Abstract
To generate functional peripheral T cells, proper gene regulation during T cell development is critical. Here, we found that histone deacetylase 3 (HDAC3) is required for T cell development. T cell development in CD2-icre HDAC3 conditional knockout mice (HDAC3-cKO) was blocked at positive selection, resulting in few CD4 and CD8 T cells, and could not be rescued by a TCR-transgene. These SP thymocytes failed to upregulate Bcl-2, leading to increased apoptosis. HDAC3-cKO mice failed to downregulate RORγt during positive selection, and phenocopied the block in positive selection in RORγt-transgenic mice. In the absence of HDAC3, the RORC promoter was hyperacetylated. In the periphery, the few CD4 T cells present were skewed towards RORγt+ IL-17-producing Th17 cells, leading to inflammatory bowel disease. Positive selection of CD8SP thymocytes was restored in RORγt-KO Bcl-xl-transgenic HDAC3-cKO mice, demonstrating that HDAC3 is required at positive selection to down-regulate RORγt.
Introduction
The generation of immune cells is dependent on epigenetic machinery that controls gene expression at each developmental stage. Lysine acetylation is a well-known post-translational modification added by histone acetyltransferases (HATs) or removed by histone deacetylases (HDACs) to regulate chromatin structure and gene expression. When HATs add acetyl groups, it neutralizes the positively charged lysine leading to the loosening of DNA around histones and gene transcription to occur (1). Removal of acetyl groups by HDACs leads to chromatin condensation and gene repression. There are eighteen HDAC enzymes grouped into four classes (I, II, III, IV) based on their domain organization and function (1). Depending on the HDAC, they are ubiquitously expressed or tissue specific.
HDAC3 belongs to class I HDAC family (1, 2). Traditionally, HDAC3 assembles with the nuclear receptor corepressor (N-CoR) and silencing mediator of retinoic and thyroid receptors (SMRT) (3–5). Together with different transcription factors, HDAC3 functions as the catalytic component of N-CoR/SMRT complexes to deacetylate histones at specific promoters to mediate gene silencing (6). Somatic deletion of HDAC3 leads to embryonic lethality, while tissue-specific deletion leads to hypertrophy in liver and heart (7), failure of hematopoietic stem cell maintenance (8), defects in peripheral T cell maturation (9), iNKT cell development (10), and regulatory T cell dysfunction (11).
Here we examined the role of HDAC3 in early T cell development (T cell development reviewed in (12, 13). Briefly, T cell precursors from the bone marrow migrate to the thymus where they commit to the T cell lineage at the double negative (DN; CD4−CD8−) stage DN2. DN3 thymocytes undergo TCRβ rearrangement and β-selection to test for proper TCRβ rearrangement and expression. Post-β-selection thymocytes (DN3b, DN4, and immature single positive (ISP)) proliferate before transitioning to the double positive (DP; CD4+CD8+) stage. After rearranging their TCRα chain, DP thymocytes that recognize self-MHC weakly are positively selected and transition to the single-positive (SP) stage (14). Thymocytes that fail to engage with MHC die by neglect (15). Thymocytes that express a TCR with strong affinity to self-peptide presented by MHC die by negative selection (14). Positively selecting thymocytes undergo CD4-versus-CD8 lineage commitment (16), maturation (17, 18), and exit the thymus to become peripheral CD4 and CD8 T cells.
Here, we demonstrated that HDAC3 is required for positive selection. HDAC3 deficient DP thymocytes fail to up-regulate Bcl-2, leading to enhanced apoptosis at the SP stage. The enhanced apoptosis was not due to a defect in TCR signaling or enhanced negative selection, but due to the failure to down-regulate RORγt during positive selection. Knocking out RORγt largely rescued this defect in HDAC3-deficient thymocytes. Thus, HDAC3 is essential for the down-regulation of RORγt during positive selection.
MATERIALS AND METHODS
Mice
HDAC3 fl/fl mice (19), and IL-7Rα-transgenic mice (20) were previously described. Human Bcl-2 transgenic mice were generated by S. Korsmeyer (Dan-Farber Cancer Institute, Boston, MA; (21)) and provided by A. Singer (National Institutes of Health, Bethesda, MD). Bcl-xl transgenic mice (22), RORγt-KO (23), and CD2-icre (24) were purchased from Jackson laboratory. OT-II mice (25) were purchased from Taconic. Mice were housed in barrier facilities and experiments were performed at Mayo Clinic with the approval of the Institutional Animal Care and Use Committee. All mice were analyzed between the ages of 6 to 14 weeks. All genetically modified mice were examined with either littermate or age-matched controls, which may include floxed only mice (no cre), CD2-icre, or WT mice, as no differences were observed between these mice. For convenience, the control mice in each experiment are termed “WT” but may represent either floxed only, CD2-icre or WT mice.
Flow Cytometry
FACS analysis was performed on a LSRII flow cytometer (BD) or Attune NxT flow cytometer (Thermo Fisher), and all experiments were analyzed using FlowJo (Tree Star). Cytoplasmic and nuclear proteins were examined via intracellular flow cytometry. Thymocytes or mesenteric lymphocytes were labeled with surface markers before being fixed and permeabilized with a FoxP3/Transcription Factor Staining Buffer Set (for nuclear protein staining; eBioscience) or an Intracellular Fixation & Permeabilization Buffer kit (for cytoplasmic protein staining; eBioscience). All analysis included size exclusion (FSC-A/SSC-A), doublet exclusion (both FSC-H/FSC-W and SSC-H/SSC-W), and dead cell exclusion (FVD; eBioscience). All other reagents for flow cytometry were purchased from Becton Dickinson, eBioscience, Biolegend, Tonbo Biosciences, or Abcam.
Cell Cycle Staining
Cell Cycle analysis was performed using a Click-iT EdU Imaging Kit (Molecular Probes) and Vybrant DyeCycle Violet stain (Molecular Probes). 10μM of EdU was added to freshly isolated thymocytes in complete media (RPMI 1640 media with 10% FCS with pen/strep/glutamine) for 2 hours at 37°C. Cells were then harvested for surface and intracellular EdU staining per manufacturer’s guidelines. Vybrant DyeCycle Violet was then used to stain for DNA content in Hanks’ Balanced Salt Solution (HBSS) for 30 minutes at 37°C.
Generation of radiation chimeras
Mixed bone marrow chimeras (BMCs) were generated by i.v. injecting 4 × 106 cells from either mixes of WT (CD45.1−/−)/B6.SJL (CD45.1+/+) or CD2-icre HDAC3-cKO (CD45.1−/−)/B6.SJL (CD45.1+/+) at a 50:50 ratio into lethally irradiated congenic B6.SJL (CD45.1+/+) recipients. Recipient mice received the antibiotic enrofloxacin in their drinking water for 3 weeks and were analyzed after 9 weeks.
Ex vivo Viability Time Course
Single cell suspensions of total thymocytes were incubated at 37°C for 0, 2, 4, 8, 16, and 24 hours in complete culture medium (RPMI 1640, 10% FCS, pen/strep/glutamine) before surface staining for thymocyte populations as well as with Annexin V (BD Biosciences) and Fixable Viability Dye (eBioscience). After staining at each time point, cells were fixed with the IC Fixation buffer from the Intracellular Fixation & Permeabilization Buffer kit (eBioscience) to allow FACS analysis of all time-points simultaneously.
Chromatin Immunoprecipitation assay (ChIP)
ChIP was performed on total thymocytes from WT or HDAC3-cKO using the SimpleChIP Enzymatic Chromatin IP kit, (Cell Signaling, #9002) per manufacturer’s guidelines. Chromatin was incubated with α-acetyl Histone 3 antibody (Millipore) and control antibodies (Cell Signaling, #9002) overnight at 4°C with rotation. Extracted DNA was analyzed by real-time PCR using previously published primer sequences spanning the RORC promoter (26).
Cytokine Stimulation
For cytokine analysis, isolated lymphocytes from mesenteric lymph nodes were stimulated or left unstimulated overnight in complete culture media (RPMI) with 200nM PMA and 1μM Ionomycin. 3 hours after stimulation, Brefeldin A/Monensin (500x) from BD was added to stop secretion of cytokines for intracellular flow cytometry analysis. Cells were harvested the following day for intracellular cytokine staining.
Histology
Mouse colon was harvested and fixed in 10% formalin for 24 hours at room temperature and then stored in 70% ethanol at 4°C until processing by paraffin embedding, sectioning, and standard H&E staining techniques. Images were viewed on a Leica DMI3000 B microscope at 10X and 20X magnification, captured using the Leica EC3 camera, and processed with the Leica Application Systems EZ software.
Statistics
Data comparing absolute cell number, percentage of Annexin V+ cells, ChIP between experimental groups were analyzed using two-tailed unpaired Student’s t test. Analysis of percentage of cells in S-phase was determined by two-tailed paired Student’s t test. Ex vivo viability time course assay was analyzed using two-way ANOVA using GraphPad Prism.
RESULTS
HDAC3 is required for normal T cell development
To investigate the role HDAC3 plays in T cell development, we generated CD2-icre HDAC3fl/fl mice (afterwards referred to as HDAC3-cKO). CD2-icre expresses Cre in a lymphoid progenitor (27), resulting in Cre-mediated deletion of HDAC3 affecting early in both T-cell and B-cell lineages. However, here we will focus only on T cell development. We used flow cytometry to identify at which stage of thymic development elimination of HDAC3 protein was complete. HDAC3 protein expression was reduced at the DN3 stage and eliminated in DN4 thymocytes onward, equivalent to an isotype control (Supplementary Fig. 1a).
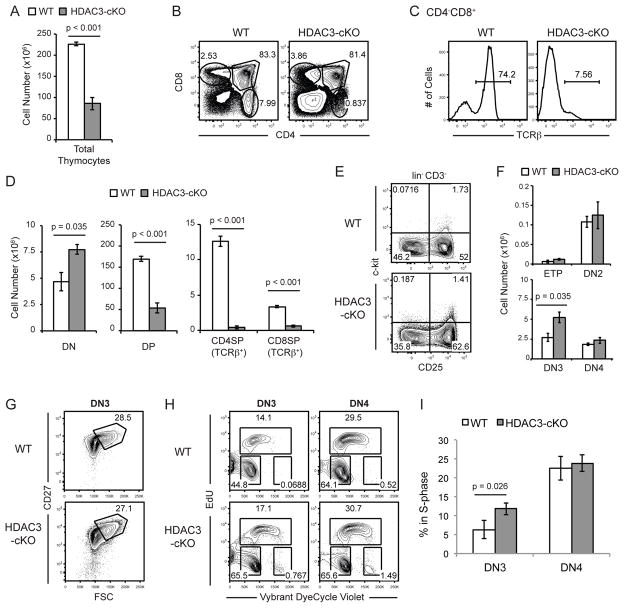
T cell development was severely blocked in HDAC3-cKO mice as compared to WT mice. There was a two-fold decrease in total thymocyte cell number in HDAC3-cKO (Fig. 1a). HDAC3-KO mice had an increased frequency of double-negative (DN) cells and a decreased frequency of CD4 single-positive (CD4SP) thymocytes as compared to WT mice (Fig. 1b). The CD4−CD8+ gate contains both CD8 single-positive (CD8SP) and immature single positive (ISP) thymocytes, which can be distinguished by TCRβ expression (28). Almost all CD8SP thymocytes in HDAC3-cKO mice were TCRβ− ISPs (Fig. 1c). Examining absolute cell numbers, there was a small increase in DN cells, a decrease in DP, and a severe defect in TCRβ+ SP thymocytes compared to WT mice (Fig. 1d).
Figure 1. HDAC3-cKO mice exhibit a block in T cell development.
(A) Quantification of total thymocytes from WT and HDAC3-cKO mice. (B) Examination of CD4 and CD8 expression in WT and HDAC3-cKO thymocytes. (C) Examination of TCRβ expression in WT and HDAC3-cKO CD8SP thymocytes. (D) Quantification of thymocyte numbers in each developmental stage (DN, DP, TCRβ+ CD4SP, and TCRβ+ CD8SP) from WT and HDAC3-cKO mice. (A–D) Data shown are representative of four WT and three HDAC3-cKO mice from at least three independent experiments. Error bars represent SEM. (E) DN thymocyte profiles using c-kit and CD25 from WT and HDAC3-cKO mice, previously gated on lin− (B220, CD11b, Gr-1, Ter119, CD19, CD11c, NK1.1, CD8α, CD4, TCRβ) CD3− thymocytes to examine ETP (c-kit+CD25−), DN2 (c-kit+CD25+), DN3 (c-kit−CD25+), and DN4 (c-kit−CD25−). (F) Quantification of cell numbers of ETP, DN2, DN3 and DN4 arising from FACS analysis in (E). (E–F) Data shown are mean ± SEM from four mice per group from four independent experiments. (G) Representative evaluation of DN3a (FSCloCD27lo) and DN3b (FSChiCD27hi) from 4 WT and 4 HDAC3-cKO mice. (H) Cell cycle analysis in DN3 and DN4 thymocytes from WT and HDAC3-cKO mice, where total thymocytes were incubated with EdU for two hours and subsequently stained for DN markers as in (E) and for DNA content (Vybrant DyeCycle Violet). (I) Quantification of frequency of DN3 and DN4 thymocytes in S-phase as gated in (H) from 5 WT and 5 HDAC3-cKO mice from 3 independent experiments. Error bars represent SEM.
HDAC3-cKO mice exhibited an increase in DN thymocytes, and a decrease in DP thymocytes, as compared to WT mice. Therefore, we analyzed DN thymocytes to determine whether there was a defect in β-selection or proliferation. After excluding lin+CD3+ cells, c-kit and CD25 delineates early thymic progenitors (ETP; c-kit+CD25−), DN2 (c-kit+CD25+), DN3 (c-kit−CD25+), and DN4 (c-kit−CD25−) thymocytes. An increased frequency of DN3 cells and a decreased frequency of DN2 and DN4 cells was observed in HDAC3-cKO mice compared to WT mice (Fig. 1e). However, absolute cell numbers revealed a two-fold increase in only DN3 cells in HDAC3-cKO mice (Fig. 1f). β-selection occurs between DN3a and DN3b stages, with cells increasing CD27 and FSC after β-selection (DN3b) (29). The frequency of DN3b cells was not altered in HDAC3-cKO mice (Fig. 1g), demonstrating that HDAC3 is not required for thymocyte β-selection. HDAC3 is required for DNA replication in hematopoietic progenitor cells (8). To determine whether the loss of HDAC3 is also required for DNA replication after β-selection, we analyzed proliferation by examining EdU incorporation. Cell cycle stages were distinguished using Vybrant DyeCycle Violet (DNA content) and EdU (G0/G1: EdU−Violetlo, S-phase: EdU+, G2/M: EdU−Violethi). Compared to WT thymocytes, HDAC3-deficient thymocytes showed a significant increase in the percentage of DN3 cells in S-phase (Fig. 1h left column, Fig. 1i) but no change in DN4 thymocytes (Fig. 1h right column, Fig. 1i). Increased proliferation of DN3 thymocytes could explain the increase in cell number of DN3 thymocytes in HDAC3-cKO mice. Thus, DN thymocytes maintain the capability to proliferate after β-selection in the absence of HDAC3. In addition, no changes in viability were observed in DN3 and DN4 cells from WT and HDAC3-cKO mice (data not shown). Therefore, HDAC3-cKO mice do not exhibit a block in T cell development at the DN stage.
HDAC3-deficient thymocytes exhibit a block in positive selection
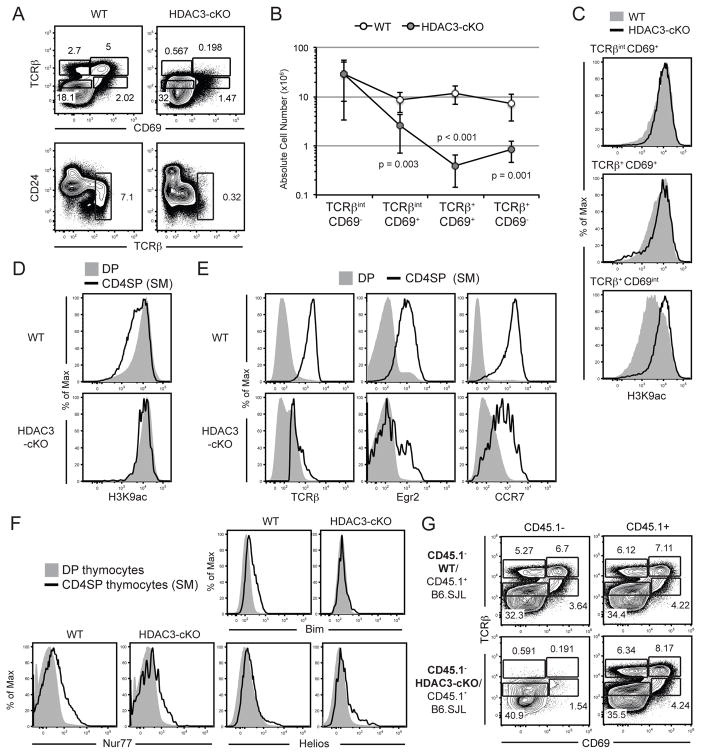
HDAC3-cKO mice showed a dramatic decrease in both the frequency and number of TCRβ+ CD4SP and TCRβ+ CD8SP thymocytes (Fig. 1a, b), suggesting that HDAC3 is required for positive selection. To analyze this, the TCRβ-versus-CD69 profile was examined in these mice. Thymocytes that successfully rearrange their TCRα chain and express TCR on the surface but have not yet received a signal through their TCR are TCRβintCD69− (immature). TCR signaling induces CD69 expression (TCRβintCD69+; selecting) and successful positive selection leads to TCRβup-regulation (TCR β+CD69+; post-positive selection) (30). Maturation of SP thymocytes leads to down-regulation of CD69 in preparation for egress (TCRβ+CD69−; mature). Compared to WT mice, HDAC3-deficient thymocytes exhibited a reduced frequency and cell number of TCRβintCD69+ thymocytes, followed by a severe reduction in the frequency and cell number of post-selection (TCRβ+CD69+) and mature thymocytes (TCRβ+CD69−) (Fig. 2a, b). Similar defects are observed using a CD24-versus-TCRβ profile, where there was a dramatic decrease in the frequency of mature CD24int/loTCRβ+ thymocytes (Fig. 2a). Although CD24 expression is lower in DP thymocytes from HDAC3-cKO mice, it would not contribute to the block in positive selection observed in these mice, as T cell development is normal in CD24-KO mice (31). Therefore, HDAC3 has a unique and critical role during positive selection that is not compensated for by any other co-expressed HDAC family member. A positive selection block was not observed in CD4-cre HDAC3-cKO mice (32), which maybe attributed to HDAC3 protein expression in DP thymocytes (Supplementary Fig. 1b). HDAC3 has a preference for acetylated Histone H3 Lys 9 (H3K9Ac) (33, 34), an epigenetic mark associated with active gene expression. Using an antibody specific for H3K9Ac in WT mice, global H3K9Ac was reduced as cells progressed from positive selection through maturation (Fig. 2c). However, in the absence of HDAC3, this mark remained high (Fig. 2c). Similarly H3K9ac decreases from DP to SP thymocytes in WT mice but was maintained at high levels in HDAC3-deficient SP thymocytes (Fig. 2d). Therefore, in the absence of HDAC3, down-regulation of H3K9Ac during thymocyte development and maturation does not occur. Examination of semi-mature SP thymocytes (TCRβ+CD24+) in HDAC3-cKO mice revealed a failure to up-regulate TCRβ, Egr2, and CCR7 as compared to WT (Fig. 2e). Semi-mature SP thymocytes were examined, as a large fraction of mature SP thymocytes (TCRβ+CD24lo) in HDAC3-cKO mice were recirculating T cells that did not express Rag1-GFP (35) (Supplementary Fig. 2). We also examined whether the lack of SP thymocytes was due to increased negative selection. Enhanced negative selection would result in increased levels of Nur77, Bim, and Helios. However, no increase in Nur77, Bim or Helios was observed in DP and semi-mature CD4SP thymocytes from HDAC3-cKO mice (Fig. 2f). Therefore, HDAC3 is required for thymocyte positive selection.
Figure 2. HDAC3-cKO mice exhibit a block in positive selection.
(A) TCRβ-versus-CD69 profile and CD24-versus-TCRβ profile of WT and HDAC3-cKO thymocytes to examine positive selection. The gating strategy delineates Immature (TCRβintCD69−), Selecting (TCRβintCD69+), Post-positive selection (TCRβ+CD69+) and Mature (TCRβ+CD69−) thymocytes. (B) Quantification of absolute cell numbers of each gate in (A), with error bars reflecting SEM. Data shown are from seven mice per group from six independent experiments. Please note that the data is presented on a logarithmic scale. (C) Examination of H3K9Ac in selecting (TCRβintCD69+), Post-positive selection (TCRβ+CD69+), and Mature (TCRβ+CD69−) thymocytes from WT and CD2-icre HDAC3-cKO mice. Data is representative of five mice per group from four independent experiments. (D) Expression of H3K9ac on DP and semi-mature CD4SP thymocytes (TCRβ+CD24+) from WT and HDAC3-cKO mice. Data is representative of five mice per group from four independent experiments. (E) Expression of TCRβ, Egr2, and CCR7 on DP and semi-mature CD4SP thymocytes (TCRβ+CD24+) from WT and HDAC3-cKO mice. Data is representative of at least four WT and at least four HDAC3-cKO mice from at least two independent experiments. (F) Intracellular expression of Nur77, Bim, and Helios on DP and semi-mature CD4SP thymocytes (TCRβ+CD24+) from WT and HDAC3-cKO mice. Data is representative of at least three mice per group from two independent experiments. (G) TCRβ-versus-CD69 expression of CD45.1− or CD45.1+ cells from 50/50 mixed BMCs from WT (CD45.1−)/B6.SJL (CD45.1+) and HDAC3-cKO (CD45.1−)/B6.SJL (CD45.1+) mice. Mice were analyzed 9–11 weeks post transfer. Plot is representative of five mice per group.
To determine whether the block in positive selection was cell intrinsic, we generated mixed BMCs with a 50:50 combination of CD45.1− WT or CD45.2+ HDAC3-cKO bone marrow cells with CD45.1+ B6.SJL bone marrow cells. The mice were analyzed 9–11 weeks post transfer. Positive selection was normal in WT mice compared to B6.SJL (Figure 2g). However, HDAC3-cKO mice still exhibited a positive selection defect that could not be rescued by with the presence of WT B6.SJL cells (Figure 2g). In addition, ~99.9% of T cells in the periphery from HDAC3-cKO/B6.SJL chimeric mice were CD45.1+, consistent with the severe block in T cell development (data not shown). Therefore, the HDAC3-dependent block in thymocyte positive selection is cell intrinsic.
OT-II TCR transgene does not rescue positive selection HDAC3-cKO mice
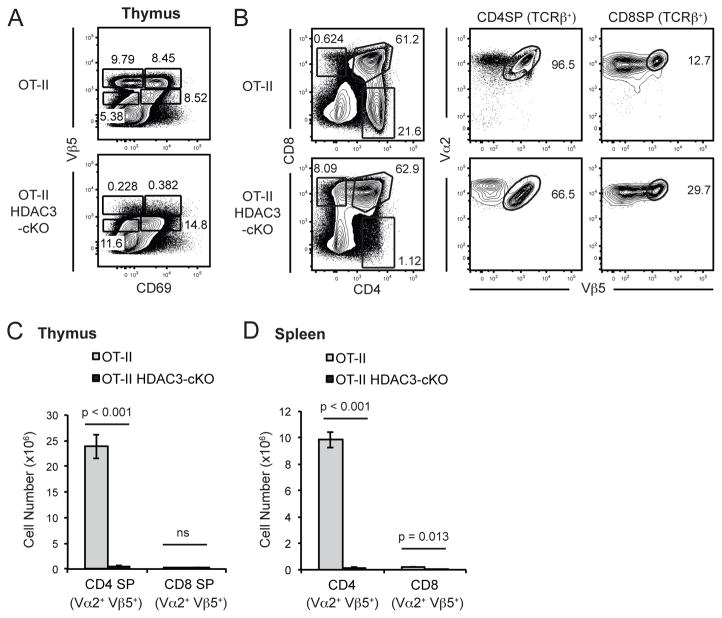
To confirm a defect in positive selection, we generated OT-II HDAC3-cKO mice. The OT-II TCR transgene recognizes OVA presented by I-Ab, leading to the generation of CD4SP but not CD8SP thymocytes (25). In contrast to OT-II thymocytes, OT-II expressing HDAC3-cKO thymocytes failed to successfully navigate positive selection even though CD69 was efficiently induced (Fig. 3a). With OT-II and OT-II HDAC3-cKO mice not being in a Rag-KO background, examination of Vβ5 and Vα2 expression to detect the OT-II transgene in SP thymocytes revealed very few Vβ5hiVα2hi CD4SP thymocytes in OT-II-HDAC3-cKO mice compared to OT-II mice (Fig. 3b, c), while the number of Vβ5hiVα2hi CD8SP thymocytes was equivalent (Fig. 3c). Furthermore, very few peripheral OT-II T cells were present in OT-II-HDAC3-cKO mice (Fig. 3d). Therefore, expression of an OT-II TCR transgene does not rescue positive selection in HDAC3-cKO mice.
Figure 3. OT-II transgene does not rescue positive selection in HDAC3-cKO mice.
(A) Examination of positive selection using Vβ5 and CD69 in OT-II and OT-II-HDAC3-cKO mice. Gating to identify stages of positive selection was performed as shown in Fig. 2a. (B) Examination of CD4 and CD8 surface expression in total thymocytes, and Vα2 and Vβ5 (OT-II) surface expression in TCRβ+ CD4SP and TCRβ+ CD8SP thymocytes from OT-II and OT-II-HDAC3-cKO mice. (C) Quantification of Vα2+ Vβ5+ CD4SP and Vα2+ Vβ5+ CD8SP thymocytes in OT-II and OT-II-HDAC3-cKO mice. (A–C) Data is representative of four mice per group from four independent experiments. (D) Quantification of Vα2+ Vβ5+ CD4 T cells and Vα2+ Vβ5+ CD8 T cells from the spleen of OT-II and OT-II-HDAC3-cKO mice. Data is representative of 3–4 mice per group from three independent experiments.
HDAC3-deficient SP thymocytes have a survival defect due to failure to up-regulate Bcl-2
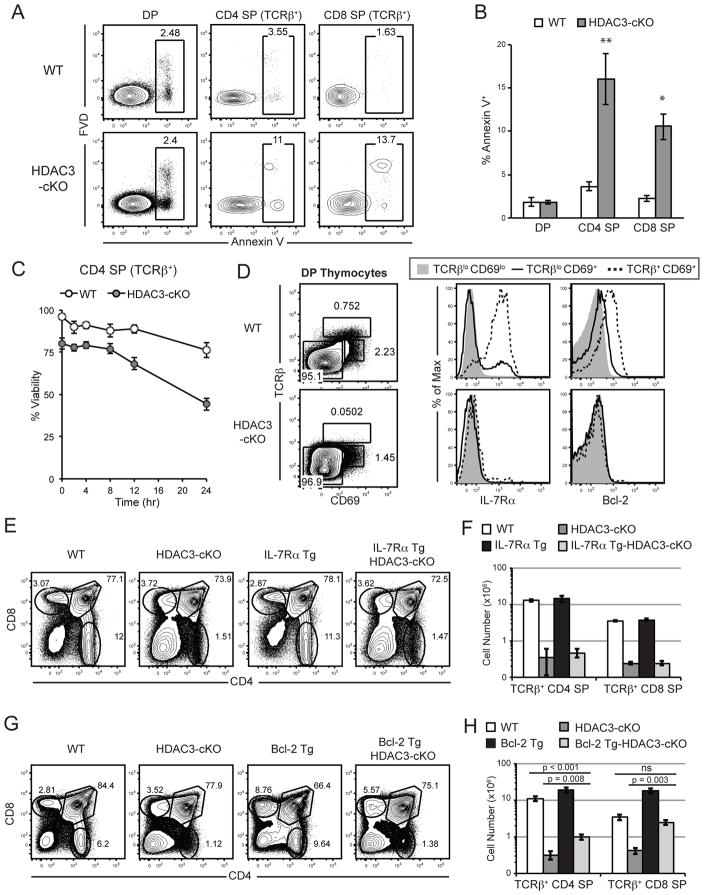
Successful positive selection leads to the induction of Bcl-2 expression and SP thymocyte survival (36). To examine whether apoptosis contributed to the lack of SP thymocytes, cell death was analyzed. CD4SP and CD8SP thymocytes from HDAC3-cKO mice exhibited about a 6-fold increase in Annexin V staining as compared to WT, indicative of enhanced apoptosis (Fig. 4a, b) independent of negative selection (Fig. 2e). The viability of HDAC3-deficient thymocytes was examined ex vivo to determine whether the enhanced cell death was cell intrinsic. After 24 hours in culture without stimulation, 76% of WT TCRβ+ CD4SP thymocytes were viable while only 44% of HDAC3-deficient TCRβ+ CD4SP thymocytes were viable (Fig. 4c). TCRβ+ CD8SP thymocytes from HDAC3-cKO mice exhibited a similar decrease in viability (data not shown). Bcl-2 is critical for SP thymocyte survival and is up-regulated after positive selection (36). IL-7Rα is also expressed after positive selection and is important for homeostasis in the periphery (37). While WT TCRβloCD69+ DP thymocytes undergoing positive selection and TCRβ+CD69+ DP thymocytes that recently completed positive selection up-regulated IL-7Rα and Bcl-2 (Fig. 4d), neither IL-7Rα or Bcl-2 expression were induced in the absence of HDAC3 (Fig. 4d). To determine the relative contribution of IL-7Rα or Bcl-2 expression to the lack of CD4SP and CD8SP thymocytes in HDAC3-cKO mice, we crossed HDAC3-cKO mice to IL-7Rα transgenic (Tg) mice or Bcl-2 Tg mice. Transgenic expression of IL-7Rα did not alter the reduced SP thymocyte frequency (Fig. 4e) or cell number (Fig. 4f) observed in HDAC3-cKO mice. Similarly, HDAC3-deficient thymocytes with the Bcl-2 transgene (Bcl-2 Tg-HDAC3-cKO) did not exhibit restored SP thymocyte development (Fig. 4g). While total SP thymocyte numbers in Bcl-2 Tg-HDAC3-cKO mice were increased as compared to HDAC3-cKO mice (Fig. 4h), they showed a similar fold decrease when compared to Bcl-2 Tg mice. Given that a large fold reduction in SP thymocytes from WT to HDAC3-cKO mice was maintained with introduction of a Bcl-2 transgene (10-fold for Bcl-2 Tg to Bcl-2 Tg-HDAC3-cKO mice), it can be concluded that Bcl-2 expression does not rescue the block in T cell development in HDAC3-cKO mice. Thus, the defects observed in HDAC3-cKO mice are not simply due to altered survival due to the inability to induce expression of IL-7Rα or Bcl-2 during positive selection.
Figure 4. HDAC3-deficient thymocytes exhibit a survival defect due to a failure to express Bcl-2.
(A) Shown is a representative analysis of apoptotic DP, TCRβ+ CD4SP and TCRβ+ CD8SP thymocytes in WT and HDAC3-cKO mice using Annexin V and fixable viability dye (FVD). (B) Quantification of frequency of apoptotic Annexin V+ DP, TCRβ+ CD4SP and TCRβ+ CD8SP thymocytes from WT and HDAC3-cKO mice as depicted in (A). Data shown are the mean ± SEM of five to seven mice per experimental group from three independent experiments. *p < 0.05 and **p < 0.01. (C) Viability of thymocytes in culture over the time period shown. Plot identifies the percentage of live (Annexin V− FVD−) TCRβ+ CD4SP thymocytes. Data shown is the mean viability ± SEM from four to five mice per group from two independent experiments. The two curves are statistically different with p<0.011 using two-way ANOVA. (D) IL-7Rα surface expression and Bcl-2 intracellular expression of pre-positive selection (TCRβloCD69lo), selecting (TCRβlo CD69+), and post-positive selection (TCRβ+CD69+) DP thymocytes. Data is representative of at least ten mice per experimental group. (E) Representative analysis of thymic development in WT, HDAC3-cKO, IL-7Rα Tg, and IL-7Rα Tg-HDAC3-cKO mice is shown. (F) Quantification of the absolute cell numbers of TCRβ+ CD4SP and TCRβ+ CD8SP thymocytes from WT, HDAC3-cKO, IL-7Rα Tg, and IL-7Rα Tg-HDAC3-cKO mice. (E–F) Data shown are the mean ± SEM from three mice per group from at least three independent experiments. Please note that the data is presented on a logarithmic scale. (G) Representative analysis of thymic development in WT, HDAC3-cKO, Bcl-2 Tg, and Bcl-2 Tg-HDAC3-cKO thymocytes is shown. (H) Quantification of the absolute cell numbers of TCRβ+ CD4SP and TCRβ+ CD8SP thymocytes from WT, HDAC3-cKO, Bcl-2 Tg, and Bcl-2 Tg-HDAC3-cKO mice. (G–H) Data shown are the mean ± SEM from five to seven mice per group from at least five independent experiments. Please note that the data is presented on a logarithmic scale.
HDAC3-deficient thymocytes fail to down-regulate RORγt during positive selection
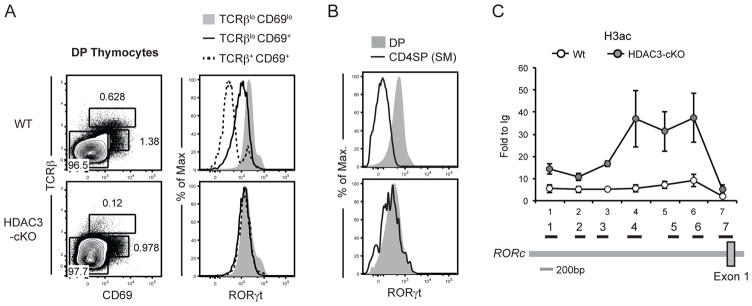
Consistent with HDAC3’s role in extinguishing rather than inducing gene expression, our focus shifted to genes required to be down-regulated during positive selection. Expression of the orphan nuclear receptor RORγt is high in DP thymocytes and then decreases upon positive selection (38). Constitutive expression of RORγt using a transgene (RORγt-Tg) causes thymocyte development to be arrested at the DP stage due to a block in positive selection, with decreased thymic cellularity and lower surface TCRβ expression on the few SP thymocytes that develop (39). This phenocopy between RORγt-Tg mice and HDAC3-cKO mice led us to examine whether RORγt expression was altered in HDAC3-cKO mice. As expected, RORγt was down-regulated in selecting (TCRβloCD69+) and post-selection (TCRβ+CD69+) DP thymocytes in WT mice (Fig. 5a). In contrast, HDAC3-deficient selecting (TCRβloCD69+) and post-selection (TCRβ+CD69+) DP thymocytes maintained high levels of RORγt expression (Fig. 5a). This high level of RORγt was also maintained at the semi-mature SP stage (Fig. 5b). We examined whether histone acetylation was enhanced at the RORC promoter using a series of previously published primers for chromatin immunoprecipitation (ChIP) (26). H3 acetylation was significantly increased (p = 0.034–0.049) at three different sites upstream of exon 1, although histone acetylation at sites that overlap with exon 1 was similar and only minor differences were observed at sites further upstream (Fig. 5c). As RORγt expression is primarily regulated at the transcriptional level at this stage (39), increased acetylation at the RORC promoter in HDAC3-cKO mice demonstrates that deacetylation of its promoter and thus the transcriptional down-regulation of RORγt during positive selection depends on HDAC3, which is not compensated by any other HDAC co-expressed during T cell development.
Figure 5. HDAC3-deficient thymocytes fail to down-regulate of RORγt during thymocyte positive selection.
(A) Intracellular expression of RORγt of pre-positive selection (TCRβloCD69lo), selecting (TCRβlo CD69+), and post-positive selection (TCRβ+CD69+) DP thymocytes. Plots are representative of at least ten mice per experimental group. (B) Intracellular expression of RORγt of DP and semi-mature CD4SP thymocytes (TCRβ+CD24+). Data is representative of at least ten mice per experimental group. (C) ChIP analysis of histone 3-acetylation (H3ac) at the RORC promoter of total thymocytes from WT and HDAC-cKO mice (numbers along x-axis correspond to primer sets in cartoon underneath plot; adapted from(26)). Results are presented as the fold-increase as compared to control ChIP with Ig alone [fold to Ig = (% input of H3ac)/(% input of Ig control)]. Data shown are the mean ± SEM of four mice per experimental group from three independent experiments.
Deletion of RORγt restores thymic cellularity and CD8SP cell number but fail to generate CD4SP thymocytes
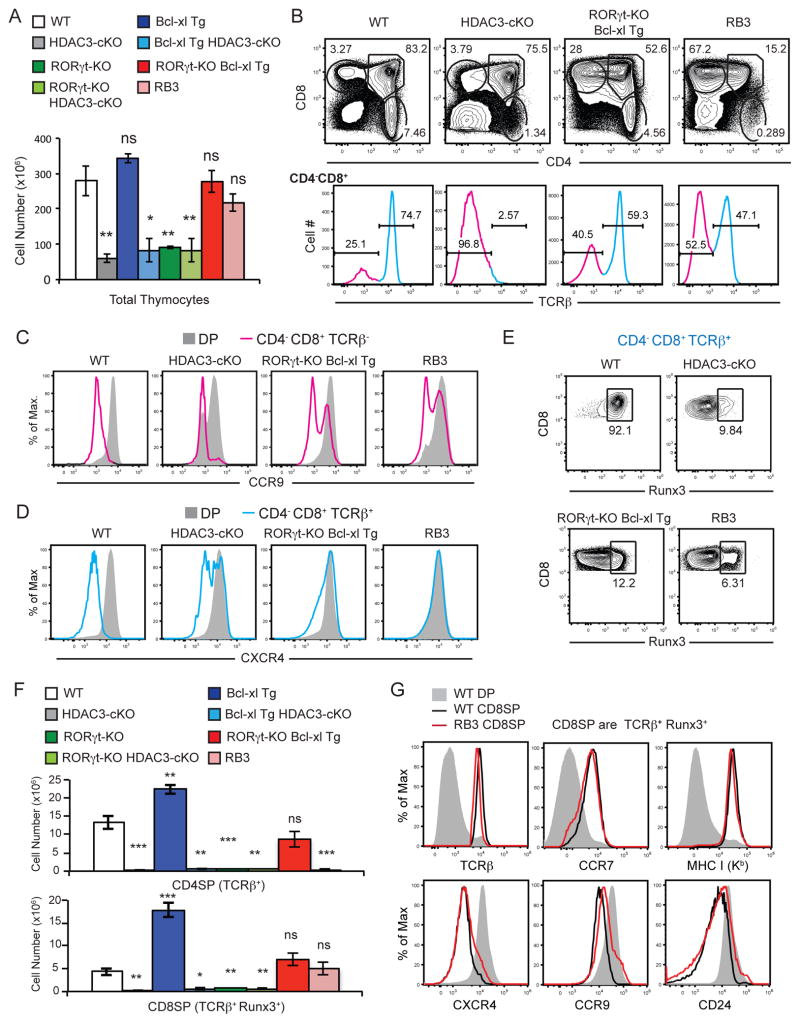
To test whether the failure to down-regulate RORγt in HDAC3-cKO mice is responsible for the block in positive selection, we interbred HDAC3-cKO mice with RORγt-KO mice to correct for the failure to down-regulate RORγt upon positive selection. However, RORγt-KO mice also have a block in T cell development, as RORγt expression is required for DP thymocyte survival through regulating Bcl-xl expression (40, 41). Transgenic expression of Bcl-xl rescues the DP thymocyte survival defect and consequently the block in T cell development in RORγt-KO mice (41). Therefore, we generated RORγt-KO Bcl-xl Tg HDAC3-cKO mice (hereafter referred to as “RB3” mice) to determine whether the deletion of RORγt (in conjunction with constitutive Bcl-xl expression) would restore positive selection in HDAC3-cKO mice. Examination of RB3 mice revealed that thymic cellularity was restored to WT and control RORγt Bcl-xl Tg levels (Fig. 6a), while interbreeding Bcl-xl Tg or RORγt-KO separately to HDAC3-cKO mice did not restore thymic development (Supplementary Fig. 3a) or thymic cellularity (Fig. 6a). In fact, HDAC3-cKO combined with RORγt-deficiency exacerbated the defect in thymic development as compared to each single KO alone, leading to a block at the ISP stage (Supplementary Fig. 3a). However, while thymic cellularity in RB3 mice was similar to WT and control RORγt-KO Bcl-xl Tg mice (Fig. 6a), the CD4-versus-CD8 profile was aberrant (Fig. 6b). Both RORγt-KO Bcl-xl Tg and RB3 mice exhibited a reduced frequency of DP thymocytes and enhanced frequency of CD4−CD8+ cells compared to WT and HDAC3-cKO mice (Fig. 6b). This pattern suggests that the CD4−CD8+ gate includes “DP” thymocytes that lack CD4. Hence, we utilized the Immunological Genome Project Consortium (www.immgen.org) to pinpoint surface markers that distinguish DP thymocytes from ISPs. We identified CCR9 as a suitable target, where ISPs express CCR9 at a much lower level than DP thymocytes, and confirmed this differential expression in WT mice by flow cytometry (Fig. 6c). Using DP expression of CCR9 as a positive control, the TCRβ− CD4−CD8+ population in RORγt-KO Bcl-xl Tg and RB3 mice was bimodal, containing both CCR9− and CCR9+ populations (Fig. 6c). This demonstrates that CCR9+ TCRβ− CD4−CD8+ thymocytes likely are CD4− “DP” thymocytes. CD4−CD8+ thymocytes that express TCRβ also contained a “DP” signature in RORγt-KO Bcl-xl Tg and RB3 mice because majority of TCRβ+ CD4−CD8+ thymocytes in these mice express CXCR4, characteristic of DP thymocytes, opposed to positively selected CD8SP thymocytes that don’t express CXCR4 (Fig. 6d). To identify positively selected CD8SP thymocytes within the TCRβ+ CD4−CD8+ cells, we examined Runx3 expression because this transcription factor is required for CD8-lineage differentiation after positive selection (42). While all WT CD8SP thymocytes were Runx3+, most TCRβ+ CD4−CD8+ cells were Runx3− in RORγt-KO Bcl-xl Tg and RB3 mice (Fig. 6e), consistent with this population being CD4− DP thymocytes. Interestingly, this population is also present in HDAC3-cKO mice (Fig. 6d, e), Bcl-xl Tg HDAC3-cKO, and RORγt-KO HDAC3-cKO mice (Supplemental Fig. 3). As a result, we used Runx3 expression to quantify positively selected CD8SP thymocytes, along with TCRβ+ CD4SP thymocytes, to determine whether deletion of RORγt (in conjunction with Bcl-xl expression) rescued positive selection in HDAC3-cKO mice. CD8SP (TCRβ+, Runx3+) cell number was restored to WT levels in RB3 mice, however there was no rescue of CD8SP thymocytes in Bcl-xl-Tg HDAC3-cKO or RORγt-KO Bcl-xl Tg mice (Fig. 6f). In addition, we performed a phenotypic analysis on CD8SP thymocytes (TCRβ+, Runx3+) from RB3 mice to determine if their expression profile was similar to WT CD8SP thymocytes. As WT DP thymocytes transition to the SP stage, TCRβ, CCR7 and MHC class I were upregulation, and CXCR4, CCR9, and CD24 were downregulated (Fig. 6g). These markers were expressed similarly on CD8SP thymocytes between WT and RB3 mice (Fig. 6g), demonstrating that CD8SP thymocytes from RB3 mice underwent an equivalent change in gene expression compared to normal CD8SP cells. Therefore, generation of CD8SP thymocytes demonstrates that deletion of RORγt rescues positive selection in HDAC3-cKO mice, but this rescue was not observed in CD4SP thymocytes.
Figure 6. Deletion of RORγt in HDAC3-deficient thymocytes restores thymic cellularity and positive selection of CD8SP thymocytes.
(A) Thymic cellularity of WT, HDAC3-cKO, Bcl-xl Tg, Bcl-xl Tg HDAC3-cKO, RORγt-KO, RORγt-KO HDAC3-cKO, RORγt-KO Bcl-xl Tg, and RB3 (aka RORγt-KO Bcl-xl Tg HDAC3-cKO) mice. Data shown are the mean ± SEM of 4–5 mice per experimental group from four independent experiments. (B) CD4-versus-CD8 profile and TCRβ surface expression of CD4−CD8+ thymocytes from WT, HDAC3-cKO, RORγt-KO-Bcl-xl Tg, and RB3 mice. Data is representative of 4–5 mice per experimental group from four independent experiments. (C) Surface expression of CCR9 on DP and CD4−CD8+ TCRβ− thymocytes from WT, HDAC3-cKO, RORγt-KO, and RB3 mice. Data is representative of at least three mice per group from three independent experiments. (D) Surface expression CXCR4 on DP and CD4−CD8+ TCRβ+ thymocytes from WT, HDAC3-cKO, RORγt-KO, and RB3 mice. Data is representative of at least three mice per group from three independent experiments. (E) Intracellular expression of Runx3 on CD4−CD8+ TCRβ+ thymocytes from WT, HDAC3-cKO, RORγt-KO, and RB3 mice. Data is representative of at least three mice per group from three independent experiments. (F) Quantification of CD4SP (TCRβ+) and CD8SP thymocytes (TCRβ+, Runx3+) from WT, HDAC3-cKO, Bcl-xl Tg, Bcl-xl Tg HDAC3-cKO, RORγt-KO, RORγt-KO HDAC3-cKO, RORγt-KO Bcl-xl Tg, and RB3 mice. Data shown are the mean ± SEM of 4–5 mice per experimental group from four independent experiments. (G) Surface expression of TCRβ, CCR7, MHC class I (Kb), CXCR4, CCR9, and CD24 on DP and CD8SP (TCRβ+, Runx3+) thymocytes from WT mice and CD8SP (TCRβ+, Runx3+) thymocytes from RB3 mice. Data is representation of at least two mice per group.
The profound alteration in the CD8SP to CD4SP ratio indicates that HDAC3 may also play an important role the development of CD4SP thymocytes. To investigate this, expression levels of transcription factors required for CD4-lineage development were examined. We examined Gata3, Tox, and ThPOK expression in TCRβintCD69+ (Selecting) and TCRβ+CD69+ (Post-Selection) populations, as these transcription factors are required for CD4SP development during these stages. Gata3 and Tox are up-regulated after DP thymocytes receive a TCR signal (43, 44), and WT thymocytes showed high levels of Gata3 and Tox within the TCRβintCD69+ population (Fig. 7). However, examination of RB3 mice revealed decreased expression of Gata3 and Tox in TCRβintCD69+ thymocytes (Fig. 7). ThPOK is the master transcription factor for CD4SP thymocyte differentiation (45, 46), and expression initiates in WT TCRβintCD69+ thymocytes (Fig. 7). Gata3 and Tox are required to induce ThPOK expression for CD4SP development (44, 47). Consequently, ThPOK was very low in TCRβintCD69+ and TCRβ+CD69+ thymocytes RB3 mice compared to WT mice (Fig. 6g), reflective of the few number of CD4SP thymocytes in RB3 mice. Therefore, transcription factors required for CD4SP development are decreased in RB3 mice.
Figure 7. Failure to upregulate Gata3, Tox, and ThPOK in RB3 mice.
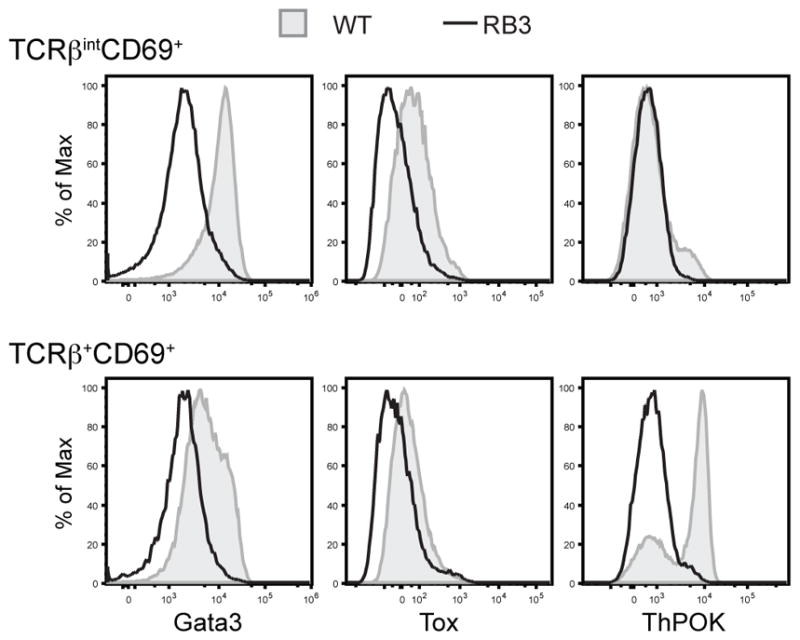
Intracellular expression of Gata3, Tox, and ThPOK in TCRβintCD69+ and TCRβ+CD69+ thymocytes from WT and RB3 mice. Data is representative of three mice per experimental group from three independent experiments.
HDAC3-cKO mice show increased frequency of RORγt+ Th17 cells and develop IBD
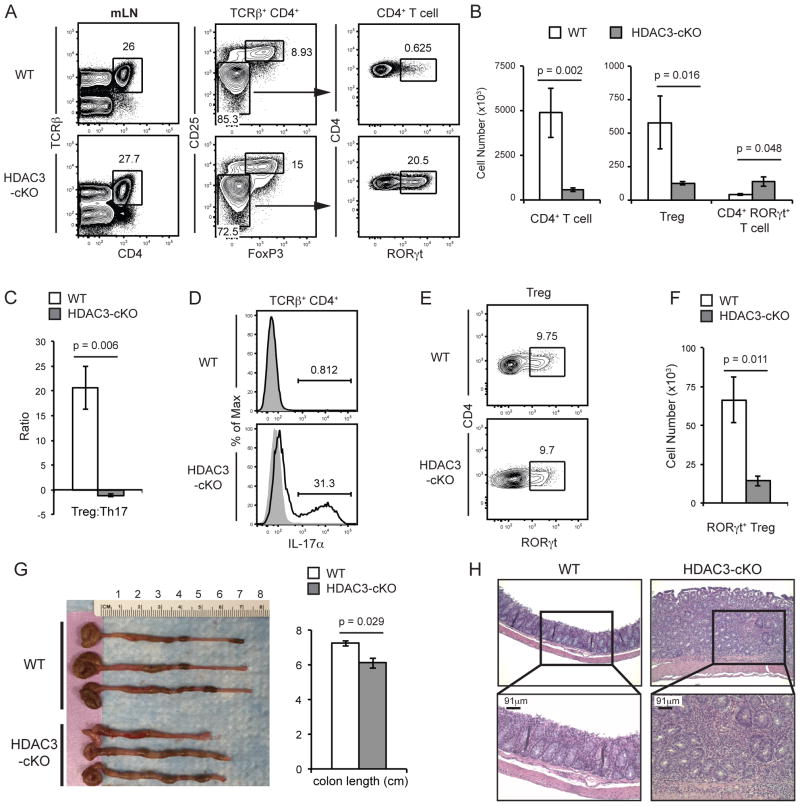
In the course of our studies, HDAC3-cKO mice developed rectal prolapse as early as 8 weeks of age. Since HDAC3-deficient DP thymocytes undergoing positive selection failed to down-regulate RORγt (Fig. 5a), the few T cells produced in these mice might skew towards the RORγt+ Th17 lineage and lead to the development of IBD. Analysis of spleens in HDAC3-cKO mice revealed very few T cells that displayed primarily a memory phenotype compared to WT mice (Supplementary Fig. 4a, b), indicative of homeostatic proliferation. Flow cytometry confirms that the majority of peripheral T cells did not express HDAC3 in HDAC3-cKO mice and are not cells that escaped deletion. Consistent with CD4-cre HDAC3-cKO mice (9), naïve T cells in CD2-icre HDAC3-cKO mice had decreased CD55 expression (Supplementary Fig. 4d), reflective of a block in peripheral T cell maturation. Analysis of mesenteric lymph nodes (mLNs) also revealed an overall decrease in T cell cellularity, but the proportion of CD4+ T cells in the mLN compared to WT mice was similar (Fig. 8a, b) as HDAC3 is also required for B cell development (R.P. and V.S.S unpublished results). Although FoxP3+ Tregs were present, there was a substantial increase in the frequency of RORγt+ Th17 cells within the Foxp3− conventional CD4+ T cell pool (Fig. 8a). A similar increase in RORγt+ Th17 cells was also present in 3-week old mice (data not shown). Comparing absolute numbers of mesenteric FoxP3+ Tregs and RORγt+ Th17 cells, Tregs vastly outnumbered RORγt+ Th17 cells in WT mice, while there were slightly more RORγt+ Th17 cells than Tregs in HDAC3-cKO mice (Fig. 8c). These RORγt+ CD4+ T cells were functional Th17 cells, as they produced IL-17 upon stimulation in culture (Fig. 8d). An enhanced frequency of RORγt+ Th17 cells was also present in the spleen of HDAC3-cKO mice, but analysis of cellularity revealed equivalent cell numbers between WT and HDAC3-cKO mice (Supplementary Fig. 4e, f). While HDAC3-cKO mice show enhanced Th17 differentiation in mLNs, there were equivalent frequencies of RORγt+ Tregs (Fig. 8g). RORγt+ Tregs have been shown to be important in constraining immunoinflammatory responses in the gut (48), therefore an overall decrease in RORγt+ Tregs cell number (Fig. 8g) along with a decrease in total Tregs and an increase in Th17 cell number (altered Treg/Th17 ratio) is consistent with the development of IBD. Examination of colon length revealed that HDAC3-cKO had a significantly shorter colon compared to WT mice (Fig. 8h). In addition, H&E staining of the colon revealed thickening and remodeling of the lamina propria as well as immune cell infiltration (Fig. 8h). Reduced colon length and abnormal gut tissue both indicate IBD, therefore, the absence of HDAC3 leads to the generation of a high proportion of RORγt+ Th17 cells in the periphery and the development of IBD.
Figure 8. HDAC3-cKO mice develop IBD.
(A) Frequency and (B) absolute cell numbers of CD4+ T cells (TCRβ+, CD4+), Tregs (TCRβ+, CD4+, CD25+, FoxP3+), and RORγt+ Th17 cells in mesenteric lymph nodes (mLN) from WT and HDAC3-cKO mice. Data shown are the mean ± SEM of six to seven mice per group from three independent experiments. (C) Ratio of Treg to RORγt+ Th17 cells in mLNs from WT and HDAC3-cKO mice. Data shown are the mean ± SEM of six to seven mice per group from three independent experiments. (D) Frequency of IL-17α+ mesenteric CD4+ T cells stimulated with PMA/Ionomycin from WT and HDAC3-cKO mice. Data is representative of four mice per group from two independent experiments. (E) Frequency and (F) absolute cell numbers of RORγt+ Tregs cells in mLN from WT and HDAC3-cKO mice. Data shown are the mean ± SEM of seven to eight mice per group from four independent experiments. (G) Image and quantification of colon length from WT and HDAC3-cKO mice. (H) H&E stained sections of colon from WT and HDAC3-cKO mice. Scale bar reflects 91μm acquired at 20x magnification. Data is representative of two WT and three HDAC3-cKO mice.
DISCUSSION
Lymphocyte development is tightly regulated, requiring successful transit of cells through several developmental stages and checkpoints prior to their release into the circulation. Each checkpoint involves induction or repression of a particular subset of genes, disruption of which causes developmental arrest. We investigated the role of histone deacetylase-3 (HDAC3) in T cell development using CD2-icre conditional knockout (HDAC3-cKO) mice. Although T cells co-express several HDAC family members during development (1, 49, 50), HDAC3 has a unique role as conditional deletion of HDAC3 led to a block in T cell development at the DP stage due to an inability to undergo positive selection. The block in T cell development could not be rescued by an OT-II TCR transgene. Successful positive selection requires down-regulation of RORγt, as mice with constitutive expression of RORγt have a similar block in T cell development at positive selection (39). In HDAC3-cKO mice, RORγt was not down-regulated upon TCR stimulation at the DP stage, demonstrating that the block in positive selection may be due to an inability to down-regulate RORγt. Consistent with this, we observed enhanced RORC promoter acetylation in the absence of HDAC3, indicating that HDAC3 may directly deacetylate histones at the RORC promoter to inhibit expression. RORγt also controls expression of Bcl-xl at the DP stage to regulate DP survival (40, 41). Therefore, to determine whether sustained expression of RORγt was responsible for the block in positive selection, RORγt-KO Bcl-xl Tg HDAC3-cKO (RB3) mice were generated, which largely restored positive selection, leading to restoration of CD8SP thymocyte (TCRβ+, Runx3+) numbers. However, CD4SP thymocytes were not generated, indicating a potential role for HDAC3 in lineage choice during T cell development. Therefore, HDAC3 is required at the DP stage for down-regulation of RORγt during positive selection.
A recent study by Stengel et al also found that HDAC3 was required for thymocyte positive selection (51). However various aspects are different from our work, including that the block in positive selection was due to a defect in gene regulation that could be bypassed by a strong TCR signaling with an OT-II TCR transgene (51). Differences observed between both studies may stem from the mouse models utilized, where Stengal et al used an Lck-cre for HDAC3 elimination compared to CD2-icre in our work. Lck-cre deletes as late as in the DN4 stage (52). While an LSL-GFP reporter was used to indicate Cre-induced deletion, the timing of the loss of HDAC3 protein expression was not examined. We demonstrate that HDAC3 protein was almost entirely eliminated by the DN3 stage in CD2-icre HDAC3-cKO mice and is absent in subsequent stages. Therefore, this could explain why CD2-icre HDAC3-cKO mice revealed a more dramatic block in thymocyte positive selection than Lck-cre HDAC3-cKO mice. In addition, as TCR transgenes are typically expressed early, at the DN stage, the rescue of T cell development in OT-II Lck-cre HDAC3-cKO mice may reflect positive selection prior to Cre deletion and loss of HDAC3 expression. Our results clearly demonstrate that the block in positive selection cannot be rescued by OT-II transgene in the absence of HDAC3 protein.
Stengal et al utilized microarray and RNA-seq to determine how gene regulation is disrupted in Lck-cre HDAC3-cKO mice and identified hundreds of genes that are differentially expressed. Based on differential gene expression, they hypothesize that TCR signaling is altered, leading to changes in positive selection. However, we demonstrate that the key gene downstream of HDAC3 required for positive selection is RORγt, as protein levels were not reduced with positive selection and CD8SP thymocyte development was restored to WT levels in RB3 mice. RORC mRNA levels were also increased in DP thymocytes from Lck-cre HDAC3-cKO mice examined by Stengel et al (51). As few CD4SP thymocytes developed in RB3 mice, HDAC3 might play an additional role in thymocyte development during CD4-lineage choice. This decision is initiated after DP thymocytes receive a positive selection signal. After positive selection, thymocytes down-regulate CD8 to become intermediate (CD4+CD8lo) thymocytes that assess TCR signal persistence, as CD4 stabilizes the TCR interaction with MHC class II-restricted thymocytes and become CD4SP thymocytes (reviewed in Singer et al (16)). This process is dependent on expression of Gata3, Tox, and ThPOK, as knockout models targeting each of these transcription factors leads to very few CD4SP thymocytes (44–47). With MHC I-restricted thymocytes, the down-regulation of CD8 in intermediate thymocytes destabilizes the TCR-MHC interaction, leading to termination of the TCR signal. IL-7R signaling then induces expression of Runx3 to suppress ThPOK and CD4 expression to become CD8SP thymocytes (53). CD69+ thymocytes in RB3 mice that had recently received a TCR signal showed decreased levels of Gata3, Tox, and ThPOK, demonstrating that the transcriptional program for the CD4-lineage was not induced. One possibility is that the lack of CD4 expression on “DP” cells in RB3 mice alters this test of MHC restriction. The absence of CD4 expression in “DP” thymocytes in RB3 as well as in RORγt-KO Bcl-xl Tg and HDAC3-cKO mice suggests that HDAC3 may work in concert with RORγt to maintain CD4 expression in DP thymocytes since the effect seen in HDAC3-cKO mice and RORγt-KO Bcl-xl Tg mice is augmented in RB3 mice, which is under investigation.
Current and previous work demonstrates HDAC3 to be required for multiple stages in T cell development, including positive selection, peripheral T cell maturation and the generation or regulatory T cells (9, 11, 51). Utilizing different mouse models that delete HDAC3 at different stages in development (CD2-icre versus CD4-cre, Supplemental Figure 1), we have shown HDAC3 to be required for thymocyte positive selection and peripheral T cell maturation (9). Positive selection and T cell maturation (both thymic maturation and post-thymic maturation) are distinct but equally important processes required to generate the peripheral naïve T cell pool (reviewed by Hogquist et al (18)). As shown by Hsu et al (9), many markers associated with thymic maturation were not altered in SP thymocytes from CD4-cre HDAC3-cKO mice, as measured by changes in CD69, CD62L, CD24, CCR4, CCR7, and CCR9 in Rag1-GFP+ SP thymocytes. Thymic egress, examined by Rag1-GFP and S1P1 expression, was also not altered in CD4-cre HDAC3-cKO mice (9). However, in the periphery, there were very few naïve CD4 and CD8 T cells, and the majority of naïve T cells present were Rag1-GFP+ recent thymic emigrants (RTEs). These HDAC3-deficient RTEs were functionally immature and were targeted for elimination by complement in the periphery (9). Thus, HDAC3 is additionally required for post-thymic T cell maturation as well as thymocyte positive selection. The peripheral T cell maturation defect was also observed in naïve T cells from CD2-icre HDAC3-cKO mice (Supplemental Figure 4), as demonstrated by low expression of CD45RB and CD55. RB3 mice, although positive selection of CD8 SP thymocytes is restored, also had a severe reduction in the number of splenic naïve CD8 T cells (Supplemental Figure 4G), consistent with a block in peripheral T cell maturation. As positive selection occurred normally in CD4-cre HDAC3-cKO mice (9) and RORγt was appropriately developmentally downregulated during positive selection (data not shown), peripheral T cell maturation in CD4-cre HDAC3-cKO mice is a process independent of altered RORγt expression. Therefore, RORγt deficiency/Bcl-xl overexpression which restores positive selection and the generation of CD8 SP thymocytes in CD2-icre HDAC3 cKO mice, would not be expected to rescue the defect in peripheral T cell maturation in the absence of HDAC3. Therefore, HDAC3 is required to downregulate RORγt during positive selection, but RB3 mice have few peripheral naïve T cells consistent with the previously characterized block in peripheral T cell maturation (9).
While there is redundancy and overlap between HDAC family members in regulating histone deacetylation, H3K9Ac deaceylation was dependent on HDAC3 in this model. H3K9 modifications are associated with activation and repression, with H3K9Ac associated with gene activation (54) and di- or tri-methylation of H3K9 (H3K9me2 or H3K9me3) associated with repression leading to condensed and transcriptionally inactive heterochromatin (55). H3K9 monomethylation (H3K9me1) is enriched at actively expressed genes indicating it is not a repressive mark (55). Using flow cytometry, global H3K9Ac was analyzed during T cell development. H3K9Ac decreases as thymocytes progress through positive selection to become mature T cells. However, in the absence of HDAC3, down-regulation of H3K9Ac remained high, demonstrating that the deacetylation of these targets is dependent on HDAC3. The identification of these targets, and the role they have in regulating T cell development, lineage commitment and maturation remain to be determined.
HDAC3-cKO mice developed rectal prolapse/inflammatory bowel disease starting at 2 months of age. Examination of T cell populations demonstrated that there were few peripheral Treg cells in these mice, consistent with the recent work of Wang et al determining that HDAC3 is required for iTreg development and function in CD4-cre HDAC3-cKO mice (11). However, a substantial number and proportion of conventional T cells in CD2-icre HDAC3-cKO mice were Th17 T cells that expressed RORγt and produced IL-17 upon stimulation. As a result, the altered Treg to Th17 ratio likely contributes to inflammatory bowel disease development. In addition, young mice (3-week old) also showed an increased frequency of RORγt+ Th17 cells before any signs of IBD appear, suggesting that this enhancement is not due to impaired Treg function but a failure to regulate RORγt expression by HDAC3. However, the frequency of RORγt+ Tregs in HDAC3-cKO mice was similar to WT levels, suggesting that additional mechanisms are required for regulating RORγt expression in Tregs. Therefore, HDAC3 is required for regulating RORγt expression in the thymus and periphery.
Currently HDAC inhibitors are in clinical trials for a variety of cancers. However, our study introduces the possibility of an unwanted side effect where inhibitors of HDAC3 would be predicted to block lymphocyte output, thus decreasing entry of new naïve T cells into the peripheral T cell pool. As tumor reactive T cells are often tolerized, cancer immunotherapy depends on a diverse naïve T cell pool for efficacy. Thus, therapies that target HDAC3 which interfere with the ability of the immune system to replenish the T cell pool with naive tumor reactive lymphocytes may not be optimal.
Supplementary Material
Acknowledgments
This work was supported by funds provided by NIH (R01 AI083279) as well as internal Mayo Clinic funds (to V.S.S.) and Mayo Graduate School (to R.P.).
We thank Dr. Al Singer for IL-7Rα tg mice, Dr. Nubuo Sakaguchi for Rag1-GFP mice, and Dr. Scott Hiebert for HDAC3 floxed mice. We thank Dr. Michael Shapiro, Dr. Fan-Chi Hsu, Puspa Thapa, and Barsha Dash for thoughtful discussions and critical reading of the manuscript.
Abbreviation
- Tg
transgenic
- cKO
conditional knockout
- HDAC
histone deacetylase
- SP
single positive
- DP
double positive
- DN
double negative
- FVD
fixable viability dye
- mLN
mesenteric lymph node
- IBD
irritable bowel syndrome
- RB3
RORγt-KO Bcl-xl-transgenic CD2-icre HDAC3-cKO
- ChIP
chromatin immunoprecipitation
- BMC
bone marrow chimera
- RTE
recent thymic emigrants
Footnotes
AUTHOR CONTRIBUTIONS
R.P. performed the experiments, M.C., D.M., P.B., and M.C. managed and genotyped the mouse colony, and R.P. and V.S. designed the experiments and wrote the paper.
The authors declare that they have no conflict of interest.
References
- 1.Shakespear MR, Halili MA, Irvine KM, Fairlie DP, Sweet MJ. Histone deacetylases as regulators of inflammation and immunity. Trends Immunol. 2011;32:335–343. doi: 10.1016/j.it.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Takami Y, Nakayama T. N-terminal region, C-terminal region, nuclear export signal, and deacetylation activity of histone deacetylase-3 are essential for the viability of the DT40 chicken B cell line. J Biol Chem. 2000;275:16191–16201. doi: 10.1074/jbc.M908066199. [DOI] [PubMed] [Google Scholar]
- 3.Wen YD, Perissi V, Staszewski LM, Yang WM, Krones A, Glass CK, Rosenfeld MG, Seto E. The histone deacetylase-3 complex contains nuclear receptor corepressors. Proc Natl Acad Sci U S A. 2000;97:7202–7207. doi: 10.1073/pnas.97.13.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li J, Wang J, Wang J, Nawaz Z, Liu JM, Qin J, Wong J. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 2000;19:4342–4350. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guenther MG, Lane WS, Fischle W, Verdin E, Lazar MA, Shiekhattar R. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 2000;14:1048–1057. [PMC free article] [PubMed] [Google Scholar]
- 6.Karagianni P, Wong J. HDAC3: taking the SMRT-N-CoRrect road to repression. Oncogene. 2007;26:5439–5449. doi: 10.1038/sj.onc.1210612. [DOI] [PubMed] [Google Scholar]
- 7.Montgomery RL, Potthoff MJ, Haberland M, Qi X, Matsuzaki S, Humphries KM, Richardson JA, Bassel-Duby R, Olson EN. Maintenance of cardiac energy metabolism by histone deacetylase 3 in mice. J Clin Invest. 2008;118:3588–3597. doi: 10.1172/JCI35847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Summers AR, Fischer MA, Stengel KR, Zhao Y, Kaiser JF, Wells CE, Hunt A, Bhaskara S, Luzwick JW, Sampathi S, Chen X, Thompson MA, Cortez D, Hiebert SW. HDAC3 is essential for DNA replication in hematopoietic progenitor cells. J Clin Invest. 2013;123:3112–3123. doi: 10.1172/JCI60806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsu FC, Belmonte PJ, Constans MM, Chen MW, McWilliams DC, Hiebert SW, Shapiro VS. Histone Deacetylase 3 Is Required for T Cell Maturation. J Immunol. 2015;195:1578–1590. doi: 10.4049/jimmunol.1500435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thapa P, Das J, McWilliams D, Shapiro M, Sundsbak R, Nelson-Holte M, Tangen S, Anderson J, Desiderio S, Hiebert S, Sant’angelo DB, Shapiro VS. The transcriptional repressor NKAP is required for the development of iNKT cells. Nat Commun. 2013;4:1582. doi: 10.1038/ncomms2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L, Liu Y, Han R, Beier UH, Bhatti TR, Akimova T, Greene MI, Hiebert SW, Hancock WW. FOXP3+ regulatory T cell development and function require histone/protein deacetylase 3. J Clin Invest. 2015;125:1111–1123. doi: 10.1172/JCI77088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rothenberg EV. Transcriptional control of early T and B cell developmental choices. Annu Rev Immunol. 2014;32:283–321. doi: 10.1146/annurev-immunol-032712-100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koch U, Radtke F. Mechanisms of T cell development and transformation. Annu Rev Cell Dev Biol. 2011;27:539–562. doi: 10.1146/annurev-cellbio-092910-154008. [DOI] [PubMed] [Google Scholar]
- 14.Klein L, Kyewski B, Allen PM, Hogquist KA. Positive and negative selection of the T cell repertoire: what thymocytes see (and don’t see) Nat Rev Immunol. 2014;14:377–391. doi: 10.1038/nri3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Surh CD, Sprent J. T-cell apoptosis detected in situ during positive and negative selection in the thymus. Nature. 1994;372:100–103. doi: 10.1038/372100a0. [DOI] [PubMed] [Google Scholar]
- 16.Singer A, Adoro S, Park JH. Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice. Nat Rev Immunol. 2008;8:788–801. doi: 10.1038/nri2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu X, Zhang S, Li P, Lu J, Xuan Q, Ge Q. Maturation and emigration of single-positive thymocytes. Clin Dev Immunol. 2013;2013:282870. doi: 10.1155/2013/282870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hogquist KA, Xing Y, Hsu FC, Shapiro VS. T Cell Adolescence: Maturation Events Beyond Positive Selection. J Immunol. 2015;195:1351–1357. doi: 10.4049/jimmunol.1501050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knutson SK, Chyla BJ, Amann JM, Bhaskara S, Huppert SS, Hiebert SW. Liver-specific deletion of histone deacetylase 3 disrupts metabolic transcriptional networks. EMBO J. 2008;27:1017–1028. doi: 10.1038/emboj.2008.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu Q, Erman B, Park JH, Feigenbaum L, Singer A. IL-7 receptor signals inhibit expression of transcription factors TCF-1, LEF-1, and RORgammat: impact on thymocyte development. J Exp Med. 2004;200:797–803. doi: 10.1084/jem.20032183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sentman CL, Shutter JR, Hockenbery D, Kanagawa O, Korsmeyer SJ. bcl-2 inhibits multiple forms of apoptosis but not negative selection in thymocytes. Cell. 1991;67:879–888. doi: 10.1016/0092-8674(91)90361-2. [DOI] [PubMed] [Google Scholar]
- 22.Chao DT, Linette GP, Boise LH, White LS, Thompson CB, Korsmeyer SJ. Bcl-XL and Bcl-2 repress a common pathway of cell death. J Exp Med. 1995;182:821–828. doi: 10.1084/jem.182.3.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivanov, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 24.Zhumabekov T, Corbella P, Tolaini M, Kioussis D. Improved version of a human CD2 minigene based vector for T cell-specific expression in transgenic mice. J Immunol Methods. 1995;185:133–140. doi: 10.1016/0022-1759(95)00124-s. [DOI] [PubMed] [Google Scholar]
- 25.Barnden MJ, Allison J, Heath WR, Carbone FR. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol Cell Biol. 1998;76:34–40. doi: 10.1046/j.1440-1711.1998.00709.x. [DOI] [PubMed] [Google Scholar]
- 26.Lazarevic V, Chen X, Shim JH, Hwang ES, Jang E, Bolm AN, Oukka M, Kuchroo VK, Glimcher LH. T-bet represses T(H)17 differentiation by preventing Runx1-mediated activation of the gene encoding RORgammat. Nat Immunol. 2011;12:96–104. doi: 10.1038/ni.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siegemund S, Shepherd J, Xiao C, Sauer K. hCD2-iCre and Vav-iCre mediated gene recombination patterns in murine hematopoietic cells. PLoS One. 2015;10:e0124661. doi: 10.1371/journal.pone.0124661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacDonald HR, Budd RC, Howe RC. A CD3- subset of CD4−8+ thymocytes: a rapidly cycling intermediate in the generation of CD4+8+ cells. Eur J Immunol. 1988;18:519–523. doi: 10.1002/eji.1830180405. [DOI] [PubMed] [Google Scholar]
- 29.Taghon T, Yui MA, Pant R, Diamond RA, Rothenberg EV. Developmental and molecular characterization of emerging beta- and gammadelta-selected pre-T cells in the adult mouse thymus. Immunity. 2006;24:53–64. doi: 10.1016/j.immuni.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 30.Yamashita I, Nagata T, Tada T, Nakayama T. CD69 cell surface expression identifies developing thymocytes which audition for T cell antigen receptor-mediated positive selection. Int Immunol. 1993;5:1139–1150. doi: 10.1093/intimm/5.9.1139. [DOI] [PubMed] [Google Scholar]
- 31.Nielsen PJ, Lorenz B, Muller AM, Wenger RH, Brombacher F, Simon M, von der Weid T, Langhorne WJ, Mossmann H, Kohler G. Altered erythrocytes and a leaky block in B-cell development in CD24/HSA-deficient mice. Blood. 1997;89:1058–1067. [PubMed] [Google Scholar]
- 32.Hsu FC, Belmonte PJ, Constans MM, Chen MW, McWilliams DC, Hiebert SW, Shapiro VS. Histone Deacetylase 3 Is Required for T Cell Maturation. J Immunol. 2015 doi: 10.4049/jimmunol.1500435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T, Liu XS, Lazar MA. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331:1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhaskara S, Knutson SK, Jiang G, Chandrasekharan MB, Wilson AJ, Zheng S, Yenamandra A, Locke K, Yuan JL, Bonine-Summers AR, Wells CE, Kaiser JF, Washington MK, Zhao Z, Wagner FF, Sun ZW, Xia F, Holson EB, Khabele D, Hiebert SW. Hdac3 is essential for the maintenance of chromatin structure and genome stability. Cancer Cell. 2010;18:436–447. doi: 10.1016/j.ccr.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boursalian TE, Golob J, Soper DM, Cooper CJ, Fink PJ. Continued maturation of thymic emigrants in the periphery. Nat Immunol. 2004;5:418–425. doi: 10.1038/ni1049. [DOI] [PubMed] [Google Scholar]
- 36.Linette GP, Grusby MJ, Hedrick SM, Hansen TH, Glimcher LH, Korsmeyer SJ. Bcl-2 is upregulated at the CD4+ CD8+ stage during positive selection and promotes thymocyte differentiation at several control points. Immunity. 1994;1:197–205. doi: 10.1016/1074-7613(94)90098-1. [DOI] [PubMed] [Google Scholar]
- 37.Tani-ichi S, Shimba A, Wagatsuma K, Miyachi H, Kitano S, Imai K, Hara T, Ikuta K. Interleukin-7 receptor controls development and maturation of late stages of thymocyte subpopulations. Proc Natl Acad Sci U S A. 2013;110:612–617. doi: 10.1073/pnas.1219242110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He YW, Deftos ML, Ojala EW, Bevan MJ. RORgamma t, a novel isoform of an orphan receptor, negatively regulates Fas ligand expression and IL-2 production in T cells. Immunity. 1998;9:797–806. doi: 10.1016/s1074-7613(00)80645-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He YW, Beers C, Deftos ML, Ojala EW, Forbush KA, Bevan MJ. Down-regulation of the orphan nuclear receptor ROR gamma t is essential for T lymphocyte maturation. J Immunol. 2000;164:5668–5674. doi: 10.4049/jimmunol.164.11.5668. [DOI] [PubMed] [Google Scholar]
- 40.Kurebayashi S, Ueda E, Sakaue M, Patel DD, Medvedev A, Zhang F, Jetten AM. Retinoid-related orphan receptor gamma (RORgamma) is essential for lymphoid organogenesis and controls apoptosis during thymopoiesis. Proc Natl Acad Sci U S A. 2000;97:10132–10137. doi: 10.1073/pnas.97.18.10132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun Z, Unutmaz D, Zou YR, Sunshine MJ, Pierani A, Brenner-Morton S, Mebius RE, Littman DR. Requirement for RORgamma in thymocyte survival and lymphoid organ development. Science. 2000;288:2369–2373. doi: 10.1126/science.288.5475.2369. [DOI] [PubMed] [Google Scholar]
- 42.Taniuchi I, Osato M, Egawa T, Sunshine MJ, Bae SC, Komori T, Ito Y, Littman DR. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002;111:621–633. doi: 10.1016/s0092-8674(02)01111-x. [DOI] [PubMed] [Google Scholar]
- 43.Hernandez-Hoyos G, Anderson MK, Wang C, Rothenberg EV, Alberola-Ila J. GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation. Immunity. 2003;19:83–94. doi: 10.1016/s1074-7613(03)00176-6. [DOI] [PubMed] [Google Scholar]
- 44.Aliahmad P, Kaye J. Development of all CD4 T lineages requires nuclear factor TOX. J Exp Med. 2008;205:245–256. doi: 10.1084/jem.20071944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun G, Liu X, Mercado P, Jenkinson SR, Kypriotou M, Feigenbaum L, Galera P, Bosselut R. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat Immunol. 2005;6:373–381. doi: 10.1038/ni1183. [DOI] [PubMed] [Google Scholar]
- 46.He X, He X, Dave VP, Zhang Y, Hua X, Nicolas E, Xu W, Roe BA, Kappes DJ. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433:826–833. doi: 10.1038/nature03338. [DOI] [PubMed] [Google Scholar]
- 47.Wang L, Wildt KF, Zhu J, Zhang X, Feigenbaum L, Tessarollo L, Paul WE, Fowlkes BJ, Bosselut R. Distinct functions for the transcription factors GATA-3 and ThPOK during intrathymic differentiation of CD4(+) T cells. Nat Immunol. 2008;9:1122–1130. doi: 10.1038/ni.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sefik E, Geva-Zatorsky N, Oh S, Konnikova L, Zemmour D, McGuire AM, Burzyn D, Ortiz-Lopez A, Lobera M, Yang J, Ghosh S, Earl A, Snapper SB, Jupp R, Kasper D, Mathis D, Benoist C. MUCOSAL IMMUNOLOGY. Individual intestinal symbionts induce a distinct population of RORgamma(+) regulatory T cells. Science. 2015;349:993–997. doi: 10.1126/science.aaa9420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dovey OM, Foster CT, Conte N, Edwards SA, Edwards JM, Singh R, Vassiliou G, Bradley A, Cowley SM. Histone deacetylase 1 and 2 are essential for normal T-cell development and genomic stability in mice. Blood. 2013;121:1335–1344. doi: 10.1182/blood-2012-07-441949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boucheron N, Tschismarov R, Goschl L, Moser MA, Lagger S, Sakaguchi S, Winter M, Lenz F, Vitko D, Breitwieser FP, Muller L, Hassan H, Bennett KL, Colinge J, Schreiner W, Egawa T, Taniuchi I, Matthias P, Seiser C, Ellmeier W. CD4(+) T cell lineage integrity is controlled by the histone deacetylases HDAC1 and HDAC2. Nat Immunol. 2014;15:439–448. doi: 10.1038/ni.2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stengel KR, Zhao Y, Klus NJ, Kaiser JF, Gordy LE, Joyce S, Hiebert SW, Summers AR. Histone Deacetylase 3 Is Required for Efficient T Cell Development. Mol Cell Biol. 2015;35:3854–3865. doi: 10.1128/MCB.00706-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mao X, Fujiwara Y, Orkin SH. Improved reporter strain for monitoring Cre recombinase-mediated DNA excisions in mice. Proc Natl Acad Sci U S A. 1999;96:5037–5042. doi: 10.1073/pnas.96.9.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park JH, Adoro S, Guinter T, Erman B, Alag AS, Catalfamo M, Kimura MY, Cui Y, Lucas PJ, Gress RE, Kubo M, Hennighausen L, Feigenbaum L, Singer A. Signaling by intrathymic cytokines, not T cell antigen receptors, specifies CD8 lineage choice and promotes the differentiation of cytotoxic-lineage T cells. Nat Immunol. 2010;11:257–264. doi: 10.1038/ni.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.