ABSTRACT
Microbial molecular hydrogen (H2) cycling is central to metabolic homeostasis and microbial composition in the human gastrointestinal tract. Molecular H2 is produced as an endproduct of carbohydrate fermentation and is reoxidised primarily by sulfate-reduction, acetogenesis, and methanogenesis. However, the enzymatic basis for these processes is incompletely understood and the hydrogenases responsible have not been investigated. In this work, we surveyed the genomic and metagenomic distribution of hydrogenase-encoding genes in the human colon to infer dominant mechanisms of H2 cycling. The data demonstrate that 70% of gastrointestinal microbial species listed in the Human Microbiome Project encode the genetic capacity to metabolise H2. A wide variety of anaerobically-adapted hydrogenases were present, with [FeFe]-hydrogenases predominant. We subsequently analyzed the hydrogenase gene content of stools from 20 healthy human subjects. The hydrogenase gene content of all samples was overwhelmingly dominated by fermentative and electron-bifurcating [FeFe]-hydrogenases emerging from the Bacteroidetes and Firmicutes. This study supports that H2 metabolism in the human gut is driven by fermentative H2 production and interspecies H2 transfer. However, it suggests that electron-bifurcation rather than respiration is the dominant mechanism of H2 reoxidation in the human colon, generating reduced ferredoxin to sustain carbon-fixation (e.g. acetogenesis) and respiration (via the Rnf complex). This work provides the first comprehensive bioinformatic insight into the mechanisms of H2 metabolism in the human colon.
KEYWORDS: electron-bifurcation, hydrogen, hydrogenase, hydrogenogen, hydrogenotroph, interspecies H2 transfer
Introduction
Harboring more than 1011 organisms per gram of content, the human colonic microbiota is a diverse ecological landscape, with impacts to health and disease that are just beginning to be elucidated. The advancement of molecular based techniques has allowed researchers to explore the residents of the colonic microenvironment, revealing specific microbes and microbial patterns associated with human health and disease.1-3 Reproduction of disease states in gnotobiotic mice upon fecal transplant implicates resident microbiota as significant contributors to disease etiology. As in the case of stomach cancer and Helicobacter pylori, some disease states are the result of specific microbial infection. However, the majority of disease appears to be determined by the compilation of host genetic and environmental impacts to microbial community structure such as diet, medication, and mode of delivery at birth. In short, it is not pathology emanating from a specific organism, but rather the accumulation of microbial metabolites from the resident community that contributes to the etiology of most colonic-related disorders. One especially abundant metabolite in the gastrointestinal tract is molecular hydrogen (H2), a diffusible gas produced and consumed by resident anaerobic microorganisms.4 The dynamics of hydrogen cycling are thought to be central to colonic metabolic homeostasis and shaping of the microbial community. As we have recently reviewed,4-6 there is growing evidence that colonic H2 metabolism in turn influences inflammatory bowel disease, colorectal cancer, gastrointestinal infections, obesity and associated metabolic disorders.
Most of our understanding of the ecology of anaerobic H2 cycling is based on studies in aquatic sediments.7 In such environments, H2 is produced predominantly by fermentative bacteria (hydrogenogens) and is thought to be reoxidised primarily by anaerobic respiratory microorganisms (hydrogenotrophs). H2 consumption is thermodynamically essential in any environment to maintain fermentative processes, and is accomplished in the sediment through interspecies hydrogen transfer or competitive hydrogenotrophy.7 Interspecies hydrogen transfer is the syntrophic evolution and consumption of hydrogen between organisms in such close proximity that hydrogen never joins the dissolved hydrogen pool.8,9 In contrast, competitive hydrogenotrophy pairs the oxidation of organic substrates by hydrogenogens with the reduction of terminal electron acceptors by hydrogenotrophs, which results in a well described ecological framework determined by H2 concentration and substrate availability. In a substrate-rich environment, competitive advantage is gained by hydrogenotrophs able to reduce substrates at low hydrogen concentrations, as the concentration is maintained too low to make growth thermodynamically favorable for other hydrogenotrophic organisms. A classic example are studies describing sulfate-reducing bacteria outcompeting methanogens.10 In a hydrogen-competitive environment, hydrogenotrophs are able to coexist through use of alternate substrates and interspecies hydrogen transfer.9
Far less is understood about the H2 economy of the human gastrointestinal tract. H2 is formed in large volumes in the colon as an end product of carbohydrate fermentation, for example by Bacteroidetes.11 There are at least 3 major pathways for disposal of the H2 produced, namely methanogenesis, sulfate reduction, and acetogenesis.12 The dominance of these pathways appears to vary among subjects, possibly as a result of competitive hydrogenotrophy.13-15 However, there are also many reports of methanogens and sulfate-reducers co-existing in the colon.12,16,17 We have hypothesized that this is due to the large spatial and temporal variations in the chemical composition of the human colon, which would enable the formation of specific microhabitats dominated by different types of hydrogenotrophs.4 Interspecies hydrogen transfer has been proposed to occur in the hydrogen-rich human colon, though microenvironmental niches in this mucosal ecosystem have not been studied and both the cellular and genetic bases for H2 cycling are underexplored.11,18
To gain a better understanding of H2 metabolism in the colon, we identified the classes of hydrogenases in genomic and metagenomic databases. Hydrogenases are metalloenzymes that reversibly oxidize dihydrogen (H2) via the reaction H2 → 2 H+ + 2 e−. These ubiquitous enzymes are found in Archaea, Bacteria, and some Eukarya, and include both anaerobically- and aerobically-adapted variants.7,19 In anoxic environments such as the human colon, hydrogenases can potentially conserve energy through 3 major processes: 1) hydrogenotrophic respiration by coupling oxidation of the high-energy fuel H2 to the reduction of exogenous oxidants such as sulfate;20,21 2) hydrogenogenic fermentation by coupling oxidation of ferredoxin or formate to the production of the dissipatable H2;22,23 or 3) hydrogenogenic respiration by coupling ferredoxin oxidation to proton reduction in cation-translocating complexes.24,25 More recently, a fourth mechanism of H2-dependent energy conservation has been described in acetogens and methanogens: flavin-based electron bifurcation.26 In this process, multimeric complexes mediate the simultaneous transfers of H2-derived electrons to high-potential (NAD+, NADP+, or heterodisulfide) and low-potential (ferredoxin) compounds, with the exergonic electron transfer driving the endergonic one.27,28 The reductant generated can be used to support biosynthetic pathways, carbon-fixation, ferredoxin respiration, and further fermentation processes.26 There is also growing evidence that H2 sensing has a role in anaerobic microorganisms.19,29
This assortment of functions is supported by the great phylogenetic diversity of the hydrogenases.19 Hydrogenases can be subdivided into 3 distinct classes based on their metal site, the [NiFe],20 [FeFe],23 and the [Fe]30 hydrogenases. Whereas the [Fe] hydrogenases form a small homogenous group,19 the [NiFe] and [FeFe] hydrogenases are highly diverse in their redox centers, associated subunits, and wider cellular integration.19,31 We recently developed a comprehensive classification scheme that correlated the primary phylogeny of these enzymes with their functions. This showed there were at least 4 groups and 22 subgroups of [NiFe]-hydrogenases, and 3 groups and 6 subtypes of [FeFe]-hydrogenases, each with distinct functions.19 This enabled the identification of the major classes of enzymes responsible for hydrogenotrophic respiration (NiFe Groups 1a, 1b, 1c), hydrogenogenic respiration (NiFe Groups 4b, 4c, 4d, 4e), hydrogenogenic fermentation (FeFe Groups A1, B; NiFe Group 4a), electron-bifurcation (FeFe Group A3, A4; NiFe Group 3c), and sensing (FeFe Group C) in anoxic environments.19 In turn, these enzymes are likely to be dominant in the human colon. The aim of this work is to use these new sequence-structure-function relationships to explore the dynamics and ecology of H2 metabolism in the human colon.
Materials and methods
This work analyzed the hydrogenase content of the public genomes and metagenomes represented in the Human Microbiome Project.3,32 We analyzed the capacity of the 343 microbial species to metabolize H2 by retrieving their hydrogenase protein sequences using BLAST searches against reference [NiFe], [FeFe], and [Fe] hydrogenases. The amino acid sequences encoding the hydrogenase catalytic subunits (for NiFe hydrogenases) or domains (for [FeFe]-hydrogenases) were aligned with ClustalW.33 The relationships between these sequences were visualized in neighbor-joining phylogenetic trees bootstrapped with 500 replicates using MEGA6.34 For metagenome data-mining, we downloaded 20 metagenomes selected at random from the Human stool microbial communities from National Health Institute, USA' project. The metagenomes were screened at a depth of 5 million reads for the presence of hydrogenase genes using a translated BLAST search referencing the 3248 sequences in our hydrogenase database.19 To remove false positives, hits within the initial screen were further sieved by removing any result with a minimum identity less than 60% and minimum query coverage less than 40 amino acids. The closest-matched BLAST hits were recorded.
Results
Two thirds of sequenced human gut microbes harbour hydrogenases
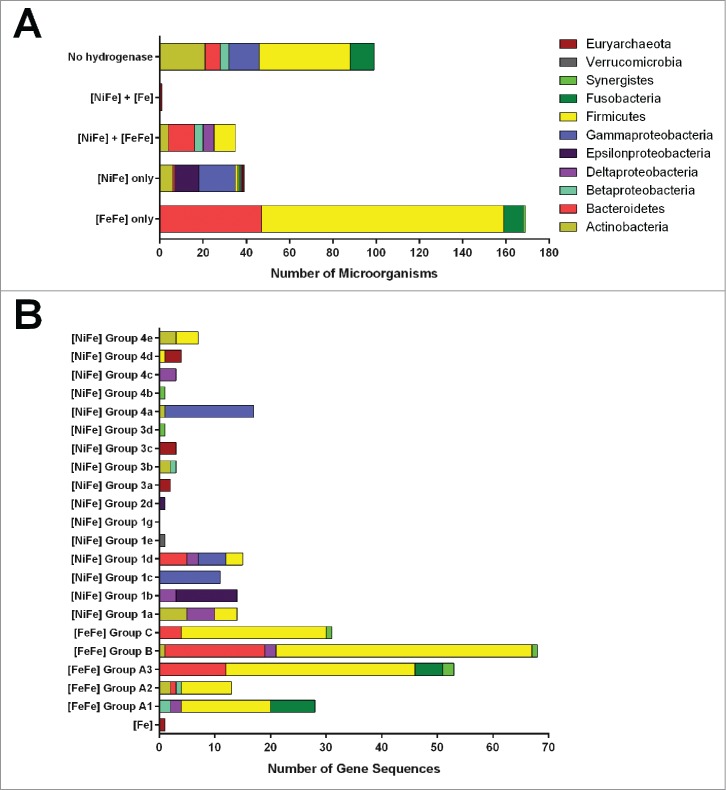
We analyzed the capacity of the 343 microbial species listed in the Human Microbiome Project Gastrointestinal Tract (HMP GI) reference genome database to metabolize H2. Some 71% of these microorganisms encoded hydrogenases. In total, 60% of organisms encoded [FeFe]-hydrogenases, 21% encoded [NiFe]-hydrogenases, and one organism (the methanogen Methanobrevibacter smithii) encoded an [Fe]-hydrogenase (Fig. 1A; Table S1). The hydrogenase sequences were distributed unevenly at the taxonomic level. All sequenced human gut representatives of Clostridiales and Bacteroidaceae harbor [FeFe]-hydrogenase genes, as do some Proteobacteria, Fusobacteria, Actinobacteria, and Synergistetes (Fig. 1A). [NiFe]-hydrogenase genes were present in all microbial phyla relevant to the human gut, with the exception Fusobacteria, but were unevenly distributed within many of these groups (Fig. 1A). Hydrogenases were entirely absent from both the Bacilli and Bifidobacteria (Table S1).
Figure 1.
Genomic distribution of hydrogenases in human colonic microorganisms. (A) The proportion of microbial species that encode hydrogenases. This was determined using the 343 microbial species represented in the Human Microbiome Project Gastrointestinal Tract (HMP GI) genome database. (B) Distribution of human colonic hydrogenase sequences by microbial phyla and hydrogenase class. Hydrogenase sequences were obtained from human colonic microorganisms represented in the NCBI Reference Sequence database and were classified according to previously-outlined criteria.19
Human gut microbiota encode a variety of anaerobe-type fermentative, respiratory, and bifurcating hydrogenases
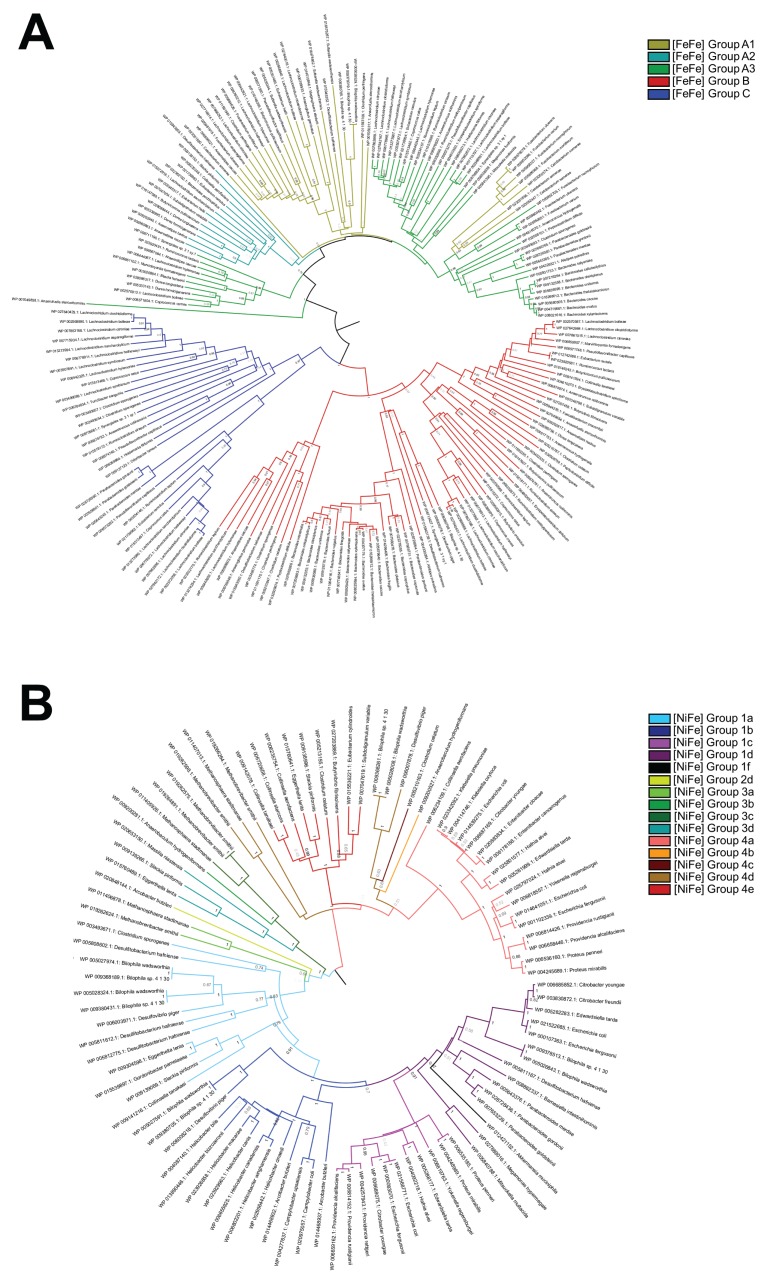
We subsequently investigated the molecular phylogeny of the hydrogenases present in these organisms using a previously-curated hydrogenase database.19 The taxonomic distribution of these enzymes is visualised in Figure 1B and their phylogenetic diversity is represented in the phylogenetic trees of Figure 2. The most abundant hydrogenases encoded in sequenced human gut genomes were [FeFe]-hydrogenases that mediate fermentative H2 production (Groups A1, B), flavin-based electron-bifurcation (Group A3), and possibly H2 sensing (Group C) (Fig. 1B; Table S2). These enzymes were especially widespread in the dominant gut phyla Firmicutes and Bacteroidetes. A diverse range of [NiFe]-hydrogenase genes were observed. The most abundant are subgroups 1a, 1b, 1c, and 1d, which couple H2 oxidation to physiologically-relevant electron acceptors such as sulfate and fumarate, primarily in enterobacteria. Also detected were enterobacteria-type formate hydrogenlyases (NiFe Group 4a), which couple formate oxidation to fermentative H2 production, and methanogenic Eha, Ehb, and Ech hydrogenases (NiFe Group 4d and 4e), which form minimalistic proton/sodium-translocating respiratory chains that reversibly couple ferredoxin oxidation to proton reduction. The other subgroups were present in one to 3 sequenced species each and hence are likely to only have a marginal impact on colonic H2 cycling.
Figure 2.
Phylogenetic trees showing the diversity of the human colonic hydrogenases. The trees represent the protein sequences of the [FeFe]-hydrogenase catalytic domains (A) and [NiFe]-hydrogenase catalytic subunits (B) derived from the genomes of human colonic microorganisms. The trees were constructed by the neighbor-joining method, are bootstrapped with 500 replicates, and are color -coded by hydrogenase class.
H2 cycling in the human gut is dominated by [FeFe]-hydrogenases from Bacteroidetes and Firmicutes
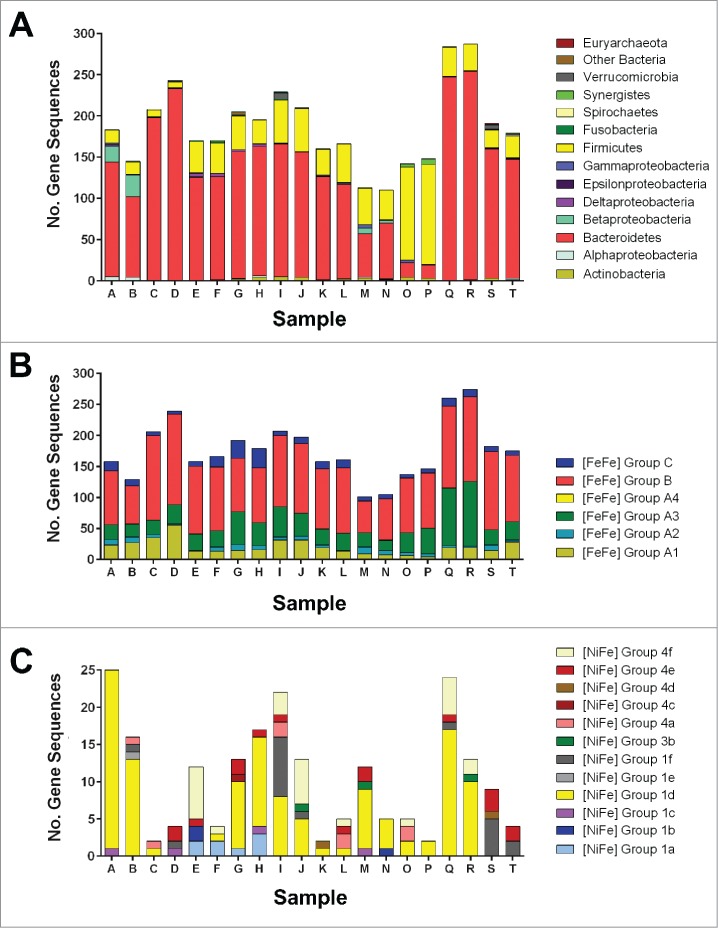
We subsequently surveyed the abundance and affiliations of hydrogenase genes using the metagenomes of stool samples from 20 healthy human subjects. There were major differences in the normalized abundance, classification, and affiliations of the hydrogenase genes among the subjects (Fig. 3; Table S3). However, the overall distribution of hydrogenases was similar, with the most abundant hydrogenase genes encoding fermentative (55% Group B, 11% Group A1), bifurcating (20% Group A3), and sensory (6% Group C) enzymes among the samples. [NiFe] enzymes accounted for just 6% of the sequences detected, with the 1d subgroup of O2-tolerant respiratory uptake enzymes proving the most abundant. Other sequences such as the methanogen-type 4d and Verrucomicrobia-type 1f categories were only found in several samples each, which is consistent with the occurrence of these organisms in only a subset of the human population.3 The vast majority of the genes were affiliated with the Bacteroidetes (73%) and Firmicutes (21%), which is consistent with their genomic hydrogenase content (Figs. 1 and 2) and known dominance in stool samples.3 While Bacteroidetes hydrogenases were generally more abundant than those in Firmicutes, this relationship was reversed in samples O and P (Fig. 3).
Figure 3.
Distribution of hydrogenase-encoding genes in human colonic metagenomes. The hydrogenase content of 20 metagenomes derived from the HMP GI metagenome database was determined at a depth of 5 million reads. (A) Probable phylum-level affiliation of hydrogenases detected. This was determined by recording the closest BLAST hit for each read against a databank of 3284 hydrogenase sequences.19 (B) Number of reads corresponding to each [FeFe]-hydrogenase class. (C) Number of reads corresponding to each [NiFe]-hydrogenase class.
Discussion
Publicly available genome and metagenome resources were used in this work to comprehensively analyze the distribution of hydrogenases in the human colon. The data indicate that H2 metabolism is more diverse and widespread on both the taxonomic and community levels than previously appreciated. Greater than 70 percent of microbial genomes in the HMP GI database harbor the capacity to synthesize hydrogenases, showing H2 cycling is a dominant mode of energy-conservation in this anaerobic ecosystem (Fig. 1). These organisms encoded a wide range of anaerobe-type fermentative, electron-bifurcating, and, to a lesser extent, respiratory [FeFe] and [NiFe] hydrogenases (Fig. 2). The capacity for H2 sensing was also inferred. The metagenome survey detected a similar diversity of hydrogenase-encoding gene sequences. However, just 3 hydrogenase classes (FeFe Groups B, A3, A1) and 2 microbial phyla (Bacteroidetes, Firmicutes) accounted for > 85% of the genes detected (Fig. 3). This reflects the well-described microbial community structure of the human colon, which is overwhelmingly dominated by Bacteroidetes and Firmicutes.3
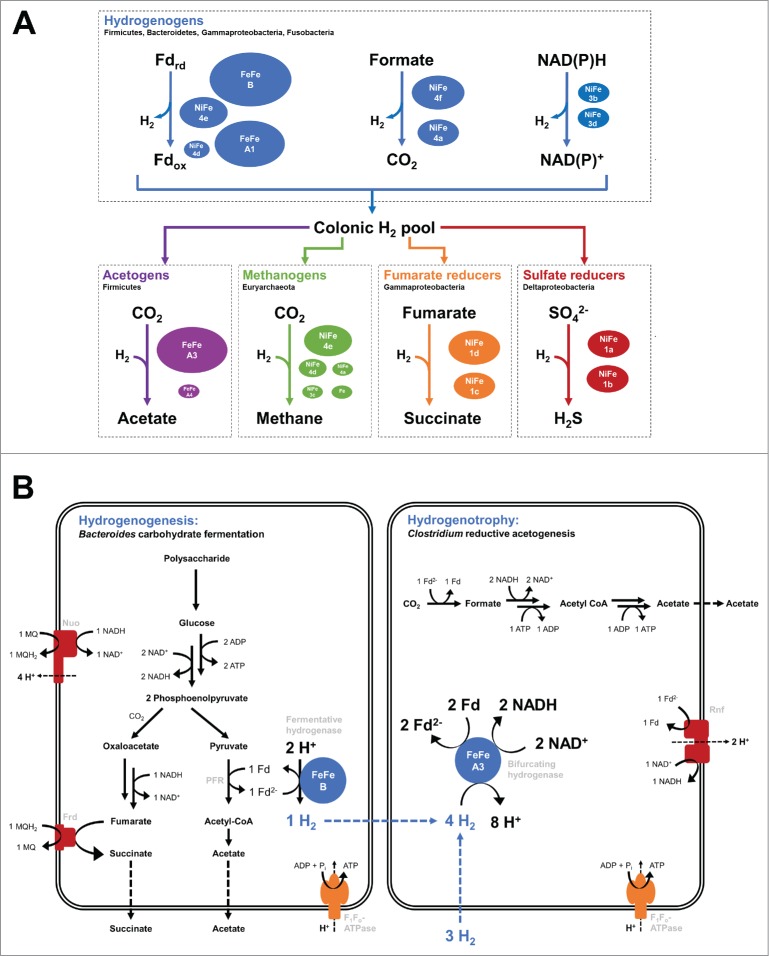
Together, the survey infers the predominant routes of H2 evolution and reoxidation in the human colon (Fig. 4). It indicates that the predominant mechanism of H2 evolution in this ecosystem is through fermentative processes mediated by Bacteroidetes and Clostridial members of the Firmicutes. Of the major pathways of hydrogenogenesis,4,19 ferredoxin-coupled H2 fermentation appears to the predominant; [FeFe]-hydrogenases responsible for this process (Group A1, likely B) were more abundant than [NiFe]-hydrogenases responsible for formate-coupled fermentation (Group 4a, likely 4f), NAD(P)H-coupled fermentation (Group 3b and 3d), and hydrogenogenic respiration (Groups 4b, 4c, 4e).
Figure 4.
Summary of human colon H2 metabolism based on the described genome and metagenome surveys. (A) Summary of the predominant known routes of H2 evolution and reoxidation in the human colon. The microbial phyla and hydrogenase classes mediating these processes are shown. The hydrogenases are sized according to the relative abundance of the genes encoding them in the 20 metagenomes surveyed. The most dominant hydrogenogenic hydrogenases were the Group A1 and Group B [FeFe]-hydrogenases that mediate ferredoxin-dependent H2 evolution. NADPH- or formate-dependent H2 evolution appears to be quantitatively less important. The electron-bifurcating Group A3 [FeFe]-hydrogenases were by far the most abundant hydrogenotrophic hydrogenases identified in our genome and metagenome surveys. These enzymes are linked to acetogenesis, though our metagenomes surveys suggest that many hydrogenotrophs are also capable of oxidising H2 without producing detectable endproducts, i.e. through H2-mediated ferredoxin reduction followed by subsequent ferredoxin respiration. The determinants of hydrogenotrophic methanogenesis, sulfate reduction, and fumarate reduction were identified, but appear to be comparatively rare.19 (B) Simplified pathways showing interspecies hydrogen transfer between the 2 most dominant H2-metabolising phyla in the human colon. The H2 evolved by a carbohydrate-fermenting Bacteroides species by the [FeFe] Group B hydrogenase. The H2 is transferred to a hydrogenotroph of the genus Clostridium and is bifurcated at [FeFe] Group A3 to reduce ferredoxin and NADH. The derived reductant sustains respiration through the Rnf complex and CO2 fixation through reductive acetogenesis. Our survey suggests alternative pathways may also occur in the human colon resulting in H2 oxidation in Firmicutes, H2 production in Bacteroidetes, and internal recycling of H2. It is probable that the [FeFe] Group A3 hydrogenase can generate reductant in Firmicutes and Bacteroidetes independently of acetogenesis. Key: Nuo = NADH dehydrogenase, Frd = fumarate reductase, Rnf = ferredoxin:NAD+ oxidoreductase. Modeled based on references.11,27
Of the mechanisms of hydrogenotrophy, the survey strongly implies that the majority of H2 is reoxidised through electron-bifurcation coupled to ferredoxin respiration. This depends on the highly abundant Group A3 [FeFe]-hydrogenases, which account for 20% of the total hydrogenase genes in the human colon. These trimeric enzymes reversibly bifurcate electrons from H2 to ferredoxin and NAD.26,35 The reductant generated can be used to sustain anabolic processes, carbon-fixation (e.g., via reductive acetogenesis), or further fermentation (via [FeFe]-hydrogenases). However, based on recent models, it is likely that a large proportion is reoxidised through the respiratory Rnf complex, which generates sodium/proton-motive force by coupling Fdred oxidation to NAD+ reduction.26,36 The only quantitatively abundant [NiFe] uptake hydrogenases detected were the oxygen-tolerant respiratory uptake hydrogenases (Group 1d [NiFe]-hydrogenases);37 such enzymes have been linked to reoxidation of fermentatively-produced H2 and might also contribute the metabolic flexibility needed for facultative anaerobes such as E. coli to transition between host-associated and free-living states.38 Between them, such processes would enable the majority of H2 produced in the human colon to be reoxidised without formation of detectable endproducts. We predict that the majority of H2 produced by fermentative processes is immediately recycled through a combination of internal reoxidation and interspecies H2 transfer without ever entering the H2 pool.
Our proposal that electron-bifurcation serves as the primary mechanism of colonic hydrogenotrophy deviates from classical literature emphasizing the roles of pathway necessitating end product secretion. It is probable that the best-characterized pathways of colonic H2 reoxidation, namely methanogenesis, sulfate-reduction, and potentially acetogenesis,4,12,14,39 serve as only fractional sinks in the colonic H2 budget. These processes have a major influence on the chemical composition of the human colon and its flatus,4,12 enhance the diversity of gut microbiota,3 and, particularly in the case of sulfate-reducers, have been extensively linked to human health and disease.40 However, they appear to be quantitatively less significant than electron-bifurcation. This likely reflects that previous studies on colonic hydrogenotrophy have focused on endproduct formation rather than molecular mechanisms of H2 oxidation. HMP studies on the community structure of the human colon indicate the abundance of sulfate-reducers and methanogens is too low and variable for these organisms to be the dominant hydrogenotrophs.3,32 Furthermore, our survey showed the key H2-oxidising enzymes responsible for methanogenesis (NiFe Groups 3a, 3c, 4d, 4e, [Fe]-hydrogenases) and sulfate-reduction (NiFe Groups 1a, 1b) were either absent or in low abundance in the 100 million sequence reads analyzed. Consistently, the levels of methane and hydrogen sulfide produced in human flatus is many orders of magnitude lower than the H2 available.5 It remains to be debated whether the energetically-constrained process of reductive acetogenesis is an important hydrogenotrophic pathway; the clostridial electron-bifurcating hydrogenases that mediate this process are abundant in the human gut, but have flexibility to support other processes such as fermentation and ferredoxin respiration.
A further surprising finding of our metagenome survey is that it emphasizes a dominant role for Bacteroidetes in H2 cycling. Genes encoding Bacteroidetes hydrogenases were more abundant than those encoding clostridial hydrogenases in 18 of the 20 samples (Fig. 3). It is established that colonic Bacteroidetes produce H2 as an end product of cellulolytic fermentation processes41,42 and that this process supports symbiotic interactions with clostridia.11,42 Given colonic Bacteroidetes genomes lack classical Group A1 [FeFe]-hydrogenases, such processes are probably supported by Group B [FeFe]-hydrogenase, a large uncharacterized class of enzymes predicted to couple Fdred oxidation to H2 evolution.19,43 The genome and metagenome surveys also detected Group A3 [FeFe]-hydrogenases from members of the genera Bacteroides, Parabacteroides, and Alistipes (Figs. 1 and 3). While the roles for these enzymes in processes such as clostridial reductive acetogenesis is well-resolved,26,44 their role in Bacteroidetes is less clear. It is probable that the majority of the reductant formed through this process respires through the Rnf complex, an example of which was recently characterized in Bacteroides fragilis.45 It is conceivable that Bacteroidetes also oxidizes internally- and externally-produced H2 to maintain redox homeostasis and support reductive processes such as CO2 fixation.46
Looking forward, there are now multiple ways to expand on these studies to develop a deeper understanding of the mechanisms of hydrogenogenesis and hydrogenotrophy in the human gut. Firstly, this study only analyses the genetic determinants of H2 metabolism; it is imperative to test the hypothesis that electron-bifurcation is the dominant mechanism of hydrogenotrophy via expression and activity studies. Secondly, fecal samples do not fully reflect the heterogeneity of the human colon over space and time.12 Biopsy studies are likely to reveal more about the diversity and distribution of H2 cycling, including why methanogens and sulfate-reducers are present and active in the human colon despite being outnumbered.12 As reflected by our findings on the demography of colonic methanogenesis,47 there is also need for clinical studies to explore how hydrogenase abundance and activity varies with factors such as subject weight, gender, race, and disease states. Our findings that over 70% of sequenced human colonic microorganisms harbor hydrogenases indicates H2 cycling may be a far more important process in human health and disease than previously recognized.4,5 Finally, there is also need for biochemical and physiological studies to resolve the physiological roles of the unexplored Group A2, B, and C [FeFe]-hydrogenases, as well as the wider roles of H2 cycling in Bacteroidetes. A deeper understanding of phylogeny as it pertains to function is necessary to fully understand the relationship among hydrogenogenic and hydrogenotrophic microbes in the human colon.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Roderick Mackie and Jason Ridlon for their review of the manuscript.
Funding
CG was supported by a CSIRO Office of the Chief Executive Postdoctoral Fellowship. PGW was supported by a predoctoral fellowship from Mayo Clinic and University of Illinois Alliance for Technology-Based Healthcare.
References
- [1].Tremaroli V, Backhed F. Functional interactions between the gut microbiota and host metabolism. Nature 2012; 489:242-9; PMID:22972297; http://dx.doi.org/ 10.1038/nature11552 [DOI] [PubMed] [Google Scholar]
- [2].Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell 2012; 148:1258-70; PMID:22424233; http://dx.doi.org/ 10.1016/j.cell.2012.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].The Human Microbiome Project Consortium . Structure, function and diversity of the healthy human microbiome. Nature 2012; 486:207-14; PMID:22699609; http://dx.doi.org/ 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Nakamura N, Lin HC, McSweeney CS, Mackie RI, Gaskins HR. Mechanisms of microbial hydrogen disposal in the human colon and implications for health and disease. Annu Rev Food Sci Technol 2010; 1:363-95; PMID:22129341; http://dx.doi.org/ 10.1146/annurev.food.102308.124101 [DOI] [PubMed] [Google Scholar]
- [5].Carbonero F, Benefiel AC, Gaskins HR. Contributions of the microbial hydrogen economy to colonic homeostasis. Nat Rev Gastroenterol Hepatol 2012; 9:504-18; PMID:22585131; http://dx.doi.org/ 10.1038/nrgastro.2012.85 [DOI] [PubMed] [Google Scholar]
- [6].Cook GM, Greening C, Hards K, Berney M. Energetics of pathogenic bacteria and opportunities for drug development In: Poole RK. editor. Advances in Bacterial Pathogen Biology. Academic Press; 2014. page 1-62 [DOI] [PubMed] [Google Scholar]
- [7].Schwartz E, Fritsch J, Friedrich B. H2-metabolizing prokaryotes. Berlin, Heidelberg: Springer Berlin Heidelberg; 2013. [Google Scholar]
- [8].Morris BEL, Henneberger R, Huber H, Moissl-Eichinger C. Microbial syntrophy: interaction for the common good. FEMS Microbiol Rev 2013; 37:384-406; PMID:23480449; http://dx.doi.org/ 10.1111/1574-6976.12019 [DOI] [PubMed] [Google Scholar]
- [9].Bryant MP, Wolin EA, Wolin MJ, Wolfe RS. Methanobacillus omelianskii, a symbiotic association of two species of bacteria. Arch Mikrobiol 1967; 59:20-31; PMID:5602458; http://dx.doi.org/ 10.1007/BF00406313 [DOI] [PubMed] [Google Scholar]
- [10].Lovley DR, Dwyer DF, Klug MJ. Kinetic analysis of competition between sulfate reducers and methanogens for hydrogen in sediments. Appl Environ Microbiol 1982; 43:1373-9; PMID:16346033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Fischbach MA, Sonnenburg JL. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe 2011; 10:336-47; PMID:22018234; http://dx.doi.org/ 10.1016/j.chom.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Nava GM, Carbonero F, Croix JA, Greenberg E, Gaskins HR. Abundance and diversity of mucosa-associated hydrogenotrophic microbes in the healthy human colon. ISME J 2012; 6:57-70; PMID:21753800; http://dx.doi.org/ 10.1038/ismej.2011.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gibson GR, Cummings JH, Macfarlane GT, Allison C, Segal I, Vorster HH, Walker AR. Alternative pathways for hydrogen disposal during fermentation in the human colon. Gut 1990; 31:679-83; PMID:2379871; http://dx.doi.org/ 10.1136/gut.31.6.679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Strocchi A, Furne JK, Ellis CJ, Levitt MD. Competition for hydrogen by human faecal bacteria: evidence for the predominance of methane producing bacteria. Gut 1991; 32:1498-501; PMID:1773956; http://dx.doi.org/ 10.1136/gut.32.12.1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Strocchi A, Levitt MD. Factors affecting hydrogen production and consumption by human fecal flora. The critical roles of hydrogen tension and methanogenesis. J Clin Invest 1992; 89:1304-11; PMID:1556190; http://dx.doi.org/ 10.1172/JCI115716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Doré J, Pochart P, Bernalier A, Goderel I, Morvan B, Rambaud JC. Enumeration of H2-utilizing methanogenic archaea, acetogenic and sulfate-reducing bacteria from human feces. FEMS Microbiol Ecol 1995; 17:279-84; http://dx.doi.org/ 10.1111/j.1574-6941.1995.tb00152.x [DOI] [Google Scholar]
- [17].Pochart P, Dore J, Lemann F, Goderel I, Rambaud JC. Interrelations between populations of methanogenic archaea and sulfate-reducing bacteria in the human colon. FEMS Microbiol Lett 1992; 77:225-8; PMID:1459413; http://dx.doi.org/ 10.1111/j.1574-6968.1992.tb05518.x [DOI] [PubMed] [Google Scholar]
- [18].Robert C, Del'Homme C, Bernalier-Donadille A. Interspecies H2 transfer in cellulose degradation between fibrolytic bacteria and H2-utilizing microorganisms from the human colon. FEMS Microbiol Lett 2001; 205:209-14; PMID:11750804; http://dx.doi.org/ 10.1111/j.1574-6968.2001.tb10949.x [DOI] [PubMed] [Google Scholar]
- [19].Greening C, Biswas A, Carere CR, Jackson CJ, Taylor MC, Stott MB, Cook GM, Morales SE. Genome and metagenome surveys of hydrogenase diversity indicate H2 is a widely-utilised energy source for microbial growth and survival. ISME J 2016; 10:761-77; PMID:26405831; http://dx.doi.org/ 10.1038/ismej.2015.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Volbeda A, Charon MH, Piras C, Hatchikian EC, Frey M, Fontecilla-Camps JC. Crystal structure of the nickel-iron hydrogenase from Desulfovibrio gigas. Nature 1995; 373:580-7; PMID:7854413; http://dx.doi.org/ 10.1038/373580a0 [DOI] [PubMed] [Google Scholar]
- [21].Caffrey SM, Park H-S, Voordouw JK, He Z, Zhou J, Voordouw G. Function of periplasmic hydrogenases in the sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. J Bacteriol 2007; 189:6159-67; PMID:17601789; http://dx.doi.org/ 10.1128/JB.00747-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].McDowall JS, Murphy BJ, Haumann M, Palmer T, Armstrong FA, Sargent F. Bacterial formate hydrogenlyase complex. Proc Natl Acad Sci U S A 2014; 111:E3948-56; PMID:25157147; http://dx.doi.org/ 10.1073/pnas.1407927111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Peters JW, Lanzilotta WN, Lemon BJ, Seefeldt LC. X-ray crystal structure of the Fe-only hydrogenase (CpI) from Clostridium pasteurianum to 1.8 Angstrom resolution. Science 1998; 282:1853-8; PMID:9836629; http://dx.doi.org/ 10.1126/science.282.5395.1853 [DOI] [PubMed] [Google Scholar]
- [24].Lie TJ, Costa KC, Lupa B, Korpole S, Whitman WB, Leigh JA. Essential anaplerotic role for the energy-converting hydrogenase Eha in hydrogenotrophic methanogenesis. Proc Natl Acad Sci U S A 2012; 109:15473-8; PMID:22872868; http://dx.doi.org/ 10.1073/pnas.1208779109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Meuer J, Kuettner H. Genetic analysis of the archaeon Methanosarcina barkeri Fusaro reveals a central role for Ech hydrogenase and ferredoxin in methanogenesis and carbon fixation. Proc Natl Acad Sci U S A 2002; 99:5632-7; PMID:11929975; http://dx.doi.org/ 10.1073/pnas.072615499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Buckel W, Thauer RK. Energy conservation via electron bifurcating ferredoxin reduction and proton/Na+ translocating ferredoxin oxidation. Biochim Biophys Acta 2013; 1827:94-113; PMID:22800682; http://dx.doi.org/ 10.1016/j.bbabio.2012.07.002 [DOI] [PubMed] [Google Scholar]
- [27].Schuchmann K, Müller V. A bacterial electron-bifurcating hydrogenase. J Biol Chem 2012; 287:31165-71; PMID:22810230; http://dx.doi.org/ 10.1074/jbc.M112.395038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kaster A-K, Moll J, Parey K, Thauer RK. Coupling of ferredoxin and heterodisulfide reduction via electron bifurcation in hydrogenotrophic methanogenic archaea. Proc Natl Acad Sci U S A 2011; 108:2981-6; PMID:21262829; http://dx.doi.org/ 10.1073/pnas.1016761108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zheng Y, Kahnt J, Kwon IH, Mackie RI, Thauer RK. Hydrogen formation and its regulation in Ruminococcus albus: involvement of an electron-bifurcating [FeFe]-hydrogenase, of a non-electron-bifurcating [FeFe]-hydrogenase, and of a putative hydrogen-sensing [FeFe]-hydrogenase. J Bacteriol 2014; 196:3840-52; PMID:25157086; http://dx.doi.org/ 10.1128/JB.02070-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Shima S, Pilak O, Vogt S, Schick M, Stagni MS, Meyer-Klaucke W, Warkentin E, Thauer RK, Ermler U. The crystal structure of [Fe]-Hydrogenase reveals the geometry of the active site. Science 2008; 321:572-5; PMID:18653896; http://dx.doi.org/ 10.1126/science.1158978 [DOI] [PubMed] [Google Scholar]
- [31].Vignais PM, Billoud B, Meyer J. Classification and phylogeny of hydrogenases. FEMS Microbiol Rev 2001; 25:455-501; PMID:11524134; http://dx.doi.org/ 10.1111/j.1574-6976.2001.tb00587.x [DOI] [PubMed] [Google Scholar]
- [32].Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett C, Knight R, Gordon JI. The human microbiome project: exploring the microbial part of ourselves in a changing world. Nature 2007; 449:804-10; PMID:17943116; http://dx.doi.org/ 10.1038/nature06244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al.. Clustal W and Clustal X version 2.0. Bioinformatics 2007; 23:2947-8; PMID:17846036; http://dx.doi.org/ 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- [34].Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol 2013; 30:2725-9; PMID:24132122; http://dx.doi.org/ 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Schut GJ, Adams MWW. The iron-hydrogenase of Thermotoga maritima utilizes ferredoxin and NADH synergistically: a new perspective on anaerobic hydrogen production. J Bacteriol 2009; 191:4451-7; PMID:19411328; http://dx.doi.org/ 10.1128/JB.01582-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Biegel E, Müller V. Bacterial Na+-translocating ferredoxin:NAD+ oxidoreductase. Proc Natl Acad Sci 2010; 107:18138-42; PMID:20921383; http://dx.doi.org/ 10.1073/pnas.1010318107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lukey MJ, Parkin A, Roessler MM, Murphy BJ, Harmer J, Palmer T, Sargent F, Armstrong FA. How Escherichia coli is equipped to oxidize hydrogen under different redox conditions. J Biol Chem 2010; 285:3928-38; PMID:19917611; http://dx.doi.org/ 10.1074/jbc.M109.067751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].van Elsas JD, Semenov AV, Costa R, Trevors JT. Survival of Escherichia coli in the environment: fundamental and public health aspects. ISME J 2011; 5:173-83; PMID:20574458; http://dx.doi.org/ 10.1038/ismej.2010.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Gibson GR, Cummings JH, Macfarlane GT, Allison C, Segal I, Vorster HH, Walker AR. Alternative pathways for hydrogen disposal during fermentation in the human colon. Gut 1990; 31:679-83; PMID:2379871; http://dx.doi.org/ 10.1136/gut.31.6.679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Carbonero F, Benefiel AC, Alizadeh-Ghamsari AH, Gaskins HR. Microbial pathways in colonic sulfur metabolism and links with health and disease. Front Physiol 2012; 3:448; PMID:23226130; http://dx.doi.org/ 10.3389/fphys.2012.00448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Betian HG, Linehan BA, Bryant MP, Holdeman LV. Isolation of a cellulolytic Bacteroides sp. from human feces. Appl Environ Microbiol 1977; 33:1009-10; PMID:869523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Murray WD. Symbiotic relationship of Bacteroides cellulosolvens and Clostridium saccharolyticum in cellulose fermentation. Appl Environ Microbiol 1986; 51:710-4; PMID:16347034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Calusinska M, Happe T, Joris B, Wilmotte A. The surprising diversity of clostridial hydrogenases: a comparative genomic perspective. Microbiology 2010; 156:1575-88; PMID:20395274; http://dx.doi.org/ 10.1099/mic.0.032771-0 [DOI] [PubMed] [Google Scholar]
- [44].Schuchmann K, Muller V. Autotrophy at the thermodynamic limit of life: a model for energy conservation in acetogenic bacteria. Nat Rev Microbiol 2014; 12:809-21; PMID:25383604; http://dx.doi.org/ 10.1038/nrmicro3365 [DOI] [PubMed] [Google Scholar]
- [45].Hess V, Gallegos R, Jones JA, Barquera B, Malamy MH, Muller V. Occurrence of ferredoxin:NAD+ oxidoreductase activity and its ion specificity in several Gram-positive and Gram-negative bacteria. PeerJ 2016; 4:e1515; PMID:26793417; http://dx.doi.org/ 10.7717/peerj.1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Macy JM, Ljungdahl LG, Gottschalk G. Pathway of succinate and propionate formation in Bacteroides fragilis. J Bacteriol 1978; 134:84-91; PMID:148460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Nava GM, Carbonero F, Ou J, Benefiel AC, O'Keefe SJ, Gaskins HR. Hydrogenotrophic microbiota distinguish native Africans from African and European Americans. Environ Microbiol Rep 2012; 4:307-15; PMID:23760794; http://dx.doi.org/ 10.1111/j.1758-2229.2012.00334.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Sieber JR, McInerney MJ, Gunsalus RP. Genomic insights into syntrophy: the paradigm for anaerobic metabolic cooperation. Annu Rev Microbiol 2012; 66:429-52; PMID:22803797; http://dx.doi.org/ 10.1146/annurev-micro-090110-102844 [DOI] [PubMed] [Google Scholar]
- [49].Buckel W, Thauer RK. Energy conservation via electron bifurcating ferredoxin reduction and proton/Na(+) translocating ferredoxin oxidation. Biochim Biophys Acta 2013; 1827:94-113; PMID:22800682; http://dx.doi.org/ 10.1016/j.bbabio.2012.07.002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.