Abstract
Identifying variants causal for complex genetic disorders is challenging. With the advent of whole exome and genome sequencing, computational tools are needed to explore and analyze the list of variants for further validation. Correlating genetic variants with subject phenotype is crucial for the interpretation of the disease causing mutations. Often such work is done by teams of researchers who need to share information and coordinate activities. To this end, we have developed a powerful, easy to use web application, ASPIREdb, which allows researchers to search, organize, analyze and visualize variants and phenotypes associated with a set of human subjects. Investigators can annotate variants using publicly available reference databases and build powerful queries to identify subjects or variants of interest. Functional information and phenotypic associations of these genes are made accessible as well. Burden analysis and additional reporting tools allow investigation of variant properties and phenotype characteristics. Projects can be shared, allowing researchers to work collaboratively to build queries and annotate the data. We demonstrate ASPIREdb's functionality using publicly available data sets, showing how the software can be used to accomplish goals that might otherwise require specialized bioinformatics expertise. ASPIREdb is available at http://aspiredb.chibi.ubc.ca.
Keywords: whole genome sequencing, WGS, whole exome sequencing, WES, computational biology, genotype-phenotype, visualization
Background
Whole genome and whole exome sequencing have enabled new genetic analysis modes in which variants across a wide range of allele frequencies are identified and considered for association with traits. At the same time there is an increasing emphasis on deep phenotyping, in which many traits (sometimes conceptualized as endophenotypes) are measured in each individual. When presented with a complex data set of variants and phenotypes across a cohort of individuals, investigators have a large number of possible questions and hypotheses to explore. A common task is prioritizing rare variants for follow-up, often requiring decisions to be made case-by-case based on the function of the gene, the phenotype of the individual, and other factors. Another task often undertaken is burden analysis, in which the prevalence of classes of variants are treated as explanatory variables for traits, as opposed to association of specific variants.
To better characterize the wide spectrum of genetic heterogeneity in genetic disorders, databases of variants and phenotypic data are being assembled in international collaborative efforts. Examples include ClinVar (Landrum et al. 2014), Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources (DECIPHER) (Bragin et al. 2013), Database of Genomic Variants (DGV) (MacDonald et al. 2014), Exome Aggregation Consortium (ExAC) (http://exac.broadinstitute.org/), and dbSNP (Sherry et al. 2001). Such data sets provide crucial background information for data interpretation and are frequently integrated into analysis workflows as filters or comparison sets.
The current work was undertaken in the context of the British Columbia Autism SPectrum Interdisciplinary REsearch (ASPIRE) Programme of the Autism Spectrum Disorders – Canadian-American Research Consortium (ASD-CARC; http://autismresearch.com). An aim of the program is to examine genetic findings in the context of deep phenotyping of patients diagnosed with an autism spectrum disorder (ASD) as well as a comorbid neurodevelopmental disorder such as intellectual disability (ID). As of August 2015, over 700 genes are associated with ASD in the Simons Foundation Autism Research Initiative (SFARI) Gene Database (Banerjee-Basu and Packer 2010), with varying degrees of confidence and associated risk burden. Many of these genes were identified using sequencing approaches. Besides gene-specific findings, genetic and phenotype studies of ASD have generated a range of more general “burden” findings. For example, genomic features such as copy number variation (CNV) size and the number of genes affected are strongly correlated with the severity of the patients phenotype (Vulto-van Silfhout et al. 2013), and 1q21.1 duplications are more prevalent in ASD patients with macrocephaly compared to deletion carriers (Bernier et al. 2015). There have also been multiple attempts to cluster, stratify or characterize ASD based on endophenotypes, such as facial features (Hammond et al. 2008). Additional phenotype assessments such as IQ, speech impairment, behaviour, and motor delay are often evaluated in conjunction with variant analysis (Bernier et al. 2014; Coe et al. 2014; Helsmoortel et al. 2014). Another family of approaches use pathway analyses, gene networks or enrichment approaches to identify patterns linking identified genes (Webber 2011; Neale et al. 2012; Sanders et al. 2012; Andrews et al. 2015). These types of analyses are highly germane to the ASPIRE Programme but also to many other genotype-phenotype studies across a wide range of traits and conditions.
We recognized that the analysis of data captured by clinical medical geneticists and cytogeneticists within the ASPIRE Programme (and projects like it) requires the involvement of bioinformaticians but must also engage clinicians and other researchers who are experts on the conditions under study but do not have bioinformatics skills. Facilitating interaction among these teams, leveraging their respective expertise, and avoiding drowning in a swarm of ad hoc Excel spreadsheets was seen as a priority. Our vision was that clinical research teams could take on the types of analyses described above without extensive bioinformatics support. The need for better tools to facilitate such analysis is especially important for smaller research teams that might not have bioinformatics experts dedicated to the project. With these requirements in mind, we developed ASPIREdb to make analysis and exploration of complex genetics and phenotype data sets accessible to clinical researchers.
In this paper we present the ASPIREdb platform. ASPIREdb is designed to sit at the stage of genome analysis after basic variant filtering and annotation has been done. At this point in a project, investigations turn to seeking patterns of variation and/or phenotypes that have statistical explanatory power (e.g., burden analysis), while also delving into the details of specific genes and variants of interest for potential follow-up studies. As we demonstrate, ASPIREdb can tackle many basic analysis and data exploration tasks in a collaborative environment.
Implementation
Software implementation
ASPIREdb is available through our public-facing instance (http://aspiredb.chibi.ubc.ca) or can be downloaded and installed on a host under the investigator's control. ASPIREdb is free software, released under the Apache 2.0 License. Source code is available at https://github.com/ppavlidis/aspiredb. The ASPIREdb website contains a user's manual with tutorials for using the web browser client, as well as instructions on how to install and run ASPIREdb on a server. Installing ASPIREdb on a server requires the assistance of a systems administrator. The ASPIREdb client works with modern web browsers but has been most extensively tested with Google Chrome.
ASPIREdb's back-end code is written in Java 1.7, using the Java Spring and Hibernate frameworks, backed by a MySQL 5.6 database. User authentication and authorization is managed using Spring Security. The web front end is developed using the ExtJS 4.2 JavaScript framework and DWR 3 for AJAX. The Subject-Phenotype heat map visualization uses the matrix2viz package (https://github.com/azub/matrix2viz/).
Data Sources
The default human genome build is hg19 / GRCh37. Gene annotations are obtained from the Ensembl BioMart API (Smedley et al. 2009). The public instance includes data from DECIPHER (accessed in February 2016) and DGV (accessed in February 2016). Disease-gene annotations are obtained from the Phenocarta database via the web service API (Portales-Casamar et al. 2013).
Analysis Methods
ASPIREdb implements several standard statistical methods for analysis and exploration of data. For burden analysis, we use the Mann-Whitney U Test (Mann and Whitney 1947), a non-parametric test for two-group comparisons, to compare features such as variant length and the number of genes overlapped by variants between subject groups. All other types of burden analyses, such as comparing variant effect type between subject groups, use the Chi-square test to test for significant association between categorical variables (Yates 1934). These statistical tests are regularly used for burden analysis of mutation data (Iossifov et al. 2012; Qiao et al. 2014). Hierarchical clustering, a commonly used data clustering methods (Kaufman and Rousseeuw 2009), is performed using the average linkage algorithm in the matrix2viz package. Compound heterozygote detection is based on the assumptions that variants are unphased, and that two variants in the same gene in the same subject are of potential interest which was inspired by the Gemini tool (Paila et al. 2013).
Results and Discussion
The main ASPIREdb user workflow is outlined in Figure 1 and a list of major features is in Table 1. Use of ASPIREdb requires registration to allow data to be kept private. Once the user has registered and logged in, they are able to create a new project, add data and explore their existing projects. The main ASPIREdb interface is divided into Subject, Variant and Phenotype panels (Figure 2). The following sections give an overview of the use and features of ASPIREdb; additional information and details are available on the web site.
Figure 1.
The main ASPIREdb workflow. Users upload subject genotype and phenotype data in comma-separated values (CSV) format using the data upload interface. Users can upload additional data anytime. Once data has been uploaded, it is displayed in the main ASPIREdb user interface. Filters can then be applied to narrow down the list of subjects and genotypes to examine. Optionally, labels are applied to the filtered subset. Analyses are performed to test hypotheses and reports are generated which can be exported as an image or a text file.
Table 1.
Summary of features in ASPIREdb.
| Feature | Description |
|---|---|
| Security | |
| Access control list | Data access is restricted to project owners, administrators and collaborators. Projects can be shared between users of the same user group. |
| Visualization | |
| Label Manager | Labels help identify subjects or variants of interest visually. Labels can also be used as a filter criteria. |
| Ideogram viewer | Quickly get an overview of where in the genome the variants are located. Variants can be colored by type and label. |
| Phenotype heatmap | Subjects and phenotype values are displayed in a heatmap with automatic clustering along rows and columns. |
| Interactive user interface | Interactive highlighting of subject, variants and phenotypes. |
| Analysis | |
| Query interface | Build simple queries such as filtering variants by location to more complex queries that involve both location and get set overlap. |
| Burden Analysis | Group patients by subject labels and compare differences between subject groups. For example, do patients in one group tend to have more variants that affect genes? Is a group more enriched for a certain phenotype? |
| Gene set manager | Upload set of genes of interest and identify which variants are located in these genes. |
| Compound heterozygote detection | Identify which variants are in the same gene of each patient. |
| Phenotype contingency table | Quickly stratify subjects based on a subset of phenotypes. |
| Gene annotation | Ensembl genes are automatically assigned to variants. |
| Integration | |
| Human Phenotype Ontology integration | Lookup HPO IDs and display the corresponding HPO terms. Parent and child HPO terms are resolved during a Phenotype filter. |
| Gemma integration | Look for genes that are coexpressed with the genes hit by a variant. |
| Phenocarta integration | Display genes associated with an HPO phenotype. |
| UCSC Genome Browser integration | View variants as UCSC tracks and highlight genomic features such as sequence conservation, regulatory regions and expression levels. |
| DECIPHER and DGV Overlap | Identify which variants are found in patients (DECIPHER) and controls (DGV). |
| Reports | |
| Variant Report | Display the frequency of variants for the selected variant characteristic as a table or barchart format. Variant lengths are displayed as a histogram which can be Log2 transformed. Barcharts are grouped by Subject Labels. |
Figure 2.
Overview of the ASPIREdb web interface. ASPIREdb is composed of three main panes. The Subject pane (A) shows the list of subjects and the total number of variants and phenotypes for each subject, as well as user-defined labels. The Variant pane has both Ideogram (B) and table (C) views as separate tabs (the table view in the figure is composed from a separate screen shot with the table tab active). The ideogram displays the location of variants along the chromosomes; variants that overlap are displayed as a pile up. Users can select a region in the ideogram of interest and subsequently display the region in the UCSC Genome Browser for more details. The tabular view shows the locations of the variants, genes overlapped, and other user-defined variant characteristics in a table format (C) The Phenotype pane (D) shows all phenotypes in the project and the number of subjects for each phenotype value. User-defined labels (E, for variants) allow for quick scanning and sorting through the list of subjects and variants and can be used as filters. In this example, variants have been labeled if they are de novo and whether they are gains or losses; samples are labeled if they contain a de novo variant.
Data upload
To upload data into ASPIREdb, users first create a project by providing a project name. The user then uploads their data into this project from simple text formatted files. The key data entities in an ASPIREdb project are subjects (study participants), variants and phenotypes. At the minimum a project requires variants and subjects, while phenotypes are optional. ASPIREdb is designed for variants at intermediate analysis stages, after variants with low quality or those considered unlikely to have functional impact have been removed: it is not meant to handle millions of raw variant calls. We have tested interactive use of ASPIREdb with data sets of up to ~20000 variants.
In the variant upload file format, each row represents a variant with a user-provided subject identifier, the variant's location, and optionally, the variant's characteristics (meta-data) which are user-defined. Variant characteristics can include inheritance, effect score, and effect type, quality measures and so forth. For phenotype inputs, the user provides a separate text file in which each row represents a subject and the phenotype values associated with that subject. Phenotype names can be entered as plain text (e.g. gender) or as Human Phenotype Ontology (HPO) IDs (Groza et al. 2015), which are automatically recognized by ASPIREdb. Since HPO terms are structured and standardized, it is possible to perform semantic inferences and comparisons with other projects, such as automatic retrieval of genes associated with the phenotype selected. Further details about the input formats are available in the online documentation.
ASPIREdb is not data management tool, so it does not provide the means for editing variant information directly. Additional variants and subjects can be appended to an existing project by using the upload tools. Updates to existing data, or adding additional metadata to existing variants will be supported in a future release.
Visualizing and analyzing variants
Once the data is loaded, users can start to explore data using various tools in ASPIREdb. Genomic hotspots can be visually identified in the Variant Ideogram pane by identifying regions where the variants are piled up next to the chromosome. There are several ways of coloring the variants in the ideogram. Selected variants can be colored by subject label, by variant label or by variant type. Users can zoom in on selected regions or focus on a single chromosome, with details of variants provided as mouse-over text. To see additional information about the highlighted region such as epigenetic marks, users can highlight the region and select the “View in UCSC Genome Browser” action. This will launch the UCSC Genome Browser in a pop-up window with the list of variants as added tracks (Rosenbloom et al. 2015). Selecting a chromosomal region in the ideogram will highlight all subjects in the subject pane having variants in that region and similarly, all variants in that region are highlighted in the variant pane. The Phenotype Heatmap tool provides a visual summary of phenotype values for selected subjects. The rows (subjects) and columns (phenotypes) are clustered based on similarity. Distributions of variant attributes such as variant type and length are available through the “Variant Report” tool. The “View compound heterozygotes” action displays the list of genes that have two variants from the same patient under the assumptions that variants are heterozygous.
Building variant and subject filters
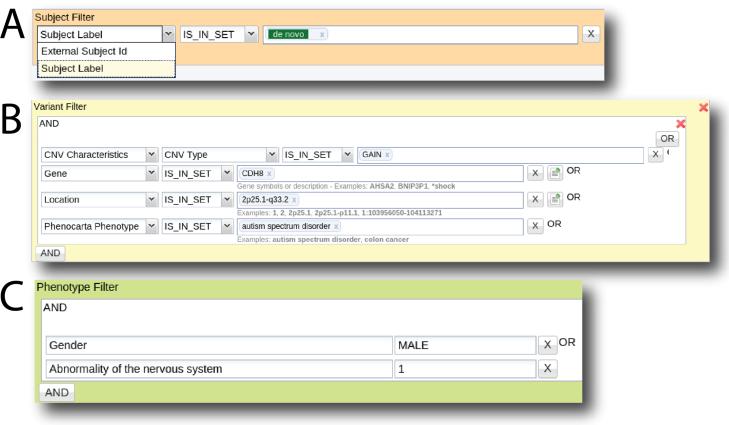
One of the most powerful features of ASPIREdb is its query builder for filtering subjects and/or variants. Filters are set up and accessed through the Filter panel (Figure 3). Filter expressions can be as simple as a single variant filter expression with just a variant location or length to more complex, involving subject, variant, phenotype, gene and project overlap filters combined using logical disjunction and conjunction operators. Users can also define Gene Sets through the “Gene Set Manager” tool and use it to filter the list of variants that overlap the gene set. To assist with the query construction, users can save queries and preview the query results to determine the number of subject and variants that will be returned. After the query is run, the filtered list of subjects and variants can be assigned labels, and these labels can then be used in other expression filters.
Figure 3.
Query builder for filtering variants and/or subjects. The filter panels are used for building simple or complex expressions. Subject filters (A) are useful for finding subjects with a specific ID or label. Variant filters (B) support queries that involve any of the required and user-defined variant characteristics such as variant type, location, effect and inheritance. Locations can be entered as base coordinates (e.g. 1:146577487-147394506) or as a cytogenetic band location (e.g. 1q21.1, 16p11.2, etc.). Genes and gene sets can also be used as a variant filter. The Phenocarta Phenotype filter allows for filtering variants that overlap genes associated with a particular phenotype from the Phenocarta database. Phenotype filters (C) are for filtering subjects which matches the specified phenotype of interest. Importantly, filters can be combined to create more complex queries.
Studying gene function
ASPIREdb provides a convenient way of accessing the gene's expression patterns and association with diseases through the Gemma (Zoubarev et al. 2012) and Phenocarta (Portales-Casamar et al. 2013) databases respectively. The “View genes” action allows users to see the list of genes overlapped by variants and see the coexpression network using data from published and curated microarray studies in Gemma. ASPIREdb queries the Phenocarta database to obtain the list of genes that are known to be associated with the phenotype's HPO ID. The list of genes can then be saved to a Gene Set and used in expression filters.
Data export
All of the ASPIREdb panels support extraction of the data either shown as tab-delimited text or shown as graphic, which can be downloaded in PNG-format. This makes it easy to move data from ASPIREdb to other tools or for preparing publication-ready materials.
Relationship to other tools
In this section we describe the relationship of ASPIREdb to other tools that operate on similar data, and highlight the extent to which they overlap with ASPIREdb's functionality and places where they differ.
As described above, ASPIREdb does not attempt to replace the entire genetics data analysis stack, and in fact it is most beneficial if applied to data that has been carefully prepared using other tools. Most importantly, many tools allow researchers to quickly annotate SNVs with information on predicted functional impact, such as SIFT (Ng and Henikoff 2003), PolyPhen (Adzhubei et al. 2010) and CADD (Kircher et al. 2014). Such variant impact calls are often included as part of commodity sequencing analysis reports. Because many potential ASPIREdb users already have impact calls, the multiplicity of approaches, and the rapid developments in methodologies, ASPIREdb does not compute variant effect scores, but they can readily be incorporated as part of the variant upload.
For more complex evaluation of variants and phenotypes, most approaches require facility with statistical computing environments and scripting. Options for interactive data analysis are limited. Family Genome Browser is a web-based tool that enables visualization and browsing of complex familial genomes (Juan et al. 2015). Aside from highlighting variants and scanning for de novo and compound heterozygotes, it does not provide any search tools, a key feature of ASPIREdb. Data in Family Genome Browser are stored locally in the browser's cache and thus cannot be worked with collaboratively. In contrast ASPIREdb requires registration and all data is associated with the user's account. MedSavant Variant Search Engine (http://genomesavant.com/p/medsavant/) is a desktop client-server variant search engine, offering annotation, filtration and variant prioritization. It enables researchers to organize patients into families, cohorts and provide phenotypes using HPO. MedSavant focuses on variant search and apparently does not provide analysis tools, though because it has a plugin framework it is extensible by third party software developers. Both Family Genome Browser and MedSavant support the representation of family structures as pedigrees, a feature currently not implemented in ASPIREdb. VariantDB is a web-based system that annotates variants with allele frequencies, functional impact, pathogenicity and pathway information (Vandeweyer et al. 2014). It allows for filtering by the chosen inheritance model and also uses the HPO for annotating phenotypes. However VariantDB does not allow for easy navigation between patients and phenotypes, nor does VariantDB provide phenotype-based filtering. Finally, ClinLabGeneticist is a software that facilitates the evaluation of pathogenic variants by providing the means to enter, annotate, review, select, report and assign candidate variants among reviewers (Wang et al. 2015). However, it is currently limited to SNVs and patient phenotypes cannot be entered into the system, while ASPIREdb supports multiple variant types as well as phenotypes. A table comparing and summarizing major features of these tools is given in the Supporting Information (Supp. Table S1). We have not attempted evaluation of commercial tools such as Golden Helix Varseq (http://goldenhelix.com/), which may offer overlapping features but differ from ASPIREdb in cost (ASPIREdb being free and open source).
Application studies
ASPIREdb is designed to address common use cases in variant and phenotype analysis. Indeed, we were motivated to create tools that would allow investigators to perform analyses that previously had been done by a dedicated bioinformatician. To demonstrate how ASPIREdb can be applied, we have developed two case studies based on published genome analyses. For each study, we show that ASPIREdb can be used to reach similar conclusions as those of the published study. Here we provide overviews, and detailed information for each application study is available on the ASPIREdb web site.
Scenario 1. Burden analyses of de novo, familial and common CNVs and phenotypes in an intellectual disability cohort
Qiao et al. (2014) identified de novo, familial and common CNVs in 78 subjects with intellectual disability using array-CGH (Qiao et al. 2014), and present a number of burden and phenotype association analyses of the data. As we show here most of the analyses presented by Qiao et al. can be conducted directly in ASPIREdb.
A total of 248 CNVs (200 of which were considered common) and 40 phenotypes from 78 subjects were uploaded to ASPIREdb. Subjects were labeled as having only common (n=40), de novo (n=18), or familial (n=20) variants based on their CNV type as provided. Qiao et al. reported significantly more genes covered by de novo than familial or common CNVs. The same analysis can be replicated in ASPIREdb, using the Burden Analysis Tool (Figure 4), comparing the number of genes per CNV between subjects with de novo CNVs (Group 1) versus those with only familial CNVs (Group 2). Consistent with the Qiao et al., ASPIREdb reports that there are more genes per CNV in the de novo group compared to the familial group (P < 0.001, Mann-Whitney U test). In a similar manner ASPIREdb can be used to replicate other findings of Qiao et al. such as the tendency for de novo CNVs to be longer than familial CNVs (P < 0.001, Mann-Whitney U test) and muscle abnormalities being more frequently seen in cases with familial CNVs than in cases with common CNVs (P < 0.05, Mann-Whitney U test).
Figure 4.
Burden analyses of the Qiao et al. data. (A) Subject group comparison between subjects with de novo CNVs (Group 1) vs. subjects with familial CNVs (Group 2). (B) Phenotype enrichment between subjects with familial vs. common CNVs.
Scenario 2. Burden analysis of de novo gene-disrupting mutations between affected and unaffected children with autism spectrum disorder
To show how ASPIREdb can be used to analyze SNVs, we take as an illustration the results of a published ASD WES study (Iossifov et al. 2012). Iossifiov et al. reported 840 quality-filtered and annotated de novo SNVs (n=754) and indels (n=86) in individuals with autism from 305 families. This resulted in a total of 475 subjects: 239 cases, 225 controls and 11 subjects from the same family with de novo SNVs that are in common between the affected child and their sibling. There are 332 males, 140 females and 3 subjects with no gender annotations. The ASPIREdb report tool shows that 88 variants are de novo loss of function as annotated by Iossifov et al. (47 frameshift, 30 nonsense, and 11 splice-site) (Supp. Figure S1).
Many of the analyses reported by Iossifov et al. can be done in ASPIREdb, such as burden analysis. For example, Iossifov et al. reported that affected children tended to have more loss of function mutations, which is readily recapitulated in ASPIREdb using the burden analysis tool (56/239 affected vs. 25/225 unaffected). As in Table 2 and 4 of Iossifov et al., it is easy in ASPIREdb to generate a summary table of de novo SNVs and indels in affected vs. unaffected subjects grouped by variant effect type (Supp. Figures S2 and S3). Additional analyses of the Iossifov et al. data are described on the ASPIREdb web site (http://aspiredb.chibi.ubc.ca).
Conclusions
We have presented a web-based platform, ASPIREdb, which allows genetics researchers to navigate, search and analyze their study genotype and phenotype data. In our groups, we are actively using ASPIREdb in studies of ASD and ID. Our use in practice has helped us identify areas for future improvements and features. Some planned enhancements include additional visualization and analysis tools. An example of one area of possible extension is in the clustering of phenotypes; currently no statistical interpretation of the clusters is offered. Integration of additional reference or disease-associated variant lists such as from ClinVar and reference databases such as ExAC would increase the ability of researchers to quickly evaluate their variants in the context of the literature. Currently such information can be incorporated as variant meta-data at upload, but direct inclusion in the system would absolve users of the need to do this step themselves. The integration of gene set enrichment analysis such as ErmineJ (Gillis et al. 2010) would provide another method for interpretation of candidate variants. We are interested in receiving suggestions, feedback, bug reports and any support requests from the community, which can be directed to aspiredb@chibi.ubc.ca.
Supplementary Material
Acknowledgments
We thank members of the Pavlidis, Lewis and Separovic labs for input on design and for additional testing. We thank Michelle Ly for additional programming contributions. The original impetus for the development ASPIREdb was the vision of the late Professor Jeanette Holden; this paper is dedicated to her memory. This study makes use of data generated by the DECIPHER community. A full list of centres that contributed to the generation of the data is available from http://decipher.sanger.ac.uk and via email from decipher@sanger.ac.uk. Supported by the Canadian Foundation for Innovation Leading Edge Fund # 19924, the British Columbia Knowledge Development Fund (UBC 11R67180, 11R67181) and the NIH grant (R01 GM076990).
Footnotes
Authors’ contributions
PP and SL conceived of the study. PP provided overall project leadership. SR led feature design and testing. PP and PPT drafted the manuscript with input from the other authors.
PPT, AZ, CM, FL, GC, MJ, MB, JL and TVR implemented software and participated in design, testing and documentation.
EPC, YQ, KC, and ERS participated in system design (use cases and requirements) and contributed to testing.
All authors read and approved the final manuscript.
Conflict of Interest
None declared.
Competing interests
None of the authors have any competing interests.
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews T, Meader S, Vulto-van Silfhout A, Taylor A, Steinberg J, Hehir-Kwa J, Pfundt R, Leeuw N de, Vries BBA de, Webber C. Gene Networks Underlying Convergent and Pleiotropic Phenotypes in a Large and Systematically-Phenotyped Cohort with Heterogeneous Developmental Disorders. PLoS Genet. 2015;11:e1005012. doi: 10.1371/journal.pgen.1005012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee-Basu S, Packer A. SFARI Gene: an evolving database for the autism research community. Dis. Model. Mech. 2010;3:133–135. doi: 10.1242/dmm.005439. [DOI] [PubMed] [Google Scholar]
- Bernier R, Golzio C, Xiong B, Stessman HA, Coe BP, Penn O, Witherspoon K, Gerdts J, Baker C, Vulto-van Silfhout AT, Schuurs-Hoeijmakers JH, Fichera M, et al. Disruptive CHD8 Mutations Define a Subtype of Autism Early in Development. Cell. 2014 doi: 10.1016/j.cell.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier R, Steinman KJ, Reilly B, Wallace AS, Sherr EH, Pojman N, Mefford HC, Gerdts J, Earl R, Hanson E, Goin-Kochel RP, Berry L, et al. Clinical phenotype of the recurrent 1q21.1 copy-number variant. Genet. Med. Off. J. Am. Coll. Med. Genet. 2015 doi: 10.1038/gim.2015.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragin E, Chatzimichali EA, Wright CF, Hurles ME, Firth HV, Bevan AP, Swaminathan GJ. DECIPHER: database for the interpretation of phenotype-linked plausibly pathogenic sequence and copy-number variation. Nucleic Acids Res. 2013;42:D993–D1000. doi: 10.1093/nar/gkt937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe BP, Witherspoon K, Rosenfeld JA, Bon BWM van, Vulto-van Silfhout AT, Bosco P, Friend KL, Baker C, Buono S, Vissers LELM, Schuurs-Hoeijmakers JH, Hoischen A, et al. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat. Genet. 2014;46:1063–1071. doi: 10.1038/ng.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis J, Mistry M, Pavlidis P. Gene function analysis in complex data sets using Ermine. J. Nat. Protoc. 2010;5:1148–1159. doi: 10.1038/nprot.2010.78. [DOI] [PubMed] [Google Scholar]
- Groza T, Köhler S, Moldenhauer D, Vasilevsky N, Baynam G, Zemojtel T, Schriml LM, Kibbe WA, Schofield PN, Beck T, Vasant D, Brookes AJ, et al. The Human Phenotype Ontology: Semantic Unification of Common and Rare Disease. Am. J. Hum. Genet. 2015 doi: 10.1016/j.ajhg.2015.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond P, Forster-Gibson C, Chudley AE, Allanson JE, Hutton TJ, Farrell SA, McKenzie J, Holden JJA, Lewis MES. Face–brain asymmetry in autism spectrum disorders. Mol. Psychiatry. 2008;13:614–623. doi: 10.1038/mp.2008.18. [DOI] [PubMed] [Google Scholar]
- Helsmoortel C, Vulto-van Silfhout AT, Coe BP, Vandeweyer G, Rooms L, Ende J van den, Schuurs-Hoeijmakers JHM, Marcelis CL, Willemsen MH, Vissers LELM, Yntema HG, Bakshi M, et al. A SWI/SNF-related autism syndrome caused by de novo mutations in ADNP. Nat. Genet. 2014;46:380–384. doi: 10.1038/ng.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, Yamrom B, Lee Y, Narzisi G, Leotta A, Kendall J, Grabowska E, et al. De Novo Gene Disruptions in Children on the Autistic Spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan L, Liu Y, Wang Y, Teng M, Zang T, Wang Y. Family genome browser: visualizing genomes with pedigree information. Bioinforma. Oxf. Engl. 2015 doi: 10.1093/bioinformatics/btv151. [DOI] [PubMed] [Google Scholar]
- Kaufman L, Rousseeuw PJ. Finding Groups in Data: An Introduction to Cluster Analysis. John Wiley & Sons; 2009. [Google Scholar]
- Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, Maglott DR. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald JR, Ziman R, Yuen RKC, Feuk L, Scherer SW. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 2014;42:D986–992. doi: 10.1093/nar/gkt958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann HB, Whitney DR. On a Test of Whether one of Two Random Variables is Stochastically Larger than the Other. Ann. Math. Stat. 1947;18:50–60. [Google Scholar]
- Neale BM, Kou Y, Liu L, Ma'ayan A, Samocha KE, Sabo A, Lin C- F, Stevens C, Wang L- S, Makarov V, Polak P, Yoon S, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012 doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paila U, Chapman BA, Kirchner R, Quinlan AR. GEMINI: Integrative Exploration of Genetic Variation and Genome Annotations. PLoS Comput Biol. 2013;9:e1003153. doi: 10.1371/journal.pcbi.1003153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portales-Casamar E, Ch'ng C, Lui F, St-Georges N, Zoubarev A, Lai AY, Lee M, Kwok C, Kwok W, Tseng L, Pavlidis P. Neurocarta: aggregating and sharing disease-gene relations for the neurosciences. BMC Genomics. 2013;14:129. doi: 10.1186/1471-2164-14-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y, Mercier E, Dastan J, Hurlburt J, McGillivray B, Chudley AE, Farrell S, Bernier FP, Lewis MS, Pavlidis P, Rajcan-Separovic E. Copy number variants (CNVs) analysis in a deeply phenotyped cohort of individuals with intellectual disability (ID). BMC Med. Genet. 2014;15:82. doi: 10.1186/1471-2350-15-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbloom KR, Armstrong J, Barber GP, Casper J, Clawson H, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L, Haeussler M, Harte RA, Heitner S, et al. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res. 2015;43:D670–681. doi: 10.1093/nar/gku1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, Ercan-Sencicek AG, DiLullo NM, Parikshak NN, Stein JL, Walker MF, Ober GT, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012 doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward M- H, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedley D, Haider S, Ballester B, Holland R, London D, Thorisson G, Kasprzyk A. BioMart--biological queries made easy. BMC Genomics. 2009;10:22. doi: 10.1186/1471-2164-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandeweyer G, Van Laer L, Loeys B, Van den Bulcke T, Kooy RF. VariantDB: a flexible annotation and filtering portal for next generation sequencing data. Genome Med. 2014;6:74. doi: 10.1186/s13073-014-0074-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulto-van Silfhout AT, Hehir-Kwa JY, Bon BWM van, Schuurs-Hoeijmakers JHM, Meader S, Hellebrekers CJM, Thoonen IJM, Brouwer APM de, Brunner HG, Webber C, Pfundt R, Leeuw N de, et al. Clinical significance of de novo and inherited copy-number variation. Hum. Mutat. 2013;34:1679–1687. doi: 10.1002/humu.22442. [DOI] [PubMed] [Google Scholar]
- Wang J, Liao J, Zhang J, Cheng W- Y, Hakenberg J, Ma M, Webb BD, Ramasamudramchakravarthi R, Karger L, Mehta L, Kornreich R, Diaz GA, et al. ClinLabGeneticist: a tool for clinical management of genetic variants from whole exome sequencing in clinical genetic laboratories. Genome Med. 2015;7:77. doi: 10.1186/s13073-015-0207-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber C. Functional enrichment analysis with structural variants: pitfalls and strategies. Cytogenet. Genome Res. 2011;135:277–285. doi: 10.1159/000331670. [DOI] [PubMed] [Google Scholar]
- Yates F. Contingency Tables Involving Small Numbers and the χ2 Test. Suppl. J. R. Stat. Soc. 1934;1:217–235. [Google Scholar]
- Zoubarev A, Hamer KM, Keshav KD, McCarthy EL, Santos JRC, Van Rossum T, McDonald C, Hall A, Wan X, Lim R, Gillis J, Pavlidis P. Gemma: a resource for the reuse, sharing and meta-analysis of expression profiling data. Bioinforma. Oxf. Engl. 2012;28:2272–2273. doi: 10.1093/bioinformatics/bts430. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.