ABSTRACT
Colorectal cancer (CRC) is one of the most frequent causes of cancer death worldwide and is associated with adoption of a diet high in animal protein and saturated fat. Saturated fat induces increased bile secretion into the intestine. Increased bile secretion selects for populations of gut microbes capable of altering the bile acid pool, generating tumor-promoting secondary bile acids such as deoxycholic acid and lithocholic acid. Epidemiological evidence suggests CRC is associated with increased levels of DCA in serum, bile, and stool. Mechanisms by which secondary bile acids promote CRC are explored. Furthermore, in humans bile acid conjugation can vary by diet. Vegetarian diets favor glycine conjugation while diets high in animal protein favor taurine conjugation. Metabolism of taurine conjugated bile acids by gut microbes generates hydrogen sulfide, a genotoxic compound. Thus, taurocholic acid has the potential to stimulate intestinal bacteria capable of converting taurine and cholic acid to hydrogen sulfide and deoxycholic acid, a genotoxin and tumor-promoter, respectively.
KEYWORDS: bile acid, colon cancer, deoxycholic acid, diet, hydrogen sulfide, taurine
Introduction
With estimated overall costs of 201.5 billion dollars in 2008, cancer is among the most significant contributors of health care spending in the United States (US).1 Colorectal cancer (CRC) is the third most frequent cancer worldwide resulting in 142,882 new CRC cases and 50,830 CRC related deaths in 2013.1 CRC is a multifactorial disease in which both nature and nurture interact in varying degrees between individuals. Only about 15% of CRC incidence can be explained by heredity alone,2 implicating environmental risk factors, particularly the sedentary, overindulgent ‘Western lifestyle’. Physical activity, smoking status, alcohol, types and quantity of dietary fiber, micronutrient intake, and diets high in animal protein and saturated fat all appear to play a role in the disease.3 The goal of much observational and mechanistic work spanning many decades is to establish how we can alter our environment so as to lower the risk for CRC. Intriguingly, each of these factors also alters the gut microbiome.4,5 Several studies have attempted to apply high-throughput sequencing followed by network correlation analysis to identify gut bacteria associated with colorectal tumors.6-11 While each study concluded that CRC is associated with gut dysbiosis, a significant shift in the gut microbiome community structure relative to healthy control populations, there has been a lack of consistently observed dysbiotic microbiota patterns.
Identification of organisms such as Fusobacterium nucleatum DNA in CRC tumors correlate with reduced survival; however, it is unclear whether F. nucleatum causes tumor formation or simply thrives in the tumor environment.12-14 In addition, toxin formation by some strains of Bacteroides fragilis and Escherichia coli provide potential mechanisms of carcinogenesis worth further exploration.15-17
Bile acids have become recognized as a particularly important class of steroid molecule regulating host and microbial physiology.18-20 The gut microbiota have the capacity to significantly alter the physicochemical properties of bile acids, generating high affinity ligands to host nuclear receptors (farnesoid X receptor, Vitamin D receptor (VDR), pregnane X receptor) and G-coupled protein receptors (TGR-5), in addition to sphingosine-1-phosphate receptor 2, and activate a range of signal transduction pathways (JNK, ERK, Akt).19,20 The levels and composition of the human bile acid pool affects gut microbiome structure, and gut microbiome structure and gene content affects bile acid pool composition. The antimicrobial nature of bile acids prevents bacterial overgrowth in the small intestine; however, microbial metabolism of bile salts in the large intestine represents long-term exposure to metabolic end-products that are implicated in CRC. The dietary context appears to be a major environmental factor determining the extent to which bile acid metabolism becomes a risk factor for the disease, as we shall demonstrate.
Here we focus on the natural history of the biliary metabolite, taurocholic acid (3α,7α,12α-trihydroxy-5β-cholan-24-oic acid N-(2-sulfoethyl)amide) (TCA), whose components, taurine and cholic acid support the growth of microbial groups of low abundance that due to their metabolic by-products, are implicated mechanistically in DNA-damage and tumor-promotion. The proportion of TCA and its toxic metabolites increases directly with a Western diet high in animal protein and fat, and low in complex carbohydrates. These observations highlight the critical link between Western diet, bile salt metabolizing microorganisms, and CRC.
Taurine
Taurine (2-aminoethanesulfonic acid) is the most abundant free amino acid in humans and plays a variety of important physiological functions. Taurine is synthesized in hepatocytes from methionine and cysteine in adults, but is considered semi-essential due to the limited synthetic capacity of neonates, and declining biosynthetic capability with age and certain diseases.21 The taurine content of various foods and food products are summarized in Table 1. Diets rich in taurine include those high in animal protein and seafood; while vegetarian diets, particularly vegan result in significantly lower levels of taurine measured in blood and urine.21 Energy drinks are a particularly modern and Western phenomenon, supplemented with roughly 8x (8 mmoles) the daily intake of taurine (∼1–1.5 mmoles, 125–188 mg) on a typical Western diet.21,22 The body maintains homeostatic levels, but is unable to metabolize taurine. Consequently, excess taurine is excreted either in urine, or through bile, conjugated to bile acids.
Table 1.
Taurine content of common foods.
| Product | Taurine content range per 100 mi in nmol | Reference |
|---|---|---|
| Energy drinks | 1600 | Reissig et al. 2009 |
| Mollusks | 2850 ± 759 – 6614 ± 123 | Reissig et al. 2009 |
| Dark Meat Poultry | 1355 ± 299 – 2445 ± 551 | Laidlaw et al. 1990 |
| Fish | 332 ± 102 – 1375 ± 428 | Laidlaw et al. 1990 |
| Beef and Pork | 307 ±78–489 ±85 | Laidlaw et al. 1990 |
| Processed Meats | 251 ± 32–981 ±42 | Laidlaw et al. 1990 |
| Light Meat Poultry | 89 ± 9–236 ±55 | Laidlaw et al. 1990 |
| Shrimp | 84 ± 11–315 ± 102 | Laidlaw et al. 1990 |
| Breast milk | 40 | Sturman 1993 |
| Infant feeding formulas | 27.2 ±0.5–60.8 | Laidlaw et al. 1990 |
| Dairy | 15 ±2–62.4 ±7.2 | Laidlaw et al. 1990 |
| Fruit | <1 | Laidlaw et al. 1990 |
| Vegetables | <1 | Laidlaw et al. 1990 |
| Grains | <1 | Laidlaw et al. 1990 |
| Nuts and Seeds | <1 | Laidlaw et al. 1990 |
| Legumes and soy milk | <1 | Laidlaw et al. 1990 |
Synthesis and enterohepatic circulation of taurocholic acid
Bile acids are water-soluble, amphipathic molecules functioning primarily to solubilize lipids in an aqueous environment. The liver is the only organ in the body expressing all 14 enzymes required for synthesis of primary bile acids from cholesterol.23 Indeed, bile acid synthesis is the principal means whereby cholesterol is removed from the body. Bile acids stimulate bile flow due to osmotic effects and further aid in removal of cholesterol by formation of mixed micelles in bile that are secreted into the small intestine. While the diversity of primary bile acids produced between vertebrates is immense,24,25 humans make primarily 2 major bile acids, cholic acid (CA; 3α,7α,12α-trihydroxy-5β-cholan-24-oic acid) and chenodeoxycholic acid (CDCA; 3α,7α-dihydroxy-5β-cholan-24-oic acid). Rodents form both CA and CDCA; however, CDCA is converted to muricholic acids by enzymatic 6α- or 6β-hydroxylation in the liver.
Free bile acids are conjugated (amidated) to the amino acids taurine and/or glycine in hepatocytes. Conjugation (amidation) of bile acids has several important biological functions. The pKa of glycine (∼4.0) and taurine (<2.0) is lower than the “free” C24 bile acid precursor (∼5.0). Consequently, conjugated bile acids are fully ionized at physiological pH and impermeable to cell membranes (hepatocytes, cholangiocytes, enterocytes). Conjugated bile acids are also soluble in aqueous solution at acidic pH and resist Ca2+ precipitation.26 The amide bond of bile acids conjugated to taurine or glycine is highly stable, and resistant to hydrolysis by pancreatic carboxypeptidases.
The N-acyl amidation of bile acids is catalyzed by 2 sequential enzymatic steps in the liver. The first reaction is catalyzed by a microsomal enzyme, cholyl-CoA synthetase (EC 6.2.1.7), resulting in an acyl-CoA thioester,27,28 followed by transfer of the bile acid moiety from the CoA-thioester to either glycine or taurine, forming N-acyl bile acid conjugate by a single cytosolic enzyme bile acid-CoA:amino acid N-acyltransferase (BAT) (EC 2.3.1.65).28,29 BAT shows high substrate specificity for both glycine (Km 5 mM) and taurine (Km 1 mM).30 A major difference between rodents and humans is that taurine conjugation in rodents is genetically determined nearly all murine bile acids are taurine-conjugated. However, in humans, bile acid taurine:glycine conjugation ratio is diet-dependent. Measurements from Native Africans consuming their traditional low fat and protein diet have taurine:glycine ratios of 1:9, while the ratio has been measured to be 10:1 in those consuming a diet high in meat and seafood.31,32 Feeding taurine, but not glycine can substantially alter the taurine:glycine ratio.32 The biochemical basis for this is likely owing to the higher affinity of taurine to BAT, and the fact that bile is a major route of excretion of excess taurine.33
The bile acid pool in healthy humans is about 2–3 g, cycling several times with each meal (4–6 g), or roughly 12 to 18 g/day circulates from liver to gallbladder through the small intestine and from ileum back to the liver.34 A recent review by Dawson describes the transport of bile acids in the intestines and the influence of the gut microbiome on this process.35
Production of deoxycholic acid
A major theme in the liver-gut microbiome axis is that the liver is conjugative and oxidative; while the gut microbiota is hydrolytic and reductive. Bile acids synthesized by the host liver are termed primary bile acids whereas bacterial metabolites of host primary bile acids are termed secondary bile acids. In general, bacterial metabolism of bile acids in the anaerobic environment of the gut is limited to oxidation-reduction reactions of the 3, 7, 12-hydroxy groups catalyzed by stereo-specific and position-specific hydroxysteroid dehydrogenase (HSDH) enzymes forming oxo-bile acids and bile acid epimers (iso or epi bile acids). In humans, oxo and iso bile acids are, as Hofmann puts it “repaired” in the liver;36 however, a small group of bacteria can remove the 7α-hydroxy group converting CA or CDCA to deoxycholic acid (3α,12α-dihydroxy-5β-cholan-24-oic acid) and lithocholic acid (LCA; 3α-hydroxy-5β-cholan-24-oic acid), respectively, which are not “repaired” in humans, but are in rodents. Some gut bacteria capable of converting CA to DCA are also capable of converting 7β-hydroxy bile acids such as ursodeoxycholic acid (UDCA; 3α, 7β, 12α-dihydroxy-5β-cholan-24-oic acid) to LCA. UDCA is a primary bile acid in some vertebrates, such as bears, but is considered a secondary bile acid (or tertiary) in humans, found in small amounts in bile, generated by the concerted action of microbial 7α-HSDH and 7β-HSDH or orally ingested as a therapy for chronic liver and biliary disorders.
While glycine and taurine conjugated bile acids are not recognized by host carboxypeptidases, bacterial bile salt hydrolases (BSH) cleave bile acid conjugates liberating taurine or glycine (reviewed recently37). To date, organisms that express BSH are not capable of converting CA to deoxycholic acid (DCA).37,38 Likewise, organisms capable of producing DCA and LCA from host primary bile acids lack BSH and are incapable of converting conjugated primary bile acids such as TCA to DCA without the presence of BSH expressing taxa capable of converting TCA to free CA.39,40
To survive in the gut environment, microorganisms have to contend with numerous host selection pressures, particularly anti-microbial agents such as lectins, defensins, and bile acids. The detergent nature of bile acids lead to membrane damage, collapse of proton motive force, and oxidative stress and DNA damage, resulting in top-down selection pressure on gut microbiome structure.37 Microbes have evolved resistance mechanisms such as expression of HSDH capable of producing iso-bile acids (3β-hydroxy) or urso-bile acids (7β-hydroxy) that are less toxic.41,42 Indeed, the therapeutic use of ursodeoxycholic acid (UDCA) (3α,7β-dihydroxy-5β-cholan-24-oic acid) is due to its hydrophilicity, it is able to dilute out toxic secondary bile acids and render the bile acid pool less damaging to the host. UDCA has been used for over a millennium in traditional Chinese medicine, and is a principle therapy for a number of GI diseases including cholestasis, and potentially a preventative treatment for patients with adenomatous polyposis.
Phillip B. Hylemon's laboratory discovered the genes involved in secondary bile acid production and worked out much of the complex biochemistry and molecular biology of bile acid 7α-dehydroxylation of CA in Clostridium scindens.38 It is interesting to note that only a small number of intestinal Clostridium spp. have evolved a multi-gene bile acid inducible operon capable of bile acid 7α-dehydroxylation of CA and CDCA and 7β-dehydroxylation of UDCA.37,38,43 While the proximal explanation of this metabolic capacity is that the bile acid 7α/β-dehydroxylation pathway yields a net 2 electron reduction, the ultimate explanation might relate to production of a toxic, anti-microbial compound.44,45 In addition, the fact that secondary bile acids (DCA, LCA and derivatives) are high affinity ligands to several host nuclear and G-coupled protein receptors suggests perhaps these metabolites are involved in interkingdom signaling.19,37,38
During enterohepatic circulation, secondary bile acids are produced in the cecum and colon and returned to the liver by passive diffusion through the gut epithelium into the portal circulation.39 Evidence is emerging that as animal-based diets stimulate more bile input into the gut the levels of DCA-producing bacteria increase.47,48 LCA is also produced from the 7α-dehydroxylation of CDCA or the 7β-dehydroxylation of UDCA and is highly toxic (potentially genotoxic and tumor-promoting). However, LCA does not tend to accumulate in the bile acid pool because it is efficiently excreted in feces by 2 mechanisms: 1) Being monohydroxylated, it is insoluble in fecal water and efficiently precipitates out in the acidic pH of the colon particularly in the presence of Ca2+ ions; and 2) binding of LCA to VDR results in enzymatic sulfation of LCA49 whose hydrophilicity prevents passive absorption in colonocytes and allows removal from the body. Indeed, LCA makes up a small proportion (<2%); whereas DCA can reach upwards of 70% of the human biliary pool.39 DCA is more hydrophobic than CA,50 but more water-soluble than LCA allowing passive diffusion through the colon into the portal circulation where it can return to the liver. Unlike LCA, DCA does not activate the signaling cascade resulting in VDR-dependent sulfation.49 The human liver cannot 7α-hydroxylate DCA and so secondary bile acids can accumulate in some individuals, and hence the reason for our focus primarily on TCA rather than taurochenodeoxycholic acid (TCDCA) or other taurine-conjugated bile acids. Importantly, diets high in animal protein and fat, and low in fiber result in greater secretion of bile, decreased transit time, and higher intestinal pH resulting in increased fecal DCA and greater accumulation of DCA in the biliary pool.
Diet and gut microbial respiration of taurine
Given that taurine is an organosulfonate common in diets high in animal protein, it is not surprising that a high meat diet significantly increases both bile acid tauro-conjugation,31,32 as well as production of fecal sulfide.51 We want to emphasize that any bile acid, not just CA, can be conjugated to taurine and thus, from the perspective of taurine metabolism, our argument extends beyond TCA. Milk fat (MF) fed mice and not lowfat (LF) or polyunsaturated fat (PUFA) fed mice increased the abundance of the taurine respiring sulfidogenic organism B. wadsworthia. Dextran sodium sulfate (DSS) treatment of MF-fed SPF C57BL/6 mice induced more severe colitis than in their LF and PUFA fed counterparts, as well as consistent observations of B. wadsworthia.52 In mono-associated germ-free Il10−/−mice, colonization of B. wadsworthia could only be obtained in MF fed mice, and abundance was greater mainly in the mucosa. B. wadsworthia grown in culture medium was selectively stimulated by gallbladder bile from MF-fed mice. When low fat (LF)-fed SPF Il10−/− mice were gavaged with either TCA or glycocholic acid, only TCA fed mice exhibited increased growth of B. wadsworthia.52 Thus, at least in a colitis-susceptible mouse model, an organosulfonate conjugated to the bile acid CA was upregulated by MF and enhanced growth of the taurine respiring bacterium B. wadsworthia, particularly in the mucosa, and this promoted colitis, a known risk factor for CRC.52 It is not known if B. wadsworthia can deconjugate TCA, and the genome sequence of B. wadsworthia ATCC 49260 appears to lack a BSH gene, suggesting the requirement of BSH expressing members of the microbiome capable of liberating free taurine.
B. wadsworthia was first identified in 1988 during a study characterizing the microbiome of gangrenous appendicitis. The organism is a common anaerobic isolate in infections,53 including ear, biliary tract and liver abscess,54 and is the third most common isolate detected in appendicitis, with abundance correlating with severity of disease. As a β-lactamase producer, B. wadsworthia is resistant to most antibiotics, with metronidazole being the only effective agent against all tested strains. The organism has endotoxic activity, and has been observed to produce intraabdominal abscess in mice when injected directly into the peritoneal cavity. Evolutionary analysis of 16s rRNA genes place B. wadsworthia within the Desulfovibrionaceae family, and interestingly Desulfovibrio spp has also been identified in pathogenic roles.55,56 B. wadsworthia is asaccharolytic, and while it is able to ferment pyruvate and reduce nitrate, the organism grows most efficiently in the presence of taurine, making it uniquely suited to thrive in the taurine and hydrogen rich environment of the colon.
Bilophila wadsworthia respires taurine in 3 enzymatic steps, resulting in the release of ammonia, acetate, carbon dioxide, and H2S (Fig. 1). In the last enzymatic step, sulfite (SO32−) reduction can be achieved using formate, lactate, pyruvate, or the taurine carbon as electron donors, but is most efficient when utilizing hydrogen. SO32−reduction is carried out by the multi-subunit enzyme known as dissimilatory-sulfite reductase (DSR), which catalyzes the 6-electron reduction of SO32− to H2S.57 Desulfoviridin type DSRs, as the type held by B. wadsworthia, were originally thought to be comprised of α, β, and γ subunits designated as dsrA, dsrB, and dsrC respectively. Subsequent evidence revealed that dsrC was not only expressed at a separate locus, but is a separate protein which interacts with the dsrAB complex with a cysteine containing C-terminal residue. dsrAB type dissimilatory (bi)sulfite reductases are highly conserved and abundant in many bacteria and archaea due to lateral gene transfer.58 The enzyme has been characterized in Salmonella enterica serovar Typhimurium59 and Clostridium pasteurianum;60 however, the majority of bacteria with this activity in humans are found in the Desulfovibrionaceae, a subdivision of the δ-Proteobacteria.61 Sequencing analysis groups B. wadsworthia's dsrAB in the Deltaproteobacteria supercluster, and similar to other Desulfoviridin type DSR harboring organisms, B. wadsworthia's DSR contains a single dsrAB transcriptional unit that is coordinately expressed. Usually, Desulfoviridin type DSRs contain an additional dsrD gene with unknown purpose at the terminal end of the dsrAB operon, but B. wadsworthia is unique in that it does not have a separate dsrD gene, but rather has a merged dsrBD unit. The dsrA gene has been a useful marker for quantification of SRB, and B. wadsworthia specific dsrA primers have proven to be most suitable for quantification of the organism in intestinal and mucosal samples.62
Figure 1.
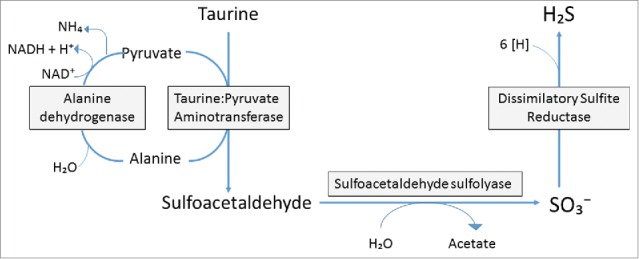
Taurine respiration by Bilophila wadsworthia. B. wadsworthia respires taurine in 3 enzymatic steps. First, taurine is transanimated from pyruvate using the enzyme taurine:pyruvate amino transferase to produce alanine and sulfoacetaldehyde. Sulfoacetaldehyde is sulfonated with water by sulfoacetaldehyde sulfolyase to produce acetate and sulfite. Sulfite is then reduced by dissimilatory sulfite reductase to hydrogen sulfide using electron donors like hydrogen and formate. Alanine is then converted back to pyruvate using alanine dehydrogenase.
Deoxycholic acid and colon cancer
During the 1970s epidemiological data correlated populations consuming diets high in fat with significantly higher rates of colon cancer.63 A sharp contrast was drawn between colon cancer rates in Japanese consuming traditional diets, and Americans consuming a “Western diet”.64 However, as Japanese immigrated to Hawaii and began consuming a ‘Western diet’, colon cancer rates rose midway between Japanese and American populations within two generations.64,65 Another population that received particular attention were the Seventh Day Adventists, who consume a vegetarian diet and have significantly lower incidence of CRC relative to age-matched and socioeconomically similar cohorts in the American population who consume a Western diet.66
Wynder et al. (1967) first proposed an association between dietary fat and CRC,67 and further proposed that dietary fat influences the fecal microbiota in a way that promotes CRC.64 Indeed, observations of high risk American populations on Western diets relative to low-risk groups (Japanese, Chinese, Seventh Day Adventists),68,69 reveal that patients with CRC and adenomas have higher total fecal bile acid levels,70 resulting from animal protein and fat.71 In developing countries, CRC rates were shown to be significantly higher in cities relative to rural areas, consistent with different dietary patterns.63 Reddy et al. (1974) conducted a small diet exchange study based on their hypothesis linking geographical observations of CRC risk and Western diet, and while they did not observe intra-individual changes in total bile acid levels, they did observe significant increases in DCA and its metabolites in subjects who consumed the animal protein and fat rich Western diet.72
The Native black African population (NA) has received considerable study due to their low CRC risk (<1 :100,000) compared to high-risk South African whites (SW) (17:100,000) and African Americans (AA) (65:100,000).73,74 Interestingly, SW consume diets higher in nutrients and vitamins, higher in insoluble fiber; however, NA consume a diet higher in resistant starch and importantly significantly lower in animal protein and fat.73 However, as NA adopt a Western diet, their incidence of CRC approaches that of SW and AA. Indeed, migrant studies have shown that one generation is sufficient for rates of CRC to elevate to that of the host western population75,76 and that dietary change is likely to be the cause. Berg (1973) observed a similar incidence of CRC in 1st and 2nd generation descendants of NA immigrants to Western countries who eschewed the traditional diet.75
The work of Wynder and Reddy during the 1970s pointed to higher fecal concentrations of bile acids in high risk populations of Americans on Western diets relative to low-risk groups (Japanese, Chinese, Seventh Day Adventists),68,75,77 that CRC and patients with adenomas have higher fecal bile acids,13 and that this was a result of animal protein and saturated fat.14 Similarly, the NA population was found to have significantly lower fecal total bile acids and significantly lower fecal DCA than AA consistent with lower meat intake.48,78
The role of DCA as a tumor promoter was established in animal models during this period. Single-dose intrarectal infusions of the carcinogen N-methyl-N′nitro- N-nitrosoguanidine (MNNG) followed by repeated rectal infusion doses of DCA significantly induced colorectal neoplasms, while DCA infusion alone was insufficient to induce tumor formation in both germ-free and conventional rats.79,80 Subsequent studies with rectal infusion of CA or CDCA in MNNG-treated rats showed significantly higher rates of tumor formation in conventional versus germ-free rats, suggesting the conversion of primary to secondary bile acids by intestinal bacteria was key to tumor promotion.81 Rats fed 20% corn oil or lard secreted significantly more fecal bile acids, and were more susceptible to colon cancer than rats fed normal levels of dietary fat (5%) in a 1,2-dimethylhydrazine-induced model.82 In the 1990s it was shown that: i) DCA was significantly elevated in serum of patients with colorectal adenomas;83-85 ii) serum DCA correlates with DCA in fecal water;83 and iii) colonic mucosal proliferation is correlated with DCA in serum.86
The activation of cellular signaling cascades and membrane perturbing effects generated by secondary bile acids, but not primary bile acids, provide mechanisms by which DCA and LCA promote CRC. The detergent properties of DCA (but not CA) cause membrane perturbations leading to the release of arachidonic acid, which is converted by the enzymes cyclooxygenase 2 (COX-2) and lipooxygenase, to pro-inflammatory and pro-angiogenic prostaglandins, and reactive oxygen species which damage DNA and inhibit DNA repair enzymes.87,88 DCA also induces COX-2 expression through transactivation of the epidermal growth factor receptor,87,89 and activates the β-catenin cell-signaling pathway resulting in colon cancer cell proliferation and invasiveness.90 Notably, a large body of published work demonstrates the effectiveness of non-steroidal anti-inflammatory drugs in significantly reducing polyp formation and reducing CRC risk by inhibiting the activity of COX-2.88 Mutations at the adenomatous polyposis coli (APC) locus are a common and early somatic event in polyp formation and CRC.25 Conspicuously, COX-2 can be downregulated by wild type but not mutant APC.91,92 APC regulates β-catenin levels in the cell, failure of which results in accumulation of transcriptionally active β-catenin, which both binds to the COX-2 promoter region activating transcription, and COX-2 mRNA increasing stabilization.93 In addition, DCA can activate proteosomal degradation of the tumor suppressor p53 selecting for cells resistant to apoptosis in spite of DNA damage.94
Taken together, diets high in animal protein and fat appear to promote colon carcinogenesis by selecting for bacteria capable of converting primary host bile acids to toxic, tumor-promoting secondary bile acids. Epidemiological, animal models, in vitro cell culture studies converge on DCA in particular as a metabolite whose serum levels correlate with disease and whose membrane-perturbing and activation of cellular signaling pathways associated with cell proliferation and apoptosis provide mechanisms for long term environmental risk of neoplasia. It is now important to identify additional bacterial metabolites than could serve genotoxic roles than might synergistically work with DCA to transform normal colonocytes into adenomas and adenocarcinomas. We will here argue that hydrogen sulfide (H2S) fits this bill.
Hydrogen sulfide is genotoxic
Sulfidogenic bacteria, including the cysteine fermenting Fusobacterium nucleatum, the taurine respiring Bilophila wadworthia, and SRB like Desulfovibrio spp., metabolize organic and inorganic sources of sulfur to produce H2S (Table 2). Several lines of evidence implicate H2S and SRB in the pathogenesis of the inflammatory bowel disease Ulcerative Colitis (UC), a risk factor for CRC. Fecal H2S is significantly elevated in UC patients experiencing active disease.95-98 However, other reports have reported no significant increase.99 It has been argued that UC is an “energy deficiency” disease due to the observation that high levels of H2S inhibits butyrate oxidation in vitro100-102 and in vivo,100 the primary energy source for colonocytes. Measurement of breath CO2 and luminal bicarbonate are significantly reduced in UC patients relative to controls after rectal infusion of butyrate.103,104 A difficulty inherent in fecal sulfide measurement may explain discrepancies between studies. It is estimated that 95% of H2S is absorbed through the colon105 owing to a natural diffusion gradient between colonic lumen (mM) and blood (μM). Furthermore, due to multiple dissociation states (HS− and H2S), which are affected by pH, roughly 2 thirds dissociates into the anionic form.
Table 2.
Sulfidogenic and 7a-dehydroxylating organisms, substrates, and end products.
| Group | Genotoxic/tumor promoting end product | Substrate | Example organism |
|---|---|---|---|
| Sulfate reducing bacteria | H2S | Inorganic sulfate Diet Organic sulfate Organosulfates Sulfomucins Sulfated bile acids Estrogen-3-sulfates | Desulfovibrio spp |
| Taurine respiring bacteria | H2s | Phenylsulfates Dietary Taurine Taurine conjugated bile acids | Bilophila wadsworthia |
| Cysteine fermenting bacteria | H2s | Dietary Cysteine Organosul fates Assimilatory sulfite reduction | Fusobacterium nucleatum |
| 7a-dehydroxylating bacteria | Deoxycholic acid | Taurocholic acid Cholic acid | Clostridium scindens |
One study showed that H2S increased proliferation in cells in the upper colonic crypts by over 54%.106 This proliferation was reversed by infusion of butyrate. Butyrate enemas are effective in treating the clinical symptoms of UC.107,108
Sulfide is thought to inhibit β-oxidation of butyrate through persulfide formation of the coenzyme A prosthetic group of the enzyme short-chain acyl CoA dehydrogenase.109 Inability to oxidize butyrate results in significant reduction in ion absorption, the formation of protective mucus, and cellular detoxification.99 Colonocytes express protective enzymes on the mucosal surface that oxidize H2S.110 At low concentration (μM), H2S is oxidized by the mitochondrial electron transport chain by H2S:quinone oxidoreductase, a gene in mitochondria, which originated in eubacteria exposed to sulfide-rich aquatic environments.111-112 Sulfide can thus increase ATP synthesis, resulting in a mix of oxidized metabolites including S2O32−, HSO3−, SO42−. At mM concentrations H2S inhibits cytochrome oxidase decreasing the electrochemical potential inhibiting basic cellular physiology. Loss of sulfide-detoxification has been reported to occur in active UC and CRC.113 Such a situation is conducive to genotoxicity leading to genetic changes and CRC.
Two studies in rats have also demonstrated that increased protein intake and increased protein fermentation were associated with genotoxicity.114,115 Fecal hydrogen sulfide levels were significantly higher in patients with neoplasm of the colon and those who have undergone surgery for sigmoid cancer.116 Animals fed DSS, but not DSS + metronidazole, had significantly higher development of dysplasia as well as adenoma and carcinoma.117 Two European studies demonstrate associations between sulfidogenic bacteria (Fusobacterium spp.) and the tumor surface in a subset of CRC118,119 Indeed, the Gaskins' lab demonstrated that sulfide is genotoxic at doses found in the colon providing a reasonable explanation for these observations.120-123 The rationale for this hypothesis is also supplemented by our long-term study of mammalian cell responses to the effects of exogenous sulfide. In a series of prior publications, we demonstrated that sulfide induces proliferative and inflammatory pathways in non-transformed intestinal crypt epithelial cells120,121 and provided unequivocal evidence that sulfide is genotoxic at concentrations less than those measured previously in human colon leading to cell-cycle arrest; and DNA damage, which is produced by oxidative stress.120-123 More recently, the genotoxic properties of sulfide have been demonstrated in non-transformed human intestinal epithelial cells with further evidence that sulfide modulates the expression of genes involved in cell-cycle progression, triggering both inflammatory and DNA repair responses.121 Together with these data, our own published work showing that sulfide is directly genotoxic at high concentrations suggests that H2S could be a significant bacterial metabolite that initiates colon cancer.
Taurocholic acid: metabolic potential for carcinogen and tumor-promoter in one
The abovementioned animal study by Devkota et al. (2012) is intriguing because it suggests a link between an animal-fat diet and a substrate that has the potential to become both a genotoxin (H2S) and a tumor-promotor (DCA) in the context of a taurine respiring immunogenic bacteria and bile acid 7α-dehydroxylating bacterial species, respectively (Fig. 2).52 Animal-based diets that favor taurine-conjugation also lead to enhanced bile acid secretion. Enhanced bile secretion provides more bile acid substrate to support a larger population of DCA-producing bacteria.19 The relationship between sulfidogenesis in the Devkota et al. (2013) model52 and bile acid 7α-dehydroxylation have not been explored. However, in vitro studies of human fecal isolates have shown that deconjugation of TCA by a sulfide-producing Bacteroides sp. strain R1 stimulated bile acid 7α-dehydroxylation of Clostridium sp strain 9/1.124 Narushima et al. (2006) established germ-free mice with a minimal microbiota and observed a significant stimulation of DCA-formation by addition of an isolate identified as closely related to B. wadsworthia.125 Higher levels of fecal secondary bile acids have been shown to worsen the severity of colitis in DSS-treated mice.126 These data suggest stimulation of secondary bile acid formation by sulfide, and exacerbation of inflammation by the combination of DCA and H2S.
Figure 2.
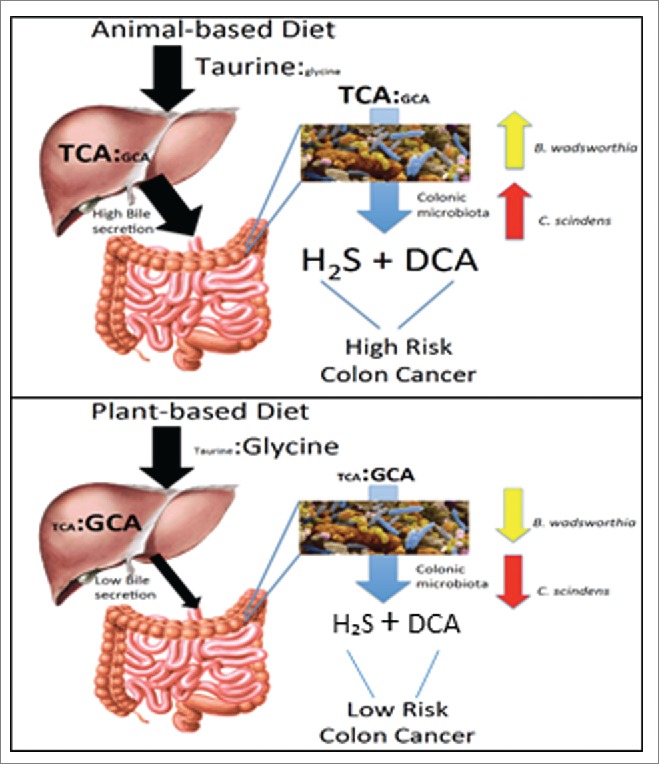
Hypothesis linking diet, TCA metabolism, and colon cancer risk. Animal-based diet high in taurine increases TCA levels in liver and gut, and results in higher levels of DCA in the gut than the plant-based diet owing to higher bile secretion due to higher fat consumption. TCA is metabolized to genotoxin H2S by B. wadsworthia and tumor-promoter, deoxycholic acid (DCA), by C. scindens. Plant-based diet, low in taurine, results in low levels of genotoxic H2S taurine metabolism, and lower fecal DCA levels.
Recent diet-exchange studies in humans have demonstrated rapid changes in the microbiota over short periods, confirming the role of diet on levels and activities of sulfidogenic, methanogenic, butyrogenic, and bile acid modifying bacteria.47,48 David et al. (2013) exchanged a plant-based and animal-based diet in healthy American volunteers and measured gut microbiome and key metabolites, including SCFA and bile acids.47 They showed that Prevotella, the major bacterial genus in NA, was reduced in subjects consuming an animal-based diet. Animal-based diet significantly increased the abundance of bile-resistant members such as B. wadsworthia, Alistipes putredinis, and Bacteroides sp., cluster 29.47 Total fecal bile acids and DCA in particular were significantly upregulated with the animal-based relative to plant-based diet. Genes involved in bile acid metabolism, including bile bsh and dsrA were significantly upregulated on animal based diet.47 SCFA associated with carbohydrate metabolism (acetate, butyrate) were significantly reduced on the animal-based diet, while SCFA (isovalerate, isobutyrate) associated with fermentation of amino acids were significantly upregulated on the animal-based diet.47
A recent 2-week food exchange study between NA and AA (NA consumed animal-based diet, AA consumed vegetarian-based diet) demonstrate rapid reciprocal changes in levels of bacteria responsible for sulfide production (B. wadsworthia, Desulfovibrio spp) and protective butyrate production, as well as levels of DCA producing bacteria (C. scindens) and DCA in stool.48 These results confirm a recent short-term diet exchange focusing on animal- and plant-based diets which found enhanced DCA formation and increased levels of taurine-utilizing, sulfide-producing B. wadsworthia.47 AA were dominated by Bacteroides while NA by Prevotella consistent with the study by David et al. (2013).47 NA had higher abundance of starch degrading gut bacteria (diet high in resistant starch), carbohydrate fermenters and butyrate producers and metabolites, while Americans had higher levels of potentially pathogenic proteobacteria (Escherichia and Acinetobacter) and bile acid deconjugators and their metabolic end products.48,78 O'Keefe et al. (2015) also measured colonic inflammation and found that it went below NA baseline when AA were placed on NA diet, while NA colonic inflammation went above AA baseline when NA were placed on AA diet.48 Butyrate production was associated with lower mucosal proliferation both in NA consuming high-fiber diet and AA switched to NA diet. NA switched to AA diet had significantly lower butyrogenesis. NA diet stimulated methanogenesis, sulfidogenesis, and acetogenesis, improving butyrate production by removing hydrogen. This study also reiterated an important connection between the gut microbiota, Western diet, and disease was the significant upregulation of choline.
Previous studies have shown that gut microbes convert choline to trimethylamine, which is absorbed and converted by the liver to the pro-atherogenic trimethylamine N-oxide (TMNO).127,128 AA had significant choline at baseline, and an observed increase in urinary TMNO in NA given the western diet. Prior studies by the same group showed that significantly higher levels of total fecal bile acids in AA than NA, and DCA in particular was significantly correlated (r2=0.65; P=0.01) with baiCD gene abundance- a gene in the bile acid-inducible operon of bile acid 7α-dehydroxylating bacteria such as Clostridium scindens and related organisms responsible for DCA production.78
In summary, we propose a novel mechanism to explain why consumption of a high red meat and saturated fat diet imparts risk for CRC development involving primary microbial risk factors (bile acid metabolizing and sulfidogenic bacteria) that are modifiable by diet (Fig. 3). The focus is on taurine, an overlooked sulfur amino acid that is abundant in red meat or provided by bacterial deconjugation of the bile salt TCA, which is increased in subjects consuming a diet high in saturated fat. The taurine provided by bacterial deconjugation of TCA is used as a substrate by B. wadsworthia for anaerobic respiration generating genotoxic H2S. And once deconjugated, free primary bile acids are further metabolized by colonic bacteria to genotoxic and proinflammatory secondary bile acids. Specifically, the production of the secondary bile acid DCA acts as a tumor promoter by causing membrane perturbations leading to the release of arachidonic acid, which is converted by the enzymes COX-2 and lipooxygenase, to pro-inflammatory and pro-angiogenic prostaglandins, and reactive oxygen species which damage DNA and inhibit DNA repair enzymes. Together these mechanisms support a compelling link among diet, levels of TCA, the metabolic end products of TCA metabolism by intestinal bacteria, and development of CRC.
Figure 3.
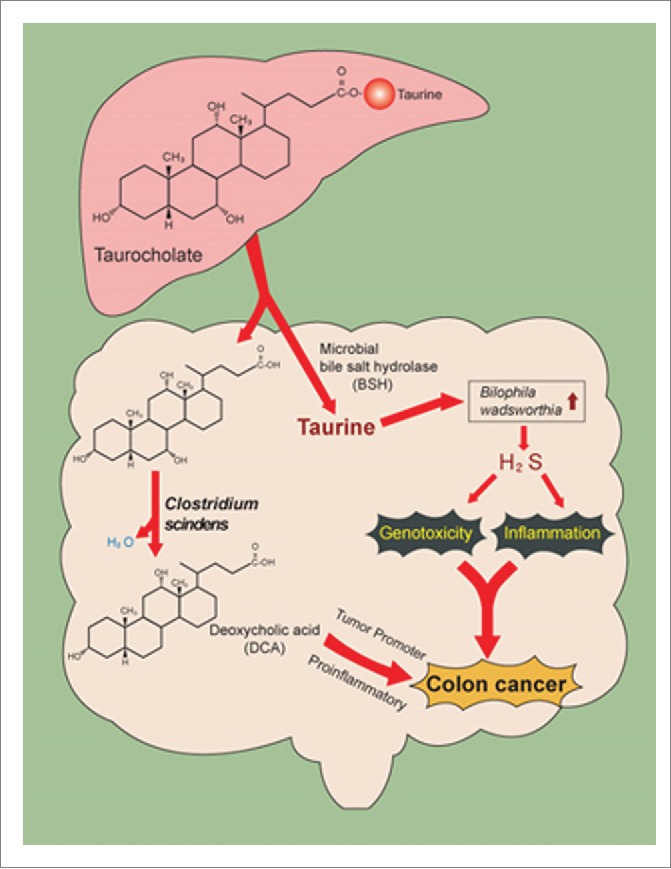
Metabolism of bile salt, taurocholic acid, by the gut microbiome. Gut microbes in the ileum and colon deconjugate bile salts to free bile acids by bile salt hydrolase. Taurine, because of sulfite moiety, can provide pathobionts in the gut with a terminal electron acceptor, allowing for their growth and expansion in the gut. High-fat diet is associated with increased taurine-conjugation in humans. Free primary bile acids are further metabolized to toxic secondary ones that can accumulate in the bile pool in humans and alter host physiology.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Reference
- [1].Howlader N, Noone AM, Krapcho M, Garshell J, Miller D, Altekruse SF, Kosary CL, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, Chen HS, Feuer EJ, Cronin KA (eds). SEER Cancer Statistics Review, 1975–2012, National Cancer Institute; Bethesda, MD, http:// seer .cancer. gov/csr/1975_2012/,-based-on-November-2014-SEER-data-submission,-posted-to-the-SEER -web-site, April 2015. [Google Scholar]
- [2].O'Shaughnessy JA, Kelloff GJ, Gordon GB, Dannenberg AJ, Hong WK, Fabian CJ, Sigman CC, Bertagnolli MM, Stratton SP, Lam S, et al.. Treatment and prevention of intraepithelial neoplasia: An important target for accelerated new agent development. Clin Cancer Res 2002; 8:314-46. [PubMed] [Google Scholar]
- [3].Durko L, Malecka-Panas E. Lifestyle modifications and colorectal cancer. Curr Colorectal Cancer Re 2014; 10:45-54; PMID:24659930; http://dx.doi.org/ 10.1007/s11888-013-0203-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Biedermann L, Zeitz J, Mwinyi J, Sutter-Minder E, Rehman A, Ott SJ, Steurer-Stey C, Frei A, Frei P, Scharl M, et al.. Smoking cessation induces profound changes in the composition of the intestinal microbiota in humans. PLoS One 2013; 8(3):e59260; PMID:23516617; http://dx.doi.org/ 10.1371/journal.pone.0059260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Mutlu EA, Gillevet PM, Rangwala H, Sikaroodi M, Naqvi A, Engen PA, Kwasny M, Lau CK, Keshavarzian A. Colonic microbiome is altered in alcoholism. Am J Physiol Gastrointest Liver Physiol 2012; 302(9):G966-78; PMID:22241860; http://dx.doi.org/ 10.1152/ajpgi.00380.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Gao Z, Guo B, Gao R, Zhu Q, Qin H. Microbiota dysbiosis is associated with colorectal cancer. Front Microbiol 2015; 6:20; PMID:25699023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Dejea CM, Wick EC, Hechenbleikner EM, White JR, Mark Welch JL, Rossetti BJ, Peterson SN, Snesrud EC, Borisy GG, Lazarev M, et al.. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc Natl Acad Sci U S A 2014; 111(51):18321-6; PMID:25489084; http://dx.doi.org/ 10.1073/pnas.1406199111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Brim H, Yooseph S, Zoetendal EG, Lee E, Torralbo M, Laiyemo AO, Shokrani B, Nelson K, Ashktorab H. Microbiome analysis of stool samples from African Americans with colon polyps. PLoS One 2013; 8(12):e81352; PMID:24376500; http://dx.doi.org/ 10.1371/journal.pone.0081352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zackular JP, Baxter NT, Iverson KD, Sadler WD, Petrosino JF, Chen GY, Schloss PD. The gut microbiome modulates colon tumorigenesis. MBio 2013; 4(6):e00692-13; PMID:24194538; http://dx.doi.org/ 10.1128/mBio.00692-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Weir TL, Manter DK, Sheflin AM, Barnett BA, Heuberger AL, Ryan EP. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS One 2013; 8(8):e70803; PMID:23940645; http://dx.doi.org/ 10.1371/journal.pone.0070803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Marchesi JR, Dutilh BE, Hall N, Peters WH, Roelofs R, Boleij A, Tjalsma H. Towards the human colorectal cancer microbiome. PLoS One 2011; 6(5):e20447; PMID:21647227; http://dx.doi.org/ 10.1371/journal.pone.0020447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, Yang J, Dou R, Masugi Y, Song M, et al.. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut 2015; pii: gutjnl-2015-310101. http://dx.doi.org/ 10.1136/gutjnl-2015-310101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Mima K, Sukawa Y, Nishihara R, Qian ZR, Yamauchi M, Inamura K, Kim SA, Masuda A, Nowak JA, Nosho K, et al.. Fusobacterium nucleatum and T cells in colorectal carcinoma. JAMA Oncol 2015; 1(5):653-61; PMID:26181352; http://dx.doi.org/ 10.1001/jamaoncol.2015.1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].McCoy AN, Araújo-Pérez F, Azcárate-Peril A, Yeh JJ, Sandler RS, Keku TO. Fusobacterium is associated with colorectal adenomas. PLoS One 2013; 8(1):e53653; PMID:23335968; http://dx.doi.org/ 10.1371/journal.pone.0053653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Sears CL, Geis AL, Housseau F. Bacteroides fragilis subverts mucosal biology: from symbiont to colon carcinogenesis. J Clin Invest 2014; 124(10):4166-72; PMID:25105360; http://dx.doi.org/ 10.1172/JCI72334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Boleij A, Hechenbleikner EM, Goodwin AC, Badani R, Stein EM, Lazarev MG, Ellis B, Carroll KC, Albesiano E, Wick EC, Platz EA, Pardoll DM, Sears CL. The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin Infect Dis 2015; 60(2):208-15; PMID:25305284; http://dx.doi.org/ 10.1093/cid/ciu787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Raisch J, Buc E, Bonnet M, Sauvanet P, Vazeille E, de Vallée A, Déchelotte P, Darcha C, Pezet D, Bonnet R, et al.. Colon cancer-associated B2 Escherichia coli colonize gut mucosa and promote cell proliferation. World J Gastroenterol 2014; 20(21):6560-72; PMID:24914378; http://dx.doi.org/ 10.3748/wjg.v20.i21.6560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ridlon JM, Alves JM, Hylemon PB, Bajaj JS. Cirrhosis, bile acids and gut microbiota: unraveling a complex relationship. Gut Microbes 2013; 4(5):382-7; PMID:23851335; http://dx.doi.org/ 10.4161/gmic.25723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ridlon JM, Kang DJ, Hylemon PB, Bajaj JS. Bile acids and the gut microbiome. Curr Opin Gastroenterol 2014; 30(3):332-8; PMID:24625896; http://dx.doi.org/ 10.1097/MOG.0000000000000057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhou H, Hylemon PB. Bile acids are nutrient signaling hormones. Steroids 2014; 86:62-8; PMID:24819989; http://dx.doi.org/ 10.1016/j.steroids.2014.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lambert IH, Kristensen DM, Holm JB, Mortensen OH. Physiological role of taurine-from organism to organelle. Acta Physiol 2015; 213:191-212; http://dx.doi.org/ 10.1111/apha.12365 [DOI] [PubMed] [Google Scholar]
- [22].Reissig CJ, Strain EC, Griffiths RR. Caffeinated energy drinks-a growing problem. Drug Alcohol Depend 2009; 99:1-10; PMID:18809264; http://dx.doi.org/ 10.1016/j.drugalcdep.2008.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Chiang JY. Bile acids: regulation of synthesis. J Lipid Res 2009; 50:1955-1966; PMID:19346330; http://dx.doi.org/ 10.1194/jlr.R900010-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wang DQ, Carey MC. Therapeutic uses of animal biles in traditional Chinese medicine: an ethnopharmacological, biophysical chemical and medicinal review. World J Gastroenterol 2014; 20(29):9952-75; PMID:25110425; http://dx.doi.org/ 10.3748/wjg.v20.i29.9952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hofmann AF, Hagey LR, Krasowski MD. Bile salts of vertebrates: structural variation and possible evolutionary significance. J Lipid Res 2010; 51(2):226-46; PMID:19638645; http://dx.doi.org/ 10.1194/jlr.R000042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hofmann AF, Roda A. Physicochemical properties of bile acids and their relationship to biological properties: an overview of the problem. J Lipid Res 1984; 25:1477-89; PMID:6397555 [PubMed] [Google Scholar]
- [27].Killenberg PG, Jordan JT. Purification and characterization of bile acid-CoA:amino acid N-acyltransferase from rat liver. J Biol Chem 1978; 253(4):1005-10; PMID:624713 [PubMed] [Google Scholar]
- [28].Shersten T. in Metabolic Conjugation and Metabolic Hydrolysis (Fisherman W.H., ed) 1971; pp. 75-121, Academic Press, New York. [Google Scholar]
- [29].Vessey DA. The co-purification and common identity of cholyl CoA:glycine- and cholyl CoA:taurine-N-acyltransferase activities from bovine liver. J Biol Chem 1979; 254(6):2059-63; PMID:422567 [PubMed] [Google Scholar]
- [30].Johnson MR, Barnes S, Sweeny DJ, Diasio RB. 2-Fluoro-beta-alanine, a previously unrecognized substrate for bile acid coenzyme A:amino acid:N-acyltransferase from human liver. Biochem Pharmacol 1990; 40(6):1241-46; PMID:2119585; http://dx.doi.org/ 10.1016/0006-2952(90)90389-3 [DOI] [PubMed] [Google Scholar]
- [31].Hardison WGM. Hepatic taurine concentration and dietary taurine as regulators of bile acid conjugation with taurine. Gastroenterology 1978; 75:71-75; PMID:401099 [PubMed] [Google Scholar]
- [32].Sjöval J. Dietary glycine and taurine on bile acid conjugation in man. Bile acids and steroids 75. Proc Soc Exp Biol Med 1959; 100(4):676-8; PMID:13645682; http://dx.doi.org/ 10.3181/00379727-100-24741 [DOI] [PubMed] [Google Scholar]
- [33].Falany CN, Johnson MR, Barnes S, Diasio RB. Glycine and taurine conjugation of bile acids by a single enzyme. Molecular cloning and expression of human liver bile acid CoA:amino acid N-acyltransferase. J Biol Chem 1994; 269(30):19375-9; PMID:8034703 [PubMed] [Google Scholar]
- [34].Hofmann AF. The continuing importance of bile acids in liver and intestinal disease. Arch Intern Med 1999; 159(22):2647-58; PMID:10597755; http://dx.doi.org/ 10.1001/archinte.159.22.2647 [DOI] [PubMed] [Google Scholar]
- [35].Dawson PA, Karpen SJ. Intestinal transport and metabolism of bile acids. J Lipid Res 2015; 56(6):1085-99; PMID:25210150; http://dx.doi.org/ 10.1194/jlr.R054114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hofmann AF, Hagey LR. Key discoveries in bile acid chemistry and biology and their clinical applications: history of the last eight decades. J Lipid Res 2014; 55:1553-1595; PMID:24838141; http://dx.doi.org/ 10.1194/jlr.R049437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ridlon JM, Harris SC, Bhowmilk S, Kang D, Hylemon PB. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes 2016; 7(1):22-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Hughes DT, Sperandio V. Inter-kingdom signaling: communication between bacteria and their hosts. Nat Rev Microbiol 2008; 6:111-120; PMID:18197168; http://dx.doi.org/ 10.1038/nrmicro1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res 2006; 47(2):241-59; PMID:16299351; http://dx.doi.org/ 10.1194/jlr.R500013-JLR200 [DOI] [PubMed] [Google Scholar]
- [40].Batta AK, Salen G, Arora R, Shefer S, Batta M, Person A. Side chain conjugation prevents bacterial 7-dehydroxylation of bile acids. J Biol Chem 1990; 265(19):10925-28; PMID:2358447 [PubMed] [Google Scholar]
- [41].Stellwag EJ, Hylemon PB. 7alpha-dehydroxylation of cholic acid and chenodeoxycholic acid by Clostridium leptum. J Lipid Res 1979; 20(3):325-333; PMID:36438 [PubMed] [Google Scholar]
- [42].Devlin AS, Fischbach MA. A biosynthetic pathway for a prominent class of microbiota-derived bile acids. Nat Chem Biol 2015; 11(9):685-90; PMID:26192599; http://dx.doi.org/ 10.1038/nchembio.1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Macdonald IA, White BA, Hylemon PB. Separation of 7 alpha- and 7 beta-hydroxysteroid dehydrogenase activities from clostridium absonum ATCC# 27555 and cellular response of this organism to bile acid inducers. J Lipid Res 1983; 24(9):1119-26; PMID:6579144 [PubMed] [Google Scholar]
- [44].Kitahara M, Takamine F, Imamura T, Benno Y. Assignment of Eubacterium sp. VPI 12708 and related strains with high bile acid 7alpha-dehydroxylating activity to Clostridium scindens and proposal of Clostridium hylemonae sp. nov., isolated from human faeces. Int J Syst Evol Microbiol 2000; 50 Pt 3:971-8; PMID:10843034; http://dx.doi.org/ 10.1099/00207713-50-3-971 [DOI] [PubMed] [Google Scholar]
- [45].Islam KB, Fukiya S, Hagio M, Fujii N, Ishizuka S, Ooka T, Ogura Y, Hayashi T, Yokota A. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology 2011; 141(5):1773-81; PMID:21839040; http://dx.doi.org/ 10.1053/j.gastro.2011.07.046 [DOI] [PubMed] [Google Scholar]
- [46].Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, et al.. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 2015; 517(7533):205-8; PMID:25337874; http://dx.doi.org/ 10.1038/nature13828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al.. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014; 505(7484):559-63; PMID:24336217; http://dx.doi.org/ 10.1038/nature12820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].O'Keefe SJ, Li JV, Lahti L, Ou J, Carbonero F, Mohammed K, Posma JM, Kinross J, Wahl E, Ruder E, et al.. Fat, fibre and cancer risk in African Americans and rural Africans. Nat Commun 2015; 6:6342; PMID:25919227; http://dx.doi.org/ 10.1038/ncomms7342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Makishima M, Lu TT, Xie W, Whitfield GK, Domoto H, Evans RM, Haussler MR, Mangelsdorf DJ. Vitamin D receptor as an intestinal bile acid sensor. Science 2002; 296(5571):1313-6; PMID:12016314; http://dx.doi.org/ 10.1126/science.1070477 [DOI] [PubMed] [Google Scholar]
- [50].Heuman DM. Quantitative estimation of the hydrophilic-hydrophobic balance of mixed bile salt solutions. J Lipid Res 1989; 30(5):719-30; PMID:2760545 [PubMed] [Google Scholar]
- [51].Magee EA, Richardson CJ, Hughes R, Cummings JH. Contribution of dietary protein to sulfide production in the large intestine: an in vitro and a controlled feeding study in humans. Am J Clin Nutr 2000; 72(6):1488-94; PMID:11101476 [DOI] [PubMed] [Google Scholar]
- [52].Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, Antonopoulos DA, Jabri B, Chang EB. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature 2012; 487(7405):104-8; PMID:22722865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Finegold S, Summanen P, Gerardo SH, Baron E. (1992) Clinical importance of Bilophila wadsworthia. Eur J Clin Microbiol Infect Dis 1992; 11:1058-63; PMID:1295759; http://dx.doi.org/ 10.1007/BF01967799 [DOI] [PubMed] [Google Scholar]
- [54].Claros MC, Schumacher UK, Jacob M, Hunt Gerardo S, Kleinkauf N, Goldstein EJC, Finegold SM, Rodloff AC. Characterization ofBilophila wadsworthia isolates using PCR fingerprinting. Anaerobe 1999; 5:589-593; http://dx.doi.org/ 10.1006/anae.1999.0307 [DOI] [Google Scholar]
- [55].Loubinoux J, Mory F, Pereira IA, Le Faou AE. Bacteremia caused by a strain of Desulfovibrio related to the provisionally named Desulfovibrio fairfieldensis. J Clin Microbiol 2000; 38(2):931-4; PMID:10655421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Tee W, Dyall-Smith M, Woods W, Eisen D. Probable new species of Desulfovibrio isolated from a pyogenic liver abscess. J Clin Microbiol 1996; 34(7):1760-64; PMID:8784584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Lengeler J, Drews G, Schlegel H. Biology of the Prokaryotes 1999. Pp. 293 Wiley-Blackwell. [Google Scholar]
- [58].Müller AL, Kjeldsen KU, Rattei T, Pester M, Loy A. Phylogenetic and environmental diversity of DsrAB-type dissimilatory (bi)sulfite reductases. The ISME Journal 2014; 9:1152-1165; PMID:25343514; http://dx.doi.org/ 10.1038/ismej.2014.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Huang CJ, Barrett EL. Sequence analysis and expression of the Salmonella typhimurium asr operon encoding production of hydrogen sulfide from sulfite. J Bacteriol 1991; 173(4):1544-53; PMID:1704886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Harrison G, Curle C, Laishley EJ. Purification and characterization of an inducible dissimilatory type sulfite reductase from Clostridium pasteurianum. Arch Microbiol 1984; 138(1):72-8; PMID:6742957; http://dx.doi.org/ 10.1007/BF00425411 [DOI] [PubMed] [Google Scholar]
- [61].Laue H, Friedrich M, Ruff J, Cook AM. Dissimilatory sulfite reductase (desulfoviridin) of the taurine-degrading, non-sulfate-reducing bacterium Bilophila wadsworthia RZATAU contains a fused DsrB-DsrD subunit. J Bacteriol 2001; 183(5):1727-33; PMID:11160104; http://dx.doi.org/ 10.1128/JB.183.5.1727-1733.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Nava GM, Carbonero F, Croix JA, Greenberg E, Gaskins HR. Abundance and diversity of mucosa-associated hydrogenotrophic microbes in the healthy human colon. ISME J 2012; 6(1):57-70; PMID:21753800; http://dx.doi.org/ 10.1038/ismej.2011.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Wynder EL. The epidemiology of large bowel cancer. Cancer Res 1975; 35(11 Pt.2):3388-94; PMID:1192406 [PubMed] [Google Scholar]
- [64].Wynder EL, Kajitani T, Ishikawa S, Dodo H, Takano A. Environmental factors of cancer of the colon and rectum. II. Japanese epidemiological data. Cancer 1969; 23(5):1210-20; PMID:5778239; http://dx.doi.org/ 10.1002/1097-0142(196905)23:5%3c1210::AID-CNCR2820230530%3e3.0.CO;2-M [DOI] [PubMed] [Google Scholar]
- [65].Haenszel W, Berg JW, Segi M, Kurihara M, Locke FB. Large-bowel cancer in Hawaiian Japanese. J Natl Cancer Inst 1973; 51(6):1765-79; PMID:4797262 [DOI] [PubMed] [Google Scholar]
- [66].WYNDER EL, LEMON FR, BROSS IJ. Cancer and coronary artery disease among Seventh-Day Adventists. Cancer 1959; 12:1016-28; PMID:13846288; http://dx.doi.org/ 10.1002/1097-0142(195909/10)12:5%3c1016::AID-CNCR2820120523%3e3.0.CO;2-2 [DOI] [PubMed] [Google Scholar]
- [67].Wynder EL, Shigematsu T. Environmental factors of cancer of the colon and rectum. Cancer 1967; 20(9):1520-61; PMID:6038396; http://dx.doi.org/ 10.1002/1097-0142(196709)20:9%3c1520::AID-CNCR2820200920%3e3.0.CO;2-3 [DOI] [PubMed] [Google Scholar]
- [68].Reddy BS, Wynder EL. Large-bowel carcinogenesis: fecal constituents of populations with diverse incidence rates of colon cancer. J Natl Cancer Inst 1973; 50(6):1437-42; PMID:4717561 [DOI] [PubMed] [Google Scholar]
- [69].Reddy BS, Mastromarino A, Wynder EL. Further leads on metabolic epidemiology of large bowel cancer. Cancer Res 1975; 35(11 Pt. 2):3403-6; PMID:1104152 [PubMed] [Google Scholar]
- [70].Reddy BS, Wynder EL. Metabolic epidemiology of colon cancer. Fecal bile acids and neutral sterols in colon cancer patients and patients with adenomatous polyps. Cancer 1977; 39(6):2533-9; PMID:872053; http://dx.doi.org/ 10.1002/1097-0142(197706)39:6%3c2533::AID-CNCR2820390634%3e3.0.CO;2-X [DOI] [PubMed] [Google Scholar]
- [71].Wynder EL, Reddy BS. Diet and cancer of the colon. Curr Concepts Nutr 1977; 6:55-71; PMID:604018 [PubMed] [Google Scholar]
- [72].Reddy BS, Weisburger JH, Wynder EL. Fecal bacterial beta-glucuronidase: control by diet. Science 1974; 183(4123):416-7; PMID:4808971; http://dx.doi.org/ 10.1126/science.183.4123.416 [DOI] [PubMed] [Google Scholar]
- [73].O'Keefe SJ, Kidd M, Espitalier-Noel G, Owira P. Rarity of colon cancer in Africans is associated with low animal product consumption, not fiber. Am J Gastroenterol 1999; 94(5):1373-80; PMID:10235221; http://dx.doi.org/ 10.1111/j.1572-0241.1999.01089.x [DOI] [PubMed] [Google Scholar]
- [74].Sharma S, O'Keefe SJ. Environmental influences on the high mortality from colorectal cancer in African Americans. Postgrad Med J 2007; 83(983):583-89; PMID:17823224; http://dx.doi.org/ 10.1136/pgmj.2007.058958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Berg A. Nutrition, development and population growth. Popul Bull 1973; 29:3-37; PMID:12309299 [PubMed] [Google Scholar]
- [76].Le Marchand L, Kolonel LN. Cancer in Japanese migrants to Hawaii: interaction between genes and environment. Rev Epidemiol Sante Publique 1992; 40(6):425-30; PMID:1287741 [PubMed] [Google Scholar]
- [77].Burkitt DP. Epidemiology of cancer of the colon and the rectum. Cancer 1971; 28(1):3-13; PMID:5165022; http://dx.doi.org/ 10.1002/1097-0142(197107)28:1%3c3::AID-CNCR2820280104%3e3.0.CO;2-N [DOI] [PubMed] [Google Scholar]
- [78].Ou J, Carbonero F, Zoetendal EG, DeLany JP, Wang M, Newton K, Gaskins HR, O'Keefe SJ. Diet, microbiota, and microbial metabolites in colon cancer risk in rural Africans and African Americans. Am J Clin Nutr 2013; 98(1):111-20; PMID:23719549; http://dx.doi.org/ 10.3945/ajcn.112.056689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Narisawa T, Magadia NE, Weisburger JH, Wynder EL. Promoting effect of bile acids on colon carcinogenesis after intrarectal instillation of N-methyl-N'-nitro-N-nitrosoguanidine in rats. J Natl Cancer Inst 1974; 53(4):1093-7; PMID:4427390 [DOI] [PubMed] [Google Scholar]
- [80].Reddy BS, Narasawa T, Weisburger JH, Wynder EL. Promoting effect of sodium deoxycholate on colon adenocarcinomas in germfree rats. J Natl Cancer Inst 1976; 56(2):441-2; PMID:1255778 [DOI] [PubMed] [Google Scholar]
- [81].Reddy BS, Watanabe K, Weisburger JH, Wynder EL. Promoting effect of bile acids in colon carcinogenesis in germ-free and conventional F344 rats. Cancer Res 1977; 37(9):3238-42; PMID:884672 [PubMed] [Google Scholar]
- [82].Reddy BS, Weisburger JH, Wynder EL. Effects of dietary fat level and dimethylhydrazine on fecal acid and neutral sterol excretion and colon carcinogenesis in rats. J Natl Cancer Inst 1974; 52(2):507-11; PMID:4816006 [DOI] [PubMed] [Google Scholar]
- [83].van Faassen A, Ochsenkühn T, Houterman S, van der Ploeg EM, Bueno-de-Mesquita BH, Ocké MC, Bayerdörffer E, Janknegt RA. Plasma deoxycholic acid is related to deoxycholic acid in faecal water. Cancer Lett 1977; 114(1-2):293-294; http://dx.doi.org/ 10.1016/S0304-3835(97)04683-1 [DOI] [PubMed] [Google Scholar]
- [84].Bayerdörffer E, Mannes GA, Ochsenkühn T, Dirschedl P, Wiebecke B, Paumgartner G. Unconjugated secondary bile acids in the serum of patients with colorectal adenomas. Gut 1995; 36(2):268-73; PMID:7883228; http://dx.doi.org/ 10.1136/gut.36.2.268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Bayerdörffer E, Mannes GA, Richter WO, Ochsenkühn T, Wiebecke B, Köpcke W, Paumgartner G. Increased serum deoxycholic acid levels in men with colorectal adenomas. Gastroenterology 1993; 104(1):145-151; PMID:8419237 [DOI] [PubMed] [Google Scholar]
- [86].Ochsenkühn T, Bayerdörffer E, Meining A, Schinkel M, Thiede C, Nüssler V, Sackmann M, Hatz R, Neubauer A, Paumgartner G. Colonic mucosal proliferation is related to serum deoxycholic acid levels. Cancer 1999; 85(8):1664-1669; PMID:10223558; http://dx.doi.org/ 10.1002/(SICI)1097-0142(19990415)85:8%3c1664::AID-CNCR4%3e3.0.CO;2-O [DOI] [PubMed] [Google Scholar]
- [87].Cheng K, Raufman JP. Bile acid-induced proliferation of a human colon cancer cell line is mediated by transactivation of epidermal growth factor receptors. Biochem Pharmacol 2005; 70(7):1035-1047; PMID:16139803; http://dx.doi.org/ 10.1016/j.bcp.2005.07.023 [DOI] [PubMed] [Google Scholar]
- [88].Brown JR, DuBois RN. COX-2: A molecular target for colorectal cancer prevention. J Clin Oncol 2005; 23:2840-2855; PMID:15837998; http://dx.doi.org/ 10.1200/JCO.2005.09.051 [DOI] [PubMed] [Google Scholar]
- [89].Qiao L, Studer E, Leach K, McKinstry R, Gupta S, Decker R, Kukreja R, Valerie K, Nagarkatti P, El Deiry W, et al.. Deoxycholic acid (DCA) causes ligand-independent activation of epidermal growth factor receptor (EGFR) and FAS receptor in primary hepatocytes: inhibition of EGFR/mitogen-activated protein kinase-signaling module enhances DCA-induced apoptosis. Mol Biol Cell 2001; 12(9):2629-2645; PMID:11553704; http://dx.doi.org/ 10.1091/mbc.12.9.2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Pai R, Tarnawski AS, Tran T. Deoxycholic acid activates β-catenin signaling pathway and increases colon cell cancer growth and invasiveness. Mol Biol Cell 2004; 15:2156-2163; PMID:15004225; http://dx.doi.org/ 10.1091/mbc.E03-12-0894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Munemitsu S, Albert I, Souza B, Rubinfeld B, Polakis P. Regulation of intracellular betacatenin levels by the adenomatous polyposis coli (APC) tumor-suppressor protein. Proc Natl Acad Sci USA 1995; 92(7):3046-3050; PMID:7708772; http://dx.doi.org/ 10.1073/pnas.92.7.3046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Oshima M, Oshima H, Kitagawa K, Kobayashi M, Itakura C, Taketo M. Loss of Apc heterozygosity and abnormal tissue building in nascent intestinal polyps in mice carrying a truncated Apcgene. Proc Natl Acad Sci USA 1995; 92:4482-4486; PMID:7753829; http://dx.doi.org/ 10.1073/pnas.92.10.4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Lee HK, Jeong S. β-catenin stabilizes cyclooxygenase-2 mRNA by interacting with AU-rich elements of 3′-UTR. Nucl Acids Res 2006; 34(19):5705-5714; PMID:17040897; http://dx.doi.org/ 10.1093/nar/gkl698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Qiao D, Gaitonde SV, Qi W, Martinez JD. Deoxycholic acid suppresses p53 by stimulating proteosome-mediated p53 protein degradation. Carcinogenesis 2001; 22(6):957-964; PMID:11375905; http://dx.doi.org/ 10.1093/carcin/22.6.957 [DOI] [PubMed] [Google Scholar]
- [95].Christl SU, Scheppach W, Kasper H. Hydrogen metabolism in the large intestine-physiology and clinical implications. Z Gastroenterol 1995; 33:408-13; PMID:7571760 [PubMed] [Google Scholar]
- [96].Pitcher MCL, Beatty ER, Cummings JH. The contribution of sulphate reducing bacteria and 5-aminosalicyclic acid to fecal sulphide in patients with ulcerative colitis. Gut 2000; 46:64-72; PMID:10601057; http://dx.doi.org/ 10.1136/gut.46.1.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Gibson GR, Cummings JH, Macfarlane GT. Growth and activities of sulphate reducing bacteria in gut contents of healthy subjects and patients with ulcerative colitis. FEMS Microbiol Ecol 1991; 86:101-112; http://dx.doi.org/ 10.1111/j.1574-6968.1991.tb04799.x [DOI] [Google Scholar]
- [98].Roediger WE, Moore J, Babidge W. Colonic sulfide in pathogenesis and treatment of ulcerative colitis. Dig Dis Sci 1997; 42:1571-1579; PMID:9286219; http://dx.doi.org/ 10.1023/A:1018851723920 [DOI] [PubMed] [Google Scholar]
- [99].Moore J, Babidge W, Millard S, Roediger WE. Colonic luminal hydrogen sulfide is not elevated in ulcerative colitis. Dig Dis Sci 1998; 43:162-65; PMID:9508519; http://dx.doi.org/ 10.1023/A:1018848709769 [DOI] [PubMed] [Google Scholar]
- [100].Roediger WE. The colonic epithelium in ulcerative colitis: An energy deficiency disease? Lancet 1980; 2:712-715; PMID:6106826; http://dx.doi.org/ 10.1016/S0140-6736(80)91934-0 [DOI] [PubMed] [Google Scholar]
- [101].Roediger WE, Nance S. Metabolic induction of experimental colitis by inhibition of fatty acid oxidation. Br J Exp Pathol 1986; 67:773-82; PMID:3099821 [PMC free article] [PubMed] [Google Scholar]
- [102].Babidge W, Millard S, Roediger WE. Sulfides impair short chain fatty acid beta-oxidation at acyl-CoA dehydrogenase level in colonocytes: Implications for ulcerative colitis. Mol Cell Biochem 1998; 181:117-124; PMID:9562248; http://dx.doi.org/ 10.1023/A:1006838231432 [DOI] [PubMed] [Google Scholar]
- [103].Roedinger WE, Lawson MJ, Kwok V, Kerr Grant A, Pannall PR. Colonic bicarbonate output as a test of disease activity in ulcerative colitis. J Clin Pathol 1984; 37:704-7; PMID:6327778; http://dx.doi.org/ 10.1136/jcp.37.6.704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Den Hond E, Hiele E, Ghoos Y, Rutgeerts P. In vivo colonic butyrate metabolism in extensive ulcerative colitis. Gastroenterology 1998; 115(3):584-90; PMID:9721155; http://dx.doi.org/ 10.1016/S0016-5085(98)70137-4 [DOI] [PubMed] [Google Scholar]
- [105].Levitt MD, Springfield J, Furne J, Koenig T, Suarez F. Physiology of sulfide in the rat colon: use of bismuth to assess colonic sulfide production. J Appl Physiol 2002; 92:L1655-60; http://dx.doi.org/ 10.1152/japplphysiol.00907.2001 [DOI] [PubMed] [Google Scholar]
- [106].Christl SU, Eisner HD, Dusel G, Kasper H, Sheppach W. Antagonistic effects of sulfide and butyrate on proliferation of colonic mucosa: A potential role for these agents in the pathogenesis of ulcerative colitis. Dig Dis Sci 1996; 41:2477-81; PMID:9011461; http://dx.doi.org/ 10.1007/BF02100146 [DOI] [PubMed] [Google Scholar]
- [107].Harig JM, Soergel KH, Komorowski RA, Wood CM. Treatment of diversion colitis with short chain fatty acid irrigation. N Engl J Med 1989; 320:23-6; PMID:2909876; http://dx.doi.org/ 10.1056/NEJM198901053200105 [DOI] [PubMed] [Google Scholar]
- [108].Sheppach W, Sommer H, Kirchner T, Paganelli GM, Bartram P, Christl S, Richter F, Dusel G, Kasper H. Effect of butyrate enemas on the colonic mucosa in distal ulcerative colitis. Gastroenterology 1992; 103:51-56; PMID:1612357 [DOI] [PubMed] [Google Scholar]
- [109].Shaw L, Engel PC. CoA-persulphide: a possible in vivo inhibitor of mammalian short-chain acyl-CoA dehydrogenase. Biochim Biophys Acta 1987; 919(2):171-4; PMID:3580384; http://dx.doi.org/ 10.1016/0005-2760(87)90204-9 [DOI] [PubMed] [Google Scholar]
- [110].Wilson K, Mudra M, Furne J, Levitt M. Differentiation of the roles of sulfide oxidase and rhodanese in the detoxification of sulfide by the colonic mucosa. Dig Dis Sci 2008; 53:277-283; PMID:17551834; http://dx.doi.org/ 10.1007/s10620-007-9854-9 [DOI] [PubMed] [Google Scholar]
- [111].Theissen U, Hoffmeister M, Grieshaber M, Martin W. Single eubacterial origin of eukaryotic sulfide:quinone oxidoreductase, a mitochondrial enzyme conserved from the early evolution of eukaryotes during anoxic and sulfidic times. Mol Biol Evol 2003; 20:1564-74; PMID:12832624; http://dx.doi.org/ 10.1093/molbev/msg174 [DOI] [PubMed] [Google Scholar]
- [112].Gouben M, Andriamihaja M, Nübel T, Blachier F, Bouillaud F. Sulfide, the first inorganic substrate for human cells. FASEB J 2007; 21:1699-1706; PMID:17314140; http://dx.doi.org/ 10.1096/fj.06-7407com [DOI] [PubMed] [Google Scholar]
- [113].Ramasamy S, Singh S, Taniere P, Langman MJS, Eggo MC. Sulfide-detoxifying enzymes in the human colon are decreased in cancer and upregulated in differentiation. Am J Physiol Gastrointest Liver Physiol 2006; 291:G288-G296; PMID:16500920; http://dx.doi.org/ 10.1152/ajpgi.00324.2005 [DOI] [PubMed] [Google Scholar]
- [114].Toden S, Bird AR, Topping DL, Conlon MA. Resistant starch prevents colonic DNA damage induced by high dietary cooked red meat or casein in rats. Cancer Biol Ther 2006; 5:267-72; PMID:16410726; http://dx.doi.org/ 10.4161/cbt.5.3.2382 [DOI] [PubMed] [Google Scholar]
- [115].Toden S, Bird AR, Topping DL, Conlon MA. High red meat diets induce greater numbers of colonic DNA double-strand breaks than white meat in rats: attenuation by high-amylose maize starch. Carcinogenesis 2007; 28:2355-62; PMID:17916911; http://dx.doi.org/ 10.1093/carcin/bgm216 [DOI] [PubMed] [Google Scholar]
- [116].Kanazawa K, Konishi F, Mitsuoka T, Terada A, Itoh K, Narushima S, Kumemura M, Kimura H. Factors influencing the development of sigmoid colon cancer. Bacteriologic and biochemical studies. Cancer 1996; 77(8 Suppl):1701-6; PMID:8608565; http://dx.doi.org/ 10.1002/(SICI)1097-0142(19960415)77:8+%3c1701::AID-CNCR18%3e3.0.CO;2-1 [DOI] [PubMed] [Google Scholar]
- [117].Yamada M, Ohkusa T, Okayasu I. Occurrence of dysplasia and adenocarcinoma after experimental chronic ulcerative colitis in hamsters induced by dextran sulphate sodium. Gut 1992; 33(11):1521-27; PMID:1333439; http://dx.doi.org/ 10.1136/gut.33.11.1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, et al.. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res 2012; 22:299-306; PMID:22009989; http://dx.doi.org/ 10.1101/gr.126516.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, et al.. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013; 14:207-15; PMID:23954159; http://dx.doi.org/ 10.1016/j.chom.2013.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Deplancke B, Gaskins HR. Hydrogen sulfide induces serum-independent cell cycle entry in nontransformed rat intestinal epithelial cells. FASEB J 2003; 17:1310-12; PMID:12738807 [DOI] [PubMed] [Google Scholar]
- [121].Attene-Ramos MS, Nava GM, Muellner MG, Wagner ED, Plewa MJ, Gaskins HR. DNA damage and toxicogenomic analyses of hydrogen sulfide in human intestinal epithelial FHs 74 Int cells. Environ Mol Mutagen 2010; 51:304-14; PMID:20120018 [DOI] [PubMed] [Google Scholar]
- [122].Attene-Ramos MS, Wagner ED, Plewa MJ, Gaskins HR. Evidence that hydrogen sulfide is a genotoxic agent. Mol Cancer Res 2006; 4:9-14; PMID:16446402; http://dx.doi.org/ 10.1158/1541-7786.MCR-05-0126 [DOI] [PubMed] [Google Scholar]
- [123].Attene-Ramos MS, Wagner ED, Gaskins HR, Plewa MJ. Hydrogen sulfide induces direct radical-associated DNA damage. Mol Cancer Res 2007; 5(5):455-59; PMID:17475672; http://dx.doi.org/ 10.1158/1541-7786.MCR-06-0439 [DOI] [PubMed] [Google Scholar]
- [124].Van Eldere J, Celis P, De Pauw G, Lesaffre E, Eyssen H. Tauroconjugation of cholic acid stimulates 7alpha-dehydroxylation by fecal bacteria. Appl Environ Microbiol 1996; 62(2):656-61; PMID:8593067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Narushima S, Itoha K, Miyamoto Y, Park SH, Nagata K, Kuruma K, Uchida K. Deoxycholic acid formation in gnotobiotic mice associated with human intestinal bacteria. Lipids 2006; 41(9):835-43; PMID:17152920; http://dx.doi.org/ 10.1007/s11745-006-5038-1 [DOI] [PubMed] [Google Scholar]
- [126].Stenman LK, Holma R, Forsgård R, Gylling H, Korpela R. Higher fecal bile acid hydrophobicity is associated with exacerbation of dextran sodium sulfate colitis in mice. J Nutr 2013; 143(11):1691-97; PMID:24047703; http://dx.doi.org/ 10.3945/jn.113.180810 [DOI] [PubMed] [Google Scholar]
- [127].Craciun S, Balskus EP. Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc Natl Acad Sci USA 2012; 109(52):21307-12; PMID:23151509; http://dx.doi.org/ 10.1073/pnas.1215689109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM, et al.. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 2011; 472(7341):57-63; PMID:21475195; http://dx.doi.org/ 10.1038/nature09922 [DOI] [PMC free article] [PubMed] [Google Scholar]


