Abstract
Evidence on whether genetic predictors of Alzheimer’s disease (AD) also predict memory decline is inconsistent and limited data are available for African ancestry populations. For 8,253 non-Hispanic white (NHW) and non-Hispanic black (NHB) Health and Retirement Study participants with memory scores measured one to eight times between 1998–2012 (average baseline age=62), we calculated weighted polygenic risk scores (AD-GRS) using the top 22 AD-associated loci, and an alternative score excluding APOE (AD-GRSexAPOE). We used generalized linear models with AD-GRS-by-age and -age2 interactions (age centered at 70) to predict memory decline. Average NHB decline was 26% faster than NHW decline (p<0.001). Among NHW, 10% higher AD-GRS predicted faster memory decline (linear β= −0.058 unit decrease over 10 years; 95% CI: −0.074, −0.043. AD-GRSexAPOE also predicted faster decline for NHW, although less strongly. Among NHB, AD-GRS predicted faster memory decline (linear β= −0.050; 95% CI: −0.106, 0.006), but AD-GRSexAPOE did not. Our non-significant estimate among NHB may reflect insufficient statistical power or a misspecified AD-GRS among NHB since an overwhelming major of GWAS studies are conducted in NHW. A polygenic score based on previously identified AD loci predicts memory loss in U.S. blacks and whites.
Keywords: Dementia, Genetics, Race, Memory, Decline, Cognition
INTRODUCTION
Genome-wide association studies (GWAS) have implicated several genetic loci in the development of Alzheimer’s disease (AD)1–5. Because AD diagnoses are potentially influenced by both pre-morbid level of cognitive function and rate of cognitive decline6, it is important to confirm associations of genetic polymorphisms with longitudinal rate of memory change, the hallmark of AD. However, apart from the apolipoprotein E ε4 allele (APOE4), the reported effect sizes of individual genetic loci associated with AD are generally small7. Polygenic risk scores can help evaluate the joint effects of multiple previously identified genetic variants, each of which may have effects too small to reliably detect in independent samples8,9.
To date, only three studies have evaluated the association of a polygenic risk score for AD and cognition in older adults, and findings were inconsistent9–11. Further, only one study has investigated the association between a polygenic risk score for AD and rate of cognitive decline11. An additional major gap in prior literature on AD-related genotypes is information on whether these loci predict outcomes for non-white populations. For example, APOE4 has not been consistently linked to rate of decline in non-white populations12. This inconsistency may be largely due to limited sample sizes in studies of non-whites, but there could be race-based genetic differences in susceptibility to AD13. Limited research has been conducted in non-whites examining associations between other genetic loci and AD, cognitive function, or decline12,14. It is important to conduct studies in racially diverse samples because genetic markers for disease discovered among primarily European ancestry populations may not be associated with disease in other racial groups for many reasons. First, individuals from different racial groups with diverse ancestry make-up are more likely to have different linkage disequilibrium (LD) patterns, e.g., the correlations among specific genetic variants may differ for blacks and whites15. Thus, if certain SNPs are in LD with a causal genetic variant among European ancestry samples, that does not guarantee that the same relationship will hold in non-European populations. Second, epistasis, or gene-gene interactions, as well as locus and allelic heterogeneity, could operate differently in diverse samples 16,17. Third, environmental modifiers – such as socioeconomic status and health behaviors – could impact SNP effects differently in European and non-European ancestry populations16,18. Fourth and finally, minor allele frequencies may vary between ancestral groups and could therefore alter the detectable effect sizes for those SNPs18.
The potential for survival bias is especially important to assess in research on determinants of cognitive decline because death rates are high in older populations and rate of cognitive decline predicts mortality19,20. However, survival bias only poses a problem if a study’s exposure of interest, in addition to the outcome, is predictive of mortality. Our exposure, genetic variants associated with AD, may impact mortality. Some research has found that carriers of the APOE4 allele have higher mortality rates than non-carriers21,22. Yet, aside from the APOE studies, few published studies assess whether genetic variants associated with dementia risk are also associated with survival. To add to this literature, we directly assess the likelihood of survival bias by testing whether the AD-GRS predicts survival and dropout after DNA collection.
We report here the associations of a 22-locus polygenic risk score (AD-GRS) with memory decline in non-Hispanic whites and blacks in the Health and Retirement Study (HRS). We report the findings for the AD-GRS both including and excluding APOE in order to test whether other loci besides APOE add to the prediction of memory decline. We hypothesized that the AD-GRS, whether constructed including or excluding the APOE gene, would predict rate of memory decline in both black and white respondents.
METHODS
Study population
HRS is a nationally representative cohort study initiated in 1992 with enrollments in 1992, 1993, 1998, 2004 and 2010. The target population is all non-institutionalized adults in the contiguous United States aged 50+ at enrollment23, but spouses of enrolled individuals are also interviewed even if aged <50. Biennial interviews (or proxy interviews for decedent or severely impaired participants) including memory assessments are available through 2012. Details of the study are provided elsewhere24–26.
Our analyses used a sub-sample with genetic data collected in 2006 or 2008, using repeated memory assessments (up to 8, from 1998–2012) on the same individuals. From 12,123 HRS participants with genetic data, we restricted to 10,728 (88.5%) who self-identified as non-Hispanic white or non-Hispanic black and contributed at least one cognitive assessment from 1998–2012. From 85,824 possible observations (10,728 participants by 8 time points) we excluded observations missing memory score due to non-response (23,565, 27.5%), death (2,705, 3.2%), or recorded when the respondent was aged <50 years (1,049, 1.2%). This translated to excluding 2,475 people (23.0%) with a resulting analytic sample of 8,253 individuals contributing 58,505 memory score observations.
Memory Score Outcomes
We used a previously developed memory composite score combining direct and proxy memory assessments for longitudinal analyses27. All HRS participants who were interviewed directly were asked to complete an immediate and delayed recall test based on a 10-word list. For individuals too impaired to directly participate in memory assessments, proxy informants, typically spouses, assessed the participants’ memory on a 5-item Likert scale and completed a 16-item version of the Informant Questionnaire for Cognitive Decline (IQCODE). We developed an algorithm to integrate direct and proxy assessments in order to retain severely impaired individuals in longitudinal studies of cognitive function. The composite score algorithm was developed in an 856-subject subsample who participated in a comprehensive neuropsychological battery as part of the Aging, Demographics, and Memory Study. We standardized the memory score by dividing each score by the 1995 standard deviation so that every unit change in memory score corresponds to one standard deviation in the population prior to baseline.
Genotyping
In 2006 and 2008, HRS invited participants to provide DNA samples (the sample was randomly split across two years, average age=68). Eligible respondents were consented and provided saliva via a mouthwash technique (2006) or an Oragene DNA self-collection kit (2008). Genotyping was completed on the Illumina Omni-2.5 chip platform and imputed using the 1000G phase 1 reference panel. Genetic information for the first 12,123 participants was filed with the Database for Genotypes and Phenotypes (dbGaP, study accession number: phs000428.v1.p1) in April 2012. Sample eigenvectors were derived from principal component analysis as implemented in the R SNPRelate package28. Exact information on the quality control procedures applied is available via HRS and dbGaP 29.
Alzheimer’s Disease Genetic Risk Score (AD-GRS)
Two SNPs (rs7412 and rs429358) are commonly used to identify APOE4 variants30. Twenty-one other genetic loci have been confirmed as genome-wide significant predictors of AD, with meta-analyzed odds ratios (ORs) reported most recently in the Lambert et al. meta-analysis1. We used proxy SNPs in LD with the SNPs reported in the Lambert et al. paper for four of the twenty-one loci. We calculated the AD-GRS by multiplying each individual’s risk allele count for each locus by the beta coefficient for that polymorphism as reported by Lambert et al. (except for APOE, where we used the beta coefficient reported by AlzGene31) and summing the products for all 22loci (for a list of SNPs and beta weights please see Table, Supplemental Digital Content 1). This step essentially weighted each polymorphism in proportion to its anticipated effect on dementia risk. Next, to convert to the odds of dementia for each individual, we exponentiated the weighted allele sum, multiplied the resulting value by 0.1 (the estimated dementia prevalence in the sample), and converted odds to probabilities.
The AD-GRS can be interpreted as the probability of dementia predicted by the 22 alleles, based on the strength of the associations estimated in previously published GWAS and meta-analyses. We also calculated an alternative AD-GRS excluding APOE to assess whether the other 21 loci contributed information in predicting memory loss. In separate analyses, we examined performance of two ABCA7 SNPs separately from the polygenic score because of evidence that rs115550680 performs better in blacks4 compared to the original ABCA7 SNP (rs3764650) used in our AD-GRS.
Age
Age was calculated as the time between self-reported date of birth and interview date, measured continuously and centered at 70 years.
Race
We used self-reported race (“What race do you consider yourself to be: White, Black or African American, American Indian, Alaska Native, Asian, Native Hawaiian, Pacific Islander, or something else?”) and ethnicity (“Do you consider yourself Hispanic or Latino?”) to restrict our sample to participants who identified as non-Hispanic white (NHW) and non-Hispanic black (NHB) only, due to small sample sizes in other groups.
Death
We used mortality information obtained via National Death Index (NDI) linkage from 2010–2012; information from proxy interviews was used for individuals without NDI information.
Dropout
Dropout was defined as the first wave a respondent was missing the composite memory score starting in 2010 (after genetic assessment in either 2006 or 2008).
Other covariates
All models were adjusted for baseline age, sex, and eigenvectors to control for population stratification32.
Statistical analysis
We used generalized linear models to estimate the association between the AD-GRS and rate of memory decline assessed from 1998–2012, using clustered standard errors to account for repeated measures on the same individual and an autoregressive covariance structure. We model a quadratic growth curve for memory decline by including AD-GRS-by-age and AD-GRS-by-age2 interaction terms. Using age as the time dimension allows us to reduce concerns about practice effects, take advantage of between-person differences that have accrued by the time of enrollment, thus improving statistical power, and is appropriate since the temporal order of exposure and outcome are clear. Next, this model was replicated using the alternative AD-GRSexAPOE which excluded APOE. We plotted memory trajectories for each racial group and each specification of the AD-GRS for a reference group of 70 year old females. When calculating these curves we used the point estimates from the models, regardless of statistical significance. In order to directly test whether there was evidence for a differential effect by race of the AD-GRS both with and without APOE on memory decline, we estimated a race-pooled model with race-by-AD-GRS-by-age and race-by-AD-GRS-by-age2 interaction terms, as well as a race-by-age interaction term. In addition, we estimate a race-pooled model adjusting only for race and age (as timescale) and their interaction terms in order to assess whether there are differences in rate of decline by race in our sample.
We examined whether the association of the AD-GRS (with and without APOE) and rate of memory declined prevailed in both middle aged and older adults by estimating the same models stratified by age (+/− 65 years). Using the alternative AD-GRS excluding APOE, we also estimated models controlling separately for APOE status to attempt to increase the precision of our estimates. We examined the two ABCA7 SNPs, rs115550680 and rs3764650, in separate models as predictors of memory change.
Finally, we conducted analyses to ascertain whether the AD-GRS predicted death and dropout and whether these associations differed by race and prior memory score. For this analysis, we used two pooled logistic models where the outcomes were death and dropout from 2008–2012 (using data from 2010 and 2012 to capture events in those four years) and the predictors were AD-GRS as well as AD-GRS-by-race and AD-GRS-by-prior memory score (2004) interaction terms.
RESULTS
Characteristics from the first wave each individual contributed an observation are shown for the 7,172 NHW and 1,081 NHB used in our models (Table 1). Average follow-up was 12.3 years for NHW and 11.3 years for NHB (of 14 possible). On average, NHW contributed 7.2 cognitive assessments and NHB contributed 6.6 (of 8 possible). Average NHB rate of decline was approximately 26% faster than the NHW rate (p<0.001 for race-by-age interaction term).
Table 1.
Baseline characteristics of the sample by race, Health and Retirement Study 1998
| Non-Hispanic white | Non-Hispanic black | |
|---|---|---|
| N=7,172 | N=1,081 | |
| Demographics | ||
| Male, No. (%) | 2,927 (40.8%) | 364 (33.7%) |
| Age, mean (SD), years | 63.0 (8.4) | 61.6 (8.0) |
| Education, mean (SD), years | 13.1 (2.5) | 11.4 (3.3) |
| Alzheimer’s Disease Genetic Risk Score | ||
| AD-GRS, mean (SD) | 1.004 (0.409) | 0.811 (0.368) |
| AD-GRS no APOE, mean (SD) | 0.946 (0.271) | 0.714 (0.226) |
| Cognitive Outcome | ||
| Memory Score, mean (SD) | 1.244 (0.310) | 0.843 (0.367) |
| Health Conditions and Behaviors | ||
| Ever diagnosed with stroke, No. (%) | 230 (3.2%) | 48 (4.4%) |
| Ever diagnosed with diabetes, No. (%) | 565 (7.9%) | 196 (18.1%) |
| Ever diagnosed with hypertension, No. (%) | 2,554 (35.7%) | 626 (58.0%) |
| Ever diagnosed with heart problems, No. (%) | 1,042 (14.5%) | 141 (13.1%) |
| Ever smoked, No. (%) | 4,105 (57.6%) | 627 (58.3%) |
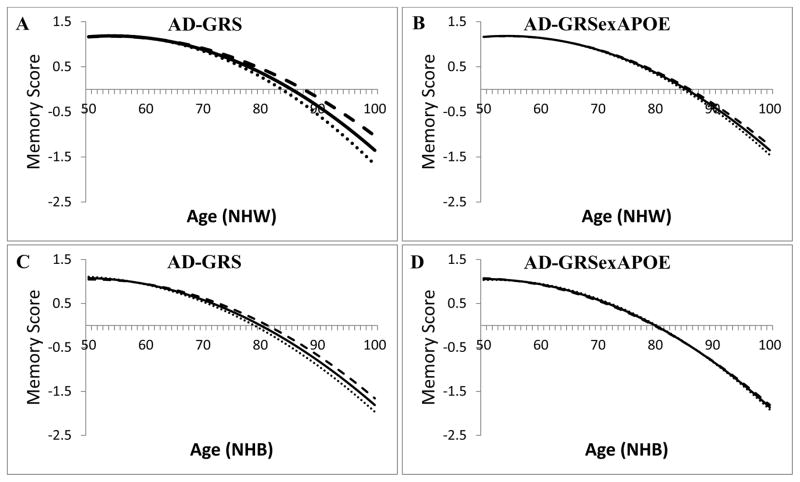
Among NHW, in the model using the AD-GRS including APOE, there was a significant negative interaction between the AD-GRS and age (β= −0.058 for a 10% higher AD-GRS and 10 year increase in age; 95% CI: −0.074, −0.043; p<0.0001) and the AD-GRS and age2 (β= −0.002; 95% CI: −0.003, −0.001; p=0.004), suggesting that an increase in the AD-GRS is associated with faster rate of memory decline, which accelerated with age (Table 2). In the model using the AD-GRS excluding APOE (AD-GRSexAPOE), effect sizes for linear (β= −0.029; 95% CI: −0.050, −0.007; p=0.008) and quadratic (β= −0.001; 95% CI: −0.003, 0.001; p=0.227) terms were smaller in absolute magnitude than effects when using the AD-GRS including APOE, and only the linear age term was significant. Models excluding the quadratic age terms were similar (see Table, Supplemental Digital Content 2). For example, a 70-year-old with an average AD-GRS score would be expected to decline 0.50 memory score units by age 80; a similar individual with a 1SD higher AD-GRS would be expected to decline 0.54 memory score units by age 80. Alternatively, while a 70-year-old with an average AD-GRSexAPOE score would also be expected to decline 0.50 memory score units by age 80; a similar individual with a 1 SD higher AD-GRSexAPOE would be expected to decline only 0.51 memory score units by age 80. Figure 1a shows the predicted memory trajectories for NHW with the average AD-GRS as well as for people 2 SD above and below the average AD-GRS. Figure 1b illustrates predicted memory decline trajectories for NHW based on the AD-GRSexAPOE.
Table 2.
Generalized Linear Models: Regression coefficients for standardized memory score by AD-GRS and age.
| Non-Hispanic White N=7,172 β (95% CI) |
Non-Hispanic Black N=1,081 β (95% CI) |
|||
|---|---|---|---|---|
| AD-GRS with APOE | AD-GRS without APOE | AD-GRS with APOE | AD-GRS without APOE | |
| AD-GRS (per 0.1 difference) | −0.035** (−0.050, −0.020) | −0.016 (−0.036, 0.005) | −0.059* (−0.101, −0.017) | −0.049 (−0.12, 0.023) |
| Current Age (in decades) | −0.325** (−0.342, −0.308) | −0.356** (−0.378, −0.334) | −0.427** (−0.475, −0.379) | −0.464** (−0.522, −0.406) |
| Current Age2 | −0.100** (−0.120, −0.090) | −0.110** (−0.130, −0.090) | −0.110** (−0.140, −0.080) | −0.130** (−0.170, −0.080) |
| AD-GRS* Current Age | −0.058** (−0.074, −0.043) | −0.029* (−0.050, −0.007) | −0.050 (−0.106, 0.006) | −0.005 (−0.080, 0.070) |
| AD-GRS* Current Age2 | −0.002* (−0.003, −0.001) | −0.001 (−0.003, 0.001) | 0.000 (−0.004, 0.004) | 0.002 (−0.005, 0.009) |
All models are additionally adjusted for: baseline age (centered at 70 years), gender and eigenvectors.
p-value <0.05;
p-value <0.001
AD-GRS = Alzheimer’s disease genetic risk score
Figure 1.
Predicted quadratic trajectories of memory function by Alzheimer’s disease genetic risk score (AD-GRS) from race-specific generalized linear models adjusted for baseline age, gender, and eigenvectors. Shown for a typical individual with an average Alzheimer’s disease genetic risk score (AD-GRS), 2 SD lower than average, and 2 SD higher than average. Note that the trajectory is predicted based on following individuals aged 50 to 100 for up to 14 years, rather than following single individuals for 50 years.
Legend:
A: NHW using an AD-GRS including APOE
B: NHW using an AD-GRS excluding APOE
C: NHB using an AD-GRS including APOE
D: NHB using an AD-GRS excluding APOE
Line styles:
Dashed: Predicted slope of memory decline by age for individuals with an AD-GRS 2 SD higher than average.
Solid: Predicted slope of memory decline by age for individuals with an average AD-GRS.
Dotted: Predicted slope of memory decline by age for individuals with an AD-GRS 2 SD lower than average.
Among NHB, there was a marginally significant interaction between the AD-GRS and age (p=0.080) but not between the AD-GRS and age2 (p=0.948), suggesting that the AD-GRS was associated with memory decline, but we do not have evidence that this association accelerated at older ages (Table 2). Among NHB, in models with linear age terms only, a 70 year old with an average AD-GRS score would be expected to decline 0.58 memory score units by age 80; a similar individual with a 1 SD higher AD-GRS would be expected to decline 0.60 memory score units by age 80. The magnitude of the association between the AD-GRS and change in rate of memory decline was smaller among NHB than among NHW. However, we found no evidence for a differential effect by race of the AD-GRS on memory decline in a race-pooled model (p=0.782 for race-by-AD-GRS-by-age interaction term; p=0.255 for race-by-AD-GRS-by-age2 interaction term). The AD-GRS excluding APOE (AD-GRSexAPOE) did not predict memory decline among NHB (β= −0.005; 95% CI: −0.080, 0.070; p=0.899). Results were similar in models without the AD-GRSexAPOE-by-age2 interaction term: the AD-GRSexAPOE-by-age term remained non-significant (p=0.30). We again found no evidence that the effect of the AD-GRSexAPOE on memory decline differed for NHW and NHB (p=0.891 for race-by-AD-GRS-by-age interaction term) in a race-pooled model. Figure 1c shows the predicted quadratic memory decline trajectories for NHB with the average AD-GRS as well as for people 2 SD above and below the average AD-GRS and Figure 1d shows the memory decline trajectories for NHB using the AD-GRSexAPOE.
Using the alternative AD-GRS excluding APOE, we estimated models controlling separately for APOE status to increase the precision of our estimates. Although APOE was predictive of memory score and decline, our standard errors for the AD-GRS beta coefficient and the AD-GRSexAPOE-by-age interaction term remained qualitatively similar to the results displayed in Table 2, regardless of separate adjustment for APOE. In addition, we tested whether new ABCA7 SNP rs115550680, recently reported as associated with AD in blacks by Reitz et al.4, was a better predictor of memory decline in our sample than the original ABCA7 SNP (rs3764650) used in our AD-GRS. We found no evidence that either ABCA7 SNP was predictive of memory decline in models restricted to NHB using ABCA7 by linear age interaction terms (β= −0.01; 95% CI: −0.08, 0.06; p=0.79 for rs115550680 and β= 0.02; 95% CI: −0.02, 0.05; p=0.34 for rs3764650).
We estimated age-stratified models and found an effect of the AD-GRS including APOE on memory decline among NHW respondents 65 and older (p<0.0001 for AD-GRS-by-age interaction), but not among older NHB (β for AD-GRS-by-age interaction=−0.056; 95% CI: −0.152, 0.040; p=0.251). We did not find an association between the AD-GRS and memory decline in younger (<65) NHW (β for AD-GRS-by-age interaction=−0.004; 95% CI: −0.095, 0.104; p=0.993), but we did in younger NHB (β for AD-GRS-by-age interaction=−0.268; 95% CI: −0.529, −0.006; p=0.045) (see Table, Supplemental Digital Content 3).
Finally, we found no evidence of survival bias. From 2008–2012, 666 individuals died and 763 individuals were lost to follow-up. The AD-GRS did not predict death (p=0.926) or dropout (p=0.890) during these four years (from 2008, the end of DNA collection when participants had to be alive, to 2012), nor did we find that the effects of race or prior memory score on death and dropout depended on the AD-GRS (Table 3). When we dropped the three-way interaction term between the AD-GRS, race, and prior memory score to boost power, results remained qualitatively similar.
Table 3.
Pooled Logistic Models: Odds Ratios for the association of the AD-GRS, race, and memory score (in 2004) with death and dropout from 2010 to 2012.
| Death OR (95% CI) |
Death OR (95% CI) |
Drop-out OR (95% CI) |
Drop-out OR (95% CI) |
|
|---|---|---|---|---|
| AD-GRS (0.1 increase) | 0.98 (0.63, 1.52) | 1.02 (0.68, 1.55) | 1.03 (0.67, 1.59) | 1.05 (0.70, 1.57) |
| Black race | 0.44 (0.096, 2.01) | 0.50 (0.12, 2.09) | 0.59 (0.14, 2.54) | 0.63 (0.16, 2.51) |
| AD-GRS* Black race | 0.83 (0.36, 1.88) | 0.73 (0.37, 1.43) | 1.04 (0.46, 2.35) | 0.98 (0.51, 1.87) |
| Memory Score | 0.42* (0.24, 0.73) | 0.44* (0.26, 0.74) | 0.48* (0.28, 0.81) | 0.49* (0.3, 0.8) |
| Black race* Memory Score | 1.53 (0.52, 4.47) | 1.20 (0.76, 1.91) | 0.89 (0.31, 2.55) | 0.79 (0.50, 1.26) |
| AD-GRS* Memory Score | 1.19 (0.77, 1.84) | 1.13 (0.76, 1.69) | 1.10 (0.73, 1.67) | 1.08 (0.74, 1.58) |
| AD-GRS* Black race* Memory Score | 0.76 (0.26, 2.18) | - | 0.88 (0.31, 2.50) | - |
All models are additionally adjusted for baseline age (centered at 70 years), current age, current age2, gender, and eigenvectors.
p-value <0.05;
p-value <0.001
AD-GRS = Alzheimer’s disease genetic risk score
DISCUSSION
We found that a 22-gene polygenic risk score for Alzheimer’s Disease (AD-GRS) predicts memory decline in nationwide samples of older NHW and NHB adults. An alternative AD-GRS excluding APOE predicted both pre-decline level of memory function8 as well as change in memory function over time among NHW, while it did not predict either among NHB. However, we were unable to find evidence that the effect of the AD-GRSexAPOE on memory decline was statistically different between NHW and NHB. This suggests that recently discovered AD related loci add to the prediction of rate of memory loss in late life among NHW, but whether they improve prediction among NHB remains to be seen.
Several studies have examined associations between individual AD polymorphisms and both cognitive level and change. Apart from APOE, evidence was mixed for other polymorphisms across different memory instruments and cohorts. CLU, PICALM, and CR1 were associated with cognitive decline (global cognition and attention) in 1,831 participants of the Cardiovascular Health Cognition Study33. When evaluating memory outcomes in black and white subjects, Pedraza et al. found a nominally significant association with CR1 in white subjects and with CLU in black subjects. CR1 was associated with episodic memory and global cognition in a combined analysis of two US cohorts of non-demented people 34. CR1 was also found to interact with APOE in predicting episodic memory decline in two US cohorts35. CLU was associated with memory scores in the Baltimore Longitudinal Study of Aging36 and with global cognitive function in two Danish cohorts37. However, these studies also report null associations between some AD polymorphisms and cognitive outcomes. For instance, PICALM was not associated with any cognitive outcomes in the study by Pedraza. Similarly, CLU was not associated with cognition in the study by Chibnik34. BIN1, CLU, ABCA7, CR1, MS4A6A, CD33, and MS4A4E were not associated with memory in the study reported by Thambisetty36. Many studies looked at genetic predictors of age-related cognitive decline beyond only AD polymorphisms and have identified APOE, COMT, BDNF and DTNBP1 as predictors of cognitive decline38.
Few studies have examined the association of a polygenic risk score for AD and cognition in older adults, and none of their samples include non-white populations. In 5,171 non demented people (age 45–99 years) from the population-based Rotterdam Study, Verhaaren et al. found that a genetic risk score constructed from APOE, EPHA1, ABCA7, BIN1, CD2AP, CLU, CR1, MS4A4E, MS4A6A, and PICALM genotypes predicted both baseline global cognition and baseline memory function9. However, after excluding APOE from the score, these associations were no longer statistically significant. In the National Institute on Aging Late-Onset Alzheimer’s Disease Family Study, a multilocus genotype, which combined information from PICALM, CR1, BIN1, and CLU, predicted episodic memory10. Conversely, a cross-sectional study across five cohorts of the GERAD1 consortium found no associations between five polygenic risk scores (created using different thresholds for p-values of AD–polymorphism associations) and cognitive ability in later life or age-related cognitive change11. Only this last study examined change in cognitive function, while the previous two estimated only cross-sectional associations. Additional research on the link between the genetic risk factors and memory change is important because the genetic predictors of memory decline could differ from those of memory level.
These contrasting results may be due to differing cognitive measures, sample differences, low statistical power, design issues (retrospective studies), and the high risk of type 1 error because of multiple testing. These methodological challenges motivated our use of a polygenic risk score, which aids in overcoming the issues of lack of power and multiple testing.
One possible explanation for the different results between NHW and NHB is that different genetic markers influence AD progression in the two racial groups. Since the GWAS studies informing the weights for the AD-GRS use predominantly NHW samples, our AD-GRS could be mis-weighted for our NHB sample. Additionally, some of the loci included in our AD-GRS may not actually be predictive of AD, pre-decline level of cognition, or decline among non-white populations. However, amidst mixed evidence of the relevance of APOE for AD prediction among NHB30,39, we found evidence that the AD-GRS including APOE strongly predicted memory decline in our sample of NHB. Moreover, we found no evidence that a new ABCA7 SNP (rs115550680) identified among blacks as predictive of AD was predictive of memory decline. We need more GWAS conducted in racially diverse samples in order to create more relevant polygenic scores for minority race groups and evaluate the performance of loci previously identified in predominantly European samples.
This large, diverse dataset with genetic information and repeated memory assessments is uniquely suited for this research question. However, it has limitations. We have no information on gene expression or epigenetic modifications. Gene expression patterns could explain differences in rate of memory decline in people with similar gene frequencies. Further, HRS does not have a clinical dementia diagnosis or assessments of many important domains of cognitive function and available measures likely have substantial measurement error. In particular, differences in the validity and reliability of the memory assessment for blacks and whites may have contributed to our findings. Memory scores were scaled based on more detailed assessments available in the predominantly white ADAMS subsample. Inequalities in quality and quantity of education may also compromise the validity and reliability of standard memory assessments for older US blacks. Both of these limitations of the memory score measure could have attenuated the observed effect among non-Hispanic blacks40. Finally, we have a smaller sample size among NHB (N=1,081), so our finding that the alternative AD-GRSexAPOE does not predict memory decline among non-Hispanic blacks could be an issue of power.
A strength of this study is that we focused on the genetic contribution to late life memory decline, not just level of functioning. Since both pre-morbid level of cognition as well as rate of decline contribute to AD development, it is critical that we understand the determinants of both. Another strength of this paper is that we directly assessed the possibly of survival bias—a potential source of bias that is widely-recognized by AD researchers but rarely addressed. For example, if a population is genotyped at age 70, those with a harmful genotype may be more likely to be omitted, due to prior mortality. The disproportionate omission of the worst performing group could make this genotype seem less harmful than it really is because survivors are not representative of other carriers. Since we found that neither APOE alone nor the AD-GRS predict death or dropout, this is evidence that survival bias in studies of the AD-GRS may not be a major concern. One limitation of our assessment was that everyone had to be alive for genotyping (in either 2006 or 2008), so we only had 4 years of mortality follow-up from 2008–2012. However, we did not find large differences in the mortality rates of NHW and NHB alive in 1998: 23.8% of NHW and 26.7% of NHB died by 2006. Whether genetic risk scores predict mortality is also of substantive interest because it may elucidate possible biological mechanisms or clinical relevance. Substantively, our findings may mean that these SNPs are causally related to memory function and rate of decline, but not to overall survival.
In conclusion, we found that a genetic risk score for AD predicts not only level but also rate of memory loss in a nationally-representative sample of older NHW and NHB. However, because of the smaller sample size of NHB, our results were consistent with an association between the AD-GRSexAPOE and memory decline among NHB of both: 1) zero; and 2) the same as the association found among NHW. While recently discovered AD related loci add to the prediction of rate of memory loss in older NHW adults, whether they contribute to our understanding of the genetic determinants of cognitive decline among NHB remains to be seen. Future studies should attempt to identify loci that are predictive of AD as well as cognitive function and decline in NHB (as well as other non-white populations) samples. Due to the heterogeneity of associations and the relatively small amount of variance in rate of cognitive decline explained by genetics to date, next steps among NHW include investigating whether gene-environment interactions exist. For example, researchers should explore whether the AD-GRS has differential effects on rate of cognitive decline among people with high versus low cognitive reserve. The AD-GRS can facilitate investigations of this and other important questions to help us better understand how genes and life experiences influence risk of developing AD.
Supplementary Material
Acknowledgments
Study Sponsors:
National Institute on Aging, grant #: NIA AG03438501; PI: Glymour
National Institute of Neurological Disorders and Stroke, grant #: NINDS T32 NS048005, PI: Betensky
National Institute of Allergy and Infectious Diseases, grant #: NIAID R01 AI104459, PI: Tchetgen Tchetgen
Sources of Funding:
Dr. Glymour received support from the National Institute on Aging (grant #: NIA AG03438501; PI: Glymour). Ms. Marden received support in the form of a training grant from the National Institute of Neurological Disorders and Stroke (grant #: NINDS T32 NS048005, PI: Betensky). Ms. Marden and Dr. Tchetgen Tchetgen received support from the National Institute of Allergy and Infectious Diseases (grant #: NIAID R01 AI104459).
Footnotes
None of the authors have any conflicts of interest or financial disclosures.
List of Supplemental Digital Content
Supplemental Digital Content 1.doc
Conflicts of Interest
No conflicts of interest for any authors were declared.
LITERATURE CITED
- 1.Lambert J-C, Ibrahim-Verbaas CA, Harold D, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nature genetics. 2013;45(12):1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Naj AC, Jun G, Beecham GW, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nature genetics. 2011;43(5):436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hollingworth P, Harold D, Sims R, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nature genetics. 2011;43(5):429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reitz C, Jun G, Naj A, et al. Variants in the ATP-binding cassette transporter (ABCA7), apolipoprotein E ε4, and the risk of late-onset Alzheimer disease in African Americans. JAMA: the journal of the American Medical Association. 2013;309(14):1483. doi: 10.1001/jama.2013.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seshadri S, Fitzpatrick AL, Ikram MA, et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. Jama. 2010;303(18):1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stern Y. Cognitive reserve in ageing and Alzheimer’s disease. The Lancet. Neurology. 2012 Nov;11(11):1006–1012. doi: 10.1016/S1474-4422(12)70191-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertram L, McQueen M, Mullin K, et al. The AlzGene Database. Alzheimer Research Forum. 2006 [Google Scholar]
- 8.Marden JR, Walter S, Tchetgen Tchetgen EJ, et al. Validation of a polygenic risk score for dementia in black and white individuals. Brain and Behavior. 2014 doi: 10.1002/brb3.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verhaaren BF, Vernooij MW, Koudstaal PJ, et al. Alzheimer’s disease genes and cognition in the nondemented general population. Biological psychiatry. 2013;73(5):429–434. doi: 10.1016/j.biopsych.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 10.Barral S, Bird T, Goate A, et al. Genotype patterns at PICALM, CR1, BIN1, CLU, and APOE genes are associated with episodic memory. Neurology. 2012 May 8;78(19):1464–1471. doi: 10.1212/WNL.0b013e3182553c48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris SE, Davies G, Luciano M, et al. Polygenic risk for Alzheimer’s disease is not associated with cognitive ability or cognitive aging in non-demented older people. Journal of Alzheimer’s disease : JAD. 2014;39(3):565–574. doi: 10.3233/JAD-131058. [DOI] [PubMed] [Google Scholar]
- 12.Plassman BL, Williams JW, Burke JR, et al. Systematic review: factors associated with risk for and possible prevention of cognitive decline in later life. Annals of internal medicine. 2010;153(3):182–193. doi: 10.7326/0003-4819-153-3-201008030-00258. [DOI] [PubMed] [Google Scholar]
- 13.Logue MW, Schu M, Vardarajan BN, et al. A comprehensive genetic association study of Alzheimer disease in African Americans. Archives of neurology. 2011;68(12):1569–1579. doi: 10.1001/archneurol.2011.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pedraza O, Allen M, Jennette K, et al. Evaluation of memory endophenotypes for association with CLU, CR1, and PICALM variants in black and white subjects. Alzheimer’s & dementia : the journal of the Alzheimer’s Association. 2014 Mar;10(2):205–213. doi: 10.1016/j.jalz.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pfaff C, Parra E, Bonilla C, et al. Population structure in admixed populations: effect of admixture dynamics on the pattern of linkage disequilibrium. The American Journal of Human Genetics. 2001;68(1):198–207. doi: 10.1086/316935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tishkoff SA, Williams SM. Genetic analysis of African populations: human evolution and complex disease. Nature Reviews Genetics. 2002;3(8):611–621. doi: 10.1038/nrg865. [DOI] [PubMed] [Google Scholar]
- 17.McClellan J, King M-C. Genetic heterogeneity in human disease. Cell. 2010;141(2):210–217. doi: 10.1016/j.cell.2010.03.032. [DOI] [PubMed] [Google Scholar]
- 18.Burchard EG, Ziv E, Coyle N, et al. The importance of race and ethnic background in biomedical research and clinical practice. New England Journal of Medicine. 2003;348(12):1170–1175. doi: 10.1056/NEJMsb025007. [DOI] [PubMed] [Google Scholar]
- 19.Yaffe K, Lindquist K, Vittinghoff E, et al. The effect of maintaining cognition on risk of disability and death. Journal of the American Geriatrics Society. 2010 May;58(5):889–894. doi: 10.1111/j.1532-5415.2010.02818.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lavery LL, Dodge HH, Snitz B, et al. Cognitive decline and mortality in a community-based cohort: the Monongahela Valley Independent Elders Survey. Journal of the American Geriatrics Society. 2009 Jan;57(1):94–100. doi: 10.1111/j.1532-5415.2008.02052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beydoun MA, Beydoun HA, Kaufman JS, et al. Apolipoprotein E epsilon4 allele interacts with sex and cognitive status to influence all-cause and cause-specific mortality in U.S. older adults. Journal of the American Geriatrics Society. 2013 Apr;61(4):525–534. doi: 10.1111/jgs.12156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosvall L, Rizzuto D, Wang HX, et al. APOE-related mortality: effect of dementia, cardiovascular disease and gender. Neurobiology of aging. 2009 Oct;30(10):1545–1551. doi: 10.1016/j.neurobiolaging.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 23.Hauser RM, Willis RJ. Survey design and methodology in the Health and Retirement Study and the Wisconsin Longitudinal Study. Population and Development Review. 2004;30:209–235. [Google Scholar]
- 24.Juster FT, Suzman R. An overview of the Health and Retirement Study. Journal of Human Resources. 1995:S7–S56. [Google Scholar]
- 25.Heeringa SG, Connor J. Technical description of the Health and Retirement Study sample design. Vol. 1995. Ann Arbor, Michigan: Survey Research Center, University of Michigan; 1995. p. DR-002. [Google Scholar]
- 26.Ofstedal MB, Fisher GF, Herzog AR. Documentation of cognitive functioning measures in the Health and Retirement Study. [Accessed March, 2005];HRS Documentation Report. 2005 :DR-006. http://hrsonline.isr.umich.edu/sitedocs/userg/dr-006.pdf.
- 27.Wu Q, Tchetgen ET, Osypuk T, et al. Combining direct and proxy assessments to reduce attrition bias in a longitudinal study. Alzheimer’s Disease and Associated Disorders. 2012 doi: 10.1097/WAD.0b013e31826cfe90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng X, Levine D, Shen J, et al. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics. 2012 Dec 15;28(24):3326–3328. doi: 10.1093/bioinformatics/bts606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Study HR. Genome Wide Association Study Overview. 2010. [Google Scholar]
- 30.Bekris LM, Yu CE, Bird TD, et al. Review article: genetics of Alzheimer disease. Journal of geriatric psychiatry and neurology. 2010;23(4):213–227. doi: 10.1177/0891988710383571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bertram L, McQueen MB, Mullin K, et al. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007 Jan;39(1):17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- 32.Weir D. Quality Control Report for Genotypic Data. Vol. 2012 University of Michigan; Mar 15, 2012. [Google Scholar]
- 33.Sweet RA, Seltman H, Emanuel JE, et al. Effect of Alzheimer’s disease risk genes on trajectories of cognitive function in the Cardiovascular Health Study. The American journal of psychiatry. 2012 Sep;169(9):954–962. doi: 10.1176/appi.ajp.2012.11121815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chibnik LB, Shulman JM, Leurgans SE, et al. CR1 is associated with amyloid plaque burden and age-related cognitive decline. Annals of neurology. 2011 Mar;69(3):560–569. doi: 10.1002/ana.22277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keenan BT, Shulman JM, Chibnik LB, et al. A coding variant in CR1 interacts with APOE-epsilon4 to influence cognitive decline. Human molecular genetics. 2012 May 15;21(10):2377–2388. doi: 10.1093/hmg/dds054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thambisetty M, Beason-Held LL, An Y, et al. Alzheimer risk variant CLU and brain function during aging. Biol Psychiatry. 2013 Mar 1;73(5):399–405. doi: 10.1016/j.biopsych.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mengel-From J, Thinggaard M, Lindahl-Jacobsen R, et al. CLU genetic variants and cognitive decline among elderly and oldest old. PloS one. 2013;8(11):e79105. doi: 10.1371/journal.pone.0079105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harris SE, Deary IJ. The genetics of cognitive ability and cognitive ageing in healthy older people. Trends in cognitive sciences. 2011 Sep;15(9):388–394. doi: 10.1016/j.tics.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 39.Barnes L, Arvanitakis Z, Yu L, et al. Apolipoprotein E and Change in Episodic Memory in Blacks and Whites. Neuroepidemiology. 2013;40(3):211–219. doi: 10.1159/000342778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manly JJ, Jacobs DM, Sano M, et al. Cognitive test performance among nondemented elderly African Americans and whites. Neurology. 1998;50(5):1238–1245. doi: 10.1212/wnl.50.5.1238. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.