake using the AUDIT questionnaire, and control individuals with an AUDIT-Score > 15 were excluded, as a higher value is suggestive for alcohol dependence.28 The SCL-90-R questionnaire was used in control individuals as well, and in addition, demographic information and health status of both—patients and control individuals—was assessed. All subjects were Caucasian and provided written informed consent prior to participation. The study was approved by the ethics committee of the University of Tuebingen and was conducted in accordance with the Declaration of Helsinki.
Ethylenediaminetetraacetic (EDTA) peripheral venous blood samples were taken from all patients immediately after hospital admission (T1). After 21 d (± 2 d) of treatment (T2), a second EDTA-blood sample was taken from the 33 patients (mean age 48.7 ± 10.92 years) who remained in the program (drop-out rate: 33%). EDTA-blood from control individuals was drawn immediately after study inclusion. Blood samples were instantly frozen and kept at −80°C until further usage. DNA was extracted using the QIAamp DNA Blood Maxi Kit (Qiagen). Genomic DNA (500 ng) was bisulfite converted using the EpiTect Fast Bisulfite Conversion Kit (Qiagen) according to the manufacturer's protocol with the following adjustments: incubation steps at 60°C were prolonged to 15 min (instead of 10 min); converted DNA was eluted in 20 µl instead of 15 µl and stored at −20°C until analysis.
Pyrosequencing was performed as follows: A 166 bp fragment covering the TSS200 region of GDAP1 and partially overlapping the transcription start site was amplified by PCR from 2 µl bisulfite-treated DNA using the PCR Primer Set from the PyroMark CpG Assay GDAP1 (PM00035399) and the PyroMark PCR Kit (both Qiagen) according to manufacturer's protocol. The CpG Assay GDAP1 covers 3 CpG sites located within chromosome 8 (site 1 located at 75,262,523, 95 bp upstream of the TSS; site 2 located at 75,262,532, 86 bp upstream of the TSS; and site 3 located at 75,262,534, 84 bp upstream of the TSS) and includes the CpG site cg23779890 (site 1) which has been previously implicated by Philibert et al.32 The 3 CpG sites are part of a larger CpG island including 48 CpG sites (chr8:75,262,522-75,263,044).
Cycling conditions were as follows: 95°C for 15 min; 94°C for 30 s, 56°C for 30 s, 72°C for 30 s (45 cycles); 72°C for 10 min. To detect potentially biased amplification of differentially methylated fragments, DNA samples with known methylation levels (0%, 25%, 50%, 75%, and 100%) were included as controls (EpiTect Control DNA, Qiagen) in the amplification and the pyrosequencing reaction.
PCR products and a no template control were visualized on a 2% agarose gel to verify successful amplification and specificity of the products. Processing of the PCR amplicons for the pyrosequencing analysis was performed in accordance with the manufacturer´s protocol and PCR products were then pyrosequenced using the PyroMark Q24 system (Qiagen) and the sequencing primer from the PyroMark CpG Assay GDAP1 (PM00035399). The percentage of methylation at each of the 3 CpG sites analyzed was quantified using the PyroMark Q24 software version 2.0.6 (Qiagen). Pyrosequencing was performed in duplicates. To avoid plate effects, samples from patients and control individuals were mixed on each plate and the samples were randomly assigned to different wells for the 2 sequencing runs. For quality control the coefficient of variance (CV) was calculated. For the 33 samples (18 control individual samples, 14 patient samples at T1 and 1 patient sample at T2) where the CV between 2 runs for any site was ≥ 0.3, a third measurement was obtained. The outlier was eliminated from further analysis, and only the 2 remaining values were used. Using this approach led to a maximum variation of 2.02%. Typically, an intra-sample variation of ≤ 3% is considered reliable.
Statistical analysis was conducted using SPSS version 21.0 (IBM). Each site was examined individually. DNA methylation levels were not normally distributed according to the Shapiro-Wilk test. Hence, non-parametric test methods were applied. Differences in the percentage of DNA methylation between the patient group and the control group were analyzed using the Mann-Whitney U test. For identifying differences in DNA methylation, GSI score, and OCDS score between the 2 time points T1 and T2 of the patients, the Wilcoxon test was used. Correlations between continuous variables were tested using the Spearman correlation test. A significance level of P ≤ 0.05 was considered significant.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Danuta Altpaß and Gisbert Farger for excellent technical assistance and expertise. We would also like to thank Sandra Eck and Dr. Ralf Brückmann for their help in recruiting control individuals. We are very grateful to all study participants. For editorial 525 assistance, we thank Drs. Nadja Freund and Daniel Bucher.
Funding
This study was supported by an intramural research grant (fortüne-program, F1331400.2) to VN and by a grant from the Wilhelm-Schuler-Stiftung to VN.
References
- 1.Agrawal A, Lynskey MT. Are there genetic influences on addiction: evidence from family, adoption and twin studies. Addiction 2008; 103:1069-81; PMID:18494843; http://dx.doi.org/ 10.1111/j.1360-0443.2008.02213.x [DOI] [PubMed] [Google Scholar]
- 2.Heath AC, Bucholz KK, Madden PA, Dinwiddie SH, Slutske WS, Bierut LJ, Statham DJ, Dunne MP, Whitfield JB, Martin NG. Genetic and environmental contributions to alcohol dependence risk in a national twin sample: consistency of findings in women and men. Psychological Med 1997; 27:1381-96; PMID:9403910; http://dx.doi.org/ 10.1017/S0033291797005643 [DOI] [PubMed] [Google Scholar]
- 3.Agrawal A, Lynskey MT, Todorov AA, Schrage AJ, Littlefield AK, Grant JD, Zhu Q, Nelson EC, Madden PA, Bucholz KK, et al.. A candidate gene association study of alcohol consumption in young women. Alcohol Clin Exp Res 2011; 35:550-8; PMID:21143251; http://dx.doi.org/ 10.1111/j.1530-0277.2010.01372.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laucht M, Treutlein J, Schmid B, Blomeyer D, Becker K, Buchmann AF, Schmidt MH, Esser G, Jennen-Steinmetz C, Rietschel M, et al.. Impact of psychosocial adversity on alcohol intake in young adults: moderation by the LL genotype of the serotonin transporter polymorphism. Biological psychiatry 2009; 66:102-9; PMID:19358979; http://dx.doi.org/ 10.1016/j.biopsych.2009.02.010 [DOI] [PubMed] [Google Scholar]
- 5.Nelson EC, Agrawal A, Pergadia ML, Wang JC, Whitfield JB, Saccone FS, Kern J, Grant JD, Schrage AJ, Rice JP, et al.. H2 haplotype at chromosome 17q21.31 protects against childhood sexual abuse-associated risk for alcohol consumption and dependence. Addict Biol 2010; 15:1-11; PMID:19878140; http://dx.doi.org/ 10.1111/j.1369-1600.2009.00181.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blomeyer D, Treutlein J, Esser G, Schmidt MH, Schumann G, Laucht M. Interaction between CRHR1 gene and stressful life events predicts adolescent heavy alcohol use. Biological psychiatry 2008; 63:146-51; PMID:17597588; http://dx.doi.org/ 10.1016/j.biopsych.2007.04.026 [DOI] [PubMed] [Google Scholar]
- 7.Nieratschker V, Batra A, Fallgatter AJ. Genetics and epigenetics of alcohol dependence. J Mol Psychiatry 2013; 1:11; PMID:25408904; http://dx.doi.org/ 10.1186/2049-9256-1-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sutherland JE, Costa M. Epigenetics and the environment. Ann N Y Acad Sci 2003; 983:151-60. [DOI] [PubMed] [Google Scholar]
- 9.Hashimoto H, Vertino PM, Cheng X. Molecular coupling of DNA methylation and histone methylation. Epigenomics 2010; 2:657-69; PMID:21339843; http://dx.doi.org/ 10.2217/epi.10.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holliday R. DNA methylation and epigenetic mechanisms. Cell biophysics 1989; 15:15-20; PMID:2476223; http://dx.doi.org/ 10.1007/BF02991575 [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Leung FC. An evaluation of new criteria for CpG islands in the human genome as gene markers. Bioinformatics 2004; 20:1170-7; PMID:14764558; http://dx.doi.org/ 10.1093/bioinformatics/bth059 [DOI] [PubMed] [Google Scholar]
- 12.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature 2004; 429:457-63; PMID:15164071; http://dx.doi.org/ 10.1038/nature02625 [DOI] [PubMed] [Google Scholar]
- 13.Doerfler W. DNA methylation and gene activity. Ann Rev Biochem 1983; 52:93-124; PMID:6311083; http://dx.doi.org/ 10.1146/annurev.bi.52.070183.000521 [DOI] [PubMed] [Google Scholar]
- 14.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 2012; 13:484-92; PMID:22641018; http://dx.doi.org/ 10.1038/nrg3230 [DOI] [PubMed] [Google Scholar]
- 15.Feil R, Fraga MF. Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet 2011; 13:97-109; PMID: 22215131; http://dx.doi.org/23817309 10.1038/nrg3142 [DOI] [PubMed] [Google Scholar]
- 16.Mill J, Heijmans BT. From promises to practical strategies in epigenetic epidemiology. Nat Rev Genet 2013; 14:585-94; PMID:23817309; http://dx.doi.org/ 10.1038/nrg3405 [DOI] [PubMed] [Google Scholar]
- 17.Januar V, Saffery R, Ryan J. Epigenetics and depressive disorders: a review of current progress and future directions. Int J Epidemiol 2015; 44 (4):1364-1387; PMID:25716985; http://dx.doi.org/ 10.1093/ije/dyu273 [DOI] [PubMed] [Google Scholar]
- 18.Abdolmaleky HM, Smith CL, Zhou JR, Thiagalingam S. Epigenetic alterations of the dopaminergic system in major psychiatric disorders. Methods Mol Biol 2008; 448:187-212; PMID:18370235; http://dx.doi.org/ 10.1007/978-1-59745-205-2_9 [DOI] [PubMed] [Google Scholar]
- 19.Plazas-Mayorca MD, Vrana KE. Proteomic investigation of epigenetics in neuropsychiatric disorders: a missing link between genetics and behavior? J Proteome Res 2011; 10:58-65; PMID:20735116; http://dx.doi.org/ 10.1021/pr100463y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bleich S, Degner D, Javaheripour K, Kurth C, Kornhuber J. Homocysteine and alcoholism. J Neural Transm Suppl 2000:187-96; PMID:11205139; http://dx.doi.org/ 10.1007/978-3-7091-6301-6_12 [DOI] [PubMed] [Google Scholar]
- 21.Bleich S, Degner D, Wiltfang J, Maler JM, Niedmann P, Cohrs S, Mangholz A, Porzig J, Sprung R, Ruther E, et al.. Elevated homocysteine levels in alcohol withdrawal. Alcohol and alcoholism 2000; 35:351-4; PMID:10905999; http://dx.doi.org/ 10.1093/alcalc/35.4.351 [DOI] [PubMed] [Google Scholar]
- 22.Heese P, Linnebank M, Semmler A, Muschler MA, Heberlein A, Frieling H, Stoffel-Wagner B, Kornhuber J, Banger M, Bleich S, et al.. Alterations of homocysteine serum levels during alcohol withdrawal are influenced by folate and riboflavin: results from the German Investigation on Neurobiology in Alcoholism (GINA). Alcohol and alcoholism 2012; 47:497-500; PMID:22645037; http://dx.doi.org/ 10.1093/alcalc/ags058 [DOI] [PubMed] [Google Scholar]
- 23.Bleich S, Hillemacher T. Homocysteine, alcoholism and its molecular networks. Pharmacopsychiatry 2009; 42 Suppl 1:S102-9; PMID:19434547; http://dx.doi.org/ 10.1055/s-0029-1214396 [DOI] [PubMed] [Google Scholar]
- 24.Bonsch D, Lenz B, Reulbach U, Kornhuber J, Bleich S. Homocysteine associated genomic DNA hypermethylation in patients with chronic alcoholism. J Neural Transm 2004; 111:1611-6; PMID:15565495; http://dx.doi.org/ 10.1007/s00702-004-0232-x [DOI] [PubMed] [Google Scholar]
- 25.French SW. Epigenetic events in liver cancer resulting from alcoholic liver disease. Alcohol Res 2013; 35:57-67; PMID:24313165 [PMC free article] [PubMed] [Google Scholar]
- 26.Varela-Rey M, Woodhoo A, Martinez-Chantar ML, Mato JM, Lu SC. Alcohol, DNA methylation, and cancer. Alcohol Res 2013; 35:25-35; PMID:24313162 [PMC free article] [PubMed] [Google Scholar]
- 27.Philibert RA, Gunter TD, Beach SR, Brody GH, Madan A. MAOA methylation is associated with nicotine and alcohol dependence in women. Am J Med Genet B Neuropsychiatr Genet 2008; 147B:565-70; PMID:18454435; http://dx.doi.org/ 10.1002/ajmg.b.30778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nieratschker V, Grosshans M, Frank J, Strohmaier J, von der Goltz C, El-Maarri O, Witt SH, Cichon S, Nothen MM, Kiefer F, et al.. Epigenetic alteration of the dopamine transporter gene in alcohol-dependent patients is associated with age. Addiction biology 2014; 19:305-11; PMID:22506971; http://dx.doi.org/ 10.1111/j.1369-1600.2012.00459.x [DOI] [PubMed] [Google Scholar]
- 29.Philibert RA, Sandhu H, Hollenbeck N, Gunter T, Adams W, Madan A. The relationship of 5HTT (SLC6A4) methylation and genotype on mRNA expression and liability to major depression and alcohol dependence in subjects from the Iowa Adoption Studies. Am J Med Genet B Neuropsychiatr Genet 2008; 147B:543-9; PMID:17987668; http://dx.doi.org/ 10.1002/ajmg.b.30657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heberlein A, Muschler M, Frieling H, Behr M, Eberlein C, Wilhelm J, Groschl M, Kornhuber J, Bleich S, Hillemacher T. Epigenetic down regulation of nerve growth factor during alcohol withdrawal. Addiction biology 2013; 18:508-10; PMID:21392176; http://dx.doi.org/ 10.1111/j.1369-1600.2010.00307.x [DOI] [PubMed] [Google Scholar]
- 31.Hillemacher T, Weinland C, Lenz B, Kraus T, Heberlein A, Glahn A, Muschler MA, Bleich S, Kornhuber J, Frieling H. DNA methylation of the LEP gene is associated with craving during alcohol withdrawal. Psychoneuroendocrinology 2015; 51:371-7; PMID:25462909; http://dx.doi.org/ 10.1016/j.psyneuen.2014.10.014 [DOI] [PubMed] [Google Scholar]
- 32.Philibert RA, Penaluna B, White T, Shires S, Gunter T, Liesveld J, Erwin C, Hollenbeck N, Osborn T. A pilot examination of the genome-wide DNA methylation signatures of subjects entering and exiting short-term alcohol dependence treatment programs. Epigenetics 2014; 9:1212-9; PMID:25147915; http://dx.doi.org/25982659 10.4161/epi.32252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruggeri B, Nymberg C, Vuoksimaa E, Lourdusamy A, Wong CP, Carvalho FM, Jia T, Cattrell A, Macare C, Banaschewski T, et al.. Association of Protein Phosphatase PPM1G With Alcohol Use sDisorder and Brain Activity During Behavioral Control in a Genome-Wide Methylation Analysis. Am J Psychiatry 2015; 172:543-52; PMID:25982659; http://dx.doi.org/ 10.1176/appi.ajp.2014.14030382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang R, Miao Q, Wang C, Zhao R, Li W, Haile CN, Hao W, Zhang XY. Genome-wide DNA methylation analysis in alcohol dependence. Addiction biology 2013; 18:392-403; PMID:23387924; http://dx.doi.org/ 10.1111/adb.12037 [DOI] [PubMed] [Google Scholar]
- 35.Clark SL, Aberg KA, Nerella S, Kumar G, McClay JL, Chen W, Xie LY, Harada A, Shabalin AA, Gao G, et al.. Combined Whole Methylome and Genomewide Association Study Implicates CNTN4 in Alcohol Use. Alcohol Clin Exp Res 2015; 39:1396-405; PMID:26146898; http://dx.doi.org/ 10.1111/acer.12790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hultberg B, Berglund M, Andersson A, Frank A. Elevated plasma homocysteine in alcoholics. Alcohol Clin Exp Res 1993; 17:687-9; PMID:8392819; http://dx.doi.org/ 10.1111/j.1530-0277.1993.tb00820.x [DOI] [PubMed] [Google Scholar]
- 37.Wedekind D, Neumann K, Falkai P, Malchow B, Engel KR, Jamrozinski K, Havemann-Reinecke U. S100B and homocysteine in the acute alcohol withdrawal syndrome. Eur Arch Psychiatry Clin Neurosci 2011; 261:133-8; PMID:20593192; http://dx.doi.org/ 10.1007/s00406-010-0121-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Biermann T, Reulbach U, Lenz B, Frieling H, Muschler M, Hillemacher T, Kornhuber J, Bleich S. N-methyl-D-aspartate 2b receptor subtype (NR2B) promoter methylation in patients during alcohol withdrawal. J Neural Transm (Vienna) 2009; 116:615-22; PMID:19350219; http://dx.doi.org/ 10.1007/s00702-009-0212-2 [DOI] [PubMed] [Google Scholar]
- 39.Barneo-Munoz M, Juarez P, Civera-Tregon A, Yndriago L, Pla-Martin D, Zenker J, Cuevas-Martin C, Estela A, Sanchez-Arago M, Forteza-Vila J, et al.. Lack of GDAP1 induces neuronal calcium and mitochondrial defects in a knockout mouse model of charcot-marie-tooth neuropathy. PLoS Genet 2015; 11:e1005115; PMID:25860513; http://dx.doi.org/ 10.1371/journal.pgen.1005115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tazir M, Hamadouche T, Nouioua S, Mathis S, Vallat JM. Hereditary motor and sensory neuropathies or Charcot-Marie-Tooth diseases: an update. J Neurological Sci 2014; 347:14-22; PMID:25454638; http://dx.doi.org/ 10.1016/j.jns.2014.10.013 [DOI] [PubMed] [Google Scholar]
- 41.Yi P, Melnyk S, Pogribna M, Pogribny IP, Hine RJ, James SJ. Increase in plasma homocysteine associated with parallel increases in plasma S-adenosylhomocysteine and lymphocyte DNA hypomethylation. J Biol Chem 2000; 275:29318-23; PMID:10884384; http://dx.doi.org/ 10.1074/jbc.M002725200 [DOI] [PubMed] [Google Scholar]
- 42.Semmler A, Heese P, Stoffel-Wagner B, Muschler M, Heberlein A, Bigler L, Prost JC, Frieling H, Kornhuber J, Banger M, et al.. Alcohol abuse and cigarette smoking are associated with global DNA hypermethylation: results from the German Investigation on Neurobiology in Alcoholism (GINA). Alcohol 2015; 49:97-101; PMID:25702197; http://dx.doi.org/ 10.1016/j.alcohol.2015.01.004 [DOI] [PubMed] [Google Scholar]
- 43.Roessler J, Ammerpohl O, Gutwein J, Hasemeier B, Anwar SL, Kreipe H, Lehmann U. Quantitative cross-validation and content analysis of the 450k DNA methylation array from Illumina, Inc. BMC Res Notes 2012; 5:210; PMID:22546179; http://dx.doi.org/ 10.1186/1756-0500-5-210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dedeurwaerder S, Defrance M, Bizet M, Calonne E, Bontempi G, Fuks F. A comprehensive overview of Infinium HumanMethylation450 data processing. Brief Bioinform 2014; 15:929-41; PMID:23990268; http://dx.doi.org/ 10.1093/bib/bbt054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dogan MV, Shields B, Cutrona C, Gao L, Gibbons FX, Simons R, Monick M, Brody GH, Tan K, Beach SR, et al.. The effect of smoking on DNA methylation of peripheral blood mononuclear cells from African American women. BMC Genom 2014; 15:151; PMID:24559495; http://dx.doi.org/ 10.1186/1471-2164-15-151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krajewski KM, Lewis RA, Fuerst DR, Turansky C, Hinderer SR, Garbern J, Kamholz J, Shy ME. Neurological dysfunction and axonal degeneration in Charcot-Marie-Tooth disease type 1A. Brain 2000; 123 (Pt 7):1516-27; PMID:10869062; http://dx.doi.org/ 10.1093/brain/123.7.1516 [DOI] [PubMed] [Google Scholar]
- 47.Watila MM, Balarabe SA. Molecular and clinical features of inherited neuropathies due to PMP22 duplication. J Neurological Sci 2015; 355:18-24; PMID:26076881; http://dx.doi.org/ 10.1016/j.jns.2015.05.037 [DOI] [PubMed] [Google Scholar]
- 48.Cartoni R, Martinou JC. Role of mitofusin 2 mutations in the physiopathology of Charcot-Marie-Tooth disease type 2A. Exp Neurol 2009; 218:268-73; PMID:19427854; http://dx.doi.org/ 10.1016/j.expneurol.2009.05.003 [DOI] [PubMed] [Google Scholar]
- 49.Kabzinska D, Kotruchow K, Cegielska J, Hausmanowa-Petrusewicz I, Kochanski A. A severe recessive and a mild dominant form of Charcot-Marie-Tooth disease associated with a newly identified Glu222Lys GDAP1 gene mutation. Acta biochimica Polonica 2014; 61:739-44; PMID:25337607 [PubMed] [Google Scholar]
- 50.Niemann A, Wagner KM, Ruegg M, Suter U. GDAP1 mutations differ in their effects on mitochondrial dynamics and apoptosis depending on the mode of inheritance. Neurobiol Dis 2009; 36:509-20; PMID:19782751; http://dx.doi.org/ 10.1016/j.nbd.2009.09.011 [DOI] [PubMed] [Google Scholar]
- 51.Noack R, Frede S, Albrecht P, Henke N, Pfeiffer A, Knoll K, Dehmel T, Meyer Zu Horste G, Stettner M, Kieseier BC, et al.. Charcot-Marie-Tooth disease CMT4A: GDAP1 increases cellular glutathione and the mitochondrial membrane potential. Hum Mol Genet 2012; 21:150-62; PMID:21965300; http://dx.doi.org/ 10.1093/hmg/ddr450 [DOI] [PubMed] [Google Scholar]
- 52.Gonzalez-Reimers E, Santolaria-Fernandez F, Martin-Gonzalez MC, Fernandez-Rodriguez CM, Quintero-Platt G. Alcoholism: a systemic proinflammatory condition. World J Gastroenterol 2014; 20:14660-71; PMID:25356029; http://dx.doi.org/25491744 10.3748/wjg.v20.i40.14660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chumakov I, Milet A, Cholet N, Primas G, Boucard A, Pereira Y, Graudens E, Mandel J, Laffaire J, Foucquier J, et al.. Polytherapy with a combination of three repurposed drugs (PXT3003) down-regulates Pmp22 over-expression and improves myelination, axonal and functional parameters in models of CMT1A neuropathy. Orphanet J Rare Dis 2014; 9:201; PMID:25491744; http://dx.doi.org/ 10.1186/s13023-014-0201-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Attarian S, Vallat JM, Magy L, Funalot B, Gonnaud PM, Lacour A, Pereon Y, Dubourg O, Pouget J, Micallef J, et al.. An exploratory randomised double-blind and placebo-controlled phase 2 study of a combination of baclofen, naltrexone and sorbitol (PXT3003) in patients with Charcot-Marie-Tooth disease type 1A. Orphanet J Rare Dis 2014; 9:199; PMID:25519680; http://dx.doi.org/ 10.1186/s13023-014-0199-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Crowley P. Long-term drug treatment of patients with alcohol dependence. Australian Prescriber 2015; 38:41-3; PMID:26648614; http://dx.doi.org/ 10.18773/austprescr.2015.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soyka M, Mutschler J. Treatment-refractory substance use disorder: Focus on alcohol, opioids, and cocaine. Prog Neuro-psychopharmacol Biol Psychiatry 2015; PMID:26577297; http://dx.doi.org/ 10.1016/j.pnpbp.2015.11.003 [DOI] [PubMed] [Google Scholar]
- 57.Ratajewski M, Pulaski L. YY1-dependent transcriptional regulation of the human GDAP1 gene. Genomics 2009; 94:407-13; PMID:19720140; http://dx.doi.org/ 10.1016/j.ygeno.2009.08.014 [DOI] [PubMed] [Google Scholar]
- 58.Mathelier A, Fornes O, Arenillas DJ, Chen CY, Denay G, Lee J, Shi W, Shyr C, Tan G, Worsley-Hunt R, et al.. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res 2016; 44:D110-5; PMID:26531826; http://dx.doi.org/ 10.1093/nar/gkv1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Donohue TM Jr., Osna NA, Trambly CS, Whitaker NP, Thomes PG, Todero SL, Davis JS. Early growth response-1 contributes to steatosis development after acute ethanol administration. Alcoholism Clin Exp Res 2012; 36:759-67; PMID:22141421; http://dx.doi.org/ 10.1111/j.1530-0277.2011.01681.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barski OA, Papusha VZ, Kunkel GR, Gabbay KH. Regulation of aldehyde reductase expression by STAF and CHOP. Genomics 2004; 83:119-29; PMID:14667815; http://dx.doi.org/ 10.1016/S0888-7543(03)00213-1 [DOI] [PubMed] [Google Scholar]
- 61.Saunders JB, Aasland OG, Babor TF, de la Fuente JR, Grant M. Development of the Alcohol Use Disorders Identification Test (AUDIT): WHO Collaborative Project on Early Detection of Persons with Harmful Alcohol Consumption–II. Addiction 1993; 88:791-804; PMID:8329970; http://dx.doi.org/ 10.1111/j.1360-0443.1993.tb02093.x [DOI] [PubMed] [Google Scholar]
- 62.Moak DH, Anton RF, Latham PK. Further validation of the Obsessive-Compulsive Drinking Scale (OCDS). Relationship to alcoholism severity. Am J Addict 1998; 7:14-23; PMID:9522003; http://dx.doi.org/ 10.1111/j.1521-0391.1998.tb00463.x [DOI] [PubMed] [Google Scholar]
- 63.Mercier C, Brochu S, Girard M, Gravel J, Ouellet R, Pare R. Profiles of alcoholics according to the SCL-90-R: a confirmative study. Int J Addict 1992; 27:1267-82; PMID:1446961; http://dx.doi.org/ 10.3109/10826089209047349 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


