Abstract
Background
Pigs are natural hosts for influenza A viruses, and the infection is widely prevalent in swine herds throughout the world. Current commercial influenza vaccines for pigs induce a narrow immune response and are not very effective against antigenically diverse viruses. To control influenza in pigs, the development of more effective swine influenza vaccines inducing broader cross-protective immune responses is needed. Previously, we have shown that a polyvalent influenza DNA vaccine using vectors containing antibiotic resistance genes induced a broadly protective immune response in pigs and ferrets using intradermal injection followed by electroporation. However, this vaccination approach is not practical in large swine herds, and DNA vaccine vectors containing antibiotic resistance genes are undesirable.
Objectives
To investigate the immunogenicity of an optimized version of our preceding polyvalent DNA vaccine, characterized by a next-generation expression vector without antibiotic resistance markers and delivered by a convenient needle-free intradermal application approach.
Methods
The humoral and cellular immune responses induced by three different doses of the optimized DNA vaccine were evaluated in groups of five to six pigs. The DNA vaccine consisted of six selected influenza genes of pandemic origin, including internally expressed matrix and nucleoprotein and externally expressed hemagglutinin and neuraminidase.
Results
Needle-free vaccination of growing pigs with the optimized DNA vaccine resulted in specific, dose-dependent immunity down to the lowest dose (200 μg DNA/vaccination). Both the antibody-mediated and the recall lymphocyte immune responses demonstrated high reactivity against vaccine-specific strains and cross-reactivity to vaccine-heterologous strains.
Conclusion
The results suggest that polyvalent DNA influenza vaccination may provide a strong tool for broad protection against swine influenza strains threatening animal as well as public health. In addition, the needle-free administration technique used for this DNA vaccine will provide an easy and practical approach for the large-scale vaccination of swine.
Keywords: Swine influenza, Polyvalent, DNA, Vaccine, Cross-reactive, Response
1. Introduction
Influenza virus is endemic in pigs and affects the majority of herds in modern swine production [1]. Reproductive problems, together with weight loss and aggravation of secondary infections, are characteristic of swine influenza and result in serious animal welfare problems and economic losses for the swine industry [2]. It is well known that pigs and humans can exchange influenza viruses, and a recent example is the triple reassortant H1N1pdm09, composed of genes from three known swine viruses, which spread rapidly among humans during the pandemic in 2009 and later transmitted from humans to pigs [3]. Protection of pigs against influenza infection by effective vaccination would provide a crucial tool to benefit swine health and reduce risks to public health.
Current vaccines against influenza virus for pigs are based on inactivated virus and only induce immunity against the virus strains included in the vaccines, thus providing limited protection against the diverse spectrum of other circulating influenza strains [1]. Thus, an effective intervention strategy for the control of influenza in pigs requires improved vaccines. DNA technology enables vaccination with versatile combinations of antigens that can simply be substituted. The DNA platform was tested early on in the influenza field with variable results [4], [5]. However, a direct comparison between early results [6], [7], [8] and more recent studies are complicated due to recent improvements in DNA vaccines as well as the improved techniques to evaluate cell-mediated immune responses. Thus, codon-optimization of genes [9], [10], [11], [12], [13], [14], improved delivery [12], [15], [16], [17] and DNA vector improvements [18] have enhanced the immunogenicity of DNA vaccines, and a number of DNA vaccine candidates have been successful in both animal and human studies [13], [14], [15], [19], [20], [21]. DNA vaccines have the advantage of inducing both cellular and humoral immunity, both of which are believed to serve important roles in protection against influenza virus infections and shedding of virus [1], [15], [22].
Previously, we and others have tested DNA vaccines against influenza in pigs in different experimental settings [6], [7], [8], [15], [20], [23], [24]. Recently, we published the optimization of a polyvalent influenza DNA vaccine using next-generation antibiotic-free vectors together with a needle-free intradermal (i.d.) application in rabbits [25]. In the present study, we conducted a DNA dose titration study in pigs to investigate the immunogenicity of our optimized influenza DNA vaccine containing pandemic genes from the 1918 H1N1-, 1968 H3N2- and pdm09 H1N1-influenza viruses. Thus, we tested the induction of both cellular and humoral immune responses directed against antigens both homologous and heterologous to the vaccine.
2. Materials and methods
2.1. Construction of DNA vaccines
The six influenza DNA vaccine genes have been described previously [25]. The NTC9385R plasmid was used as an expression vector [18], [25].
2.2. Animals and experimental design
Twenty-two five-week-old, recently weaned pigs obtained from a Danish specific pathogen free (SPF) herd were randomly assigned to four groups of five or six animals. The pigs were housed without contact to other animals in separate isolation facilities at the Department of Animal Science, Aarhus University. The pigs were allowed to acclimatize for 1 week before the initiation of the experiment. With an interval of 3 weeks, three groups of pigs were vaccinated twice on the dorsal site of the back using the needle-free IntraDermal Application of Liquids (IDAL®) device (Henke Sass Wolf). Six pigs were vaccinated with 200 μg of DNA each (one injection site on the back), another six pigs received 800 μg of DNA each (distributed into four injection sites) and five pigs received 1972 μg of DNA (distributed into 10 injection sites). For use of the IDAL® device, the vaccine constructs were premixed at a 1:1 volume ratio with an α-tocopherol-based aqueous solution (Diluvac Forte®, MSD Animal Health). Two pigs remained unvaccinated, and three additional pigs received the Diluvac Forte® solution without any DNA vaccine. The latter five pigs displayed similar immune profiles in the analyses and were thus combined into the non-treated control group. All pigs were monitored daily for clinical signs of disease or any adverse vaccination-related effects. Rectal body temperatures were recorded 2 days before and 2 days after each vaccination. Whole-blood samples were collected from the anterior vena cava of all pigs on days 0, 7, 14, 21, 28 and 35 post-first vaccination (pv1). Serum was isolated and stored at -20 °C for subsequent examination. On day 35pv1, peripheral blood mononuclear cells (PBMC) were isolated from freshly collected heparinized blood samples by density gradient centrifugation and cryopreserved until use. On a weekly basis starting from day 0pv1, nasal swab (MicroRheologics) samples were collected in virus transport medium from all pigs to test for potential accidental influenza infection during the experiment. Upon termination of the experiment, on day 35pv1, the pigs were euthanized by i.v. injection of a lethal dose of pentobarbital. All animal handling and experimentation procedures were approved by the Danish Animal Experiments Inspectorate (2014-15-0201-00251).
2.3. Influenza detection
Nasal swab samples (day 0, 7, 14, 21, 28 and 35pv1) were examined for influenza A virus RNA using an in-house real-time reverse transcription (RT)-PCR assay. Primers and probes for the matrix gene of influenza A virus, the NA gene of H1N1pdm09 and the HA gene of human seasonal H3N2 were used.
2.4. Enzyme-linked immunosorbent assay (ELISA)
ELISA was conducted to measure influenza-specific IgG responses in the sera as previously described [25]. The influenza virus proteins used for coating were HA from A/California/04/09(H1N1)pdm09, A/Aichi/2/1968(H3N2), A/swine/Guangxi/13/2006(H1N2) or A/Brisbane/59/07(H1N1); NA from A/Aichi/2/1968(H3N2); NP from A/California/07/09(H1N1)pdm09; M1 protein from A/Brevig Mission/1/1918(H1N1) (all from Sino Biological Inc.); or M2e polypeptide (GenScript). A horseradish peroxidase-conjugated anti-pig-IgG antibody (AbD Serotec) was used for detection.
2.5. Hemagglutination inhibition (HI) assay
The HI assay was performed according to the protocols of the WHO [26] as previously described [25]. The virus isolates tested were two swine strains, A/swine/Denmark (DK)/10409/2013(H1N1pdm) and A/swine/DK/10525/2008(H1N2).
2.6. Microneutralization assay (MN)
Development of neutralizing antibodies was determined according to the protocols of the WHO [27]. Viruses used were A/California/07/09(H1N1pdm09), A/NewCaledonia/20/99(H1N1), and A/swine/DK/10409/2013(H1N1pdm), with 100 TCID50 as the inoculum.
2.7. PBMC stimulation and cell-mediated immune assays
Prior to stimulation, the cryopreserved PBMC were thawed and rested overnight in R10 (RPMI, Gibco) supplemented with 10% heat-inactivated FBS (Gibco) and 1% penicillin–streptomycin (Gibco) (culture medium) at 37 °C with 5% CO2. During stimulation, the R10 was supplemented with 50 ng/ml porcine IL-18 (R&D). The PBMC were stimulated with 5 μg/ml recombinant influenza proteins, including NP from A/California/07/09(H1N1)pdm09 and A/Brevig Mission/1/1918(H1N1), HA from A/California/04/09(H1N1)pdm09 or matrix 1 (M1) from A/Brevig Mission/1/1918(H1N1) (all from Sino Biological Inc.). One microgram per milliliter Staphylococcus Enterotoxin B (SEB, Sigma) served as a positive control and media alone served as a negative control. After 18 h of stimulation, 10 μg/ml Brefeldin A (Sigma) was added, followed by an additional 6 h of incubation. The stimulation was halted by 2 mM EDTA. The cells were stained with anti-CD3 PE-Cy7 (BD Pharmingen), anti-CD4 FITC (Serotec), anti-CD8 PE (Serotec) and a violet dead cell staining kit (Invitrogen), fixed and permeabilized with Cytofix/Cytoperm (BD) and stained with anti-IFN-γ AF647 (Serotec). The stained cells were acquired using a BD LSRII and analyzed using FlowJo (Tree Star). The background level of cytokine staining in the non-stimulated samples was subtracted for each individual animal. For the assessment of cell proliferation, in combination with the IFN-γ response, PBMC were labeled with 5 μM CellTrace Violet (Molecular probes), as described by the manufacturer, prior to stimulation. The cells were suspended in R10 supplemented with IL-18 and 50 μM 2-mercaptoethanol (Sigma) and stimulated for 5 days with 2 μg/ml of recombinant influenza proteins. At day 5, the PBMC were re-stimulated with the same amount of proteins for an additional 18 h. Next, 10 μg/ml Brefeldin A was added, followed by an additional 6 h of incubation. The cells were stained and acquired as described above but with the near IR dead cell staining kit (Invitrogen).
2.8. Statistical analysis
Differences between the groups were calculated using two-way ANOVA and Bonferroni multiple comparison test (GraphPad Prism v.6, GraphPad software).
3. Results
3.1. Clinical observations
None of the pigs displayed any signs of clinical disease or side effects of vaccination during the experiment. In addition, influenza virus could not be identified in any of the weekly collected nasal secretions.
3.2. Induction of cross-reactive antibodies
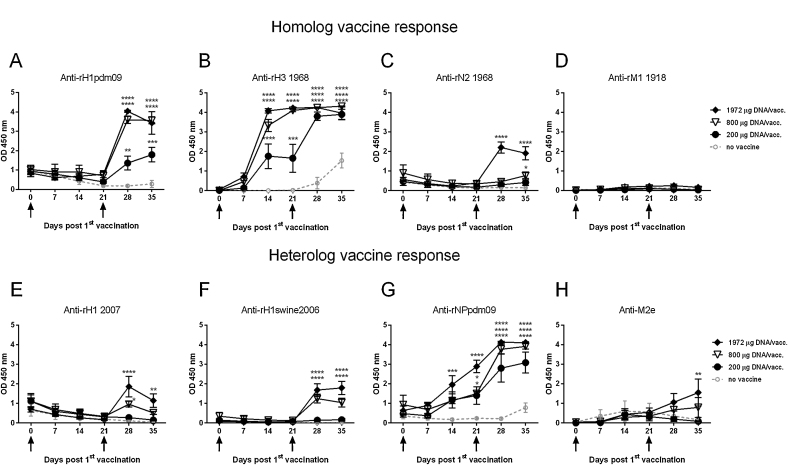
Antibody responses against three out of the four tested different influenza proteins, homologous to the vaccine genes, could be detected in the vaccinated pigs (Fig. 1a−d). In particular, the HA-specific antibodies were found to be present at high titers after day 28pv1, and anti-H3 antibodies were detected at day 14pv1. The antibody response levels correlated well with the applied DNA doses. In addition, antibody responses against influenza proteins not corresponding to the vaccine genes were detected (Fig. 1e−h). Antibodies against recombinant HA of both human and swine origin (Fig. 1e,f) were seen after day 28pv1 in the two pig groups receiving the highest DNA doses. A high antibody response was detected against NP originating from H1N1pdm09 in all vaccinated groups. Both vaccinated and control pigs had low levels of influenza-specific IgG against several different antigens at day 0pv1 (Fig. 1a H1pdm09, 1C N2 1968, 1E H1 2007 and 1G NPpdm09). This low level detected at day 0 gradually deceased over time in the control group, thus indicating that these antibodies represent maternally derived antibodies (MDA).
Fig. 1.
Influenza-specific antibody response following DNA vaccination. Pigs were vaccinated twice (arrows) i.d. with needle-free delivery with 200 μg (n = 6), 800 μg (n = 6) or 1972 μg (n = 5) DNA, or not DNA vaccinated at all (n = 5). Levels of IgG in the sera were measured by ELISA. Recombinant influenza proteins that were (a-d) homologous to the vaccine or (e-h) heterologous to the vaccine were used as the coating antigens. All serum samples were tested using a fixed 1:100 or 1:125 serum dilution. Error bars indicate the mean ± SEM, and significant differences from the no-vaccine control group are indicated by: ****: p < 0.0001; ***: p < 0.001; **: p < 0.01; *: p < 0.05.
3.3. Induction of HI antibodies
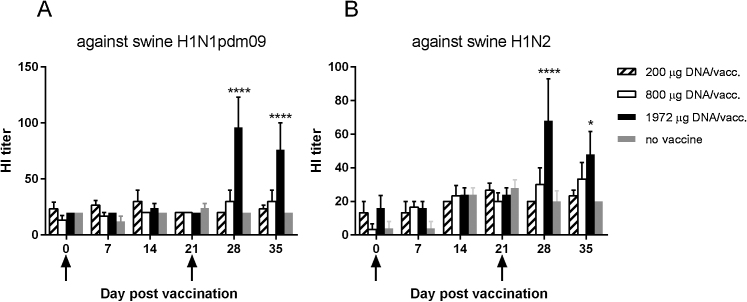
Vaccinated pigs had vaccine-induced serum HI antibodies that were cross-reactive against two swine virus strains, H1N1pdm09 and H1N2, which were heterologous to the vaccine genes (Fig. 2). The HI antibody levels were significantly higher in the group vaccinated with the highest dose of DNA than in the control group after day 28pv1. The HI titers obtained with the two different virus strains correlated significantly with each other (Spearman correlation, r = 0.50, p < 0.0001).
Fig. 2.
Serum HI antibody titers in vaccinated pigs. Pigs were vaccinated twice (arrows) i.d. with needle-free delivery with 200 μg (n = 6), 800 μg (n = 6) or 1972 μg (n = 5) DNA, or not DNA vaccinated at all (n = 5). Vaccine-induced hemagglutination inhibition antibody responses in pig sera against (a) swine H1N1pdm09 and (b) swine H1N2 isolates were measured. The data presented are from one representative experiment out of two performed. Error bars indicate the mean ± SEM, and significant differences from the no-vaccine control group are indicated by: ****: p < 0.0001; *: p < 0.05.
3.4. Induction of neutralizing activity
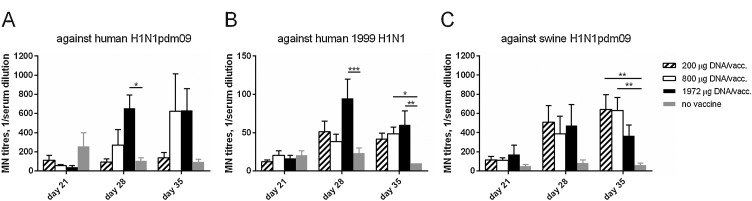
Neutralizing activity against H1N1 virus strains, both homologous and heterologous to the vaccine genes, developed in the vaccinated pigs (Fig. 3, only day 21 and beyond are shown). Neutralization could not be detected at time points earlier than day 28pv1, i.e. 1 week after the second vaccination. At this stage, the pigs receiving the highest dose of DNA had developed significantly higher MN titers against the homologous influenza virus H1N1pdm09 (Fig. 3a) and a heterologous human isolate (Fig. 3b) than the control group. At day 35pv1, the pigs given the middle dose of DNA, 800 μg, also had elevated MN titers against both human H1N1 isolates. Neutralization of a heterologous swine virus, H1N1pdm09, was also detected in the vaccinated pigs on day 35pv1 (Fig. 3c). Notably, only the lower- and middle-dose DNA groups demonstrated significant levels of neutralization compared to the control group. Significant correlations were observed for the MN titers derived from the three virus strains tested (Spearman correlations for all combinations between the three virus strains, r-range 0.45−0.64, p range 0.0001 to < 0.0001).
Fig. 3.
Neutralizing activity in vaccinated pig sera. Pigs were vaccinated twice (day 0 and 21 pv1) i.d. with needle-free delivery with 200 μg (n = 6), 800 μg (n = 6) or 1972 μg (n = 5) DNA, or not DNA vaccinated at all (n = 5). The pig sera were tested in a microneutralization assay on days 21, 28 and 35 pv1. Neutralizing antibody titers, MN titers, were evaluated by the capacity of the sera to prevent the infection of MDCK cells by (a) H1N1pdm09, (b) 1999 H1N1 and (c) swine H1N1 isolates. The MN titer was defined as the reciprocal dilution giving 50% infection inhibition and calculated with a linear interpolation method [28]. Serum samples with a titer below the detectable limit of the assay (lowest serum dilution tested was 1:20) were assigned a value of 10 for graphical representation and statistical analyses. Error bars indicate the mean ± SEM, and significant differences from the no-vaccine control group are indicated by: ***: p < 0.001; **: p < 0.01; *: p < 0.05.
3.5. Induction of antigen-specific T cell responses
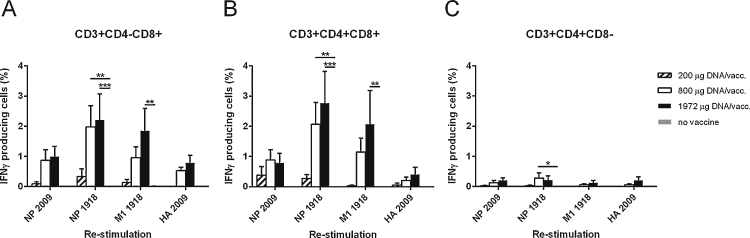
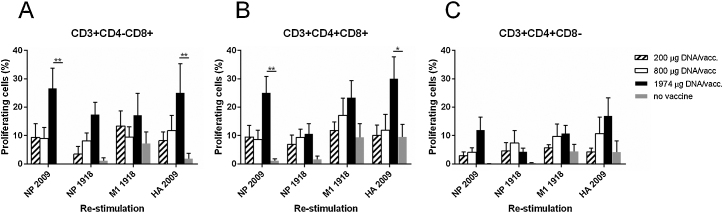
The DNA vaccine elicited NP-, M1- and HA-specific CD4-CD8+ T cells and CD4+CD8+T cells producing IFN-γ (Fig. 4a and b). The IFN-γ response levels correlated with the DNA vaccine doses. Both vaccine-homologous and vaccine-heterologous NP (1918 and 2009, respectively) could re-stimulate the PBMC, and their respective IFN-γ responses correlated significantly (r = 0.78, p < 0.0001 (Spearman correlation) for CD4-CD8+ T cells and r = 0.86, p < 0.0001 for CD4+CD8+T cells.) CD4+CD8-T cells contained lower levels of re-stimulated cells (Fig. 4c). A similar pattern was observed when the proliferation level of re-stimulated PBMC was assessed (Fig. 5). Pigs receiving the highest dose of the vaccine had a proliferating recall response significantly higher than the control group. The expression of IFN-γ coincided with proliferating cells; the mean for all groups was 64.4% (standard deviation, 14.8) of proliferating cells also expressing IFN-γ.
Fig. 4.
T cell sub-population IFN-γ response to in vitro re-stimulation with influenza proteins. Pigs were vaccinated twice (day 0 and 21pv1) i.d. with needle-free delivery with 200 μg (n = 6), 800 μg (n = 6) or 1972 μg (n = 5) DNA, or not DNA vaccinated at all (n = 5). PBMC from the vaccinated pigs on day 35pv1 were cultured in vitro in the presence of recombinant influenza NP 2009, NP 1918, M1 1918 and HA 2009. After 24 h, the cells were stained with anti-CD3, -CD4, -CD8 and -IFN-γ monoclonal antibodies and analyzed by flow cytometry. Three T cell subsets were identified based on their CD4 and CD8 expression: (a) CD4-CD8+, (b) CD4+CD8+ and (c) CD4+CD8- cells. Error bars indicate the mean ± SEM, and significant differences from the no-vaccine control group are indicated by: ***: p < 0.001; **: p < 0.01; *: p < 0.05.
Fig. 5.
T cell sub-population proliferation response to in vitro re-stimulation with influenza proteins. Pigs were vaccinated twice (day 0 and 21pv1) i.d. with needle-free delivery with 200 μg (n = 6), 800 μg (n = 6) or 1972 μg (n = 5) DNA, or not DNA vaccinated at all (n = 5). PBMC from the vaccinated pigs on day 35pv1 were cultured in vitro in the presence of recombinant influenza NP 2009, NP 1918, M1 1918 and HA 2009. After 6 days, the cells were stained with anti-CD3, -CD4, and -CD8 monoclonal antibodies and analyzed by flow cytometry. Three T cell subsets were identified based on their CD4 and CD8 expression: (a) CD4-CD8+, (b) CD4+CD8+ and (c) CD4+CD8- cells. Error bars indicate the mean ± SEM, and significant differences from the no-vaccine control group are indicated by: **: p < 0.01; *: p < 0.05.
4. Discussion
In the present study, we demonstrated that our polyvalent DNA influenza vaccine delivered via needle-free i.d. application induced a significant immune response against homologous as well as heterologous influenza antigens, represented by both antibody- and cell-mediated reactivity. The vaccine responses correlated with the DNA doses, i.e. higher responses with increasing DNA doses. The intermediate dose of 800 μg DNA/vaccination seemed to be almost as effective as the highest dose of 1972 μg DNA, and even the lowest dose of 200 μg DNA/vaccination could induce a detectable response. However, the protective effect of the various doses requires further challenge studies.
The highest dose of DNA in this study was chosen to be equimolar to the dosage used in our previous pig challenge study [20], where pigs vaccinated via i.d. needle and electroporation demonstrated protective immunity, as measured by reduced viral shedding after challenge. Similar to our previous study [20], Gorres et al. [15] also demonstrated reduced virus shedding with a comparable amount of DNA in a pig challenge study. The results of the present study suggest that the DNA dose can be reduced to less than half amount of plasmid compared to our previous challenge study [20] and still maintain immunogenicity. Moreover, we have removed the antibiotic resistance selection in the plasmid to avoid interference with antibiotics used in the pig industry [18].
Humoral immune response analyses revealed an antibody response after the first vaccination that was boosted after re-vaccination. In addition to a homologous response, vaccination with our polyvalent DNA vaccine appears to induce broadly reactive antibodies against multiple H1N1 and H1N2 strains of human and swine origin. Because our DNA vaccine encodes four different influenzas surface-exposed glycoproteins, H1, N1, H3 and N2, we expected to detect a heterogeneous antibody response against several influenza strains. HA-specific antibodies are of importance for vaccine efficacy because they can provide protection by blocking virus attachment and entry [28]. In addition, NA-specific antibodies have been shown to reduce virus shedding and decrease severe illness (reviewed by Marcelin et al. [29]). The NP plasmid was included to induce cellular immunity to NP proteins, which are relatively constant among different influenza virus strains [30]. However, the in vivo-encoded NP also gave rise to high antibody titers that are not expected to prevent viral entry because NP is not exposed on the surface of the virion [31], [32]. The M protein is a relatively constant protein with an extracellular surface loop, M2e, which is exposed to antibodies [33]. Thus, antibodies against M may be cross-protective against different influenza strains [34]. Indeed, we did observe anti-M2e-specific antibodies in vaccinated pigs, which potentially can serve a protective role during infection. The combination of antibodies against both conserved (NP and M2e) and more diverse influenza antigens (HA and NA) suggest a potent cross-reactive IgG response generated by our polyvalent DNA vaccine. Indeed, the cross-reactive response was reflected in the functional humoral hemagglutination inhibition and neutralization assays. Both of these assays demonstrated a vaccine response against circulating swine influenza isolates that were heterologous to our vaccine, which is promising for a future swine vaccine. In addition, the HI and MN titers were correlated between the different virus strains tested, indicating that the induced antibody response is cross-reactive.
Cellular immune responses play an important role during influenza infection by contributing to eliminate infected cells and reduce virus shedding [35], and DNA vaccines have the advantage of inducing both a humoral and a cellular immune response [36], [37]. While Gorres et al. [15] could not detect significant levels of IFN-γ secreting cells after needle-free (subcutaneous (s.c.)/i.m.) vaccination with a DNA influenza vaccine, we demonstrated significant dose-related levels of IFN-γ-producing T cells against influenza virus specific proteins 2 weeks after the second needle-free i.d. vaccination. A number of factors related to the different analysis methods may explain this discrepancy, but it may also underline the superiority of i.d. compared to s.c./i.m. application. In our assay, both the NP 1918 and M1 1918 proteins, homologous to the DNA vaccine, could re-stimulate the T cells. In addition, a NP protein from the pdm09 virus strain could also stimulate the T cell response, which indicates that the NP-specific T cell response is cross-reactive. Moreover, there also seems to be a T cell response against the externally expressed proteins, represented by HA protein in our assay. The functionality of an influenza-specific T cell response, including proliferation and degranulation, has previously been published to correlate with IFN-γ-production [38], which was also confirmed herein. Although the humoral response, especially the HA-specific response, may be the primary factor in influenza protection, other studies have suggested the importance of cell-mediated immunity in pigs and ferrets upon DNA vaccination followed by challenge [15], [23], [39]. Thus, the present confirmation of the vaccine-induced IFN-γ response in T cells to coincide with proliferation indicates that our i.d.-applied DNA vaccine provokes a specific cell-mediated response, which may provide strong contribute to heterotypic influenza immunity [40], [41], [42].
Vaccination of piglets with MDA using a conventional influenza vaccine may have potential complications such as the suppression of the immune response [43], [44], [45] and the induction of vaccine-associated enhanced respiratory disease (VAERD) [45], [46]. Our results demonstrating the development of antibody responses in pigs with MDA support previous findings indicating the ability of the DNA vaccine to function in the presence of MDA [47]. This ability of DNA vaccines represents a major advantage of using a DNA vaccine approach in the influenza vaccination of swine herds.
We believe that our approach of using pandemic-derived surface influenza antigens with conserved internal antigens has the ability to confer broad protection against both homologous and heterologous virus strains, with both antibody and T cell responses as contributors. Furthermore, the lack of adverse reactions to vaccination indicates that the vaccine is safe for the animals. The possibility to deliver the DNA vaccine using a needle-free approach is a realistic and attractive alternative for convenient, safe and animal welfare-friendly mass vaccination in swine herds, a method already in use for traditional protein vaccines in pigs. The present results encourage influenza challenge studies with a lower dose of DNA than used previously [20] to demonstrate the efficacy of the polyvalent DNA vaccine, thus paving the way for an improved intervention strategy in the fight against influenza.
Conflicts of interest
James Williams has an equity interest in Nature Technology Corporation. The other authors declare that there are no other conflicts of interest.
Acknowledgments
The technical assistance of Lene Rosborg Dal, Birgit Knudsen, Randi Thøgersen, Bente Andersen and the animal care staff at Aarhus University is gratefully acknowledged. This project has received funding from the European Union's Seventh Framework Programme for research, technological development and demonstration under grant agreement no. 602012.
References
- 1.Sandbulte M.R., Spickler A.R., Zaabel P.K., Roth J.A. Optimal use of vaccines for control of influenza A virus in swine. Vaccines (Basel) 2015;3:22–73. doi: 10.3390/vaccines3010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olsen C.W., Reeth K.V., Brown I., Easterday B.C. Swine influenza. In: Straw J.J., Zimmerman S., D’Allaire D.J., Taylor, editors. Diseases of swine. 9th ed. 2006. pp. 469–482. [Google Scholar]
- 3.Neumann G., Noda T., Kawaoka Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature. 2009;459:931–939. doi: 10.1038/nature08157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robinson H.L., Hunt L.A., Webster R.G. Protection against a lethal influenza virus challenge by immunization with a haemagglutinin-expressing plasmid DNA. Vaccine. 1993;11:957–960. doi: 10.1016/0264-410x(93)90385-b. [DOI] [PubMed] [Google Scholar]
- 5.Ulmer J.B., Donnelly J.J., Parker S.E., Rhodes G.H., Felgner P.L., Dwarki V.J. Heterologous protection against influenza by injection of DNA encoding a viral protein. Science. 1993;259:1745–1749. doi: 10.1126/science.8456302. [DOI] [PubMed] [Google Scholar]
- 6.Heinen P.P., Rijsewijk F.A., de Boer-Luijtze E.A., Bianchi A.T. Vaccination of pigs with a DNA construct expressing an influenza virus M2-nucleoprotein fusion protein exacerbates disease after challenge with influenza A virus. J Gen Virol. 2002;83:1851–1859. doi: 10.1099/0022-1317-83-8-1851. [DOI] [PubMed] [Google Scholar]
- 7.Eriksson E., Yao F., Svensjo T., Winkler T., Slama J., Macklin M.D. In vivo gene transfer to skin and wound by microseeding. J Surg Res. 1998;78:85–91. doi: 10.1006/jsre.1998.5325. [DOI] [PubMed] [Google Scholar]
- 8.Macklin M.D., McCabe D., McGregor M.W., Neumann V., Meyer T., Callan R. Immunization of pigs with a particle-mediated DNA vaccine to influenza A virus protects against challenge with homologous virus. J Virol. 1998;72:1491–1496. doi: 10.1128/jvi.72.2.1491-1496.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shan S., Jiang Y., Bu Z., Ellis T., Zeng X., Edwards J. Strategies for improving the efficacy of a H6 subtype avian influenza DNA vaccine in chickens. J Virol Methods. 2011;173:220–226. doi: 10.1016/j.jviromet.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 10.Li J., Jiang Y., Zhao S., Chang X., Liu J., Zeng X. Protective efficacy of an H5N1 DNA vaccine against challenge with a lethal H5N1 virus in quail. Avian Dis. 2012;56:937–939. doi: 10.1637/10150-040812-ResNote.1. [DOI] [PubMed] [Google Scholar]
- 11.Wang S., Hackett A., Jia N., Zhang C., Zhang L., Parker C. Polyvalent DNA vaccines expressing HA antigens of H5N1 influenza viruses with an optimized leader sequence elicit cross-protective antibody responses. PLoS One. 2011;6:e28757. doi: 10.1371/journal.pone.0028757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ault A., Zajac A.M., Kong W.P., Gorres J.P., Royals M., Wei C.J. Immunogenicity and clinical protection against equine influenza by DNA vaccination of ponies. Vaccine. 2012;30:3965–3974. doi: 10.1016/j.vaccine.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones S., Evans K., McElwaine-Johnn H., Sharpe M., Oxford J., Lambkin-Williams R. DNA vaccination protects against an influenza challenge in a double-blind randomised placebo-controlled phase 1b clinical trial. Vaccine. 2009;27:2506–2512. doi: 10.1016/j.vaccine.2009.02.061. [DOI] [PubMed] [Google Scholar]
- 14.Smith L.R., Wloch M.K., Ye M., Reyes L.R., Boutsaboualoy S., Dunne C.E. Phase 1 clinical trials of the safety and immunogenicity of adjuvanted plasmid DNA vaccines encoding influenza A virus H5 hemagglutinin. Vaccine. 2010;28:2565–2572. doi: 10.1016/j.vaccine.2010.01.029. [DOI] [PubMed] [Google Scholar]
- 15.Gorres J.P., Lager K.M., Kong W.P., Royals M., Todd J.P., Vincent A.L. DNA vaccination elicits protective immune responses against pandemic and classic swine influenza viruses in pigs. Clin Vaccine Immunol. 2011;18:1987–1995. doi: 10.1128/CVI.05171-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Otten G., Schaefer M., Doe B., Liu H., Srivastava I., zur Megede J. Enhancement of DNA vaccine potency in rhesus macaques by electroporation. Vaccine. 2004;22:2489–2493. doi: 10.1016/j.vaccine.2003.11.073. [DOI] [PubMed] [Google Scholar]
- 17.Widera G., Austin M., Rabussay D., Goldbeck C., Barnett S.W., Chen M. Increased DNA vaccine delivery and immunogenicity by electroporation in vivo. J Immunol. 2000;164:4635–4640. doi: 10.4049/jimmunol.164.9.4635. [DOI] [PubMed] [Google Scholar]
- 18.Williams J.A. Vector design for improved DNA vaccine efficacy, safety and production. Vaccines. 2013;1:225–249. doi: 10.3390/vaccines1030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ledgerwood J.E., Wei C.J., Hu Z., Gordon I.J., Enama M.E., Hendel C.S. DNA priming and influenza vaccine immunogenicity: two phase 1 open label randomised clinical trials. Lancet Infect Dis. 2011;11:916–924. doi: 10.1016/S1473-3099(11)70240-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bragstad K., Vinner L., Hansen M.S., Nielsen J., Fomsgaard A. A polyvalent influenza A DNA vaccine induces heterologous immunity and protects pigs against pandemic A(H1N1)pdm09 virus infection. Vaccine. 2013;31:2281–2288. doi: 10.1016/j.vaccine.2013.02.061. [DOI] [PubMed] [Google Scholar]
- 21.Yan J., Villarreal D.O., Racine T., Chu J.S., Walters J.N., Morrow M.P. Protective immunity to H7N9 influenza viruses elicited by synthetic DNA vaccine. Vaccine. 2014;32:2833–2842. doi: 10.1016/j.vaccine.2014.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ulmer J.B., Fu T.M., Deck R.R., Friedman A., Guan L., DeWitt C. Protective CD4+ and CD8+ T cells against influenza virus induced by vaccination with nucleoprotein DNA. J Virol. 1998;72:5648–5653. doi: 10.1128/jvi.72.7.5648-5653.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larsen D.L., Karasin A., Olsen C.W. Immunization of pigs against influenza virus infection by DNA vaccine priming followed by killed-virus vaccine boosting. Vaccine. 2001;19:2842–2853. doi: 10.1016/s0264-410x(01)00014-7. [DOI] [PubMed] [Google Scholar]
- 24.Olsen C.W. DNA vaccination against influenza viruses: a review with emphasis on equine and swine influenza. Vet Microbiol. 2000;74:149–164. doi: 10.1016/s0378-1135(00)00175-9. [DOI] [PubMed] [Google Scholar]
- 25.Borggren M., Nielsen J., Bragstad K., Karlsson I., Krog J.S., Williams J.A. Vector optimization and needle-free intradermal application of a broadly protective polyvalent influenza A DNA vaccine for pigs and humans. Hum Vaccines Immunother. 2015;11(8):1983–1990. doi: 10.1080/21645515.2015.1011987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.WHO Recommendations for Influenza Vaccine Composition [http://www.who.int/influenza/vaccines/virus/en/].
- 27.World Health Organization Global Influenza Surveillance Network. Manual for the laboratory diagnosis and virological surveillance of influenza http://wwwwhoint/influenza/gisrs_laboratory/manual_diagnosis_surveillance_influenza/en/.2011.
- 28.Wong S.S., Webby R.J. Traditional and new influenza vaccines. Clin Microbiol Rev. 2013;26:476–492. doi: 10.1128/CMR.00097-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marcelin G., Sandbulte M.R., Webby R.J. Contribution of antibody production against neuraminidase to the protection afforded by influenza vaccines. Rev Med Virol. 2012;22:267–279. doi: 10.1002/rmv.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gorman O.T., Bean W.J., Kawaoka Y., Webster R.G. Evolution of the nucleoprotein gene of influenza A virus. J Virol. 1990;64:1487–1497. doi: 10.1128/jvi.64.4.1487-1497.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carragher D.M., Kaminski D.A., Moquin A., Hartson L., Randall T.D. A novel role for non-neutralizing antibodies against nucleoprotein in facilitating resistance to influenza virus. J Immunol. 2008;181:4168–4176. doi: 10.4049/jimmunol.181.6.4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lamere M.W., Moquin A., Lee F.E., Misra R.S., Blair P.J., Haynes L. Regulation of antinucleoprotein IgG by systemic vaccination and its effect on influenza virus clearance. J Virol. 2011;85:5027–5035. doi: 10.1128/JVI.00150-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zebedee S.L., Richardson C.D., Lamb R.A. Characterization of the influenza virus M2 integral membrane protein and expression at the infected-cell surface from cloned cDNA. J Virol. 1985;56:502–511. doi: 10.1128/jvi.56.2.502-511.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heinen P.P., de Boer-Luijtze E.A., Bianchi A.T. Respiratory and systemic humoral and cellular immune responses of pigs to a heterosubtypic influenza A virus infection. J Gen Virol. 2001;82:2697–2707. doi: 10.1099/0022-1317-82-11-2697. [DOI] [PubMed] [Google Scholar]
- 35.Thomas P.G., Keating R., Hulse-Post D.J., Doherty P.C. Cell-mediated protection in influenza infection. Emer Infect Dis. 2006;12:48–54. doi: 10.3201/eid1201.051237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu M.A., McClements W., Ulmer J.B., Shiver J., Donnelly J. Immunization of non-human primates with DNA vaccines. Vaccine. 1997;15:909–912. doi: 10.1016/s0264-410x(96)00280-0. [DOI] [PubMed] [Google Scholar]
- 37.Okuda K., Ihata A., Watabe S., Okada E., Yamakawa T., Hamajima K. Protective immunity against influenza A virus induced by immunization with DNA plasmid containing influenza M gene. Vaccine. 2001;19:3681–3691. doi: 10.1016/s0264-410x(01)00078-0. [DOI] [PubMed] [Google Scholar]
- 38.Talker S.C., Koinig H.C., Stadler M., Graage R., Klingler E., Ladinig A. Magnitude and kinetics of multifunctional CD4+ and CD8beta+ T cells in pigs infected with swine influenza A virus. Vet Res. 2015;46:52. doi: 10.1186/s13567-015-0182-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bragstad K., Martel C.J., Thomsen J.S., Jensen K.L., Nielsen L.P., Aasted B. Pandemic influenza 1918 H1N1 and 1968 H3N2 DNA vaccines induce cross-reactive immunity in ferrets against infection with viruses drifted for decades. Influenza Other Respir Viruses. 2011;5:13–23. doi: 10.1111/j.1750-2659.2010.00177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma W., Richt J.A. Swine influenza vaccines: current status and future perspectives. Anim Health Res Rev/Conf Res Workers Anim Dis. 2010;11:81–96. doi: 10.1017/S146625231000006X. [DOI] [PubMed] [Google Scholar]
- 41.Reeth K.V., Brown I., Essen S., Pensaert M. Genetic relationships, serological cross-reaction and cross-protection between H1N2 and other influenza A virus subtypes endemic in European pigs. Virus Res. 2004;103:115–124. doi: 10.1016/j.virusres.2004.02.023. [DOI] [PubMed] [Google Scholar]
- 42.Van Reeth K., Gregory V., Hay A., Pensaert M. Protection against a European H1N2 swine influenza virus in pigs previously infected with H1N1 and/or H3N2 subtypes. Vaccine. 2003;21:1375–1381. doi: 10.1016/s0264-410x(02)00688-6. [DOI] [PubMed] [Google Scholar]
- 43.Loeffen W.L., Heinen P.P., Bianchi A.T., Hunneman W.A., Verheijden J.H. Effect of maternally derived antibodies on the clinical signs and immune response in pigs after primary and secondary infection with an influenza H1N1 virus. Vet Immunol Immunopathol. 2003;92:23–35. doi: 10.1016/s0165-2427(03)00019-9. [DOI] [PubMed] [Google Scholar]
- 44.Markowska-Daniel I., Pomorska-Mol M., Pejsak Z. The influence of age and maternal antibodies on the postvaccinal response against swine influenza viruses in pigs. Vet Immunol Immunopathol. 2011;142:81–86. doi: 10.1016/j.vetimm.2011.03.019. [DOI] [PubMed] [Google Scholar]
- 45.Kitikoon P., Nilubol D., Erickson B.J., Janke B.H., Hoover T.C., Sornsen S.A. The immune response and maternal antibody interference to a heterologous H1N1 swine influenza virus infection following vaccination. Vet Immunol Immunopathol. 2006;112:117–128. doi: 10.1016/j.vetimm.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 46.Vincent A.L., Lager K.M., Janke B.H., Gramer M.R., Richt J.A. Failure of protection and enhanced pneumonia with a US H1N2 swine influenza virus in pigs vaccinated with an inactivated classical swine H1N1 vaccine. Vet Microbiol. 2008;126:310–323. doi: 10.1016/j.vetmic.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 47.Fischer L., Barzu S., Andreoni C., Buisson N., Brun A., Audonnet J.C. DNA vaccination of neonate piglets in the face of maternal immunity induces humoral memory and protection against a virulent pseudorabies virus challenge. Vaccine. 2003;21:1732–1741. doi: 10.1016/s0264-410x(02)00736-3. [DOI] [PubMed] [Google Scholar]