ABSTRACT
Non-steroidal anti-inflammatory drugs (NSAIDs) are one of the most frequently used classes of medications in the world. Unfortunately, NSAIDs induce an enteropathy associated with high morbidity and mortality. Although the pathophysiology of this condition involves the interaction of the gut epithelium, microbiota, and NSAIDs, the precise mechanisms by which microbiota influence NSAID enteropathy are unclear. One possible mechanism is that the microbiota may attenuate the severity of disease by specific metabolite-mediated regulation of host inflammation and injury. The microbiota-derived tryptophan-metabolite indole is abundant in the healthy mammalian gut and positively influences intestinal health. We thus examined the effects of indole administration on NSAID enteropathy. Mice (n = 5 per group) were treated once daily for 7 days with an NSAID (indomethacin; 5 mg/kg), indole (20 mg/kg), indomethacin plus indole, or vehicle only (control). Outcomes compared among groups included: microscopic pathology; fecal calprotectin concentration; proportion of neutrophils in the spleen and mesenteric lymph nodes; fecal microbiota composition and diversity; small intestinal mucosal transcriptome; and, fecal tryptophan metabolites. Co-administration of indole with indomethacin: significantly reduced mucosal pathology scores, fecal calprotectin concentrations, and neutrophilic infiltration of the spleen and mesenteric lymph nodes induced by indomethacin; modulated NSAID-induced perturbation of the microbiota, fecal metabolites, and inferred metagenome; and, abrogated a pro-inflammatory gene expression profile in the small intestinal mucosa induced by indomethacin. The microbiota-derived metabolite indole attenuated multiple deleterious effects of NSAID enteropathy, including modulating inflammation mediated by innate immune responses and altering indomethacin-induced shift of the microbiota.
KEYWORDS: dysbiosis, indole, microbiota, microbial metabolites, non-steroidal antiinflammatory drugs (NSAIDs), transcriptome
Introduction
Non-steroidal anti-inflammatory drugs (NSAIDs) are among the most frequently used medications worldwide for routine relief of pain or fever, to manage various forms of arthritis and inflammatory intestinal disorders, and to prevent or treat alimentary cancers.1-3 Despite their effectiveness for managing these varied and highly prevalent conditions, NSAIDs cause damage to the gastrointestinal (GI) tract. Although methods for diagnosis and effective treatment of NSAID-induced lesions of the proximal GI tract (i.e., gastropathy) have been well documented, the pathogenesis, diagnosis, and treatment of NSAID-induced damage of the GI tract distal to the duodenum (known as NSAID enteropathy, primarily affecting the distal jejunum and ileum) remain unclear.4,5 The magnitude of the problem of NSAID enteropathy is alarmingly high. In the United States, NSAID enteropathy results in approximately 100,000 hospitalizations and 16,500 deaths each year.6 Additionally, 2-thirds of both short- and long-term NSAID users develop distal small intestinal lesions.7,8 Although the use of either NSAIDs considered to be safer for the GI tract or other ancillary therapies have reduced the incidence and severity of NSAID-induced gastropathy, the incidence of NSAID enteropathy has remained constant or has increased.9
The pathophysiology of NSAID enteropathy is complex and incompletely understood.10 It appears to involve deleterious effects of NSAIDs on the intestinal mucosa including enterocyte cell death, increased mucosal permeability, and interaction of the damaged mucosa with luminal contents including bacteria (GI microbiota) and bacterial products or components such as lipopolysaccharide (LPS).5,11 The GI microbiota has been implicated as an important contributor to NSAID enteropathy.12-16 Administration of NSAIDs causes a dysbiosis characterized by a reduction of the predominately gram-positive phylum Firmicutes and a corresponding increase of gram-negative bacteria.13,14 Germ-free rats lacking intestinal microbiota do not develop NSAID enteropathy, whereas they develop NSAID-induced intestinal lesions when colonized with gram-negative bacteria.15 Concurrent administration of NSAIDs and antimicrobials targeting gram-negative bacteria reduces the severity of NSAID-induced gastrointestinal lesions in rats.16
A mechanism by which the microbiota might influence NSAID-induced intestinal mucosal damage is by producing metabolites that protect intestinal epithelial cells.17,18 Previously, we identified tryptophan metabolites, including indole, as an important class of GI microbiota-derived compounds.19 Indole is a quorum-sensing molecule produced by bacterial metabolism of L-tryptophan that mediates communication among bacterial population and inter-kingdom signaling between the host and microbe.17,20,21 Indole improves barrier function and decreases intestinal inflammation in vitro and in vivo.17,18 Moreover, several other tryptophan metabolites reportedly exert similar salutary effects on the intestinal epithelium.22,23 Therefore, we hypothesized that indole would mitigate the severity of NSAID enteropathy. To investigate this hypothesis, we co-administered indole with the NSAID indomethacin and demonstrated a reduction in severity of mucosal injury caused by indomethacin alone. To determine whether the protective effects of indole were associated with alterations in the GI microbiota, we characterized the effects of administration of NSAIDs, indole, and their co-administration on the composition and diversity of the fecal microbiota. We observed that the co-administration of indole with indomethacin resulted in maintenance of or even an increase of an important member of the Firmicutes phylum. Finally, to better understand the mechanisms of NSAID enteropathy and the effects of indole on these processes, we performed RNA-Seq of the distal small intestinal mucosa in mice that were untreated or treated with indomethacin, indole alone, or the combination of indomethacin and indole. Pro-inflammatory pathways associated with innate immune responses were up-regulated by indomethacin administration relative to control mice, and co-administration of indole significantly mitigated the up-regulation of these pathways concomitant with reduced GI pathology.
Results
Indole reduced the severity of NSAID enteropathy
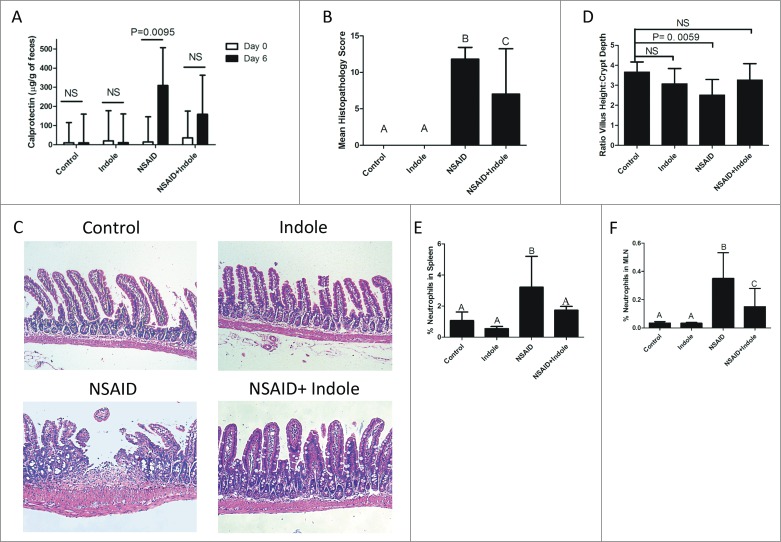
Calprotectin ELISA of fecal samples collected on days 0 and 6 revealed that co-administration of indole with indomethacin significantly decreased fecal calprotectin levels (Fig. 1A). Moreover, attenuation of NSAID-induced small intestinal damage was confirmed by microscopic pathology scores (Fig. 1B and C) of mice in which indole was co-administered with indomethacin. The reduction in mean microscopic pathology scores were corroborated by morphological parameters which revealed maintenance of villus height:crypt depth ratio (Fig. 1D) and thickness of the submucosal layers (Supplementary Fig. 1) in mice in which indole was co-administered. Indomethacin had a negligible effect on the large intestine (data not shown), consistent with evidence that indomethacin induces small intestinal ulcerations in mice in a location similar to where NSAIDs injure people.24-28
Figure 1.
Indole attenuates severity of NSAID enteropathy. A) Fecal calprotectin levels were determined by ELISA on fecal samples on Day 0 (white bar) and then again after 6 days of therapy (black bar) with indomethacin, indole, or the combination. Day 6 fecal calprotectin values were significantly different from day 0 calprotectin only for the animals that received indomethacin alone (P=0.0095). B) Microscopic pathology scores from analysis of Swiss-rolled H&E stained small intestinal sections (n = 5/group). Error bars represent 95% confidence intervals about the mean score. Groups with different letters differed significantly (P < 0.05): control and indole-treated mice are not different from one another; NSAID alone results in a significant difference from control, indole alone, and from NSAID+indole; the NSAID + indole group was significantly higher than control and indole alone groups, but significantly lower than the NSAID group). C) Representative H&E stained sections of small intestine. The microscopic appearance of both the control and indole appear normal, whereas the small intestine of NSAID-treated mice showed ulceration, inflammation in the lamina propria and epithelium, and thickening and blunting of the villi. These pathological findings were ameliorated by co-administration of indole with NSAID (i.e., NSAID + indole). D) Ratio of villus height to crypt depth taken from small intestinal mucosa; relative to the controls, this ratio was only significantly decreased for mice in the NSAID group. E) % CD11b- and GR-1-positive cells (i.e., neutrophils) in the spleen after 7 days of treatment. Groups with different letters differed significantly. Indole significantly attenuated the NSAID-induced increase in splenic neutrophils. F) % CD11b positive and GR-1-positive cells (i.e., neutrophils) in the mesenteric lymph nodes (MLN) after 7 days of treatment. Groups with different letters differed significantly. Indole significantly attenuated the NSAID-induced increase in MLN neutrophils.
Indole reduced indomethacin-induced neutrophilic infiltration of spleen and MLN
Neutrophilic inflammation is primarily responsible for NSAID enteropathy, therefore, we quantified the abundance of neutrophils in the spleen and MLNs as measures of systemic neutrophilic response and trafficking of neutrophils though the GI tract, respectively.29-30 Indomethacin treatment resulted in a significant increase in neutrophils (defined as both CD11b- and GR-1 double-positive; Supplementary Fig. 2) in both the spleen (Fig. 1E) and MLNs (Fig. 1F), and co-administration of indole significantly decreased neutrophilic infiltration of these tissues.31 Taken together, these data demonstrate that indomethacin induced small intestinal injury in mice that was accompanied by neutrophilic infiltration of the spleen and mesenteric lymph node and that co-administration of indole attenuated the small intestinal injury and limited neutrophilic infiltration of the spleen and mesenteric lymph nodes caused by indomethacin.
Indole prevented indomethacin-induced fecal microbiota shift and alterations of the inferred metagenome
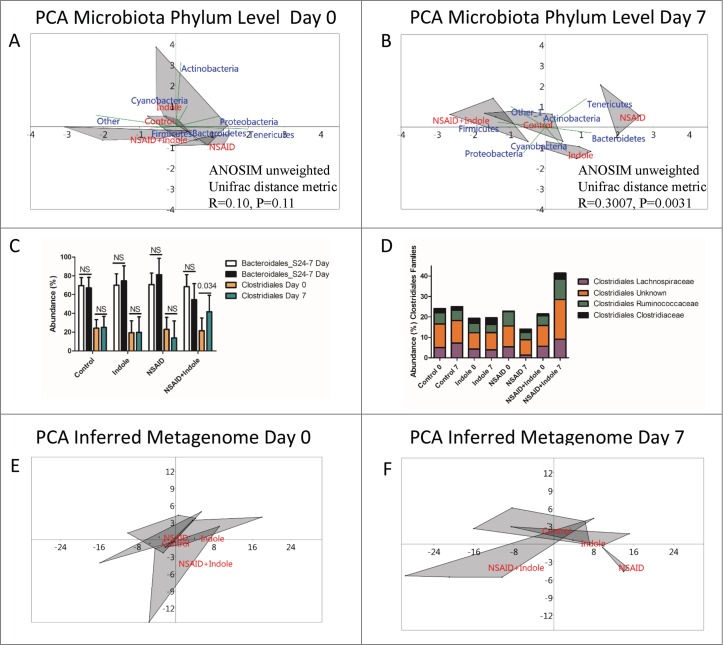
Because NSAIDs alter the intestinal microbiota, we evaluated the composition and diversity of the fecal microbiota in the 4 groups of mice. To adjust for uneven sequencing depth among the samples, each sample was rarefied to an even sequencing depth of 10,000 reads per sample prior to analysis. Alpha rarefaction curves and Good's coverage index estimates indicated that over 90% of the species were represented across all samples at this sequencing depth (Supplementary Fig. 3). Using analysis of similarities (ANOSIM), no significant difference in the unweighted Unifrac distance metric among the groups was observed on day 0 (R = 0.10; P = 0.11); however, by day 7 there was a significant difference among the groups (R = 0.3007; P = 0.0031). Pairwise comparisons between groups revealed that a significant difference in the Unifrac distance metric existed only between NSAID and control animals (Table 1) and that co-administration of indole with indomethacin attenuated this change in the β diversity of the fecal microbiota.
Table 1.
Pairwise ANOSIM R-values based on the unweighted Unifrac distance metric at day 7. Only the difference between control and NSAIDs was significantly different than 0.
| Pairwise AOSIM R-values | Control | NSAID | Indole | NSAID+Indole |
|---|---|---|---|---|
| Control | 0.5487* | 0.2 | 0.2813 | |
| NSAID | 0.5487* | 0.2615 | 0.4444 | |
| Indole | 0.2 | 0.2615 | 0.1938 | |
| NSAID+Indole | 0.2813 | 0.4444 | 0.1938 |
Note.
R value is significantly (P < 0.05) different than a value of 0.
The primary gram-positive and gram-negative phyla found in murine feces are Firmicutes and Bacteroidetes, respectively.32 At the phylum level, PCA revealed a separation of NSAID-treated mice from the other groups characterized by increase in members of the phyla Bacteroidetes in the NSAID-treated animals between day 0 and day 7 (Fig. 2A and 2B). Interestingly, PCA revealed qualitatively that co-administration of indole counteracted the increase in Bacteroidetes and instead appeared to shift the group closer to the phylum Firmicutes. Based on the results of PCA, we compared the abundance of members of the phyla Firmicutes and Bacteroidetes and found no significant difference after treatment with NSAIDs, indole, or their combination (Supplementary Fig. 4). At lower taxonomic levels, however, significant differences were observed. For example, co-administration of indole with indomethacin prevented a decrease in Clostridiales and instead this group had a significant increase in several members of the Clostridales order (Fig. 2C). Furthermore, similarity percentage (SIMPER) based on the Bray Curtis dissimilarity measure at phylum, order, and family levels further confirmed that indomethacin treatment resulted in the gain of members of the Bacteroidales S24-7 family, a major family of Bacteroidetes found in murine feces, indicating that gain of this family contributed to the dissimilarity between NSAID and the other groups.33 Co-administration of indole prevented this increase in Bacteroidales and instead resulted in an increase in Clostridiales with marginal increases in several other members of the Firmicutes phyla (Supplemental Tables 1 to 3 and Fig. 2D).
Figure 2.
Indole increases abundance of Clostridiales and prevents NSAID-induced shift of the microbiota and inferred metagenome. A) Principal component analysis (PCA) plots of 16S rRNA sequencing of the fecal microbiota at the phylum level with biplot overlay and ANOSIM based on the unweighted Unifrac distance metric (located in the lower right quadrant) of the OTU table from day 0 revealed no significant differences among the groups at day 0.The area of the gray shaded shapes reflects the variation among individuals in a group: the larger the shaded area, the more variation among the individuals in that group. B) Principal component analysis (PCA) plots of 16S rRNA sequencing of the fecal microbiota at the phylum level with biplot overlay and ANOSIM based on the unweighted Unifrac distance metric (located in the lower right quadrant) of the OTU table from day 7 reveal that there is a significant difference among the groups at day 7. After 7 days, the NSAID group is significantly shifted from the other 3 groups, and this shift was associated with a qualitative increase in the phylum Bacteroidetes and loss of Firmicutes. C) Abundance of Clostridales from Day 0 (orange) and Day 7 (green) and Bacteroidales Day 0 (white) and Day 7 (black). Error bars represent the upper bound of the symmetrical 95% confidence intervals about the mean of the 5 animals in each group and P values displayed on the graph are corrected for multiplicity of comparisons by the method of Sidak. Clostridiales were significantly increased on Day 7 relative to Day 0 by co-administration of indole with the NSAID indomethacin. D) Mean abundance of several families of Clostridiales at Day 0 and Day 7 for each of the treatment groups depicting reduction of these families following NSAID therapy but expansion indole is co-administered. E) PCA plots of inferred metagenome of the fecal microbiota from Day 0. ANOSIM based on the Bray Curtis distance measure revealed no significant difference among the groups at day 0. F) PCA plots of inferred metagenome of the fecal microbiota from Day 7. ANOSIM based on the Bray Curtis distance measure revealed that, consistent with the microbiota data, the NSAID group visually separated from the other groups; however, this apparent difference was not significant (P > 0.05 by ANOSIM based on Bray Curtis dissimilarity measure).
PCA of the inferred metagenome revealed clustering and separation of the NSAID-treated mice from the other groups (Fig. 2E and 2F), although ANOSIM based on the Bray Curtis dissimilarity metric indicated this difference was not significant (P > 0.05). Consistent with the microbiota data, the distance among groups was greatest between the NSAID group and the NSAID + indole group. The major up- and down-regulated inferred functional pathways between NSAID and NSAID + indole groups were tabulated (Supplementary Table 4).
Co-administration of indole prevents the indomethacin-induced tryptophan-derived metabolite disruption in feces
We have identified multiple tryptophan-derived metabolites produced by the microbiota predicted to be bioactive and exert effects on the host.19 Given the importance of the microbiota in NSAID enteropathy, we chose to examine whether the effects of indole on the intestinal epithelium and microbiota (Fig. 2) were correlated with tryptophan metabolites. No single tryptophan metabolite was significantly correlated with fecal calprotectin (data not shown). Only the metabolite tyramine was significantly (P < 0.01) correlated with microscopic pathology scores (Supplemental Table 5). Examination of the fecal profile of all tryptophan metabolites using PCA revealed a visible separation of the NSAID-treated mice from the remaining groups on day 7, while co-administration of indole appeared to prevent this separation (Supplementary Fig. 5); however, ANOSIM of the Bray Curtis dissimilarity measure indicated this apparent difference was not significant (P = 0.55). Examination of tryptophan metabolite concentrations by treatment group revealed a significant increase in tyramine in the NSAID group that was attenuated by co-administration of indole (Table 2); moreover, several other tryptophan metabolites tended to be increased only in the feces of mice treated exclusively with indomethacin, indicating that NSAID treatment caused differences that were attenuated by co-administration of indole (Table 2). Relative to controls, mice in the indole-only and indomethacin-only groups had significantly more indole; although fecal indole concentrations were generally higher in the NSAID + indole mice, this difference was not significant.
Table 2.
Group effects of tryptophan-derived metabolites identifying the direction and magnitude of change in fecal concentration between Days 0 and 7. Several tryptophan-derived metabolites tended to be increased in the NSAID treated mice (5-hydroxytryptamine, 5-hydroxyindole, indole, serotonin, tyramine). Fecal concentrations of indole increased in the groups in which indole was administered.
| Metabolite | Group effect | Magnitude (95%CI) | P valué |
|---|---|---|---|
| 5 - Hv d roxy try pt a mi ne | t NSAID onlv | 13.9 (−1.4 to 29.3) | 0.1029 |
| 5-Hydroxvindole | t NSAID ünly | 27.2 (−2.2 to 56.6) | 0.0657 |
| Arginine | O | ||
| Glutamine | <-> | ||
| lndole-3-acetamide | <-> | ||
| lndole-3-acetate | |||
| lndole-3-carboxaldehvde | |||
| Indole | T NSAID | 6.7 {1.6 to 28.2) | 0.0255 |
| ^Indole | 5.3 (1.1 to 24.7) | 0.0364 | |
| t NSAID + Indole | 4.7 {0.9 to 24.7) | 0.0641 | |
| Kyneurine | O | ||
| Ornithine | O | ||
| Phenvlalanine | O | ||
| Serotonin | T NSAID only | 1.6 (−0.3 to 3.5) | 0.0895 |
| Tryptamine | O | ||
| Tryptophan | O | ||
| Tyramine | T NSAID onlv | 6.7 (1.3 to 33.6) | 0.0024 |
| Tyrosine | O |
Co-administration of indole attenuates NSAID-induced pro-inflammatory mucosal transcriptomic changes
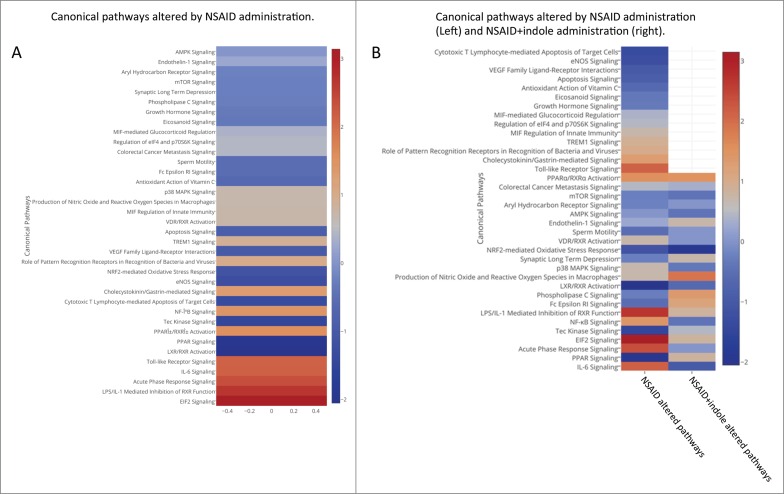
We performed RNA-Seq of the distal small intestinal mucosa to examine the in vivo transcriptomic changes associated with NSAID enteropathy and to gain insight into how the co-administration of indole altered gene expression. The top genes that were significantly up- or downregulated (fold change ≥ 2) in NSAID-treated mice relative to control mice, indole-treated mice relative to control mice, and NSAID + indole-treated mice relative to control mice were tabulated (Supplemental Table 6). The Ingenuity Pathway Analysis software package was used to identify pathways represented by differentially expressed genes. Several canonical pathways were altered in NSAID-treated mice relative to controls (Fig. 3A). Several of the pathways that were modulated by NSAID administration were either shifted to the opposite direction (i.e., inhibited or activated, respectively), or the degree of activation or inhibition was markedly attenuated by co-administration of indole (Fig. 3B). Moreover, transcription of specific pro-inflammatory cytokines (interleukin [IL]-1α, IL-1β, TNF, IL-6) and chemokines (CXCL1, CXCL3, CXCL2, CXCL5, CCL2, CCL7) was significantly upregulated in NSAID-treated mice; however, when indole was co-administered the degree of upregulation was not significantly different than control mice (Supplementary Fig. 6). Based on our results, a proposed schema of the interaction of NSAIDs, the host mucosal epithelium, the microbiota, and indole is presented (Fig. 4).
Figure 3.
NSAID enteropathy results in upregulation of innate immune response pathways and the co-administration of indole regulates this response. A) Heatmap showing upregulated (red) and down regulated (blue) pathways in the small intestinal mucosa from NSAID-treated mice compared with control animals. B) 2-column heatmap showing upregulated (red) and down-regulated (blue) pathways in the small intestinal mucosa from NSAID-treated mice compared with control mice (left column) and NSAID + indole versus control mice (right column).
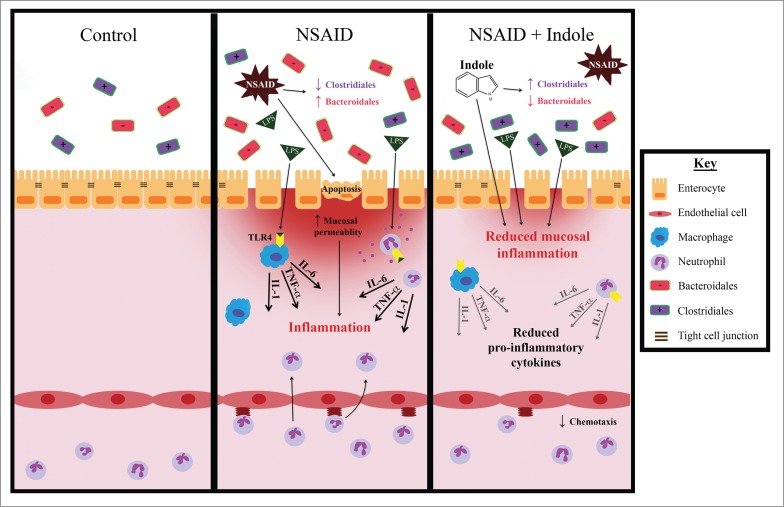
Figure 4.
Co-administration of indole with indomethacin attenuates NSAID enteropathy and indole may exert these beneficial effects through several possible mechanisms. NSAID enteropathy is characterized by a loss of barrier function resulting in an influx of luminal contents and a massive innate immune response. Indole is known to have direct effects on intestinal epithelial cells including upregulation of tight-cell junctional proteins, effects on immune responses including inhibition of NF-κB, and regulating the overall innate immune response. Moreover, co-administration of indole with NSAID administration resulted in an increase in the abundance of Firmicutes, principally Costridales, known to be important in maintaining intestinal homeostasis, and appeared to prevent any increase in Bacteroidales.
Discussion
Co-administration of indole attenuated small intestinal mucosal damage induced by administration of indomethacin in mice as manifested by reduced microscopic pathology and fecal calprotectin concentration. Fecal calprotectin is a well-established, non-invasive indicator of intestinal mucosal injury induced by NSAIDs in human patients and animal models, and correlates well with 4-day fecal excretion of 111Indium-labeled leukocytes.34,35 The findings of decreased fecal calprotectin and decreased microscopic pathology have important clinical implications. A variety of NSAIDs are used widely for an array of clinical conditions ranging from pain-relief for minor injuries to management of rheumatoid arthritis or cancer. The relatively low cost, high effectiveness, and lack of alternatives to NSAIDs indicate that their use will continue to be highly prevalent. Consequently, agents that might be co-administered with NSAIDs to diminish NSAID enteropathy would be clinically important. Further evaluation of indole to ameliorate NSAID enteropathy in animal models and naturally-occurring disease is warranted by our findings.
Administration of NSAIDs increases the proportion of gram-negative organisms at the expense of gram-positive organisms in the intestinal microbiota, and this shift has been shown to contribute to NSAID-induced intestinal injury.14,36,37 Specifically, NSAID administration decreases various members of the class Clostridia and increases members of the class Bacteroidia.38 Mice treated with indole and indomethacin did not have a change in the abundance of Bacteroidia but did have an increase in several members of the gram-positive family Clostridiales in concert with diminution of the severity of intestinal mucosal damage. Evidence exists that the microbiota plays an important role in the development of NSAID enteropathy. Germ-free rats treated with NSAIDs develop less severe enteropathy than specific-pathogen-free rats or germ-free rats that have been colonized with gram-negative bacteria.39 Toll-like receptor (TLR) 4-deficient mice develop less severe lesions than isogeneic TLR4-competent strains.37 Dramatic NSAID-induced alterations of the gut microbiota are well-documented and most often characterized by a loss of gram-positive bacteria with a concurrent increase in gram-negative bacteria.14,40 Moreover, this particular shift in the microbiota has been associated with increased severity of intestinal mucosal injury, and preventing this shift can reduce mucosal injury.15 The classic indomethacin-induced increase in types of Bacteroidia we observed in our study were not significant, but this was likely attributable to limited power to detect a statistically significant difference resulting from our small sample size. It is not clear why increased abundance of gram-negative bacteria worsens the severity of NSAID enteropathy, but direct effects of LPS and the host innate immune response to LPS appear to be important.14,16 It is also possible that loss of beneficial gram-positive bacteria is important. Commensal Clostridia have been shown to be critically important in gut homeostasis, specifically members of Clostridium cluster XIVa and Clostridium cluster IV.41 Interestingly, several members of these 2 clostridial clusters were increased in the NSAID-treated animals in which indole was co-administered.
The host response to the microbiota might be more important than the microbiota itself in the pathogenesis of NSAID enteropathy. Neutrophils are key effector cells of innate immunity and are critically important in the pathogenesis of NSAID enteropathy.29,42 Neutrophils are recruited to the site of injury by the influx of luminal contents following increased mucosal permeability. The resident innate immune cells present in the epithelium and lamina propria release cytokines and chemokines that attract circulating neutrophils. These neutrophils, along with other innate immune cells, then release pro-inflammatory cytokines, typically characterized by an abundance of the IL-1 super family, TNF-α, IL-6, and others, that are responsible for the damage to the lower GI tract.43,44 This critical role for neutrophils in NSAID enteropathy is supported by our findings of neutrophilic infiltration of both the spleen and MLN following NSAID administration and reduced neutrophil concentrations in these tissues with co-administration of indole. Our data also affirm the importance of the innate immune response in the pathophysiology of NSAID enteropathy because many of the most up-regulated pathways identified by RNA-Seq reflected the innate immune response (viz., NF-κB pathway, TLR signaling pathways, and the LPS/IL-1 response). Moreover, at the individual gene level, several of the classic pro-inflammatory cytokines associated with neutrophil activation and known to be important in NSAID enteropathy (e.g., TNF-α, IL-1, and IL-6) were upregulated among NSAID-treated mice.43,45,46 Co-administration of indole, however, attenuated or reversed up-regulation of genes associated with innate immunity and inflammation that contribute to the pathogenesis of NSAID enteropathy. In addition, several chemokines that attract neutrophils were up-regulated in the NSAID-treated mice and this upregulation was dramatically attenuated by the co-administration of indole. These data indicate that indole can mitigate the host innate immune response to the influx of luminal contents across injured epithelia. Indole has been shown to inhibit NF-κB signaling in intestinal epithelial cells.17 It is thus plausible that indole exerts similar effects on neutrophils and other innate immune cells, thereby reducing cytokines dependent upon NF-κB signaling. Although NF-κB signaling was activated by administration of both NSAID and NSAID + indole, activation of this pathway was greatly diminished by co-administration of indole with NSAIDs, suggesting indole might mitigate NSAID damage in part by inhibiting NF-κB. It is important to note that the mRNA for RNA-seq was isolated from mucosal scrapings. These scrapings likely contain epithelial cells as well as cells residing in the lamina propria (i.e., immune cells recruited to the inflamed gut due to loss of barrier function) and therefore we cannot be sure the exact source of gene expression profiles. NSAID enteropathy is characterized as a mucosal injury and we felt examining the transcriptome of this location would be more informative than examining the transcriptome of whole tissue.11 The observed anti-inflammatory effects of indole may be due to indole's effects on epithelial cells, immune cells in the lamina propria, or both.
It is unclear how indole prevented the characteristic shifts in the composition of the microbiota associated with NSAID administration. Indole has long been recognized as a quorum-sensing molecule so it is possible that indole directly affected the microbial community by acting as an intra-kingdom signaling molecule.20,21 It is, however, also plausible that changes in the microbiota associated with NSAID use occur secondary to mucosal inflammation because mucosal inflammation has been shown to alter the luminal environment.47 Indole might have attenuated mucosal damage, which in-turn prevented the expected changes in the microbiota. Finally, it has been shown that GI microbiota can vary with environment including cage-dependent variation and cage-dependent clustering.48 In order to mitigate this phenomenon, after acclimation and immediately prior to starting the study (Day 0), we randomly assigned mice into treatment groups and then moved them into cages based on the group to which they were randomly assigned. There were no significant differences in the fecal β diversity among the groups at Day 0. It is possible, however, that were some cage-dependent microbiota changes that occurred over the 7 days of treatment that might have influenced the composition of the microbiota after treatment.
Indole is present in the GI tract of humans and animals at relatively high concentrations (∼250–1,100 µM).49,50 It has been speculated that, because intestinal epithelial cells are continually exposed to indole, indole may act as an interkingdom signaling molecule. Indeed we have shown, in vitro, that indole does behave in this manner and has anti-inflammatory effects on intestinal epithelial cells and upregulates expression of genes associated with tight cell junctions.17 We therefore expected increasing the concentration of indole within the lumen of the GI tract might mitigate NSAID enteropathy because of the in vitro effects of indole on intestinal epithelial cells. As expected, we observed that mice gavaged with indole (alone or in combination with indomethacin) had increased fecal concentration of indole. Interestingly, the NSAID-only treated mice also had increased fecal concentrations of indole. Although both gram-negative and gram-positive bacteria produce indole, the list of gram-negative bacteria known to produce indole is much larger than gram-positive bacteria.20 Thus, the observed increase in gram-negative bacteria in the NSAID treated mice might explain their increased fecal concentrations of indole. The beneficial effects of indole in this study were observed at the distal small intestine, but the concentration of indole and the microbiota diversity were determined using fecal samples. Fecal metabolite and microbiota characterization do not always correlate well with those of more proximal mucosal locations in the GI tract.51,52 Therefore, it is possible that indole was present in higher concentrations in the distal small intestine of mice treated with NSAID combined with indole compared with mice treated with the NSAID alone, thus contributing to the attenuation of NSAID-induced injury by indole administration. Interestingly, several tryptophan metabolites that act as neurotransmitters including serotonin and 5-hydroxytryptamine were also increased in NSAID-treated mice. Hypermotility of the GI tract is induced by NSAIDs and contributes to the pathophysiology of NSAID enteropathy.53,54 The microbiota shift induced by NSAIDs might result in production of prokinetic metabolites that contribute to GI hypermotility.
In summary, indole supplementation of mice attenuates the deleterious effects of NSAIDs on the distal small intestine and modulates NSAID-induced alterations in the composition of the fecal microbiota. The major events in the onset of NSAID enteropathy are intestinal epithelial cell death, increased mucosal permeability, influx of luminal contents, and host innate immune response to the microbiota.36,55,56 Indole likely reduces intestinal injury induced by NSAIDs at multiple levels, including neutrophilic infiltration, NSAID-induced dysbiosis, and pro-inflammatory pathways in the distal small intestine. Future work will focus on interrogating the potential mechanisms by which indole exerts this beneficial effect in order to further elucidate means to control or prevent NSAID enteropathy.
Materials and methods
Animal protocols were approved by the Texas A&M Institutional Animal Care and Use Committee in accordance with appropriate institutional and regulatory bodies' guidelines.
Mice and treatments
Eight- to 10-week-old specific-pathogen-free C57BL/6J mice were purchased and allowed to acclimate for 2 weeks. Mice were fed standardized laboratory rodent diet and sterile water ad libitum. Mice were randomly divided into the following 4 groups (n = 5 mice/group): 1) NSAID (indomethacin); 2) indole; 3) NSAID + indole; and, 4) untreated controls. Mice were then rehoused on the basis of treatment group assignment, with 5 animals/group-cage. To induce NSAID enteropathy, mice in group 1 were gavaged once daily with indomethacin (5 mg/kg; for 7 days; Sigma Aldrich, St. Louis, MO) dissolved in DMSO (Sigma Aldrich, St. Louis, MO) and further diluted in phosphate buffered saline (PBS). Mice in group 2 received indole by gavage (20 mg/kg; once daily for 7 days; Sigma Aldrich, St. Louis, MO) dissolved in sterile water warmed to 55°C. Mice in group 3 received indole co-administered with indomethacin by gavage at the dosages described above. All mice were gavaged with equal volumes (200 μL) and equal concentrations of DMSO (0.001%).
Sample collection
Feces were collected daily by placing individual animals in sterile plastic cups that were RNase- and DNase-free until they passed feces. The mice were immediately returned to their home cages and the feces immediately flash frozen at −80°C. All animals were euthanized via CO2 asphyxiation on day 8 (i.e., after 7 days of treatment). The small intestine was harvested, opened longitudinally, rinsed with ice-cold PBS, and the distal 1/3 of the intestinal mucosa was scraped for tissue gene expression analysis. The remaining small intestine was fixed in 4% paraformaldehyde, Swiss-rolled, paraffin-embedded, and stained with hematoxylin and eosin. The spleen and mesenteric lymph nodes (MLN) were harvested, and immediately placed in ice cold RPMI-1640-c + 10% fetal calf serum (FCS: Life Technologies, Carlsbad, CA), homogenized, and prepared as a single cell suspension for flow cytometric analysis as previously described.57
Fecal calprotectin ELISA
A murine calprotectin ELISA kit (HK214, Hycult Biotech, Plymouth Meeting, PA) was used according to the manufacturer's protocol with slight modifications. Briefly, 100 mg of feces was homogenized in extraction buffer (0.1 M Tris, 0.15 M NaCl, 1.0 M urea, 10 mM CaCl2, 0.1 M citric acid monohydrate, 5 g/l bovine serum albumin (BSA) and 0.25 mM thimerosal [pH 8.0]). The homogenate was centrifuged at 10,000 x g at 4°C for 20 minutes and the supernatant used as directed in manufacturer's protocol.
Tissue RNA extraction, sequencing, and processing
RNA was extracted from the mucosal scrapings using an RNeasy mini kit (QIAGEN, Redwood City, CA) following the manufacturer's instructions and including on-column DNase treatment. RNA quantity was determined using a Nanodrop spectrophotometer (Fisher Thermoscientific) and the quality was assessed using the Nano6000 chip on a Bioanalyzer 2100(Agilent Technologies). Only RNA with an integrity number (RIN) > 8 was used. The samples were randomized before beginning the RNA-Seq library preparation. Sequencing libraries were made using 250 ng of RNA and the TruSeq RNA Sample Preparation kit (Illumina) following the manufacturer's instructions. A volume of 2.5 µl of ERCC spike-in RNA control mix (Life Technologies) was added to the starting RNA at a dilution of 1:1000. The libraries were pooled and sequenced on an Illumina HiSeq 2500 at the Texas AgriLife Genomics and Bioinformatics Services Core Facility (College Station, TX). Sequencing data were provided in a de-multiplexed format and aligned using Spliced Transcripts Alignment to a Reference (STAR) software with default parameters and referenced against the genome of Mus musculus (Ensembl version GRCm38).58 Differentially expressed genes were determined using EdgeR based on the matrix of gene counts.59 Gene lists were analyzed through use of QIAGEN's Ingenuity ® Pathway Analysis (IPA, QIAGEN, Redwood City, CA http://www.qiagen.com/ingenuity). Sequence data were uploaded into NCBI small reads archive (Accession number PRJNA290483).
Microbiota DNA extraction, sequencing, and processing
Microbiota 16S rRNA gene sequencing methods were adapted from the methods developed for the NIH-Human Microbiome Project.60,61 Briefly, bacterial genomic DNA was extracted using MO BIO PowerSoil DNA Isolation Kit (MO BIO Laboratories) according to the manufacturer's protocol. The 16S rDNA V4 region was amplified by PCR and sequenced in the MiSeq platform (Illumina) using the 2 x 250-bp paired-end protocol yielding paired-end reads that overlap almost completely.62 The primers used for amplification contain adapters for MiSeq sequencing and dual-index barcodes so that the PCR products can be pooled and sequenced directly. The software suite Quantitative Insights Into Microbial Ecology (QIIME v1.9 [http://qiime.sourceforge.net]) was used for data processing and analysis.63 The raw sequence data were de-multiplexed, and low-quality reads were filtered using the database's default parameters. Chimeric sequences were detected using Uchime and removed prior to further analysis.64 Sequences were then assigned to operational taxonomic units (OTUs) using an open-reference OTU picking protocol [http://qiime.org/scripts/pick_open_reference_otus.html] with UCLUST software in QIIME based on 97% identity with the Greengenes database (v13_5).65-67 To adjust for uneven sequencing depth among the samples, each sample was rarefied to an even sequencing depth (10,512 reads/sample) prior to further analysis.
Alpha rarefaction, β diversity measures, richness, taxonomic summaries, and tests for significance were calculated and plotted using QIIME. The weighted and unweighted Unifrac distances were calculated for comparison of β diversity. Differences in microbial communities among the treatment groups were investigated by visual assessment of clustering on principal component analysis (PCA) and by analysis of similarity (ANOSIM) calculated on unweighted UniFrac distance metrics.68-70 ANOSIM is a non-parametric test of difference between 2 or more groups based on a distance metric. This test gives an R value between −1 and 1 where large positive R values indicate a large magnitude of dissimilarity between the groups and small R values indicate small magnitudes of dissimilarity; the P value provides statistical significance.68 When ANOSIM identified significant differences among groups, then pairwise ANOSIM was performed to determine which groups differed significantly and similarity percentage (SIMPER) was used to examine which features contributed to the differences among groups. ANOSIM, SIMPER, and PCA plots were performed with PAST v3.05.71
The software Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) was used to predict the metagenome.72 Sequencing data were prepared as described above, but sequences were then clustered into OTUs using a closed-reference OTU picking protocol at the 97% sequencing identity level [http://qiime.org/scripts/pick_closed_reference_otus.html]. The resulting OTU table was normalized by the expected copy number(s) of the 16s rRNA gene in each OTU. PICRUSt was then used to predict the metagenome [https://picrust.github.io/picrust/tutorials/metagenome_prediction.html#metagenome-prediction-tutorial]. Each sample was rarefied to an even sequencing depth to adjust for uneven sequencing depth prior to further analysis. Differences in the metagenomes among the groups were investigated by visual assessment of clustering on PCA and by analysis of similarity (ANOSIM) calculated on Bray Curtis dissimilarity metric.
Metabolite extraction from fecal samples
Metabolites from the fecal contents were extracted using a solvent-based method as previously described.19 Briefly, fecal pellets were homogenized using a homogenizer (Omni International) with equal volume of cold methanol and half volume of chloroform. The samples were then centrifuged at 10,000 g at 4°C (Thermo Fisher Scientific) for 10 min. Supernatant was passed through a 70-µm sterile nylon cell strainer (Falcon) and 0.6 ml of ice cold water was added. The samples were vortex and centrifuged again at 10,000 g for 5 minutes. The upper phase and lower phase were collected and 400 µl of upper phase was dried to a pellet using a vacufuge (Eppendorf, Hauppauge, NY), and then reconstituted in 50 μl of methanol/water (1:1, v/v). The samples were stored at −80°C until analysis. Tryptophan metabolites in the samples were detected and quantified on a triple quadrupole linear ion trap mass spectrometer (3200 QTRAP, AB SCIEX, Foster City, CA) coupled to a binary pump HPLC (Prominence LC-20, Shimazu, Concord, Ontario, Canada).
Flow cytometry
Spleens and MLNs were processed individually to single-cell suspensions with frosted glass slides in RPMI-c + 10% FCS, and spleen cells underwent red blood cell lysis.73 Cell suspensions were plated in individual wells, washed with 0.5% BSA in PBS, surface-stained for CD11b-AlexaFluor488 (eBioscience cat. #53-0112-82) and Gr-1-biotin (BD cat. #553125), followed by streptavidin-PE (eBioscience cat. #12-4317), fixed with 0.4% paraformaldehyde, and samples were acquired on a BD FACS Aria II in the College of Medicine Cell Analysis Facility (COM-CAF) at the Texas A&M Health Science Center.
Histology and small intestinal morphometric measurements
The stained sections of the small intestine were analyzed by a board-certified veterinary pathologist (BRW) blinded to treatment group. The slides were scored as previously described for intestinal inflammation.74 Briefly, mucosal injury was determined by the following parameters scored from 0 (no evidence) to 3 (marked): mucosal ulceration, mucosal erosion, and presence of squamified epithelium. Inflammatory changes were scored similarly based on the following parameters: lymphocytic infiltration, plasma cell infiltration, and neutrophilic infiltration. Finally, an overall evidence of injury score was used to document total injury graded from 0 (none) to 4 (marked). Morphological parameters were obtained from digitally scanned slides using SPOT vr 5.0 software. Three sets of measurements from 3 separate sections were recorded for each animal by an observer blinded to treatment group. Measurements consisted of villus height, crypt depth, and submucosal mural thickness. The ratio of the villus height to crypt depth was calculated (Supplementary Fig. 1)
Data analysis
Results were expressed as mean ± 95% confidence interval unless indicated otherwise. For all analyses, significance was set P ≤ 0.05. Data were analyzed using S-PLUS statistical software (Version 8.2, TIBCO Inc., Seattle, WA) unless otherwise noted. Histology scores, the proportion of neutrophils in the spleen and MLNs, ratios of villus height to crypt depth, submucosal thicknesses, paired differences between Day 7 and Day 0 in phyla and families, and fecal tryptophan metabolites were compared among treatment groups using a generalized linear model with post hoc testing for pairwise differences among groups using the method of Sidak.75 To meet statistical assumptions underlying the generalized linear model, the histology scores were converted to ranks and the neutrophil data were log10 transformed prior to analysis. Ratios of villus height to crypt depth and submucosal thicknesses were compared among groups using a generalized linear model with post hoc testing for pairwise differences among groups using the method of Sidak. Fecal calprotectin concentrations were analyzed as a function of treatment group, time (Day 0 [baseline] and Day 7), and their interaction using linear mixed-effects modeling with treatment group and time modeled as fixed, categorical effects and individual mouse modeled as a random effect to account for repeated measures on individual mice. Paired differences between Day 0 and Day 7 in phyla and families were compared among groups using a generalized linear model and post hoc testing for pairwise differences among groups using the method of Sidak.
Supplementary Material
Abbreviations
- ANOSIM
Analysis of similarity
- CXCL
Chemokine (C-X-C motif)
- DMSO
Dimethylysulfoxide
- ELISA
Enzyme-linked immunosorbent assay
- FDR
False discovery rate
- GI
Gastrointestinal
- IL
Interleukin
- LPS
Lipopolysaccharide
- MLN
Mesenteric lymph node
- NF-κB
Nuclear factor kappa-light-chain-enhancer of activated B cells
- NSAID
Non-steroidal anti-inflammatory drug
- OTU –
Operational taxonomic unit
- PBS
Phosphate buffered saline
- PCA
Principal component analysis
- PICRUSt
Phylogenetic investigation of communities by reconstruction of unobserved states
- QIIME
Quantitative insights into microbial ecology
- RNA-Seq
RNA sequencing
- SIMPER
Similarity percentage analysis
- TLR
Toll-like receptor
- TNF
Tumor necrosis factor
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by grants from the Department of Large Animal Clinical Sciences, College of Veterinary Medicine & Biomedical Sciences, Texas A&M University, College Station, Texas, United States of America and a post-doctoral trainee grant from the College of Veterinary Medicine & Biomedical Sciences, Texas A&M University, College Station, Texas, United States of America. Additional support was provided by the Link Equine Research Endowment at Texas A&M University and the Center for Translational Environmental Health Research (NIH P30ES023512). Dr. Chapkin is supported by a grant from the National Institutes of Health (NIH R35CA197707).
References
- [1].Thun MJ, Henley SJ, Patrono C. Nonsteroidal anti-inflammatory drugs as anticancer agents: mechanistic, pharmacologic, and clinical issues. J Natl Cancer Inst 2002; 94:252-66; PMID:11854387; http://dx.doi.org/ 10.1093/jnci/94.4.252 [DOI] [PubMed] [Google Scholar]
- [2].Garcia Rodriguez LA, Huerta-Alvarez C. Reduced incidence of colorectal adenoma among long-term users of nonsteroidal antiinflammatory drugs: a pooled analysis of published studies and a new population-based study. Epidemiol (Cambridge, Mass) 2000; 11:376-81; http://dx.doi.org/ 10.1097/00001648-200007000-00003 [DOI] [PubMed] [Google Scholar]
- [3].The use of medicines in the United States: review of 2011. IMS Institute Healthcare Informatics 2012 [Google Scholar]
- [4].Becker JC, Domschke W, Pohle T. Current approaches to prevent NSAID-induced gastropathy–COX selectivity and beyond. Br J Clin Pharmacol 2004; 58:587-600; PMID:15563357; http://dx.doi.org/ 10.1111/j.1365-2125.2004.02198.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wallace JL. Mechanisms, prevention and clinical implications of nonsteroidal anti-inflammatory drug-enteropathy. World J Gastroenterol 2013; 19:1861-76; PMID:23569332; http://dx.doi.org/ 10.3748/wjg.v19.i12.1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wolfe MM, Lichtenstein DR, Singh G. Gastrointestinal toxicity of nonsteroidal antiinflammatory drugs. N Eng J Med 1999; 340:1888-99; http://dx.doi.org/ 10.1056/NEJM199906173402407 [DOI] [PubMed] [Google Scholar]
- [7].Graham DY, Opekun AR, Willingham FF, Qureshi WA. Visible small-intestinal mucosal injury in chronic NSAID users. Clin Gastroenterol Hepatol 2005; 3:55-9; PMID:15645405; http://dx.doi.org/ 10.1016/S1542-3565(04)00603-2 [DOI] [PubMed] [Google Scholar]
- [8].Maiden L. Capsule endoscopic diagnosis of nonsteroidal antiinflammatory drug-induced enteropathy. J Gastroenterol 2009; 44(Suppl 19):64-71; PMID:19148796; http://dx.doi.org/ 10.1007/s00535-008-2248-8 [DOI] [PubMed] [Google Scholar]
- [9].Lanas A, Garcia-Rodriguez LA, Polo-Tomas M, Ponce M, Alonso-Abreu I, Perez-Aisa MA, Perez-Gisbert J, Bujanda L, Castro M, Munoz M, et al.. Time trends and impact of upper and lower gastrointestinal bleeding and perforation in clinical practice. Am J Gastroenterol 2009; 104:1633-41; PMID:19574968; http://dx.doi.org/ 10.1038/ajg.2009.164 [DOI] [PubMed] [Google Scholar]
- [10].Wallace JL. NSAID gastropathy and enteropathy: distinct pathogenesis likely necessitates distinct prevention strategies. Br J Pharmacol 2012; 165:67-74; PMID:21627632; http://dx.doi.org/ 10.1111/j.1476-5381.2011.01509.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Handa O, Naito Y, Fukui A, Omatsu T, Yoshikawa T. The impact of non-steroidal anti-inflammatory drugs on the small intestinal epithelium. J Clin Biochem Nutri 2014; 54:2-6; http://dx.doi.org/ 10.3164/jcbn.13-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wallace JL, Syer S, Denou E, de Palma G, Vong L, McKnight W, Jury J, Bolla M, Bercik P, Collins SM, et al.. Proton pump inhibitors exacerbate NSAID-induced small intestinal injury by inducing dysbiosis. Gastroenterol 2011; 141(1314-22):22.e1-5 [DOI] [PubMed] [Google Scholar]
- [13].Makivuokko H, Tiihonen K, Tynkkynen S, Paulin L, Rautonen N. The effect of age and non-steroidal anti-inflammatory drugs on human intestinal microbiota composition. Br J Nutri 2010; 103:227-34; http://dx.doi.org/ 10.1017/S0007114509991553 [DOI] [PubMed] [Google Scholar]
- [14].Hagiwara M, Kataoka K, Arimochi H, Kuwahara T, Ohnishi Y. Role of unbalanced growth of gram-negative bacteria in ileal ulcer formation in rats treated with a nonsteroidal anti-inflammatory drug. J Medical Invest 2004; 51:43-51; http://dx.doi.org/ 10.2152/jmi.51.43 [DOI] [PubMed] [Google Scholar]
- [15].Uejima M, Kinouchi T, Kataoka K, Hiraoka I, Ohnishi Y. Role of intestinal bacteria in ileal ulcer formation in rats treated with a nonsteroidal antiinflammatory drug. Microbiol Immunol 1996; 40:553-60; PMID:8887349; http://dx.doi.org/ 10.1111/j.1348-0421.1996.tb01108.x [DOI] [PubMed] [Google Scholar]
- [16].Koga H, Aoyagi K, Matsumoto T, Iida M, Fujishima M. Experimental enteropathy in athymic and euthymic rats: synergistic role of lipopolysaccharide and indomethacin. Am J Physiol 1999; 276:G576-82; PMID:10070032 [DOI] [PubMed] [Google Scholar]
- [17].Bansal T, Alaniz RC, Wood TK, Jayaraman A. The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc Natl Acad Sci U S A 2010; 107:228-33; PMID:19966295; http://dx.doi.org/ 10.1073/pnas.0906112107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Shimada Y, Kinoshita M, Harada K, Mizutani M, Masahata K, Kayama H, Takeda K. Commensal bacteria-dependent indole production enhances epithelial barrier function in the colon. PloS One 2013; 8:e80604; PMID:24278294; http://dx.doi.org/ 10.1371/journal.pone.0080604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sridharan GV, Choi K, Klemashevich C, Wu C, Prabakaran D, Pan LB, Steinmeyer S, Mueller C, Yousofshahi M, Alaniz RC, et al.. Prediction and quantification of bioactive microbiota metabolites in the mouse gut. Nat Communications 2014; 5:5492; http://dx.doi.org/ 10.1038/ncomms6492 [DOI] [PubMed] [Google Scholar]
- [20].Lee JH, Lee J. Indole as an intercellular signal in microbial communities. FEMS Microbiol Rev 2010; 34:426-44; PMID:20070374; http://dx.doi.org/ 10.1111/j.1574-6976.2009.00204.x [DOI] [PubMed] [Google Scholar]
- [21].Lee J, Jayaraman A, Wood TK. Indole is an inter-species biofilm signal mediated by SdiA. BMC Microbiol 2007; 7:42; PMID:17511876; http://dx.doi.org/ 10.1186/1471-2180-7-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, Zecchi R, D'Angelo C, Massi-Benedetti C, Fallarino F, et al.. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 2013; 39:372-85; PMID:23973224; http://dx.doi.org/ 10.1016/j.immuni.2013.08.003 [DOI] [PubMed] [Google Scholar]
- [23].Venkatesh M, Mukherjee S, Wang H, Li H, Sun K, Benechet AP, Qiu Z, Maher L, Redinbo MR, Phillips RS, et al.. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity 2014; 41:296-310; PMID:25065623; http://dx.doi.org/ 10.1016/j.immuni.2014.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Fukumoto K, Naito Y, Takagi T, Yamada S, Horie R, Inoue K, Harusato A, Hirata I, Omatsu T, Mizushima K, et al.. Role of tumor necrosis factor-α in the pathogenesis of indomethacin-induced small intestinal injury in mice. Int J Mol Med 2011; 27:353-9; PMID:21249312 [DOI] [PubMed] [Google Scholar]
- [25].Narimatsu K, Higashiyama M, Kurihara C, Takajo T, Maruta K, Yasutake Y, Sato H, Okada Y, Watanabe C, Komoto S, et al.. Toll-like receptor (TLR) 2 agonists ameliorate indomethacin-induced murine ileitis by suppressing the TLR4 signaling. J Gastroenterol Hepatol 2015; 30:1610-7; PMID:25867219; http://dx.doi.org/ 10.1111/jgh.12980 [DOI] [PubMed] [Google Scholar]
- [26].Adebayo D, Bjarnason I. Is non-steroidal anti-inflammaory drug (NSAID) enteropathy clinically more important than NSAID gastropathy? Postgraduate Med J 2006; 82:186-91; http://dx.doi.org/ 10.1136/pgmj.2005.039586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Caunedo-Alvarez A, Gomez-Rodriguez BJ, Romero-Vazquez J, Arguelles-Arias F, Romero-Castro R, Garcia-Montes JM, Pellicer-Bautista FJ, Herrerias-Gutierrez JM. Macroscopic small bowel mucosal injury caused by chronic nonsteroidal anti-inflammatory drugs (NSAID) use as assessed by capsule endoscopy. Revista Espanola De Enfermedades Digestivas 2010; 102:80-5; PMID:20361843 [DOI] [PubMed] [Google Scholar]
- [28].Tacheci I, Bradna P, Douda T, Bastecka D, Kopacova M, Rejchrt S, Bures J. NSAID-Induced Enteropathy in Rheumatoid Arthritis Patients with Chronic Occult Gastrointestinal Bleeding: A Prospective Capsule Endoscopy Study. Gastroenterol Res Practice 2013; 2013:268382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Stadnyk AW, Dollard C, Issekutz TB, Issekutz AC. Neutrophil migration into indomethacin induced rat small intestinal injury is CD11a/CD18 and CD11b/CD18 co-dependent. Gut 2002; 50:629-35; PMID:11950807; http://dx.doi.org/ 10.1136/gut.50.5.629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wallace JL, Keenan CM, Granger DN. Gastric ulceration induced by nonsteroidal anti-inflammatory drugs is a neutrophil-dependent process. The Am J Physiol 1990; 259:G462-7; PMID:2169206 [DOI] [PubMed] [Google Scholar]
- [31].Kim HJ, Alonzo ES, Dorothee G, Pollard JW, Sant'Angelo DB. Selective depletion of eosinophils or neutrophils in mice impacts the efficiency of apoptotic cell clearance in the thymus. PloS One 2010; 5:e11439; PMID:20625428; http://dx.doi.org/ 10.1371/journal.pone.0011439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Jones-Hall YL, Kozik A, Nakatsu C. Ablation of tumor necrosis factor is associated with decreased inflammation and alterations of the microbiota in a mouse model of inflammatory bowel disease. PloS One 2015; 10:e0119441; PMID:25775453; http://dx.doi.org/ 10.1371/journal.pone.0119441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Sands SA, Tsau S, Yankee TM, Parker BL, Ericsson AC, LeVine SM. The effect of omeprazole on the development of experimental autoimmune encephalomyelitis in C57BL/6J and SJL/J mice. BMC Res Notes 2014; 7:605; PMID:25190469; http://dx.doi.org/ 10.1186/1756-0500-7-605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Jacob M, Foster R, Sigthorsson G, Simpson R, Bjarnason I. Role of bile in pathogenesis of indomethacin-induced enteropathy. Arch Toxicol 2007; 81:291-8; PMID:17151867; http://dx.doi.org/ 10.1007/s00204-006-0149-2 [DOI] [PubMed] [Google Scholar]
- [35].Tibble JA, Sigthorsson G, Foster R, Scott D, Fagerhol MK, Roseth A, Bjarnason I. High prevalence of NSAID enteropathy as shown by a simple faecal test. Gut 1999; 45:362-6; PMID:10446103; http://dx.doi.org/ 10.1136/gut.45.3.362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Reuter BK, Davies NM, Wallace JL. Nonsteroidal anti-inflammatory drug enteropathy in rats: role of permeability, bacteria, and enterohepatic circulation. Gastroenterol 1997; 112:109-17; http://dx.doi.org/ 10.1016/S0016-5085(97)70225-7 [DOI] [PubMed] [Google Scholar]
- [37].Watanabe T, Higuchi K, Kobata A, Nishio H, Tanigawa T, Shiba M, Tominaga K, Fujiwara Y, Oshitani N, Asahara T, et al.. Non-steroidal anti-inflammatory drug-induced small intestinal damage is Toll-like receptor 4 dependent. Gut 2008; 57:181-7; PMID:17639086; http://dx.doi.org/ 10.1136/gut.2007.125963 [DOI] [PubMed] [Google Scholar]
- [38].Blackler RW, De Palma G, Manko A, Da Silva GJ, Flannigan KL, Bercik P, Surette MG, Buret AG, Wallace JL. Deciphering the pathogenesis of NSAID enteropathy using proton pump inhibitors and a hydrogen sulfide-releasing NSAID. Am J Physiol Gastrointestinal Liver Physiol 2015; 308:G994-1003; http://dx.doi.org/ 10.1152/ajpgi.00066.2015 [DOI] [PubMed] [Google Scholar]
- [39].Robert A, Asano T. Resistance of germfree rats to indomethacin-induced intestinal lesions. Prostaglandins 1977; 14:333-41; PMID:331401; http://dx.doi.org/ 10.1016/0090-6980(77)90178-2 [DOI] [PubMed] [Google Scholar]
- [40].Dalby AB, Frank DN, St Amand AL, Bendele AM, Pace NR. Culture-independent analysis of indomethacin-induced alterations in the rat gastrointestinal microbiota. Applied Environmental Microbiol 2006; 72:6707-15; http://dx.doi.org/ 10.1128/AEM.00378-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lopetuso LR, Scaldaferri F, Petito V, Gasbarrini A. Commensal Clostridia: leading players in the maintenance of gut homeostasis. Gut Pathogens 2013; 5:23; PMID:23941657; http://dx.doi.org/ 10.1186/1757-4749-5-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zeino Z, Sisson G, Bjarnason I. Adverse effects of drugs on small intestine and colon. Best Practice Res Clin Gastroenterol 2010; 24:133-41; http://dx.doi.org/ 10.1016/j.bpg.2010.02.008 [DOI] [PubMed] [Google Scholar]
- [43].Bertrand V, Guimbaud R, Tulliez M, Mauprivez C, Sogni P, Couturier D, Giroud JP, Chaussade S, Chauvelot-Moachon L. Increase in tumor necrosis factor-α production linked to the toxicity of indomethacin for the rat small intestine. Br J Pharmacol 1998; 124:1385-94; PMID:9723949; http://dx.doi.org/ 10.1038/sj.bjp.0701968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Konaka A, Kato S, Tanaka A, Kunikata T, Korolkiewicz R, Takeuchi K. Roles of enterobacteria, nitric oxide and neutrophil in pathogenesis of indomethacin-induced small intestinal lesions in rats. Pharmacological Res 1999; 40:517-24; http://dx.doi.org/ 10.1006/phrs.1999.0550 [DOI] [PubMed] [Google Scholar]
- [45].Saud B, Nandi J, Ong G, Finocchiaro S, Levine RA. Inhibition of TNF-α improves indomethacin-induced enteropathy in rats by modulating iNOS expression. Digestive Dis Sci 2005; 50:1677-83; http://dx.doi.org/ 10.1007/s10620-005-2914-0 [DOI] [PubMed] [Google Scholar]
- [46].Al-Sadi RM, Ma TY. IL-1beta causes an increase in intestinal epithelial tight junction permeability. J Immunol 2007; 178:4641-9; PMID:17372023; http://dx.doi.org/ 10.4049/jimmunol.178.7.4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, et al.. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 2010; 467:426-9; PMID:20864996; http://dx.doi.org/ 10.1038/nature09415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Teran-Ventura E, Roca M, Martin MT, Abarca ML, Martinez V, Vergara P. Characterization of housing-related spontaneous variations of gut microbiota and expression of toll-like receptors 2 and 4 in rats. Microb Ecol 2010; 60:691-702; PMID:20717659; http://dx.doi.org/ 10.1007/s00248-010-9737-z [DOI] [PubMed] [Google Scholar]
- [49].Karlin DA, Mastromarino AJ, Jones RD, Stroehlein JR, Lorentz O. Fecal skatole and indole and breath methane and hydrogen in patients with large bowel polyps or cancer. J Cancer Res Clin Oncol 1985; 109:135-41; PMID:3980562; http://dx.doi.org/ 10.1007/BF00391888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Zuccato E, Venturi M, Di Leo G, Colombo L, Bertolo C, Doldi SB, Mussini E. Role of bile acids and metabolic activity of colonic bacteria in increased risk of colon cancer after cholecystectomy. Dig Dis Sci 1993; 38:514-9; PMID:8444084; http://dx.doi.org/ 10.1007/BF01316508 [DOI] [PubMed] [Google Scholar]
- [51].Sartor RB. Gut microbiota: Optimal sampling of the intestinal microbiota for research. Nat Rev Gastroenterol Hepatol 2015; 12:253-4 [DOI] [PubMed] [Google Scholar]
- [52].Lavelle A, Lennon G, O'Sullivan O, Docherty N, Balfe A, Maguire A, Mulcahy HE, Doherty G, O'Donoghue D, Hyland J, et al.. Spatial variation of the colonic microbiota in patients with ulcerative colitis and control volunteers. Gut 2015; 64:1553-61; PMID:25596182; http://dx.doi.org/ 10.1136/gutjnl-2014-307873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Takeuchi K, Miyazawa T, Tanaka A, Kato S, Kunikata T. Pathogenic importance of intestinal hypermotility in NSAID-induced small intestinal damage in rats. Digestion 2002; 66:30-41; PMID:12379813; http://dx.doi.org/ 10.1159/000064419 [DOI] [PubMed] [Google Scholar]
- [54].Satoh H, Shiotani S, Otsuka N, Hatao K, Nishimura S. Role of dietary fibres, intestinal hypermotility and leukotrienes in the pathogenesis of NSAID-induced small intestinal ulcers in cats. Gut 2009; 58:1590-6; PMID:19060018; http://dx.doi.org/ 10.1136/gut.2008.156596 [DOI] [PubMed] [Google Scholar]
- [55].Bjarnason I, Fehilly B, Smethurst P, Menzies IS, Levi AJ. Importance of local versus systemic effects of non-steroidal anti-inflammatory drugs in increasing small intestinal permeability in man. Gut 1991; 32:275-7; PMID:1901563; http://dx.doi.org/ 10.1136/gut.32.3.275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Bjarnason I, Hayllar J, MacPherson AJ, Russell AS. Side effects of nonsteroidal anti-inflammatory drugs on the small and large intestine in humans. Gastroenterol 1993; 104:1832-47 [DOI] [PubMed] [Google Scholar]
- [57].Wang N, Strugnell R, Wijburg O, Brodnicki T. Measuring bacterial load and immune responses in mice infected with Listeria monocytogenes. J Visualized Experiments 2011; 54(e3076):1–10; http://dx.doi.org/ 10.3791/3076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England) 2013; 29:15-21; PMID:23104886; http://dx.doi.org/ 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics (Oxford, England) 2010; 26:139-40; PMID:19910308; http://dx.doi.org/ 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].A framework for human microbiome research. Nature 2012; 486:215-21; PMID:22699610; http://dx.doi.org/ 10.1038/nature11209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Structure, function and diversity of the healthy human microbiome. Nature 2012; 486:207-14; PMID:22699609; http://dx.doi.org/ 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, et al.. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 2012; 6:1621-4; PMID:22402401; http://dx.doi.org/ 10.1038/ismej.2012.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al.. QIIME allows analysis of high-throughput community sequencing data. Nature Methods 2010; 7:335-6; PMID:20383131; http://dx.doi.org/ 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics (Oxford, England) 2011; 27:2194-200; PMID:21700674; http://dx.doi.org/ 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics (Oxford, England) 2010; 26:2460-1; PMID:20709691; http://dx.doi.org/ 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- [66].DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Applied Environmental Microbiol 2006; 72:5069-72; http://dx.doi.org/ 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 2012; 6:610-8; PMID:22134646; http://dx.doi.org/ 10.1038/ismej.2011.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Clark K. Non-parametric multivariate analyses of changes in community structure. Australian J Ecol 1993; 18:117-43; http://dx.doi.org/ 10.1111/j.1442-9993.1993.tb00438.x [DOI] [Google Scholar]
- [69].Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Applied Environmental Microbiol 2005; 71:8228-35; http://dx.doi.org/ 10.1128/AEM.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Bray JR, Curtis JT. An ordination of upland forest communities of southern Wisconsin. Ecological Monographs 1957; 27:325-49; http://dx.doi.org/ 10.2307/1942268 [DOI] [Google Scholar]
- [71].Hammer O, Harper D, Ryan PD. PAST: Paleon Statictics Software Package for Education and Data Analysis. Palaeontologica Electronica 2001; 4:9 [Google Scholar]
- [72].Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, et al.. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 2013; 31:814-21; PMID:23975157; http://dx.doi.org/ 10.1038/nbt.2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Charles N, Hardwick D, Daugas E, Illei GG, Rivera J. Basophils and the T helper 2 environment can promote the development of lupus nephritis. Nature Med 2010; 16:701-7; PMID:20512127; http://dx.doi.org/ 10.1038/nm.2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Jia Q, Lupton JR, Smith R, Weeks BR, Callaway E, Davidson LA, Kim W, Fan YY, Yang P, Newman RA, et al.. Reduced colitis-associated colon cancer in Fat-1 (n-3 fatty acid desaturase) transgenic mice. Cancer Res 2008; 68:3985-91; PMID:18483285; http://dx.doi.org/ 10.1158/0008-5472.CAN-07-6251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Šidák ZK. Rectangular confidence regions for the means of multivariate normal distributions. J Am Stat Assoc 1967; 62:626-33 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.