Abstract
The aim of the present was to evaluate the effects of DNA methylation and histone acetylation on 2,3,7,8-tetrachlo-rodibenzo-p-dioxin (TCDD)-induced cleft palate in fetal mice. Pregnant mice (n=10) were randomly divided into two groups: i) TCDD group, mice were treated with 28 µg/kg TCDD on gestation day (GD) 10 by oral gavage; ii) control group, mice were treated with an equal volume of corn oil. On GD 16.5, the fetal mice were evaluated for the presence of a cleft palate. An additional 36 pregnant mice were divided into the control and TCDD groups, and palate samples were collected on GD 13.5, GD 14.5 and GD 15.5, respectively. Transforming growth factor-β3 (TGF-β3) mRNA expression, TGF-β3 promoter methylation, histone acetyltransferase (HAT) activity and histone H3 (H3) acetylation in the palates were evaluated in the two groups. The incidence of a cleft palate in the TCDD group was 93.55%, and no cases of cleft palate were identified in the control group. On GD 13.5 and GD 14.5, TGF-β3 mRNA expression, HAT activity and acetylated H3 levels were significantly increased in the TCDD group compared with the control. Methylated bands were not observed in the TCDD or control groups. In conclusion, at the critical period of palate fusion (GD 13.5–14.5), TCDD significantly increased TGF-β3 gene expression, HAT activity and H3 acetylation. Therefore, histone acetylation may be involved in TCDD-induced cleft palate formation in fetal mice.
Keywords: 2,3,7,8-tetrachlorodibenzo-p-dioxin; cleft palate; transforming growth factor-β3; histone acetyltransferases; histone H3
Introduction
Clefts of the lip and palate (CLP) are common birth defects. CLP can be corrected to varying degrees by surgery, dental treatment, speech therapy and psychosocial intervention. CLP is not a major cause of mortality in developed countries, however, it does cause considerable morbidity in affected children and imposes a societal and financial burden for families (1). Thus, preventing the occurrence of CLP has important practical significance. In order for prevention strategies to be effective, the etiology and pathogenesis of CLP requires further elucidation.
Previous studies have demonstrated that various environmental risk factors are associated with the incidence of a cleft lip and palate, including exposure to dioxin-like chemicals during pregnancy (2,3). In addition, mice exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) during organo-genesis was shown to result in the formation of a cleft palate (4). It has been established that the transforming growth factor-β (TGF-β) superfamily is important in palate development (5). TGF-β3 is considered to be an important regulator of palatal fusion in mice, and TGF-β3-null mice exhibited cleft palates without any other major deformities (5). Congenital cleft palate patients usually have aberrant expression of the TGF-β3 gene as a result of epigenetic regulation rather than genetic mutations (6,7). Our previous study determined that the expression of TGF-β3 was gradually upregulated in TCDD-induced cleft palates in fetal mice (8); however, the underlying mechanism for elevated expression of TGF-β3 in the TCDD-induced cleft palate remains to be determined.
A previous study demonstrated that changes in DNA methylation caused by environmental factors, such as maternal exposure to all-trans-retinoic acid may be important in cleft palate formation (9). Additionally, treatment with lipopolysaccharide or TCDD may alter DNA methylation in splenocytes (10). Histones are globular proteins that undergo post-translational modifications and interact with DNA and other nuclear proteins (11). H3 and H4 histones have long tails protruding from the nucleosome which may be covalently modified by acetylation, methylation, ubiquitination, phosphorylation, sumoylation, citrullination and ADP-ribosylation, thus, modifying chromatin structure and gene expression. Histone acetylation is a dynamic process regulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs). HATs activate gene transcription via histone acetylation, while HDACs inhibit gene transcription via histone deacetylation. Our previous study revealed that the acetylation of histone H3 (H3) is involved in TCDD-induced cleft palate development in C57BL/6J mice (12). Therefore, it is possible that an epigenetic mechanism may be involved in the TCDD-induced cleft palate formation in fetal mice and may be one of the factors contributing to the upregulation of TGF-β3 expression. The present study aimed to elucidate the effects of epigenetic regulation, including DNA methylation and histone acetylation, on TCDD-induced cleft palate in fetal mice.
Materials and methods
Animals and drugs
The present study was approved by the Institutional Review Board of the Children's Hospital of Chongqing Medical University (Chongqing, China). C57BL/6J mice (46 female experimental mice; 23 male mice to impregnate the females; age, 8–10 weeks) were obtained from the Experimental Animal Center of Chongqing Medical University and treated in accordance with the Guide for the Care and Use of Laboratory Animals, US National Institutes of Health (13). Mice were caged under controlled conditions of 22–24°C, 55±5% humidity, a 12 h light/dark cycle and access to food and water ad libitum. Two female mice were housed overnight with one male mouse after one week of acclimation, and the presence of vaginal plugs on the next morning was defined as gestation day 0 (GD 0).
TCDD and corn oil were purchased from Sigma-Aldrich (St. Louis, MO, USA). TCDD was diluted with corn oil to 4 µg/ml.
Animal treatment and sample collection
A total of 10 pregnant mice were randomly divided into two groups (n=5). The TCDD group was administered a single dose of 28 µg/kg TCDD by oral gavage on GD 10. The control group was given an equal volume of corn oil by oral gavage on GD 10. All of these mice were sacrificed by cervical dislocation on GD 16.5. The body weight of the pregnant and fetal mice, and the numbers of live and total fetal mice were recorded. The cleft palate and other malformations were examined under a stereomicroscope (SMZ1500; Nikon Corporation, Tokyo, Japan). The palate was quickly isolated from each embryo and fixed overnight in 4% paraformaldehyde (Shanghai Chemical Reagent Co., Ltd., Shanghai, China) for histological analysis. Fixed palates were dehydrated in alcohol (80% ethanol for 60 min; 95% ethanol for 60 min, twice; 100% ethanol for 60 min, three times), embedded in paraffin (Shanghai Chemical Reagent Co., Ltd.), and cut into 4-µm sections. The palate sections were stained with hematoxylin and eosin (Shanghai Chemical Reagent Co., Ltd.) and examined by light microscopy (Eclipse 55i; Nikon Corporation).
An additional 36 pregnant mice were randomly divided into TCDD and control groups (n=18) as described, and sacrificed on GD 13.5 (pre-palate fusion), GD 14.5 (period of palate fusion), and GD 15.5 (post-palate fusion). A total of 6 mice were sacrificed at each time point. The palate was quickly removed from every embryo following sacrifice of the mother and stored at −80°C for molecular analysis.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
The mRNA expression of TGF-β3 in the palates of fetal mice was quantified by RT-qPCR. Total RNA was extracted from the frozen palates using RNApure High-purity Total RNA Rapid Extraction kit (BioTeke Corporation, Beijing, China) and 1,000 ng was reverse transcribed into cDNA using PrimeScript™ RT reagent kit (Takara Biotechnology Co., Ltd., Dalian, China). qPCR was performed with a total reaction volume of 20 µl, containing 2 µl cDNA, 10 µl SYBR® Fast qPCR mix (Takara Biotechnology Co., Ltd.), 0.8 µl of sense and antisense primers, and 6.4 µl ddH2O by using StepOnePlus™ real-time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The primer sequences (Beijing SBS Genetech Co., Ltd., Beijing, China) for TGF-β3 and β-actin were as follows: Sense: 5′-CCTGGCCCTGCTGAACTTG-3′ and antisense: 5′-TTGATGTGGCCGAAGTCCAAC-3′ for TGF-β3; and sense: 5′-CCAGCCTTCCTTCTTGGGTAT-3′ and antisense: 5′-TTGGCATAGAGGTCTTTACGG-3′ for β-actin. The PCR was performed at 95°C for 30 sec, followed by 39 cycles of 95°C for 5 sec, 60°C for 30 sec, and 63°C for 30 sec. The quantification cycle indicated the fractional cycle number at which the PCR product was first detected above a fixed threshold. The relative mRNA expression levels were determined by normalization to β-actin using the 2−ΔΔCq method (14).
Methylation analysis
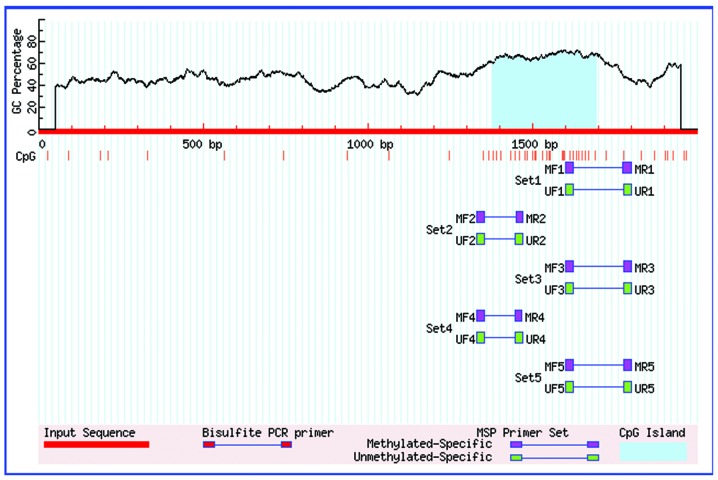
Methylation of the TGF-β3 gene promoter was examined using a DNA methylation kit (Zymo Research Corporation, Irvine, CA, USA). The TGF-β3 gene promoter sequences were obtained from the UCSC Genome Browser Home and inputted into MethPrimer software (15). The location of the CpG island in the TGF-β3 gene promoter is shown in Fig. 1, and the primers for methylation analysis were synthesized by Shanghai Biological Engineering Co., Ltd. (Shanghai, China) and are presented in Table I. Total DNA was extracted from the frozen palates using a Cell/Tissue DNA Extraction kit (BioTeke Corporation). PCR was performed in a reaction volume of 25 µl containing 2 µl cDNA, 12.5 µl Zymo Taq PreMix (Zymo Research Corporation), 1 µl of forward and reverse primers (Beijing SBS Genetech Co., Ltd.), and 8.5 µl ddH2O. The PCR parameters were as follows: Denaturation at 95°C for 10 min; 35 cycles of 95°C for 30 sec, 56°C for 40 sec, and 72°C for 60 sec; and a final extension at 72°C for 7 min. Equal quantities (5 µl) of PCR product from each group were analyzed on a 1.5% agarose gel containing ethidium bromide (Shanghai Chemical Reagent), and the target bands were analyzed densitometrically on a Vistra Fluor Imager SI (Molecular Dynamics Inc., Albuquerque, NM, USA).
Figure 1.
Transforming growth factor-β3 CpG island locations.
Table I.
Sequences of primers used for methylation analysis.
| Primer | Primer sequence | Product length (bp) |
|---|---|---|
| Methylated TGF-β3 primer | F: 5′-TATTTAATTAGATTGGAAAGGAGCG-3′ R: 5′-AACCGAAAATACCCTCTAACGA-3′ |
138 |
| Non-methylated TGF-β3 primer | F: 5′-ATTTAATTAGATTGGAAAGGAGTGG-3′ F: 5′-AAAAAACCAAAAATACCCTCTAACAA-3′ |
141 |
TGF-β3, transforming growth factor β3; F, forward; R, reverse.
Detection of HAT activity
Palatal tissues (0.05–0.1 g) were homogenized in 300 µl of ice-cold lysis buffer (50 mM NaCl, 0.5% Triton X-100, 10 mM HEPES, 1 mM EDTA, 0.05% 2-mercaptoethanol, 0.1 mM polymethyl sulphonate and 10 µg/ml aprotinin). After centrifugation at 22 x g and 4°C for 10 min, the supernatant lysates were collected and stored at −80°C. The palatal protein concentration was quantified using the bicinchoninic acid assay reagent (BioTeke Corporation) and adjusted to 1.25 µg/µl. Next, 68 µl lysate was mixed with HAT activity analysis liquid (BioVision, Inc., Milpitas, CA, USA), added to a 96-well plate, and incubated at 37°C for 4 h. The optical density was measured at a wavelength of 440 nm using a microplate reader (Varioskan™ Flash; Thermo Fisher Scientific, Inc.). The HAT activity was determined by the optical density of the experimental group minus that of the blank control.
Western blotting
A total of 50 µg of whole cell lysate was separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Beyotime Institute of Biotechnology, Haimen, China) at 60 V for 30 min and 110 V for 1 h, and transferred onto a polyvinylidene difluoride membrane (Immobilon; EMD Millipore, Billerica, MA, USA). After blocking in 5% non-fat dry milk, the membranes were incubated with primary antibodies overnight at 4°C, as follows: Rabbit polyclonal anti-acetylated H3 (Ac-H3, 1:10,000, EMD Millipore; cat. no. 382158) and rat monoclonal anti-laminin β1 (1:500, Abcam, Cambridge, MA, USA; cat. no. ab44941) overnight at 4°C. Polyclonal horseradish peroxidase-conjugated goat anti-rabbit IgG (1:5,000; OriGene Technologies, Beijing, China; cat. no. TA130023) was used as the secondary antibody, and the bands were detected using an Enhanced Chemiluminescence Detection kit (Nanjing KeyGen Biotech Co., Ltd. Nanjing, China) and analyzed by Quantity One Software (version 4.62; Bio-Rad Laboratories, Inc.). Protein expression was determined relative to expression of laminin β1.
Statistical analysis
Data are expressed as the mean ± standard deviation and analyzed using SPSS 17.0 (SPSS, Inc., Chicago, IL, USA). Wilcoxon rank sum test was used to compare count data and the Student's t-test was used to compare the data between two groups. P<0.05 was considered to indicate a statistically significant difference.
Results
Effects of TCDD on pregnant and fetal mice
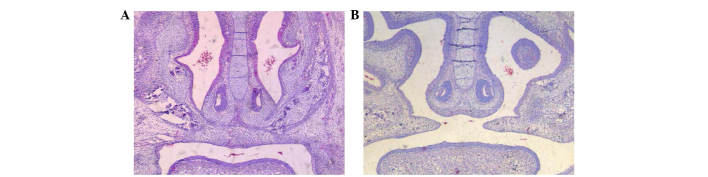
The appearance, feeding, drinking, and activity of the mice were not affected by TCDD treatment. There was no significant difference in the weight gain of pregnant mice, body weights of live fetal mice, or the numbers of live fetuses per litter in the TCDD group compared with the control group (P>0.05, Table II). No cleft palates were observed in fetal mice in the control group, while 93.55% of fetal mice in the TCDD group had a cleft palate. The incidence of a cleft palate was significantly different between the two groups (P<0.01, Table II). Additionally, on GD 16.5, the palatine process fused to the opposite side and formed a complete palate in the control group, whereas the palatine process in the TCDD group was not complete and a cleft was observed in the middle (Fig. 2).
Table II.
Development of pregnant mice and fetal mice and the incidence of cleft palate in each group.
| Group (n=5) | Pregnant mouse weight gain (g) | Live fetus body weight (g) | Number of fetal mice
|
|||
|---|---|---|---|---|---|---|
| Total | Cleft palate | Stillbirth or absorbed fetus | Incidence of cleft palate (%) | |||
| Control | 2.05±0.43 | 1.08±0.10 | 33 | 0 | 0 | 0.00 |
| TCDD (28 µg/kg) | 1.79±0.57 | 1.06±0.11 | 31 | 29 | 1 | 93.55a |
Data are presented as the mean ± standard deviation. Pregnant mouse weight gain represents the weight increase from GD 0 to GD 16.5.
P<0.01 compared with the control group. TCDD, 2,3,7,8-tetrachlorodibenzo-p-dioxin; GD, gestation day.
Figure 2.
Histology of the palate on GD 16.5. (A) Control group and (B) TCDD group. Stained with hematoxylin and eosin. Magnification, ×40. GD, gestation day; TCDD, 2,3,7,8-tetrachlorodibenzo-p-dioxin.
Effect of TCDD on TGF-β3 mRNA expression levels in the palate and on methylation in the TGF-β3 gene promoter
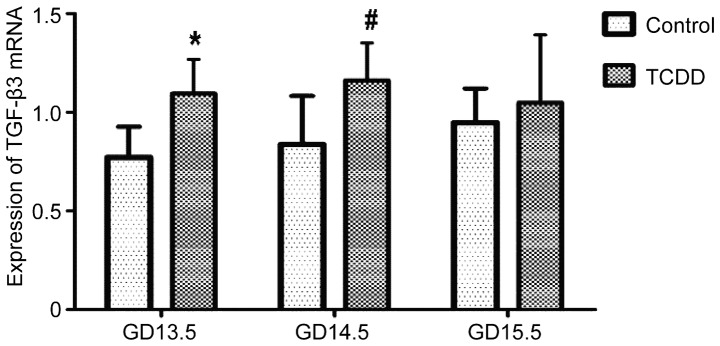
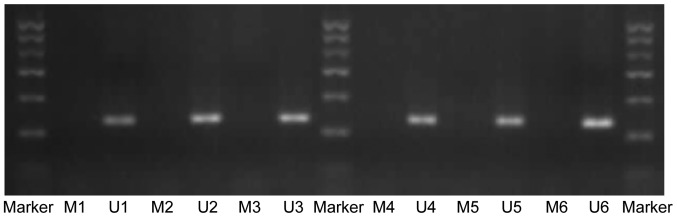
The relative expression of TGF-β3 mRNA in the palate of fetal mice was normalized to that of β-actin. On GD 13.5 and GD 14.5, TGF-β3 mRNA expression was significantly greater in the TCDD group compared with the control (P<0.01 and P<0.05, respectively; Fig. 3); however, the mRNA expression level was not significantly different on GD 15.5 between the two groups (P>0.05, Fig. 3). In addition, the effect of TCDD on methylation of the TGF-β3 gene promoter was examined using PCR. Notably, on GD 13.5, GD 14.5 and GD 15.5, only unmethylated bands were observed in the TCDD and control groups (Fig. 4).
Figure 3.
TGF-β3 mRNA expression on GD 13.5, GD 14.5 and GD 15.5 in the TCDD and control groups. Data are presented as the mean ± standard deviation. *P<0.01 and #P<0.05 vs. the control group. TGF-β3, transforming growth factor-β3; GD, gestation day; TCDD, 2,3,7,8-tetrachlorodibenzo-p-dioxin.
Figure 4.
Detection of methylation status in the TGF-β3 gene promoter by methylation-specific polymerase chain reaction. Marker, 100–600 bp; M, methylation band (138 bp); U, nonmethylated band (141 bp); 1–3, GD13.5, GD14.5, and GD15.5, respectively, in the control group; 4–6, GD13.5, GD14.5 and GD15.5, respectively, in the TCDD group. TGF-β3, transforming growth factor-β3; GD, gestation day; TCDD, 2,3,7,8-tetrachlorodibenzo-p-dioxin.
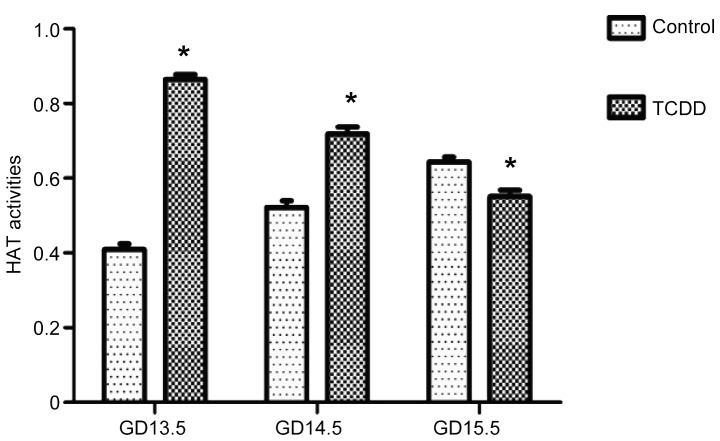
Effect of TCDD on HAT activity
On GD 13.5 and GD 14.5, HAT activity in the TCDD group was significantly higher compared with the control group (P<0.01). Conversely, on GD 15.5, it was significantly lower than that in the control group (P<0.01, Fig. 5). HAT activity in the control group was gradually increased from GD 13.5 to GD 15.5 by contrast, its activity in the TCDD group was significantly reduced (P<0.05, Fig. 5).
Figure 5.
HAT activity in the cleft palate on GD 13.5, GD 14.5 and GD v15.5 in the control and TCDD groups. Data are presented as the mean ± standard deviation. *P<0.01 vs. the control group. HAT, histone acetyltransferase; GD, gestation day; TCDD, 2,3,7,8-tetrachlorodibenzo-p-dioxin.
Effect of TCDD on H3 acetylation
The level of H3 acetylation in the TCDD group was significantly increased on GD 13.5 and GD 14.5 compared with the control group (P<0.01 and P<0.05, respectively; Fig. 6). On GD 15.5, it was significantly reduced (P<0.05; Fig. 6). H3 acetylation level in the control group was increased from GD 13.5 to GD 15.5; however, in the TCDD group, it was not significantly affected from GD 13.5 to GD 15.5 (Fig. 6).
Figure 6.
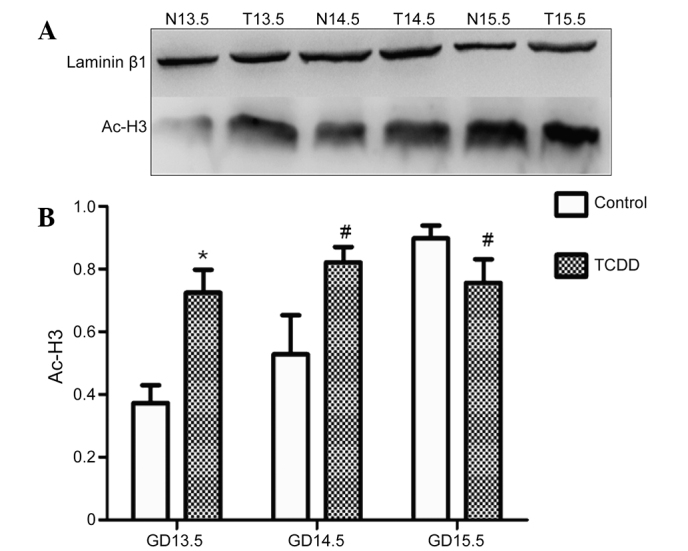
Detection of Ac-H3 in the TCDD and control groups by western blotting. (A) Representative blot. (B) Statistical analysis of the acetylated histone H3 level. Data are presented as the mean ± standard deviation. *P<0.01 and #P<0.05 vs. the control group. N, control group, T, TCDD group. Ac-H3, acetylated histone 3; GD, gestation day; TCDD, 2,3,7,8-tetrachlorodibenzo-p-dioxin.
Discussion
The combination of gene mutations and environmental effects contribute to the complex etiology of cleft palate formation. Although progress has been made in screening mutant genes in the cells of the cleft palate, it is unclear how environmental factors can result in this defect. Environmental influences can adversely affect the development of the secondary palate through genetic variants or epigenetic alterations. Single nucleotide polymorphisms (SNPs) in specific genes confer an increased risk for adverse developmental outcomes subsequent to exposure to certain environmental hazards. For example, infants with a specific transforming growth factor-α polymorphism whose mothers smoked cigarettes during their pregnancy exhibit an increased risk for cleft palate (16–18). Although SNPs are easily identifiable due to the change in DNA sequence, epigenetic alterations may result in gene expression changes without altering the DNA sequence (19), and so cannot be identified through genome-wide association studies that determine nucleotide changes. Epigenetics include, but are not limited to, DNA methylation, histone modification, and microRNA function (19). The pollutant TCDD, a highly toxic halogenated aromatic hydrocarbon, has been implicated in the etiology of various human diseases and birth defects, including the cleft palate. However, the exact mechanisms of TCDD-induced cleft palate formation remain to be fully elucidated.
TGF-β3 is considered to be an important regulator of palatal fusion in mice as TGF-β3-null mice were shown to exhibit a cleft palate but did not have any other major deformities (5). Exposure to dioxin in vivo and in vitro, leads to a reduction in the number of filopodial extensions at the medial epithelial edge of the developing palate, similar to what was observed in TGF-β3-null mice (20). The addition of TGF-β3 to an in vitro palate culture model prevented the dioxin-induced reduction in filopodial density (20). TGF-β3 exposure completely prevented the dioxin-induced block of palatal fusion (20). Our previous study revealed that the expression of TGF-β3 was gradually upregulated in the TCDD-treated cleft palate (32 and 64 µg/kg/day TCDD on GD 10) (8). In the present study, the optimal dose of TCDD was reduced to 28 µg/kg to induce a cleft palate (21) and the epigenetic mechanisms were investigated. Consistent with our previous study, TGF-β3 mRNA expression was significantly increased in the TCDD-induced cleft palate on GD 13.5 and GD 14.5. However, when the methylation status of the TGF-β3 promoter was examined, only unmethylated bands were observed in the cleft palate from the TCDD and control groups. In human and mouse genomes, ~50% of all CpG islands are remote from annotated promoters but often have promoter-like features (22). In a previous study, dioxin exposure was associated with changes in DNA methylation (23). TCDD may induce CpG methylation of the breast cancer-1 (BRCA-1) gene and reduce BRCA-1 expression in breast cancer cell lines (24). Additionally, when a pregnant female was exposed to TCDD epigenetic transgenerational inheritance of adult-onset disease and sperm epimutations through altering DNA methylated regions in gene promoters occurred (25). Therefore, TCDD may affect the methylation status of other gene promoters, global DNA methylation, and promoter-like CpG island methylation in the TCDD-induced cleft palate. However, these mechanisms require further investigation.
Histones are globular proteins that undergo posttranslational modifications and interact with DNA and other nuclear proteins (11). H3 and H4 histones have long tails protruding from the nucleosome, which may be covalently modified by acetylation, methylation, ubiquitination, phosphorylation, sumoylation, citrullination and ADP-ribosylation, thus influencing chromatin structure and gene expression. Histone acetylation is a dynamic process regulated by HATs and HDACs. HATs activate gene transcription through histone acetylation, while HDACs inhibit gene transcription through histone deacetylation. The key period for palate development is between GD 13.5 and GD 14.5; therefore, TCDD may regulate HAT activity and H3 acetylation at the critical period of palate fusion. HAT activity and the acetylated H3 level in the cleft palates from the TCDD group was observed to be significantly higher in the present study on GD 13.5 and GD 14.5 than those from the control group. Since palate fusion completes on GD 15.5, the HAT activity and acetylated H3 levels in the TCDD group were determined to be significantly lower than those in the control group of the current study. The HAT activity and acetylated H3 levels in the control group were increased from GD 13.5 to GD 15.5; however, in the TCDD group, the HAT activity was gradually reduced and the acetylated histone H3 level was not significantly affected. These data indicate that histone acetylation is important in cleft palate development and that the TCDD-induced cleft palate may be the result of the deregulation of HAT activity and histone H3 acetylation. However, the underlying mechanisms for the deregulation of histone acetylation remain to be determined. A previous study reported that the exposure of pregnant women to valproic acid, an antiepileptic drug and an established human teratogen, led to neural tube defects and orofacial clefts, including a cleft palate (26). The teratogenicity of this drug was likely due to its ability to inhibit histone deacetylases, thereby altering the chromatin conformation and regulating gene transcription (27). In addition, the increased deacetylation of histones was important in the TCDD-induced reduction in luteinizing hormone β in the fetal pituitary (28). It has been determined that the acetylation of H3 and H4 was markedly increased at the CYP1B1 promoter in the MCF-7 human breast cancer cell line following dioxin treatment, which is dependent on p300 (29). In conjunction, these studies indicate that histone acetylation is involved in dioxin-induced teratogenic and pathogenic mechanisms.
In conclusion, 28 µg/kg TCDD successfully induced cleft palate development in fetal mice, and upregulated TGF-β3 mRNA expression, HAT activity and H3 levels in the cleft palate. Therefore, TCDD-induced cleft palate development may be associated with the deregulation of histone acetylation. Other epigenetic mechanisms may be involved in TCDD-induced cleft palate and require further research, which may contribute towards strategies of cleft palate prevention.
Acknowledgments
The present study was funded by the National Natural Science Youth Foundation (grant no. 81202167), the National Key Clinical Specialist Construction Project Foundation [(2013) 544] and the Yuzhong District Science and technology planning project (grant nos. 20130121 and 20150112).
References
- 1.Wehby GL, Cassell CH. The impact of orofacial clefts on quality of life and healthcare use and costs. Oral Dis. 2010;16:3–10. doi: 10.1111/j.1601-0825.2009.01588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leite IC, Paumgartten FJ, Koifman S. Chemical exposure during pregnancy and oral clefts in newborns. Cad Saude Publica. 2002;18:17–31. doi: 10.1590/S0102-311X2002000100003. [DOI] [PubMed] [Google Scholar]
- 3.Murray JC. Gene/environment causes of cleft lip and/or palate. Clin Genet. 2002;61:248–256. doi: 10.1034/j.1399-0004.2002.610402.x. [DOI] [PubMed] [Google Scholar]
- 4.Yamada T, Hirata A, Sasabe E, Yoshimura T, Ohno S, Kitamura N, Yamamoto T. TCDD disrupts posterior palato-genesis and causes cleft palate. J Craniomaxillofac Surg. 2014;42:1–6. doi: 10.1016/j.jcms.2013.01.024. [DOI] [PubMed] [Google Scholar]
- 5.Proetzel G, Pawlowski SA, Wiles MV, Yin M, Boivin GP, Howles PN, Ding J, Ferguson MW, Doetschman T. Transforming growth factor-beta 3 is required for secondary palate fusion. Nat Genet. 1995;11:409–414. doi: 10.1038/ng1295-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marazita ML, Lidral AC, Murray JC, Field LL, Maher BS, Goldstein McHenry T, Cooper ME, Govil M, Daack-Hirsch S, Riley B, et al. Genome scan, fine-mapping and candidate gene analysis of non-syndromic cleft lip with or without cleft palate reveals phenotype-specific differences in linkage and association results. Hum Hered. 2009;68:151–170. doi: 10.1159/000224636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jenuwein T. The epigenetic magic of histone lysine methylation. FEBS J. 2006;273:3121–3135. doi: 10.1111/j.1742-4658.2006.05343.x. [DOI] [PubMed] [Google Scholar]
- 8.Gan LQ, Fu YX, Liu X, Qiu L, Wu SD, Tian XF, Liu Y, Wei GH. Transforming growth factor-beta3 expression up-regulates on cleft palates induced by 2,3,7,8-tetrachloro-dibenzo-p-dioxin in mice. Toxicol Ind Health. 2009;25:473–478. doi: 10.1177/0748233709345940. [DOI] [PubMed] [Google Scholar]
- 9.Kuriyama M, Udagawa A, Yoshimoto S, Ichinose M, Sato K, Yamazaki K, Matsuno Y, Shiota K, Mori C. DNA meth-ylation changes during cleft palate formation induced by retinoic acid in mice. Cleft Palate Craniofac J. 2008;45:545–551. doi: 10.1597/07-134.1. [DOI] [PubMed] [Google Scholar]
- 10.McClure EA, North CM, Kaminski NE, Goodman JI. Changes in DNA methylation and gene expression during 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced suppression of the lipopolysaccharide-stimulated IgM response in splenocytes. Toxicol Sci. 2011;120:339–348. doi: 10.1093/toxsci/kfq396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 12.Cuiping L, Xingang Y, Yuexian F, Lin Q, Xiaofei T, Yan L, Guanghui W. The role of histone H3 acetylation on cleft palate in mice induced by 2,3,7,8-tetrachlorodibenzopdioxin. Zhonghua Zheng Xing Wai Ke Za Zhi. 2014;30:369–372. In Chinese. [PubMed] [Google Scholar]
- 13.National Research Council . Guide for the Care and Use of Laboratory Animals. 8th edition. National Academy Press; Washington, DC: 2011. [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Li LC, Dahiya R. MethPrimer: Designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 16.Beaty TH, Maestri NE, Hetmanski JB, Wyszynski DF, Vanderkolk CA, Simpson JC, McIntosh I, Smith EA, Zeiger JS, Raymond GV, et al. Testing for interaction between maternal smoking and TGFA genotype among oral cleft cases born in Maryland 1992–1996. Cleft Palate Craniofac J. 1997;34:447–454. doi: 10.1597/1545-1569(1997)034<0447:TFIBMS>2.3.CO;2. [DOI] [PubMed] [Google Scholar]
- 17.Hwang SJ, Beaty TH, Panny SR, Street NA, Joseph JM, Gordon S, McIntosh I, Francomano CA. Association study of transforming growth factor alpha (TGF alpha) TaqI polymorphism and oral clefts: Indication of gene-environment interaction in a population-based sample of infants with birth defects. Am J Epidemiol. 1995;141:629–636. doi: 10.1093/oxfordjournals.aje.a117478. [DOI] [PubMed] [Google Scholar]
- 18.Shaw GM, Wasserman CR, Lammer EJ, O'Malley CD, Murray JC, Basart AM, Tolarova MM. Orofacial clefts, parental cigarette smoking, and transforming growth factor-alpha gene variants. Am J Hum Genet. 1996;58:551–561. [PMC free article] [PubMed] [Google Scholar]
- 19.Seelan RS, Mukhopadhyay P, Pisano MM, Greene RM. Developmental epigenetics of the murine secondary palate. ILAR J. 2012;53:240–252. doi: 10.1093/ilar.53.3-4.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomae TL, Stevens EA, Bradfield CA. Transforming growth factor-beta3 restores fusion in palatal shelves exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin. J Biol Chem. 2005;280:12742–12746. doi: 10.1074/jbc.M410780200. [DOI] [PubMed] [Google Scholar]
- 21.He X, Liu C, Pu Y, Gan L, Yuan X, Wei G, Fu Y. Be based on the morphological and histological changes to study optimal dose of TCDD induced cleft palate in mice embryo. Wei Sheng Yan Jiu. 2013;42:277–281. In Chinese. [PubMed] [Google Scholar]
- 22.Deaton AM, Webb S, Kerr AR, Illingworth RS, Guy J, Andrews R, Bird A. Cell type-specific DNA methylation at intragenic CpG islands in the immune system. Genome Res. 2011;21:1074–1086. doi: 10.1101/gr.118703.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lind L, Penell J, Luttropp K, Nordfors L, Syvänen AC, Axelsson T, Salihovic S, van Bavel B, Fall T, Ingelsson E, Lind PM. Global DNA hypermethylation is associated with high serum levels of persistent organic pollutants in an elderly population. Environ Int. 2013;59:456–461. doi: 10.1016/j.envint.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 24.Papoutsis AJ, Borg JL, Selmin OI, Romagnolo DF. BRCA-1 promoter hypermethylation and silencing induced by the aromatic hydrocarbon receptor-ligand TCDD are prevented by resveratrol in MCF-7 cells. J Nutr Biochem. 2012;23:1324–1332. doi: 10.1016/j.jnutbio.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 25.Manikkam M, Tracey R, Guerrero-Bosagna C, Skinner MK. Dioxin (TCDD) induces epigenetic transgenerational inheritance of adult onset disease and sperm epimutations. PLoS One. 2012;7:e46249. doi: 10.1371/journal.pone.0046249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ornoy A. Valproic acid in pregnancy: How much are we endangering the embryo and fetus? Reprod Toxicol. 2009;28:1–10. doi: 10.1016/j.reprotox.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 27.Phiel CJ, Zhang F, Huang EY, Guenther MG, Lazar MA, Klein PS. Histone deacetylase is a direct target of valproic acid, a potent anticonvulsant, mood stabilizer and teratogen. J Biol Chem. 2001;276:36734–36741. doi: 10.1074/jbc.M101287200. [DOI] [PubMed] [Google Scholar]
- 28.Takeda T, Fujii M, Taura J, Ishii Y, Yamada H. Dioxin silences gonadotropin expression in perinatal pups by inducing histone deacetylases: A new insight into the mechanism for the imprinting of sexual immaturity by dioxin. J Biol Chem. 2012;287:18440–18450. doi: 10.1074/jbc.M111.335158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ingelman-Sundberg M, Zhong XB, Hankinson O, Beedanagari S, Yu AM, Peng L, Osawa Y. Potential role of epigenetic mechanisms in the regulation of drug metabolism and transport. Drug Metab Dispos. 2013;41:1725–1731. doi: 10.1124/dmd.113.053157. [DOI] [PMC free article] [PubMed] [Google Scholar]