Abstract
In silico drug design using virtual screening, absorption, distribution, metabolism and excretion (ADME)/Tox data analysis, automated docking and molecular dynamics simulations for the determination of lead compounds for further in vitro analysis is a cost effective strategy. The present study used this strategy to discover novel lead compounds from an in-house database of Traditional Chinese Medicinal (TCM) compounds against epithelial growth factor receptor (EGFR) protein for targeting non-small cell lung cancer (NSCLC). After virtual screening of an initial dataset of 2,242 TCM compounds, leads were identified based on binding energy and ADME/Tox data and subjected to automated docking followed by molecular dynamics simulation. Triptolide, a top compound identified by this vigorous in silico screening, was then tested in vitro on the H2347 cell line carrying wild-type EGFR, revealing an anti-proliferative potency similar to that of known drugs against NSCLC.
Keywords: lung cancer, drug design, automated docking, simulation, Traditional Chinese Medicine
Introduction
Lung cancer leads to the greatest number of cancer-associated mortalities in China and worldwide (1–3). In 2010, nearly 600,000 newly diagnosed cases of lung cancer were reported in China. The crude incidence rate of lung cancer was estimated to be 46 per ten thousand individuals with a gender-specific incidence of 62 and 30 per ten thousand individuals for men and women, respectively (4). In China, 480,000 mortalities with a crude mortality rate of 37 per ten thousand individuals were estimated, which is higher by ten individuals per ten thousand compared to world-wide statistics (5).
The etiology of lung cancer has been linked to factors including tobacco smoking (6), genetic factors (7), diet and obesity (8,9) as well as certain environmental factors such as air pollution (10–14). Lung cancers are broadly classified as small cell lung cancer and non-small cell lung cancer (NSCLC); the latter constitutes 85% of total cases (15) and 40% of NSCLCs are adenocarcinoma (16). Targeting of epidermal growth factor receptor (EGFR) is currently considered to be the most suitable way to treat NSCLC adenocarcinoma (17). Erlotinib (18), Afatinib (19) and Gefitinib (20) are the most common drugs in use that target EGFR.
In China, the wisdom of Traditional Chinese Medicine has been recently sourced for use in modern medicine (21), and has been applied in numerous cancer centers in china (22). In silico approaches are a tool for drug discovery (23), which may be utilized for lead identification from large pools of drugs for further in vitro analysis. Molecular dynamics simulation is a tool that can be used to assess the stability of drug-target complexes (24). The present study used a computer-aided drug design (CADD) approach to identify compounds from an in-house pool of Chinese medicinal compounds for targeting EGFR for NSCLC treatment. The initial dataset was reduced to three compounds and then a final lead compound using CADD.
Materials and methods
Data set preparation
The crystallographic structure of the kinase domain of EGFR protein was retrieved from the Protein Databank repository (www.rcsb.org/pdb; ID: 2ITY) (25), representing the crystal structure of EGFR kinase domain with gefitinib (Iressa). After removing the drug Iressa from the crystal structure, the Gromos force field was used for energy minimization using Swiss-PDB viewer (http://spdbv.vital-it.ch/).
Virtual screening
The AutoDock Vina (26) platform using the Pymol interface (27) was used for the study. A total of 2,242 Traditional Chinese Medicinal compounds of plant origin were docked into the active site of the kinase domain of the EGFR protein. A uniform grid of 60 Å in each plane was created for the run. The top compounds based on the cutoff of Gibbs free energy (ΔG) of −9 kcal/mol were selected for further analysis.
Molecular docking
The Lamarckian algorithm of the AutoDock 4.2 (28) tool was used for automated docking of the top compounds with the adenosine triphosphate (ATP) binding pocket of EGFR. Based on the binding energy in kcal/mol, the screening was limited to top three compounds. The sub-set of the top three compounds was limited to one based on binding energy and the Lipinski rule of five (29).
Molecular dynamics simulation
The lead compound identified was subjected to the computational chemistry engine within GROMACS 4.6.5 (http://www.gromacs.org/) for understanding the stability and nature of the ligand protein interaction over time. The protocol used by Chikan et al (23) was used for this study.
Cell growth inhibition assay
H2347 cells (Hybridoma Lab., Ningbo No. 2 Hospital, Ningbo, China), which are known to carry the wild-type EGFR gene with a relative copy number of 4.18, were selected for the study. The cells were grown in Gibco RPMI-1640 medium (Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with Gibco 10% fetal bovine serum (Thermo Fisher Scientific, Inc.) and were seeded into a 96-well plate. Following attachment for 24 h, the cells were incubated with erlotinib, afatinib, gefitinib, as well as the lead compound triptolide (all purchased from Sigma-Aldrich, St. Louis, MO, USA) at concentrations of 0.001, 0.005, 0.01, 0.05, 0.1, 0.5, 1, 5 and 10 μM for a total of 48 h. The Cell Titer-Blue cell viability kit (Promega Corp., Madison, WI, USA) was used for a growth inhibition assay, which was performed as five independent experiments. Growth inhibition was calculated according to the CellTiter-Blue-specific fluorescence at 530 nm (excitation)/590 nm (emission) using a Fluoroskan Ascent FL plate reader (Thermo Fisher Scientific, Inc.).
Results and Discussion
Protein preparation and virtual screening
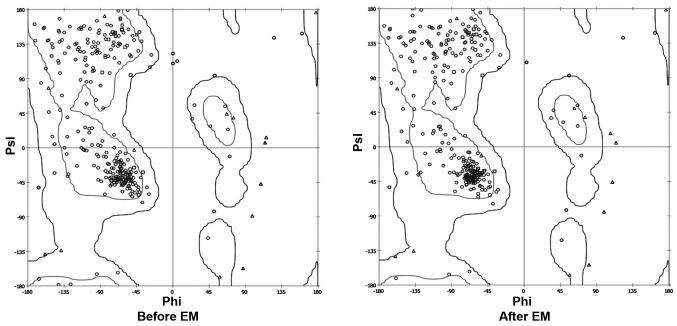
The crystal structure of the kinase domain of the EGFR protein, ranging from amino acids 652–974, was observed using the SPDB viewer to identify missing amino acids and perform energy minimization. The Ramachandran plot before and after energy minimization shows the improved acceptability of the overall structure (Fig. 1), where points represent residues inside and outside the acceptable region, and the contours represent the hardsphere and overlap. The minimized structure of the domain was then processed using the VINA plugin of Pymol, where the grid around the ATP binding site was created. The grid box was subjected to screening of the Traditional Chinese Medicinal compound database (Ningbo No. 2 Hospital), which was useful for narrowing down the number of compounds based on their binding energy. The cutoff value of the ΔG used for initial screening was −9 kcal/mol.
Figure 1.
Psi/phi distribution before and after EM. EM, energy minimization.
Absorption, distribution, metabolism and excretion (ADME)/Tox prediction and molecular docking
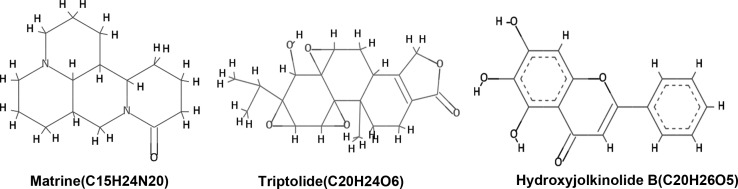
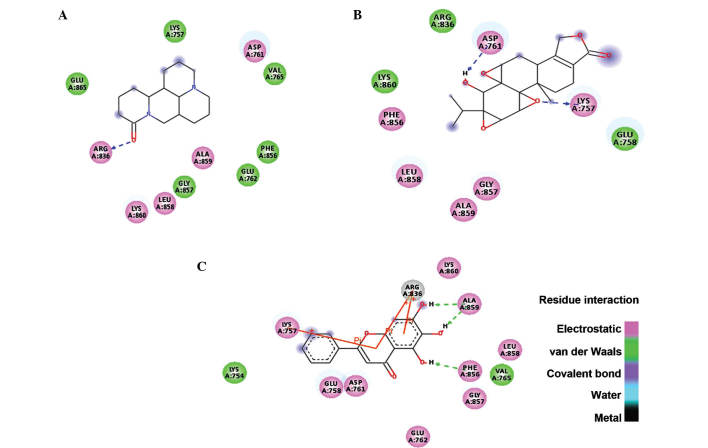
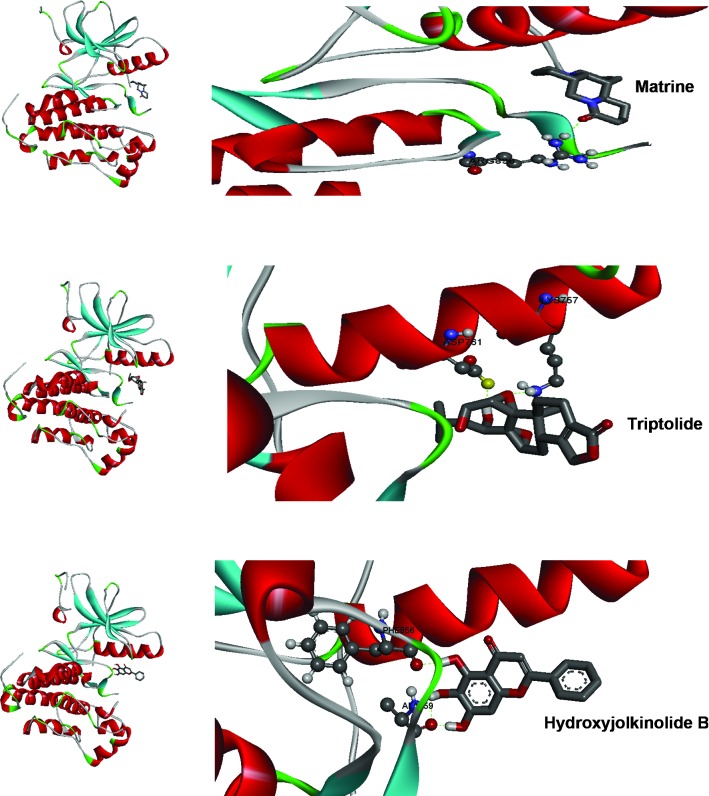
The identified lead compounds were further screened using the Lamarkian algorithm-based AutoDock 4.2 suit. The flexible docking approach of 50 GA runs of maximum evaluations was performed for the top 67 compounds. The 67 compounds were screened from the second dataset of 337 compounds using ADME/Tox analysis, with cutoff based on the parameters set by Lipinski. Out of these 67 compounds, three candidates showed physical interaction with the ATP binding pocket of the kinase domain of EGFR (Fig. 2). In Table I, the top three compounds were ranked on the basis of their binding free energy. The top ranked compound based on docking score was matrine, an alkaloid present in Sophora flarescens. The compound showed a binding energy of −6.19 kcal/mol and was indicated to form single hydrogen bonds with ARG836 of the crystal structure of EGFR as shown in Fig. 3A. The oxygen at the first position of matrine engages in a hydrogen bond with the hydrogen at the 11th position of ARG836 over a distance of 1.91 Å. The second ranked compound was triptolide, a diterpenoid epoxide present in Tripterygium wilfordii, also known as thunder god vine. The compound formed two hydrogen bonds with the ATP binding pocket of EGFR with a binding free energy of −5.69 kcal/mol (Fig. 3B). The hydrogen bonds formed between oxygen in the first position and hydrogen in position 40 of triptolide with HZ2 of LYS757 and OD2 of ASP761 had a bond distance of 2.03 and 1.71 Å, respectively. The compound ranked third regarding interaction with the ATP binding domain of EGFR was hydroxyjolkinolide B, found in Euphorbia fischeriana, with a binding free energy of −3.19 kcal/mol. Two hydroxy groups engaged in hydrogen bonds with ALA859 and one hydrogen bond was present between another hydroxy group and PHE856 over distances of 1.93 and 1.81 Å, respectively. Furthermore, pi-pi interactions of the aromatic core with LYS757 and ARG836 were observed (Fig. 3C). In Table II, the Lipinski rule of five properties of the top three compounds are listed. Three-dimensional representations of the binding interactions at the atomic level are shown in Fig. 4.
Figure 2.
Chemical structures of the top three Traditional Chinese Medicinal lead compounds against epidermal growth factor receptor protein.
Table I.
Binding mode of the top three Traditional Chinese Medicinal compounds.
| Lead | Chem ID | ΔG (kcal/mol) | Binding pocket | Physical interaction |
|---|---|---|---|---|
| Matrine | 91466 | −6.19 | GLU865, LYS757, ASP761, VAL765, PHE856, GLU762, ALA859, GLY857, LEU858, LYS860, ARG863 | Single H-bond |
| Triptolide | 107985 | −5.69 | PHE856, LYS860, ARG836, ASP761, LYS757, GLU758, GLY857, ALA859, LEU858 | Two H-bonds |
| Hydroxyjolkinolide B | 6712607 | −3.19 | LYS754, LYS757, ARG836, LYS860, ALA859, LEU858, VAL765, PHE856, GLY857, GLU762, ASP761, GLU758 | Three H-bonds Three pi-pi |
Figure 3.
Two-dimensional representation of the binding modes of (A) matrine, (B) triptolide and (C) hydoxyjolkinolide B.
Table II.
Lipinski rule of five properties of the top three Traditional Chinese Medicinal compounds.
| Lead | Hydrogen bond donor | Hydrogen bond acceptor | TPSA (Å) | Log P | Molecular weight (Da) | Lipinski rule violations |
|---|---|---|---|---|---|---|
| Matrine | 0 | 3 | 24 | 1.44 | 248.36 | 0 |
| Triptolide | 1 | 6 | 84 | 1.27 | 360.41 | 0 |
| Hydroxyjolkinolide B | 1 | 5 | 72 | 3.07 | 332.43 | 0 |
TPSA, topological polar-surface area; Log P, partition coefficient.
Figure 4.
Top binding modes of the three lead Traditional Chinese Medicinal compounds with the active site of epidermal growth factor receptor.
Molecular dynamics simulation
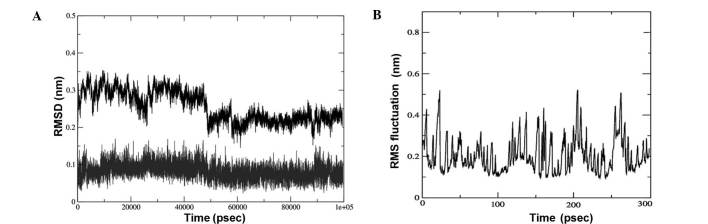
The lead compound triptolide was selected based on the binding energy and number of hydrogen bonds. Its complex with EGFR was subjected to root-mean-square (RMS) simulation over 100 nsec. In a neutral aqueous environment, all trajectories converged within the first five nsec. All calculations were based on the backbone atoms of the protein. RMS deviation (RMSD), RMS fluctuation, radius of gyration (Rg) and hydrogen bond analysis of the complex were performed. The stability of the complex of EGFR with triptolide using RMSD calculations (Fig. 5A) revealed that the binding of the drug was stable. The fluctuations observed in the plot are the attributes of the changing position of the drug in the binding pocket. The fluctuations at each amino acid position were also calculated for the complex using the Grms tool, revealing that the fluctuation of the amino acids in the binding pocket of the complex was reduced, indicating a stable interaction of the drug with EGFR (Fig. 5B).
Figure 5.
(A) RMSD of the EGFR - triptolide complex. The top graph represents EGFR and the bottom graph represents triptolide - RMSD. (B) RMS fluctuation of the amino acids of the EGFR over time. RMSD, root-mean-square deviation; EGFR, epidermal growth factor receptor.
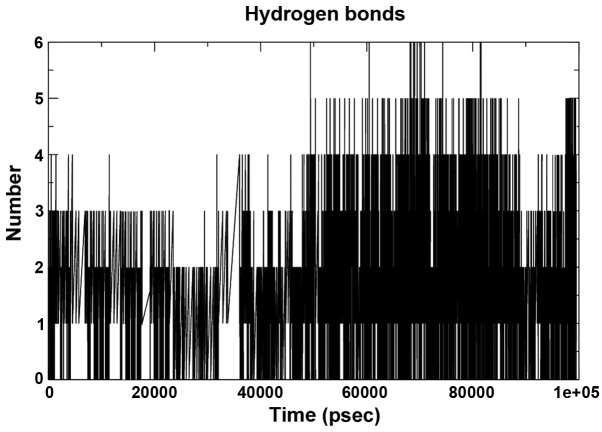
The hydrogen bond interaction between 280 potential donors and 280 potential acceptors present in the two systems were found to comprise 1.1 out of 15,000 possible interactions. The pattern throughout the run is depicted in Fig. 6. The grid that was used to calculate the hydrogen bond using the ghbond tool of gromacs was 16×16×11 and the cutoff was set at 0.35 Å. The Rg of the complex, which is indicative of its fluctuation, showed a decrease in the area of the complex between 0 and 20,000 psec and subsequently increased until 90,000 psec, remaining static until 100 nsec, indicating a maximum amplitude at this time-point, with an average Rg of ~1.32 Å (Fig. 7).
Figure 6.
Hydrogen bond pattern of the complex over a 100-nsec run.
Figure 7.
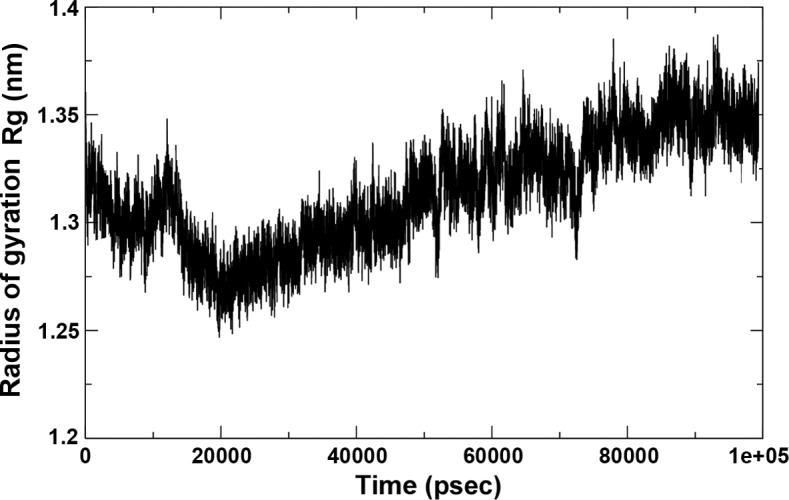
Radius of gyration of the complex over time.
Lead compound triptolide shows potency against lung adenocarcinoma cells
To assess the potency of triptolide in comparison with other known anti-EGFR drugs (erlotinib, afatinib and gefitinib) against lung adenocarcinoma cells in vitro, the H2347 cell line was treated it with either of these drugs at 0.001–10 μM for 48 h, followed by a viability assay. As shown in Fig. 8, the potency of triptolide was at par with afitinib and erlotinib at concentrations <1 μM, while being at par with gefitinib at 5 and 10 μM. The IC50 value for triptolide on H2347 cells was 350±0.658 mg/ml (mean ± standard deviation).
Figure 8.
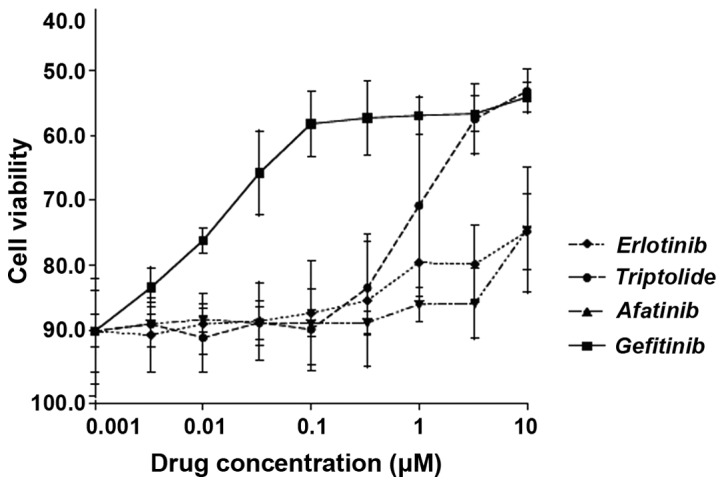
Effects of triptolide and three known drugs on the viability of H2347 lung adenocarcinoma cell line, which is known to carry wild-type epidermal growth factor receptor, following 48 h of incubation.
Conclusions
The present study performed a computer-aided screening approach of a database of 2,242 Chinese Traditional Medicinal compounds with regard to their binding to EGFR for identifying novel drugs for the treatment of lung adenocarcinoma. This virtual screening, ADME/Tox prediction, molecular docking and molecular dynamics simulation provided triptolide as a lead compound. Furthermore, an in vitro study was performed to validate the effects of triptolide on the H2347 NSCLC cell line, which expresses wild-type EGFR. Following 48 h of treatment, triptolide produced growth inhibition, which was comparable with that of known EGFR-targeting drugs. Triptolide has previously been reported to show anti-tumor effects (30–33); the compound is reported to inhibit cell proliferation, induce cell apoptosis, inhibit tumor metastasis and enhance the effect of other therapeutic methods in various cancer cell lines (31). In lung cancer cell lines, the mechanism of action of triptolide remains to be fully elucidated; however it has been indicated that triptolide causes hypomethylation (32) and sensitizes lung cancer cells to cisplatin-induced apoptosis (33). A previous study revealed that triptolide demonstrated synergistic anti-tumor effects with celastrol, another Chinese medicinal compound (30). Out of the two other compounds reported in the present study, matrine has also been reported to exert anti-tumor effects (34). Hydroxyjolkinolide B, is a novel type of lead compound and, to the best of our knowledge, has not undergone ample investigation, therefore further analysis is required. The present study provided an atomic-scale analysis of the interaction of triptolide with EGFR, indicating the highest stability of this complex among all compounds screened. The in silico and in vitro investigations performed in the present study suggest the suitability of triptolide for the treatment of NSCLC, which warrants further in vitro and in vivo analysis.
Acknowledgments
The present study was supported by Programs Supported by the Ningbo Science and Technology Innovation Team Project (grant no. 2011B82016).
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 3.Chen Z. The 3th National Death Cause Survey Report. Beijing: Chinese Academy of Medical Sciences & Peking Union Medical College Press; 2008. [Google Scholar]
- 4.Chen W, Zhang S, Zou X. Evaluation on the incidence, mortality and tendency of lung cancer in China. Thoracic Cancer. 2010;1:48–53. doi: 10.1111/j.1759-7714.2010.00011.x. [DOI] [PubMed] [Google Scholar]
- 5.Chen W, Zheng R, Zeng H, Zhang S. The Epidemiology of Lung Cancer in China. J Cancer Biol Res. 2014;2:1043. doi: 10.1111/1759-7714.12169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guindon GE, Boisclair D. Past, current and future trend in tobacco. Washington, DC: International Bank for Reconstruction and Development. The World Bank; 2009. pp. 13–16. [Google Scholar]
- 7.Cassidy A, Myles JP, van Tongeren M, Page RD, Liloglou T, Duffy SW, Field JK. The LLP risk model: An individual risk prediction model for lung cancer. Br J Cancer. 2008;98:270–276. doi: 10.1038/sj.bjc.6604158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Willett WC, Trichopoulos D. Nutrition and cancer: A summary of the evidence. Cancer Causes Control. 1996;7:178–180. doi: 10.1007/BF00115648. [DOI] [PubMed] [Google Scholar]
- 9.Ruano-Ravina A, Figueiras A, Barros-Dios JM. Diet and lung cancer: A new approach. Eur J Cancer Prev. 2000;9:395–400. doi: 10.1097/00008469-200012000-00004. [DOI] [PubMed] [Google Scholar]
- 10.Rudin CM, Avila-Tang E, Samet JM. Lung cancer in never smokers: A call to action. Clin Cancer Res. 2009;15:5622–5625. doi: 10.1158/1078-0432.CCR-09-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skillrud DM, Offord KP, Miller RD. Higher risk of lung cancer in chronic obstructive pulmonary disease. A prospective, matched, controlled study. Ann Intern Med. 1986;105:503–507. doi: 10.7326/0003-4819-105-4-503. [DOI] [PubMed] [Google Scholar]
- 12.Luo RX, Wu B, Yi YN, Huang ZW, Lin RT. Indoor burning coal air pollution and lung cancer-a case-control study in Fuzhou, China. Lung Cancer. 1996;14(Suppl 1):S113–S119. doi: 10.1016/S0169-5002(96)90217-2. [DOI] [PubMed] [Google Scholar]
- 13.Pershagen G. Air pollution and cancer. IARC Sci Publ. 1990;104:240–251. [PubMed] [Google Scholar]
- 14.Cohen BL. How dangerous is low level radiation? Risk Anal. 1995;15:645–653. doi: 10.1111/j.1539-6924.1995.tb01336.x. [DOI] [PubMed] [Google Scholar]
- 15.Devesa SS, Bray F, Vizcaino AP, Parkin DM. International lung cancer trends by histologic type: Male: Female differences diminishing and adenocarcinoma rates rising. Int J Cancer. 2005;117:294–299. doi: 10.1002/ijc.21183. [DOI] [PubMed] [Google Scholar]
- 16.Morgensztern D, Ng SH, Gao F, Govindan R. Trends in stage distribution for patients with non-small cell lung cancer: A National Cancer Database survey. J Thorac Oncol. 2010;5:29–33. doi: 10.1097/JTO.0b013e3181c5920c. [DOI] [PubMed] [Google Scholar]
- 17.Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, Palmero R, Garcia-Gomez R, Pallares C, Sanchez JM, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): A multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 18.Zhou C, Wu YL, Chen G, Feng J, Liu XQ, Wang C, Zhang S, Wang J, Zhou S, Ren S, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): A multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12:735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 19.Yang JC, Schuler MH, Yamamoto N, et al. LUX-Lung 3: A randomized, open-label, phase III study of afatinib versus pemetrexed and cisplatin as first-line treatment for patients with advanced adenocarcinoma of the lung harboring EGFR-activating mutations. J Clin Oncol. 2012;30(Suppl) Abstract no. LBA7500. [Google Scholar]
- 20.Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 21.Nestler G. Traditional Chinese medicine. Med Clin North Am. 2002;86:63–73. doi: 10.1016/S0025-7125(03)00072-5. [DOI] [PubMed] [Google Scholar]
- 22.Xu W, Towers AD, Li P, Collet JP. Traditional Chinese medicine in cancer care: Perspectives and experiences of patients and professionals in China. Eur J Cancer Care (Engl) 2006;15:397–403. doi: 10.1111/j.1365-2354.2006.00685.x. [DOI] [PubMed] [Google Scholar]
- 23.Chikan NA, Bhavaniprasad V, Anbarasu K, Shabir N, Patel TN. From natural products to drugs for epimutation computer-aided drug design. Appl Biochem Biotechnol. 2013;170:164–175. doi: 10.1007/s12010-013-0158-6. [DOI] [PubMed] [Google Scholar]
- 24.Chikan NA, Vipperla B. KAISO inhibition: An atomic insight. J Biomol Struct Dyn. 2015;33:1794–1804. doi: 10.1080/07391102.2014.974072. [DOI] [PubMed] [Google Scholar]
- 25.Yun CH, Boggon TJ, Li Y, Woo MS, Greulich H, Meyerson M, Eck MJ. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: Mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11:217–227. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J comput chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seeliger D, de Groot BL. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J Comput Aided Mol Des. 2010;24:417–422. doi: 10.1007/s10822-010-9352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem. 1998;19:1639–1662. doi: 10.1002/(SICI)1096-987X(19981115)19:14<1639::AID-JCC10>3.0.CO;2-B. [DOI] [Google Scholar]
- 29.Lipinski CA. Lead-and drug-like compounds: The rule-of-five revolution. Drug Discov Today Technol. 2004;1:337–341. doi: 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 30.Jiang QW, Cheng KJ, Mei XL, et al. Synergistic anticancer effects of triptolide with celastrol, two main compounds from thunder god vine. Oncotarget. 2015;6:32790–32804. doi: 10.18632/oncotarget.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meng C, Zhu H, Song H, Wang Z, Huang G, Li D, Ma Z, Ma J, Qin Q, Sun X, Ma J. Targets and molecular mechanisms of triptolide in cancer therapy. Chin J Cancer Res. 2014;26:622–626. doi: 10.3978/j.issn.1000-9604.2014.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Shen B, Kim J, Raz D. Triptolide inhibits Wnt signaling due to DNA methylation alteration that is determined by dynamic histone 3 K79 lysine methylation in NSCLC. Cancer Res. 2015;75(15 Suppl) Abstract no. 4777. [Google Scholar]
- 33.Wang G, Xu XS, Zhang Y. The role of triptolide in sensitizing lung tumor cells to cisplatin-induced apoptosis. Cancer Res. 2014;74(19 Suppl) Abstract no. 5110. [Google Scholar]
- 34.Ma L, Wen S, Zhan Y, He Y, Liu X, Jiang J. Anticancer effects of the Chinese medicine matrine on murine hepatocellular carcinoma cells. Planta Med. 2008;74:245–251. doi: 10.1055/s-2008-1034304. [DOI] [PubMed] [Google Scholar]