Abstract
Alloreactive T lymphocytes are the primary mediators of allograft rejection. The size and diversity of the HLA-alloreactive T cell repertoire has thus far precluded the ability to follow these T cells and thereby to understand their fate in human transplant recipients. This review summarizes the history, challenges, and recent advances in the study of alloreactive T cells. We highlight the historical development of assays to measure alloreactivity and discuss how high-throughput T cell receptor (TCR) sequencing-based assays can provide a new window into the fate of alloreactive T cells in human transplant recipients. A specific approach combining a classical in vitro assay, the mixed lymphocyte reaction, with deep T cell receptor sequencing is described as a tool to track the donor-reactive T cell repertoire for any specific HLA-mismatched donor-recipient pair. This assay can provide mechanistic insights and has potential as a non-invasive, highly specific biomarker for rejection and tolerance.
Introduction
When transplant immunologists began to study and quantify alloreactive lymphocytes, the cells presumed responsible for organ rejection, the number of cells was described as “inordinate,”1 as the number of lymphocytes responding to the cells of another individual was found to be more than an order of magnitude larger than that of previously characterized antigen-specific responses. Now, more than half a century later, the clinical importance of alloreactive T cells and their fundamental role in transplantation are clear; however, the size and diversity of the alloreactive T cell repertoire have rendered a full understanding of this response somewhat elusive. In this review, we summarize the history, challenges, and recent advances in the study of alloreactive T cells. We highlight the emergence of fundamental concepts and discuss how high-throughput T cell receptor (TCR) sequencing-based assays may provide a new window into tolerance and rejection in human transplant recipients.
The Mixed Lymphocyte Response
The need for an in vitro surrogate of the transplant rejection response has existed since transplantation entered clinical practice. The hope for such an assay is that it could predict rejection episodes and identify tolerant patients. The oldest and most widely used in vitro functional assay in transplantation immunology is the mixed lymphocyte reaction (MLR). The MLR largely measures proliferation of T cells activated by the direct pathway of allorecognition, in which T cells are directly activated by allogeneic antigen-presenting cells (APCs). This is in contrast to the indirect pathway, in which T cells are activated by autologous APCs presenting peptides derived from polymorphic proteins of an allogeneic donor in the groove of their major histocompatibility complex (MHC) heterodimers. The magnitude of the direct alloresponse is unusually strong, whereas the magnitude of the indirect response more closely resembles that of the response to other polymorphic proteins. In contrast to most types of antigen-specific responses, direct MLR responses do not require priming in order to be measurable, reflecting their markedly greater magnitude.
The clinical importance of alloreactive T cells activated directly by the presence of allogeneic APCs transplanted in the graft is obvious in the immediate post-transplant period, but the endothelial and parenchymal cells of the allograft may express donor human leukocyte antigen (HLA) molecules that could activate directly alloreactive T cells at any time.2,3 Another more recently-described allorecognition pathway is the semi-direct pathway,4 in which recipient cells can present donor-HLA molecules directly on their surface that are acquired via a process known as trogocytosis, thereby possibly triggering T cells that are directly alloreactive.5 Taken together, there is compelling support for the importance of directly alloreactive lymphocytes in the immunologic response in transplantation.
The first MLR documented in the literature appeared in 1963 in an abstract from Bain et al. in the Federation Proceedings. 6 Inspired by the incidental observation via time-lapse cinematography of mitosis induced by the co-culture of blood samples from two different patients,7 the investigators set up a co-culture of cells from two individuals and after five days pulsed the samples with radioactive thymidine, enabling the identification of allo-activated cells undergoing DNA synthesis. The finding of human lymphocytes reacting against each other in culture was published shortly thereafter in 1963 by Hirschhorn et al., demonstrating enlargement and cell division of the alloreactive lymphocytes.8 The potential use of the MLR for studying the immune reaction or histocompatibility between donor and recipient in the context of transplantation was immediately recognized.9 Furthermore, Bain at al. showed that the extent of cell division occurring in MLRs of monozygotic twins was markedly reduced compared to unrelated individuals, suggesting a possible genetic underpinning to histocompatibility.10 Shortly thereafter, studies in rodents and humans with known histoincompatibility supported the notion that MLR proliferation depends, at least partially, upon MHC differences.11,12
Concurrently, extensive work was performed to illuminate fundamental features of the cellular response in the MLR.13,14 The difficulty in accurately quantifying alloreactive T cells has been recognized since the publication of the mixed lymphocyte reaction15, as specific culture conditions and methodologies markedly affected the outcome. Consistent with earlier studies,16 however, the finding that arose again and again was the large number of lymphocytes of one person responding to those of another.1,17–19 Despite the acknowledged limitations of these early estimates, a range of 1–10% of the entire T cell repertoire20 is often described as alloreactive, though the evidence from the early MLR studies themselves point to a range of 0.5–3%. Several additional studies using complementary approaches supported this approximation: 4.5–12% in an in vivo graft-versus-host model in mice; 21–23 1–2% alloreactive cytotoxic T lymphocyte precursors via limiting dilution assays in mice.24
On the origin and diversity of alloreactive T cells
A myriad of hypotheses arose to explain why and how there might be such a large population of alloreactive cells.1,25–27 While much remains to investigate, there is compelling evidence for the role of both the foreign MHC molecule and the peptide presented, though the relative contribution of each for different clones may not be equivalent.20,28–30 Although humans certainly did not evolve to mount an immune response in the context of organ transplantation, the germline T cell repertoire has been shown to be strongly enriched for MHC recognition.31 Because the processes of positive and negative selection in the thymus take place after TCR α and β chain rearrangements have occurred, T cells developing in the thymus have the potential to recognize a diversity of potential HLA types; given the inherent cross-reactivity/degeneracy of TCR recognition due to the rotational flexibility of the TCR/MHC-peptide interface,32–35 alloreactivity can be thought of as the consequence of selection for weak recognition of self MHC-peptide complexes in the thymus in combination with selection against strong recognition of those same specificities36–39 Some clones exiting the thymus as naïve T cells may in fact be more strongly reactive to allogeneic HLA alleles, but have sufficient cross-reactivity to self-HLA/peptide complexes to survive positive selection.40 While this inherent MHC-reactivity may help explain the large proportion of naïve alloreactive T cells, alloreactive memory cells may reflect a combination of previous exposure of an individual to alloantigens, such as during pregnancy, as well as cross-reactivity of pathogen-specific memory T cells that emerge in response to a viral or bacterial infection.30,41–43 The connection between alloreactivity and viral-reactive T cells began with experiments focused on T cells reactive to Epstein Barr virus (EBV); such studies have demonstrated that there are even public TCRs shared across individuals of the same HLA-type that cross-react to EBV and specific foreign HLA molecules.41 A more recent hypothesis to help explain the large size of the alloreactive T cell population is that T cells expressing two different TCRs simultaneously—specifically two distinct alpha chains paired with the same beta chain—may have a particular propensity for allorecognition, with support from studies in mice and humans.44,45
The large number and diversity of T cell clones that recognize allogeneic MHC has presented a challenge for the identification of alloreactive T cells. The antigen specificity of the millions of unique T cells circulating in a healthy human adult at any given time arises primarily from the amino acid sequence of the third complementarity-determining region (CDR3) of the variable regions of the α and β chains that form the heterodimer of the αβ TCR: the extensive variation in the precise CDR3 region emerges from somatic recombination of variable (Vβ), diversity (Dβ), and joining (Jβ) genes encoding the β-chain and Vα and Jα of the α-chain, with even further variation created by the insertion of additional nucleotides within these regions.46,47
Early studies investigating alloreactive repertoire diversity in mice failed to identify dominant usage of a particular TCR Vβ gene in alloreactive T cell populations, even when directed against only a three amino acid difference in one MHC molecule.48–50 Spectratyping, which examined distribution of CDR3 lengths within each V-Jβ family51, suggested the activation of many TCRs, with a near-Gaussian distribution among multiple Vβ families in human and rat cells responding in MLRs,52 consistent with previous studies focusing on smaller, more targeted alloreactive populations53. Thus, it was surmised that the alloreactive repertoire is highly diverse. Only recent advances in gene sequencing capabilities have made the study of this enormously diverse T cell population technically feasible.
Utility of the CFSE MLR
The original MLR technique measured cell division in the MLR via the incorporation of radioactive thymidine. This assay measures the amount of total cell division at any given time, but does not indicate how many times a cell has divided and, most importantly, only provides a snapshot of cell division over a short period of time. Since kinetics of division may differ for different T cell clones, it is reasonable to assume that not all cell division is captured with this assay. In 1994, Lyons and Parish published the use of the fluorescent dye carboxyfluorescein diacetate succinimidyl ester (CFSE) to study cell division:54 because CFSE binds covalently to intracellular amines, when a cell divides in half the CFSE dye divides in half with it and thus the fluorescence intensity of the daughter cells are half that of the original parent cell.55 With histograms of CFSE fluorescence intensity, individual CFSE peaks correspond to a given number of cell divisions. This information can be used to quantify the precursor frequency of the T cells that divide in an MLR.56 The CFSE MLR enabled the phenotypic characterization and physical isolation of alloreactive cells by flow cytometry and fluorescence-activated cell sorting (FACS), respectively. The assay has the advantage of capturing divided cells regardless of the kinetics of cell division, in contrast to radioactive thymidine assays, which only capture cells dividing during the period of the thymidine pulse.
Suchin et al. used CFSE MLR approach to compare alloreactive T cell frequencies obtained in in vitro MLRs versus in vivo adoptive cell transfer in mice. While their in vitro experiments yielded a frequency of 4.61 +/- 2.22%, three different in vivo models gave a range of 0.71% to 21.05%, depending on how they defined the denominator of the total number of potential responder cells.57 Macedo et al. used the CFSE MLR with other phenotypic markers to confirm a more than decade-old observation that both naïve and memory cells contribute to the alloresponse.58 Furthermore, they demonstrated that known EBV-reactive CD8 T cells were cross-reactive to alloantigens in healthy controls. While the findings in this work are valuable, the specific precursor frequency values must be interpreted cautiously, as the calculations did not account for the potentially extensive cell death among non-dividing cells in culture and thus may result in overestimations.
Existing tools for tracking alloresponses in vivo
The first surrogate for alloreactive clones that could be tracked in vivo in mice was built upon the observation that certain Vβ families were deleted in the thymus of specific strains of mice due to the presence in their genomes of endogenous superantigens that are the products of endogenous retroviruses and bind outside the binding groove of distinct MHC class II molecules.59,60 This approach was used to demonstrate intra-thymic deletion as a major mechanism of tolerance in full chimeras prepared with myeloablative conditioning61 and in mixed chimeras prepared with non-myeloablative conditioning.62 Subsequently, the development of alloreactive transgenic TCRs allowed tracking of individual allospecific clones in vivo, demonstrating intrathymic deletion of T cells expressing the TCR and its MHC ligand.63 This approach has allowed the kinetics of the graft-versus-host alloresponses to be delineated 64 and to confirm an intrathymic deletional mechanism of tolerance in mixed allogeneic chimeras65 These studies are most informative when TCR transgenic alloreactive cells represent a measurable fraction of T cells on the backdrop of a normal polyclonal T cell repertoire.66–68
Humans present a far greater challenge for investigating alloreactive T cells than animal models because the in vivo techniques described above cannot be employed in humans, for obvious reasons. The main strategies, therefore, have been: (1) to study a small population of donor-reactive T cells with known HLA-peptide complex reactivity via tetramers, soluble MHC-peptide multimers with a fluorescent tag69 or (2) to use in vitro functional assays.
The use of tetramers in the study of transplant patients is most relevant in the setting of HLA-identical transplantation, where peptides from minor histocompatibility antigens (HA) are used to form tetramers with HLA molecules that are shared between the donor and recipient. Because minor HA are of clearest relevance in the absence of HLA disparity, these tetramers have been most useful in hematopoietic cell transplantation, which is commonly performed in the HLA-identical or closely-matched setting 70–72 Using tetramers specific for the HLA-type of the patient and the known minor histocompatibility antigen, specific alloreactive T cells have been tracked in the transplant setting.73 The greatest limitation in the use of tetramers to study alloreactive clones is that it requires knowing exactly the HLA-peptide specificities of interest. Given the enormous number of potential allogeneic MHC-peptide combinations in HLA-mismatched transplantation and the inability to predict immunodominant responses, such a strategy is not feasible.
Efforts to correlate in vitro assays and transplant outcomes
Soon after development of the MLR, studies began in an effort to correlate in vitro results with outcomes in skin transplantation in humans 74 and non-human primates75 and ultimately to evaluate the role of the MLR as a possible tool for predicting immunological compatibility pre-transplant for kidney transplantation.76 However, the correlation of clinical outcomes with in vitro functional assays in general has been disappointing.77–86 In addition to the MLR, these traditional assays include the cell-mediated lympholysis assay and limiting dilution assays (LDA) to quantify cytotoxic T lymphocyte precursors and interleukin-2-producing helper T lymphocytes. The LDA was evaluated as a tool to quantify directly and indirectly allospecific T cells in association with outcomes of bone marrow and solid organ transplantation.87–89 Although a significant correlation was observed between graft-versus-host (GVH)-reactive IL-2-producing cell frequencies in the donor and the subsequent development of GVH disease (GVHD) in the HLA-identical transplant setting, the LDA results overlapped considerably between individuals who did and did not develop GVHD.90 While correlations with solid organ transplant outcomes have not been consistent,91,92 the assay provided evidence for a role for the indirect pathway in chronic rejection.93 Heeger et al. developed the cytokine enzyme linked immunosorbent spot (ELISPOT) assay,94,95 in which recipient and donor cells are co-cultured just as in the MLR and alloreactivity is measured as a function of interferon-γ expression. While initial single-center and retrospective studies focused on correlations with clinical outcomes in kidney transplantation, predictive value remains low for the individual patient96–98 and the most recent clinical trial results show no clear correlation between ELISPOT reactivity and early post-transplant outcomes.99 Nevertheless, the assay may ultimately prove useful in helping to personalize immunosupressive therapy.100 ELISPOT assays measure the activity of memory cells, which is a limitation given that the alloreactive repertoire is a combination of naïve and memory cells. A somewhat technically challenging assay known as the trans vivo delayed-type hypersensitivity assay involves the injection of recipient cells and donor antigens into the footpads of immunodeficient mice with donor-reactivity assessed via swelling of the mouse footpad; its results have been used to implicate regulatory or suppressive mechanisms of tolerance in transplant patients.83,101–104 A recent approach identifies alloreactive T cells via upregulation of CD154 (CD40 ligand) in a modified MLR in which donor and recipient cells are mixed in the presence of anti-CD40 antibody to stabilize surface CD154 expression.105 Results of this measurement of cytotoxic memory T cells pre- or post-transplant have been correlated with outcomes and with proliferative responses in pediatric liver transplant studies;106 however, similar to the other assays discussed, this approach has not been shown to predict outcomes for the individual patient.
The functional studies described are all limited by the particular culture conditions provided, which can never replicate the in vivo setting. Unresponsiveness of donor-reactive clones in vitro may be due to deletion, anergy, or suppression of the anti-donor response. While suppression can be teased out in limiting dilution assays or by removal of putative suppressive populations, there has not previously been a good way of distinguishing deletion from anergy as mechanisms of unresponsiveness of human T cells in vitro. Given that anergy is a relative term that may in fact reflect the failure of the culture conditions to provide critical factors such as important cytokines, it is hardly surprising that in vitro assays have been poorly predictive of in vivo outcomes for the individual.
An impressive amount of research has aimed to develop biomarkers of tolerance and rejection. Though beyond the scope of this paper, these approaches have included, but are not limited to, phenotypic studies via flow cytometry particularly focused on regulatory markers,107–110 assays measuring T cell activation,111,112 and gene expression and proteomic signatures in blood and urine.113–124 While many of these tests show promise, with one exception showing strong ability to predict acute cellular rejection,125 most have not yet been shown to accurately predict tolerance or rejection for the individual patient. All of these biomarkers measure correlations with pathogenic processes, such as inflammation or T cell activation in general, rather than assessing the specific anti-donor T cell response.
Before the high-throughput sequencing era, more standard clonotyping methods were used in efforts to identify alloreactive TCRs in the setting of HLA-identical hematopoietic cell transplantation. These weak MLRs did not reproducibly identify immunodominant clones that could be expected to contribute to GVHD.126 Michalek et al. used an MLR to identify a single alloreactive clone of unknown peptide-specificity in a patient who underwent allogeneic hematopoietic stem cell transplantation for acute myelogenous leukemia with only one HLA allele mismatch from the graft donor; tracking of this this clone via quantitative PCR post-transplant revealed a marked increase in frequency that correlated with clinical GVHD.127 Ex vivo “pruning” or removal of alloreactive CD4 cells, defined by those T cells proliferating in a CFSE-MLR, delayed rejection of skin and cardiac allografts in an adoptive transfer model in mice,128,129 demonstrating the relevance of the CFSE MLR for allograft rejection in this model.
The use of high-throughput TCR CDR3 sequencing to track the specific anti-donor response in humans
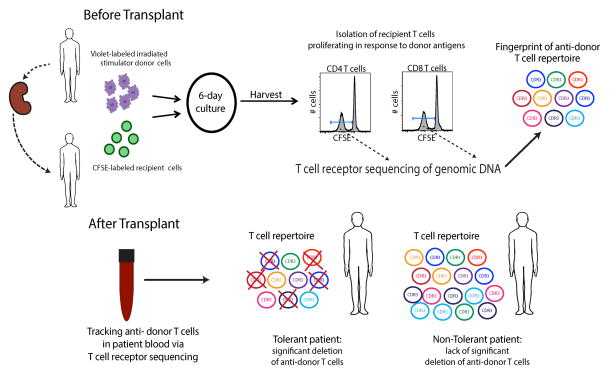
We have recently developed a high-throughput TCR sequencing-based approach that overcomes some of the limitations discussed above by specifically identifying the donor-reactive T cell repertoire before transplant and then tracking those clones following the transplant, without relying on any functional readout. A pre-transplant CFSE MLR is performed with FACS sorting of the recipient T cells dividing in response to the donor. Genomic DNA is extracted from this divided cell population and from unstimulated recipient T cells at the same time. High throughput sequencing of TCR β chain CDR3 regions in both populations is then used to generate a “fingerprint” of the alloreactive T cell repertoire specific for that patient’s donor. The fate of thousands of clones identified as donor-reactive is traced in the peripheral circulation over time after transplantation, without the requirement for any post-transplant functional assay.78
The need for such an approach arose in our efforts to understand the mechanisms of tolerance in recipients of combined kidney and bone marrow transplantation at the Massachusetts General Hospital: 7 of 10 patients achieved tolerance, as defined by the successful withdrawal of immunosuppression without organ rejection for periods of years.130–132 The results of mechanistic studies performed on cells from these patients implicated a possible role for regulatory T cells early post-transplant, but not long-term.133,134 In all of the tolerant patients, in vitro assays revealed unresponsiveness or hypo-responsiveness of the donor-reactive T cells post-transplant, with recovery of third party responses; however, these results could not differentiate between two key mechanistic possibilities: either the donor-reactive T cells that persisted were rendered inactive or anergic, resulting in the observed lack of anti-donor reactivity, or the anti-donor T cells had been deleted from the recipient T cell repertoire. Given that chimerism was very transient, lasting at most a few weeks in these patients,130,131,133 straightforward central deletion could not readily explain tolerance on a long-term basis as it did in mouse studies.135 We therefore sought to develop an assay that could specifically distinguish anergy from deletion.
The advent of high-throughput T cell receptor sequencing provided the opportunity to develop such an assay. TCR deep sequencing enabled the generation of catalogs of the thousands of individual clones proliferating specifically in response to donor stimulation. These clones were defined by the unique nucleotide sequence of the CDR3 region of their TCR β chains. For the first time, what had been presumed to be an enormous number of T cell clones reactive against allogeneic HLA-peptide complexes in humans could be individually delineated. Figure 1 schematically describes our assay for defining a fingerprint of the alloreactive T cell repertoire and the general findings from our studies in six transplant patients, including four recipients of combined kidney and bone marrow transplants (CKBMT), three of whom achieved tolerance, and two conventional kidney transplant patients. The findings in this small series of patients validated the in vivo biological significance of the T cell clones defined as alloreactive by this new method and provided mechanistic insights into the tolerance achieved in three of the CKBMT recipients. The evidence for their significance includes the post-transplant expansion of the number of clones defined as donor-reactive circulating in two conventional transplant patients, despite extensive repertoire turnover, demonstrating that the same clones that were defined as donor-reactive in the MLR respond in vivo to donor antigens.78. Additionally, we observed a lack of expansion and statistically significant reductions in the number of circulating donor-reactive clones compared to pre-transplant over time only in the three tolerant patients. In the context of the studies already performed to investigate the mechanisms of tolerance in this unique cohort of patients, the TCR sequencing approach provided an unparalleled insight into the mechanisms at play. In view of the very transient nature of the chimerism detected in these patients, the role of the kidney itself in promoting tolerance, the possible early role of regulatory cells suggested by functional studies134 and the enrichment for FOXP3+ cells in the graft,130 we speculate that repeated encounter of donor-reactive T cells with donor antigen in the rich capillary bed of the immunologically quiescent environment established in the kidney following CKBMT ultimately leads to deletion of these T cells.
Figure 1.
Schematic of the technique and findings presented in “Tracking donor-reactive T cells: Evidence for clonal deletion in tolerant kidney transplant patients.”78
One of the unique aspects of this TCR sequencing approach is that it specifically addresses the donor-specific T cell repertoire. Other human TCR sequencing studies performed to date have focused only on changes within the entire peripheral blood repertoire.136,137 The TCR sequencing results also allowed us to study the extent of T cell turnover and compare the relative number of donor-reactive and non-donor-reactive clones persisting over time post-transplantation. This analysis permitted assessment of the contributions of: (1) the T cell depleting conditioning regimen that non-specifically eliminated many T cells; (2) antigen-driven expansion of the donor-reactive clones in the early post-transplant setting of lymphopenia-induced proliferation (LIP); and (3) the progressive antigen-driven deletion of donor-reactive clones. We interpret the outcome of our study to implicate a role for all three factors in determining the number of donor-reactive clones detected after transplantation.
Figure 2 presents an example of the marked difference in the post-transplant changes in the CD4 anti-donor T cell repertoire between a tolerant and non-tolerant subject. It is visually obvious from this presentation that there is a “thinning” of donor-reactive clones in the tolerant patient, whereas there is an increase in the number of circulating donor-reactive clones in the non-tolerant subject at the same time about one and a half years post-transplant. While the tolerant subjects showed a loss of donor-reactive clones over time post-transplant,, the allograft seemed to provide strong antigenic pressure for expansion of pre-existing donor-reactive clones in conventional transplant recipients, despite considerable repertoire turnover induced by their immunosuppressive regimens.78 These contrasting results implicate a role for the allograft itself rather than simply non-specific T cell depletion in the decline of donor-reactive clones in the tolerant patients. Most importantly, the loss of donor-reactive clones aligned more closely with tolerance induction than any other assay. This finding is particularly striking in Subject 5, who failed to achieve tolerance despite receiving the CKBMT regimen. Remarkably, this patient showed robust donor-specific unresponsiveness in all in vitro functional assays, including MLR, CML and LDA, clearly illustrating the unreliability of post-transplant functional assays in specifying tolerance.78 The post-transplant in vitro unresponsiveness in this subject suggests that the donor-reactive clones in this subject may have been anergic, raising the possibility that the infection preceding the rejection in this patient triggered the re-activation of these alloreactive cells.
Figure 2.
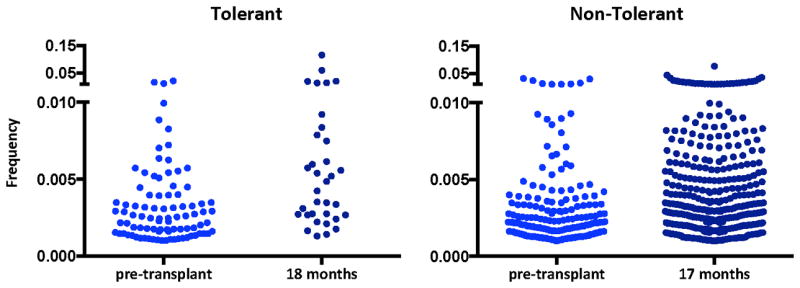
Comparison of the CD4 donor-reactive T cells as defined by the pre-transplant donor-reactive fingerprint from a tolerant (Subject 1) and non-tolerant (IS #1) transplant patient (minimum frequency threshold of dection in unstimulated samples 10^-5).
On the other hand, in one tolerant patient who, like the others, showed a significant post-transplant decline over time in clones defined pre-transplant as donor-reactive, a measurable post-transplant MLR persisted. This MLR permitted clonal analysis of post-transplant donor-reactive T cells. The clones contributing to this post-transplant alloresponse showed very little overlap with the donor-reactive clones identified in the pre-transplant MLR and showed no significant overall increase or decrease in number post-transplant. 78 Since this patient accepted the donor kidney without long-term immunosuppression, these results suggest that, in contrast to the pre-transplant MLR, the post-transplant MLR did not identify sufficient numbers of biologically relevant clones in the context of the tolerance mechanisms that had been established in the patient.
It can be informative to examine both the number of donor-reactive clones and the frequency of those clones using the alloreactive TCR tracking approach. A comparison of the frequency distribution of donor-reactive clones pre- and post-transplant can be seen in Figure 2: interestingly, in the non-tolerant patient there is an increase in the proportion of relatively abundant CD4 donor-reactive T cell clones post-transplant, a trend that is not seen for the persisting donor-reactive clones in the tolerant patient, for which the frequency distribution is similar compared to pre-transplant. This increase in frequency of circulating donor-reactive clones in a non-tolerant kidney transplant recipient provides further support for the biological relevance of the clones identified in the pre-transplant MLR and the potential clinical utility of tracking them.
Strengths and limitations of the TCR sequencing approach
Many questions remain to be explored in terms of understanding the assay’s potential and limitations. Considerable overlap was detected in alloreactive repertoire from normal donors against the same stimulator tested at times two weeks or one year apart. However, the level of this overlap was generally greater for CD4 clones than CD8 clones.78 Overall, the clonality of the unstimulated CD8 T cell repertoire was greater than that of CD4 cells in our studies78, and this may reflect occupation of larger amounts of CD8 repertoire “space” by oligoclonal expansions of CD8 cells, resulting in a smaller effective CD8 repertoire size in a given sample and hence greater sampling error.
The background frequency of a given clone and the number of T cells in each sample determines the chance of detecting a clone in two different blood samples. Since any given blood draw captures only a fraction of the T cells actively in circulation at any given time, large cell numbers are necessary not only in the set-up of the pre-transplant MLR to identify the greatest possible number of donor-reactive clones, but perhaps more importantly in the sequencing of unstimulated peripheral blood sample populations, particularly when deletion of alloreactive clones is suspected. For the MLR, although a large starting pool of clones is essential, the antigen-driven expansion permits repeated detection of the same top clones. We have demonstrated considerable overlap in the alloreactive repertoires in MLRs from the same responder-stimulator pair performed as far as one year apart as well as relative stability of the overall TCR repertoire in this period.78 The overlap in alloreactive repertoires observed in independent blood samples is consistent with estimates of repertoire size and T cell pool size that suggest there are many thousands of copies of each T cell clone in the circulating repertoire at a given time.47,138 In the unstimulated setting, proof of deletion necessitates analysis of enough cells to provide statistical power to say that a clone is truly not there, rather than merely not detected. In addition to setting a frequency “threshold” of detection based on these power calculations, our studies required a particular fold expansion in frequency in MLR compared to unstimulated T cells in order to avoid assignment as donor-reactive of highly abundant non-donor-specific clones sorted into the CFSE-low pool. Emerson et al. used a similar approach to study TCR usage in human MLRs.139 We also required a minimum frequency threshold in the alloreactive T cell pool to help ensure that the clones we define as alloreactive are not proliferating primarily due to a bystander effect. As the sequencing protocols improve and the number of patients studied increases, we expect the precise fold-expansion and minimum frequency cut-offs to evolve.
While the requirement for only peripheral blood is a benefit in terms of minimizing invasiveness, conclusions are currently limited to the repertoire of circulating T cells. Identification of these alloreactive clones within the allograft is likely to provide further evidence for the biological relevance of these clones and expand the potential utility of this approach as a biomarker of tolerance and rejection. It should also be noted that another potential improvement to the assay would be the distinction of indirectly donor-reactive cells from those that are directly alloreactive. The current assay is likely to primarily capture directly alloreactive T cells clones, which are expected to dominate the MLR even when recipient APCs are present in addition to those of the donor. Separate identification and tracking of indirectly reactive clones would allow assessment of this critical component of the alloresponse, which is thought to play a major role in chronic rejection and alloantibody responses. Specific tracking of donor-reactive regulatory T cells is an additional area of interest for future development of this assay. While the current assay does not distinguish between functionally distinct subsets of T cells, additional FACS sorting parameters to capture alloreactive T cell subsets with cytotoxic function and particular cytokine production patterns could be developed.
There has been considerable interest in the possibility that infection shapes the alloreactive repertoire both acutely and in the post-infectious setting. While there are many examples in the literature of T cells that cross-react on viral antigens and alloantigens,36 the remarkable stability we have observed of the alloreactive repertoires of normal donors over time argues that major shifts in the alloreactive repertoire may not occur following common infections and vaccinations. In this regard, the “fold expansion” criterion for defining alloreactive clones in the MLR is important in excluding clonally expanded T cells that accumulate in the normal adult repertoire, presumably in response to infection, and that may appear in the CFSE-low population by virtue of their abundance, with minimal overall increase in frequency compared to unstimulated T cells. In general, the frequencies of donor-reactive clones detected in our transplant recipients do not reach levels that can be considered “immunodominant”, either pre- or post-transplant (e.g. Figure 2),78 arguing against changes resulting from cross-reactivity against infectious agents. It is theoretically possible that during an acute infection, a dominant T cell population might dilute alloreactive clones, rendering the detection of alloreactive clones less statistically likely in that setting. On the other hand, the specificity for the alloresponse of the alloreactive TCR tracking assay may help to differentiate infection from rejection, which can create a diagnostic challenge and may be indistinguishable using other types of biomarkers.
From a logistical perspective, the alloreactive TCR tracking approach is quite simple, as only functional assay necessary is the pre-transplant MLR. The availability of pre-transplant donor and recipient lymphocytes is critical for this assay. All post-transplant studies require only peripheral blood, from which fresh or cryopreserved PBMCs can be stained with three antibodies and sorted for genomic DNA extraction from the relevant cell subsets. The cost of deep sequencing is currently the most prohibitive aspect of the assay, though this is expected to diminish over time. Although it markedly increases the cost, we have elected to sort and sequence CD4 and CD8 cells separately in our studies, as this allows additional mechanistic information about tolerance and rejection to be obtained. Because changes in the CD4 and CD8 T cell repertoires do not always occur exactly in parallel, we would have missed key differences if we had not studied the subsets independently.
An alloreactive TCR tracking system that does not rely on post-transplant functional assays is advantageous in avoiding the interpretation of donor-specific unresponsiveness as tolerance. Indeed, the unresponsiveness observed in functional assays in a patient who failed to achieve tolerance78 suggests that such donor-specific unresponsiveness may reflect a fragile state of T cell “anergy” that may even be an artifact of the in vitro culture conditions. However, since new alloreactive clones likely are likely to emerge post-transplant, identification and tracking of those T cells could also provide important insights and assistance in managing immunosuppression.140 New anti-viral clones may emerge over the post-transplant period as the post-transplant immune system adapts to viral antigens that themselves may be changing,141 and new anti-donor clones may also develop post-transplant, However, our data suggest that donor-reactive clones detected in the pre-transplant repertoire, which encounter the allograft early, often under lymphopenic conditions, expand under the antigenic pressure of the graft and thereby remain important throughout the post-transplant course, despite eventual turnover of the non-donor-reactive repertoire.142
A limitation to consider at this time, as we found in our study and others have noted,126 is that a weak pre-transplant MLR may result in the identification of too few donor-reactive clones, thus limiting the power for statistical comparisons. Furthermore, CDR3 sequencing is currently restricted to the β chain of the TCR, thereby risking that two T cell clones with different α chains might not be differentiated because of the same TCRβ sequence. Improved technologies are already being developed that could overcome this shortcoming.
A new tool for understanding the alloreactive T cell repertoire
The combination of the CFSE MLR with TCR sequencing provides an entirely new approach to tackle the original questions of Bain, Bach, and Wilson in assessing the size and diversity of the alloreactive T cell repertoire. While many technical challenges remain as key assay parameters continue to be investigated, the great potential of this approach in readily apparent. In our study,78 we quantified the alloreactive repertoire size and diversity of healthy control responder-stimulator pairs. The cumulative frequencies of T cells reacting to an allogeneic donor were in the 0.3–2.3% range in unstimulated blood, and these increased into the 47.9–80.4% range in the allostimulated population. Since we have found that donor-reactive clones are, overall, not particularly abundant (i.e. not clonally expanded) in the unstimulated repertoire, their likelihood of being detected in a small unstimulated T cell sample is quite small, necessitating the use of larger samples, ideally containing at least one million CD4 and CD8 cells and the use of statistical analyses to compare the chances of detecting pre-existing donor-reactive versus non-donor-reactive clones over time.
The TCR sequencing approach also provides an unprecedented level of resolution for describing alloreactive T cell repertoire diversity. As discussed earlier in this paper, previous studies of TCR Vβ gene usage48–50 and CDR3 length distributions of each TCR V-Jβ family 51–53 suggested a high level of diversity in the alloresponse, but did not permit more detailed analyses. The high throughput CDR3 sequencing assay provides an opportunity to define the nature of the alloresponse in more detail.
Several different quantitative approaches can be used to quantify T cell repertoire diversity with robustness towards varying sample size. These include entropy, clonality (normalized entropy), and Simpson’s Index, all of which investigate different aspects of T cell repertoire composition. Both the number of unique clones and the frequency of those clones are integral components of repertoire diversity, and several types of analysis are needed to tease out the distinct contributions of these two elements. The Simpson’s Index is sensitive to changes in frequency of dominant clones as clones are weighted by their frequency, rather than their log frequency as they are in entropy and clonality. When the repertoire is dominated by highly frequent clones, Simpson’s Index may help show key aspects of the data that are not apparent in comparisons of clonality.
Aside from measuring diversity within a given T cell population, comparisons across populations are of considerable interest with respect to the overlap in both the identity and frequency. Scatter plots comparing two repertoires may qualitatively help compare repertoires. The Jaccard Index has been used as a type of quantitative Venn diagram, purely comparing the number of unique clones overlapping between two populations.143 More advanced tools include the Jensen Shannon Divergence and Morisita-Horn similarity index,140 both of which take into account the frequency of clones in a population to help assess the overall divergence or similarity, respectively, between two repertoires. As the use of high-throughput TCR sequencing expands, so will the analytic tools for understanding and comparing the different aspect of T cell repertoire composition.
On the future of TCR sequencing in transplantation
There is great interest in generating a tool to identify those transplant patients who have developed operational tolerance to their allografts, either intentionally or spontaneously. Such patients could be spared the panoply of deleterious side-effects of long-term immunosuppression if a reliable assay of this nature existed. Indeed among long-term liver transplant recipients who are free of rejection, as many as 20% (and a higher percentage of children) may be operationally tolerant, and yet there is currently no definitive tool to detect those individuals.144,145 TCR sequencing and tracking of the donor-reactive repertoire may prove useful for distinguishing operationally tolerant from non-tolerant patients and prospective trials to address this hypothesis are warranted.
Another key avenue for future investigation is the use of TCR sequencing in the diagnosis and/or prediction of rejection. Across the field of transplantation, strategies are lacking to successfully anticipate and recognize rejection episodes. Before a rejection episode there might be an increase in donor-specific clones in the circulation and/or within the allograft itself. Evaluation of this hypothesis in clinical studies might not only lead to the development of less invasive diagnostic tools and predictors of rejection but might also provide novel mechanistic insights into the role of T cells in acute and chronic rejection.
Conclusion
Although the formidable size and diversity of the directly alloreactive T cell repertoire has been a major challenge for transplant immunologists and physicians, the development of high-throughout TCR sequencing has transformed our ability to understand this response. Combining the classical MLR with T cell sequencing to define a fingerprint of the donor-reactive T cell repertoire for any specific HLA-mismatched donor-recipient pair has provided an unprecedented level of resolution for studying the fate of alloreactive clones in association with clinical outcomes. This approach has yielded mechanistic insights in a tolerant kidney transplant cohort and shows potential as a tool for differentiating tolerant and non-tolerant patients, meriting evaluation in additional cohorts of transplant recipients. Though much remains to be understood about how to use this new assay for identifying and tracking alloreactive clones, including both graft-versus-host and host-versus-graft, in the appropriate settings, its tremendous promise for elucidating mechanisms of tolerance and rejection are clear, particularly as the sequencing technology and accessibility improves. With this new window into the human alloreactive T cell repertoire, many of the fundamental questions about the nature of this enigmatic population may soon have a more definitive answer.
Acknowledgments
Funding: NIH awards: N01AI15416, P01AI106697, S10RR027050.
We thank Dr. Julien Zuber for his guidance and suggestions.
Abbreviations
- MLR
mixed lymphocyte reaction
- TCR
T cell receptor
- APC
antigen-presenting cells
- MHC
major histocompatibility complex
- HLA
human leukocyte antigen
- EBV
Epstein Barr virus
- CDR3
third complementarity-determining region
- BrdU
bromodeoxyuridine
- CFSE
carboxyfluorescein diacetate succinimidyl ester
- FACS
fluorescence-activated cell sorting
- HA
histocompatibility antigens
- GVH
graft-versus-host
- GVHD
graft-versus-host-disease
- ELISPOT
enzyme linked immunosorbent spot
- CKBMT
combined kidney bone marrow transplantation
- LIP
lymphopenia-induced proliferation
Footnotes
Author Contributions: S.D., Y.S., and M.S. interpreted data and wrote the review.
Disclosures: The authors declare no conflicts of interest.
References
- 1.Wilson DB, Blyth JNP. Quantitative Studies on the Mixed Lymphocyte Interaction in Rats. 3. Kinetics of the Response. J Exp Med. 1968 Nov 1;128(5):1157–1181. doi: 10.1084/jem.128.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lakkis FG, Lechler RI. Origin and Biology of the Allogeneic Response. Cold Spring Harb Perspect Med. 2013 Aug;3(8) doi: 10.1101/cshperspect.a014993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ali JM, Bolton EM, Bradley JA, Pettigrew GJ. Allorecognition Pathways in Transplant Rejection and Tolerance. Transplantation. 2013 Oct 27;96(8):681–688. doi: 10.1097/TP.0b013e31829853ce. [DOI] [PubMed] [Google Scholar]
- 4.Herrera OB, Golshayan D, Tibbott R, et al. A Novel Pathway of Alloantigen Presentation by Dendritic Cells. J Immunol. 2004 Oct 15;173(8):4828–4837. doi: 10.4049/jimmunol.173.8.4828. [DOI] [PubMed] [Google Scholar]
- 5.Smyth LA, Afzali B, Tsang J, Lombardi G, Lechler RI. Intercellular Transfer of Mhc and Immunological Molecules: Molecular Mechanisms and Biological Significance. Am J Transplant. 2007 Jun;7(6):1442–1449. doi: 10.1111/j.1600-6143.2007.01816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bain B, Vas M, Lowenstein L. Reaction between Leukocytes in Mixed Peripheral Blood Cultures. Federation Proceedings; April 1963. [Google Scholar]
- 7.Schrek R, Donnelly WJ. Differences between Lymphocytes of Leukemic and Non-Leukemic Patients with Respect to Morphologic Features, Motility, and Sensitivity to Guinea Pig Serum. Blood. 1961 Nov;18:561–571. [PubMed] [Google Scholar]
- 8.Hirschhorn K, Bach F, Kolodny RL, Firschein IL, Hashem N. Immune Response and Mitosis of Human Peripheral Blood Lymphocytes in Vitro. Science. 1963 Nov 29;142(3596):1185–1187. doi: 10.1126/science.142.3596.1185. [DOI] [PubMed] [Google Scholar]
- 9.Bach F, Hirschhorn K, Schreibman RR, Ripps C. Immunological Responses of Human Lymphocytes in Vitro. Ann N Y Acad Sci. 1964 Nov 30;120:299–302. doi: 10.1111/j.1749-6632.1964.tb34728.x. [DOI] [PubMed] [Google Scholar]
- 10.Bain B, Vas MR, Lowenstein L. The Development of Large Immature Mononuclear Cells in Mixed Leukocyte Cultures. Blood. 1964 Jan;23:108–116. [PubMed] [Google Scholar]
- 11.Silvers WK, Wilson DB, Palm J. Mixed Leukocyte Reactions and Histocompatibility in Rats. Science. 1967 Feb 10;155(3763):703–704. doi: 10.1126/science.155.3763.703. [DOI] [PubMed] [Google Scholar]
- 12.Amos DB, Bach FH. Phenotypic Expressions of the Major Histocompatibility Locus in Man (Hl-a): Leukocyte Antigens and Mixed Leukocyte Culture Reactivity. J Exp Med. 1968 Oct 1;128(4):623–637. doi: 10.1084/jem.128.4.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elves MW, Israuels MC. Lymphocyte Transformation in Cultures of Mixed Leucocytes: A Possible Test of Histocompatibility. Lancet. 1965 Jun 5;1(7397):1184–1186. doi: 10.1016/s0140-6736(65)92719-4. [DOI] [PubMed] [Google Scholar]
- 14.Marshall WH, Valentine FT, Lawrence HS. Cellular Immunity in Vitro. Clonal Proliferation of Antigen-Stimulated Lymphocytes. J Exp Med. 1969 Aug 1;130(2):327–343. doi: 10.1084/jem.130.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bach FH, Bock H, Graupner K, Day E, Klostermann H. Cell Kinetic Studies in Mixed Leukocyte Cultures: An in Vitro Model of Homograft Reactivity. Proc Natl Acad Sci U S A. 1969 Feb;62(2):377–384. doi: 10.1073/pnas.62.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simonsen M. The Clonal Selection Hypothesis Evaluated by Grafted Cells Reacting against Their Host. Cold Spring Harbor symposia on quantitative biology. 1967;32:517–523. [Google Scholar]
- 17.Wilson DB. Quantitative Studies on the Mixed Lymphocyte Interaction in Rats. I. Conditions and Parameters of Response. J Exp Med. 1967 Oct 1;126(4):625–654. doi: 10.1084/jem.126.4.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson DB, Silvers WK, Nowell PC. Quantitative Studies on the Mixed Lymphocyte Interaction in Rats. Ii. Relationship of the Proliferative Response to the Immunologic Status of the Donors. J Exp Med. 1967 Oct 1;126(4):655–665. doi: 10.1084/jem.126.4.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones G. The Number of Reactive Cells in Mouse Lymphocyte Cultures Stimulated by Phytohemagglutinin, Concanavalin a or Histocompatibility Antigen. J Immunol. 1973 Sep;111(3):914–920. [PubMed] [Google Scholar]
- 20.Sherman LA, Chattopadhyay S. The Molecular Basis of Allorecognition. Annu Rev Immunol. 1993;11:385–402. doi: 10.1146/annurev.iy.11.040193.002125. [DOI] [PubMed] [Google Scholar]
- 21.Ford WL, Atkins RC. The Proportion of Lymphocytes Capable of Recognizing Strong Transplantation Antigens in Vivo. Adv Exp Med Biol. 1973;29(0):255–262. doi: 10.1007/978-1-4615-9017-0_37. [DOI] [PubMed] [Google Scholar]
- 22.Atkins RC, Ford WL. Early Cellular Events in a Systemic Graft-Vs.-Host Reaction. I. The Migration of Responding and Nonresponding Donor Lymphocytes. J Exp Med. 1975 Mar 1;141(3):664–680. doi: 10.1084/jem.141.3.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ford WL, Simmonds SJ, Atkins RC. Early Cellular Events in a Systemic Graft-Vs.-Host Reaction. Ii. Autoradiographic Estimates of the Frequency of Donor Lymphocytes Which Respond to Each Ag-B-Determined Antigenic Complex. J Exp Med. 1975 Mar 1;141(3):681–696. doi: 10.1084/jem.141.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindahl KF, Wilson DB. Histocompatibility Antigen-Activated Cytotoxic T Lymphocytes. Ii. Estimates of the Frequency and Specificity of Precursors. J Exp Med. 1977 Mar 1;145(3):508–522. doi: 10.1084/jem.145.3.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jerne NK. The Somatic Generation of Immune Recognition. 1971. Eur J Immunol. 2004 May;34(5):1234–1242. doi: 10.1002/eji.200425132. [DOI] [PubMed] [Google Scholar]
- 26.Matzinger P, Bevan MJ. Hypothesis: Why Do So Many Lymphocytes Respond to Major Histocompatibility Antigens? Cell Immunol. 1977 Mar 1;29(1):1–5. doi: 10.1016/0008-8749(77)90269-6. [DOI] [PubMed] [Google Scholar]
- 27.Bevan MJ. High Determinant Density May Explain the Phenomenon of Alloreactivity. Immunol Today. 1984 May;5(5):128–130. doi: 10.1016/0167-5699(84)90233-0. [DOI] [PubMed] [Google Scholar]
- 28.Whitelegg AM, Oosten LE, Jordan S, et al. Investigation of Peptide Involvement in T Cell Allorecognition Using Recombinant Hla Class I Multimers. J Immunol. 2005 Aug 1;175(3):1706–1714. doi: 10.4049/jimmunol.175.3.1706. [DOI] [PubMed] [Google Scholar]
- 29.Felix NJ, Donermeyer DL, Horvath S, et al. Alloreactive T Cells Respond Specifically to Multiple Distinct Peptide-Mhc Complexes. Nat Immunol. 2007 Apr;8(4):388–397. doi: 10.1038/ni1446. [DOI] [PubMed] [Google Scholar]
- 30.Archbold JK, Macdonald WA, Burrows SR, Rossjohn J, McCluskey J. T-Cell Allorecognition: A Case of Mistaken Identity or Deja Vu? Trends Immunol. 2008 May;29(5):220–226. doi: 10.1016/j.it.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 31.Zerrahn J, Held W, Raulet DH. The Mhc Reactivity of the T Cell Repertoire Prior to Positive and Negative Selection. Cell. 1997 Mar 7;88(5):627–636. doi: 10.1016/s0092-8674(00)81905-4. [DOI] [PubMed] [Google Scholar]
- 32.Mason D. A Very High Level of Crossreactivity Is an Essential Feature of the T-Cell Receptor. Immunol Today. 1998 Sep;19(9):395–404. doi: 10.1016/s0167-5699(98)01299-7. [DOI] [PubMed] [Google Scholar]
- 33.Wilson DB, Wilson DH, Schroder K, et al. Specificity and Degeneracy of T Cells. Mol Immunol. 2004 Feb;40(14–15):1047–1055. doi: 10.1016/j.molimm.2003.11.022. [DOI] [PubMed] [Google Scholar]
- 34.Wucherpfennig KW, Allen PM, Celada F, et al. Polyspecificity of T Cell and B Cell Receptor Recognition. Semin Immunol. 2007 Aug;19(4):216–224. doi: 10.1016/j.smim.2007.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maverakis E, van den Elzen P, Sercarz EE. Self-Reactive T Cells and Degeneracy of T Cell Recognition: Evolving Concepts-from Sequence Homology to Shape Mimicry and Tcr Flexibility. J Autoimmun. 2001 May;16(3):201–209. doi: 10.1006/jaut.2000.0493. [DOI] [PubMed] [Google Scholar]
- 36.D’Orsogna LJ, Roelen DL, Doxiadis II, Claas FH. Tcr Cross-Reactivity and Allorecognition: New Insights into the Immunogenetics of Allorecognition. Immunogenetics. 2012 Feb;64(2):77–85. doi: 10.1007/s00251-011-0590-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Griesemer AD, Sorenson EC, Hardy MA. The Role of the Thymus in Tolerance. Transplantation. 2010 Sep 15;90(5):465–474. doi: 10.1097/TP.0b013e3181e7e54f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robey E, Fowlkes BJ. Selective Events in T Cell Development. Annu Rev Immunol. 1994;12:675–705. doi: 10.1146/annurev.iy.12.040194.003331. [DOI] [PubMed] [Google Scholar]
- 39.Jameson SC, Hogquist KA, Bevan MJ. Positive Selection of Thymocytes. Annu Rev Immunol. 1995;13:93–126. doi: 10.1146/annurev.iy.13.040195.000521. [DOI] [PubMed] [Google Scholar]
- 40.Marrack P, Kappler J. T Cells Can Distinguish between Allogeneic Major Histocompatibility Complex Products on Different Cell Types. Nature. 1988 Apr 28;332(6167):840–843. doi: 10.1038/332840a0. [DOI] [PubMed] [Google Scholar]
- 41.Burrows SR, Khanna R, Burrows JM, Moss DJ. An Alloresponse in Humans Is Dominated by Cytotoxic T Lymphocytes (Ctl) Cross-Reactive with a Single Epstein-Barr Virus Ctl Epitope: Implications for Graft-Versus-Host Disease. J Exp Med. 1994 Apr 1;179(4):1155–1161. doi: 10.1084/jem.179.4.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amir AL, D’Orsogna LJ, Roelen DL, et al. Allo-Hla Reactivity of Virus-Specific Memory T Cells Is Common. Blood. 2010 Apr 15;115(15):3146–3157. doi: 10.1182/blood-2009-07-234906. [DOI] [PubMed] [Google Scholar]
- 43.D’Orsogna LJ, Roelen DL, Doxiadis II, Claas FH. Alloreactivity from Human Viral Specific Memory T-Cells. Transpl Immunol. 2010 Aug;23(4):149–155. doi: 10.1016/j.trim.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 44.Morris GP, Allen PM. Cutting Edge: Highly Alloreactive Dual Tcr T Cells Play a Dominant Role in Graft-Versus-Host Disease. J Immunol. 2009 Jun 1;182(11):6639–6643. doi: 10.4049/jimmunol.0900638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morris GP, Uy GL, Donermeyer D, Dipersio JF, Allen PM. Dual Receptor T Cells Mediate Pathologic Alloreactivity in Patients with Acute Graft-Versus-Host Disease. Sci Transl Med. 2013 Jun 5;5(188):188ra174. doi: 10.1126/scitranslmed.3005452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davis MM, Bjorkman PJ. T-Cell Antigen Receptor Genes and T-Cell Recognition. Nature. 1988 Aug 4;334(6181):395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- 47.Robins HS, Campregher PV, Srivastava SK, et al. Comprehensive Assessment of T-Cell Receptor Beta-Chain Diversity in Alphabeta T Cells. Blood. 2009 Nov 5;114(19):4099–4107. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sherman LA. Dissection of the B10.D2 Anti-H-2kb Cytolytic T Lymphocyte Receptor Repertoire. J Exp Med. 1980 Jun 1;151(6):1386–1397. doi: 10.1084/jem.151.6.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bill J, Yague J, Appel VB, et al. Molecular Genetic Analysis of 178 I-Abm12-Reactive T Cells. J Exp Med. 1989 Jan 1;169(1):115–133. doi: 10.1084/jem.169.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rotzschke O, Falk K, Faath S, Rammensee HG. On the Nature of Peptides Involved in T Cell Alloreactivity. J Exp Med. 1991 Nov 1;174(5):1059–1071. doi: 10.1084/jem.174.5.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pannetier C, Even J, Kourilsky P. T-Cell Repertoire Diversity and Clonal Expansions in Normal and Clinical Samples. Immunol Today. 1995 Apr;16(4):176–181. doi: 10.1016/0167-5699(95)80117-0. [DOI] [PubMed] [Google Scholar]
- 52.Sebille F, Gagne K, Guillet M, et al. Direct Recognition of Foreign Mhc Determinants by Naive T Cells Mobilizes Specific Vbeta Families without Skewing of the Complementarity-Determining Region 3 Length Distribution. J Immunol. 2001 Sep 15;167(6):3082–3088. doi: 10.4049/jimmunol.167.6.3082. [DOI] [PubMed] [Google Scholar]
- 53.Bragado R, Lauzurica P, Lopez D, Lopez de Castro JA. T Cell Receptor V Beta Gene Usage in a Human Alloreactive Response. Shared Structural Features among Hla-B27-Specific T Cell Clones. J Exp Med. 1990 Apr 1;171(4):1189–1204. doi: 10.1084/jem.171.4.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lyons AB, Parish CR. Determination of Lymphocyte Division by Flow Cytometry. J Immunol Methods. 1994 May 2;171(1):131–137. doi: 10.1016/0022-1759(94)90236-4. [DOI] [PubMed] [Google Scholar]
- 55.Parish CR. Fluorescent Dyes for Lymphocyte Migration and Proliferation Studies. Immunol Cell Biol. 1999 Dec;77(6):499–508. doi: 10.1046/j.1440-1711.1999.00877.x. [DOI] [PubMed] [Google Scholar]
- 56.Noorchashm H, Lieu YK, Rostami SY, et al. A Direct Method for the Calculation of Alloreactive Cd4+ T Cell Precursor Frequency. Transplantation. 1999 May 15;67(9):1281–1284. doi: 10.1097/00007890-199905150-00015. [DOI] [PubMed] [Google Scholar]
- 57.Suchin EJ, Langmuir PB, Palmer E, Sayegh MH, Wells AD, Turka LA. Quantifying the Frequency of Alloreactive T Cells in Vivo: New Answers to an Old Question. J Immunol. 2001 Jan 15;166(2):973–981. doi: 10.4049/jimmunol.166.2.973. [DOI] [PubMed] [Google Scholar]
- 58.Macedo C, Orkis EA, Popescu I, et al. Contribution of Naive and Memory T-Cell Populations to the Human Alloimmune Response. Am J Transplant. 2009 Sep;9(9):2057–2066. doi: 10.1111/j.1600-6143.2009.02742.x. [DOI] [PubMed] [Google Scholar]
- 59.Choi Y, Kappler JW, Marrack P. A Superantigen Encoded in the Open Reading Frame of the 3′ Long Terminal Repeat of Mouse Mammary Tumour Virus. Nature. 1991 Mar 21;350(6315):203–207. doi: 10.1038/350203a0. [DOI] [PubMed] [Google Scholar]
- 60.Woodland DL, Happ MP, Gollob KJ, Palmer E. An Endogenous Retrovirus Mediating Deletion of Alpha Beta T Cells? Nature. 1991 Feb 7;349(6309):529–530. doi: 10.1038/349529a0. [DOI] [PubMed] [Google Scholar]
- 61.Yoshikai Y, Ogimoto M, Matsuzaki G, Nomoto K. Bone Marrow-Derived Cells Are Essential for Intrathymic Deletion of Self-Reactive T Cells in Both the Host- and Donor-Derived Thymocytes of Fully Allogeneic Bone Marrow Chimeras. J Immunol. 1990 Jul 15;145(2):505–509. [PubMed] [Google Scholar]
- 62.Tomita Y, Khan A, Sykes M. Role of Intrathymic Clonal Deletion and Peripheral Anergy in Transplantation Tolerance Induced by Bone Marrow Transplantation in Mice Conditioned with a Nonmyeloablative Regimen. J Immunol. 1994 Aug 1;153(3):1087–1098. [PubMed] [Google Scholar]
- 63.Sha WC, Nelson CA, Newberry RD, Kranz DM, Russell JH, Loh DY. Positive and Negative Selection of an Antigen Receptor on T Cells in Transgenic Mice. Nature. 1988 Nov 3;336(6194):73–76. doi: 10.1038/336073a0. [DOI] [PubMed] [Google Scholar]
- 64.Dey B, Yang YG, Preffer F, et al. The Fate of Donor T-Cell Receptor Transgenic T Cells with Known Host Antigen Specificity in a Graft-Versus-Host Disease Model. Transplantation. 1999 Jul 15;68(1):141–149. doi: 10.1097/00007890-199907150-00026. [DOI] [PubMed] [Google Scholar]
- 65.Manilay JO, Pearson DA, Sergio JJ, Swenson KG, Sykes M. Intrathymic Deletion of Alloreactive T Cells in Mixed Bone Marrow Chimeras Prepared with a Nonmyeloablative Conditioning Regimen. Transplantation. 1998 Jul 15;66(1):96–102. doi: 10.1097/00007890-199807150-00015. [DOI] [PubMed] [Google Scholar]
- 66.Kurtz J, Shaffer J, Lie A, Anosova N, Benichou G, Sykes M. Mechanisms of Early Peripheral Cd4 T-Cell Tolerance Induction by Anti-Cd154 Monoclonal Antibody and Allogeneic Bone Marrow Transplantation: Evidence for Anergy and Deletion but Not Regulatory Cells. Blood. 2004 Jun 1;103(11):4336–4343. doi: 10.1182/blood-2003-08-2642. [DOI] [PubMed] [Google Scholar]
- 67.Fehr T, Takeuchi Y, Kurtz J, Wekerle T, Sykes M. Early Regulation of Cd8 T Cell Alloreactivity by Cd4+Cd25- T Cells in Recipients of Anti-Cd154 Antibody and Allogeneic Bmt Is Followed by Rapid Peripheral Deletion of Donor-Reactive Cd8+ T Cells, Precluding a Role for Sustained Regulation. Eur J Immunol. 2005 Sep;35(9):2679–2690. doi: 10.1002/eji.200526190. [DOI] [PubMed] [Google Scholar]
- 68.Gibbons C, Sykes M. Manipulating the Immune System for Anti-Tumor Responses and Transplant Tolerance Via Mixed Hematopoietic Chimerism. Immunol Rev. 2008 Jun;223:334–360. doi: 10.1111/j.1600-065X.2008.00636.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Altman JD, Moss PA, Goulder PJ, et al. Phenotypic Analysis of Antigen-Specific T Lymphocytes. Science. 1996 Oct 4;274(5284):94–96. doi: 10.1126/science.274.5284.94. [DOI] [PubMed] [Google Scholar]
- 70.Tsoi MS, Storb R, Dobbs S, Medill L, Thomas ED. Cell-Mediated Immunity to Non-Hla Antigens of the Host by Donor Lymphocytes in Patients with Chronic Graft-Vs-Host Disease. J Immunol. 1980 Nov;125(5):2258–2262. [PubMed] [Google Scholar]
- 71.Goulmy E, Gratama JW, Blokland E, Zwaan FE, van Rood JJ. A Minor Transplantation Antigen Detected by Mhc-Restricted Cytotoxic T Lymphocytes During Graft-Versus-Host Disease. Nature. 1983 Mar 10;302(5904):159–161. doi: 10.1038/302159a0. [DOI] [PubMed] [Google Scholar]
- 72.Goulmy E, Termijtelen A, Bradley BA, van Rood JJ. Alloimmunity to Human H-Y. Lancet. 1976 Nov 27;2(7996):1206. doi: 10.1016/s0140-6736(76)91727-x. [DOI] [PubMed] [Google Scholar]
- 73.Mutis T, Gillespie G, Schrama E, Falkenburg JH, Moss P, Goulmy E. Tetrameric Hla Class I-Minor Histocompatibility Antigen Peptide Complexes Demonstrate Minor Histocompatibility Antigen-Specific Cytotoxic T Lymphocytes in Patients with Graft-Versus-Host Disease. Nat Med. 1999 Jul;5(7):839–842. doi: 10.1038/10563. [DOI] [PubMed] [Google Scholar]
- 74.Bach F, Hirschhorn K. Lymphocyte Interaction: A Potential Histocompatibility Test in Vitro. Science. 1964 Feb 21;143(3608):813–814. doi: 10.1126/science.143.3608.813. [DOI] [PubMed] [Google Scholar]
- 75.Moynihan PC, Jackson JF, Hardy JD. Lymphocyte Transformation as an in-Vitro Histocompatibility Test. Lancet. 1965 Feb 27;1(7383):453–455. doi: 10.1016/s0140-6736(65)91587-4. [DOI] [PubMed] [Google Scholar]
- 76.Rubin AL, Stenzel KH, Hirschhorn K, Bach F. Histocompatibility and Immunologic Competence in Renal Homotransplantation. Science. 1964 Feb 21;143(3608):815–816. doi: 10.1126/science.143.3608.815. [DOI] [PubMed] [Google Scholar]
- 77.Goulmy E, Stijnen T, Groenewoud AF, Groenewoud Af, Persijn GG, et al. Renal Transplant Patients Monitored by the Cell-Mediated Lympholysis Assay. Evaluation of Its Clinical Value. Transplantation. 1989 Oct;48(4):559–563. doi: 10.1097/00007890-198910000-00004. [DOI] [PubMed] [Google Scholar]
- 78.Morris H, DeWolf S, Robins H, et al. Tracking Donor-Reactive T Cells: Evidence for Clonal Deletion in Tolerant Kidney Transplant Patients. Sci Transl Med. 2015 Jan 28;7(272):272ra210. doi: 10.1126/scitranslmed.3010760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dinavahi R, Heeger PS. T-Cell Immune Monitoring in Organ Transplantation. Transplantation. 2009 Nov 27;88(10):1157–1158. doi: 10.1097/TP.0b013e3181bdbf92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Streilein JW, Strome P, Wood PJ. Failure of in Vitro Assays to Predict Accurately the Existence of Neonatally Induced H-2 Tolerance. Transplantation. 1989 Oct;48(4):630–634. [PubMed] [Google Scholar]
- 81.Cullen PR, Lester S, Rouch J, Morris PJ. Mixed Lymphocyte Reaction and Graft Survival in Forty Cadaveric Renal Transplants. Clin Exp Immunol. 1977 May;28(2):218–222. [PMC free article] [PubMed] [Google Scholar]
- 82.Goulmy E, Persijn G, Blokland E, D’Amaro J, van Rood JJ. Cell-Mediated Lympholysis Studies in Renal Allograft Recipients. Transplantation. 1981 Mar;31(3):210–217. doi: 10.1097/00007890-198103000-00014. [DOI] [PubMed] [Google Scholar]
- 83.Najafian N, Albin MJ, Newell KA. How Can We Measure Immunologic Tolerance in Humans? J Am Soc Nephrol. 2006 Oct;17(10):2652–2663. doi: 10.1681/ASN.2005070707. [DOI] [PubMed] [Google Scholar]
- 84.Reinsmoen NL, Jackson AM, DeOliveira A, Matas AJ, Gillingham K, Ward FE. Cellular Immunology Markers Postransplantation Predictive of Long-Term Graft Outcome. Ann Transplant. 2000;5(2):50–60. [PubMed] [Google Scholar]
- 85.Steinmann J, Kaden J, May G, Schroder K, Herwartz C, Muller-Ruchholtz W. Failure of in Vitro T-Cell Assays to Predict Clinical Outcome after Human Kidney Transplantation. J Clin Lab Anal. 1994;8(3):157–162. doi: 10.1002/jcla.1860080308. [DOI] [PubMed] [Google Scholar]
- 86.Segall M, Noreen H, Edwins L, Haake R, Shu XO, Kersey J. Lack of Correlation of Mlc Reactivity with Acute Graft-Versus-Host Disease and Mortality in Unrelated Donor Bone Marrow Transplantation. Hum Immunol. 1996 Aug;49(1):49–55. doi: 10.1016/0198-8859(96)00055-9. [DOI] [PubMed] [Google Scholar]
- 87.Lefkovits I, Waldmann H. Limiting Dilution Analysis of the Cells of Immune System I. The Clonal Basis of the Immune Response. Immunol Today. 1984 Sep;5(9):265–268. doi: 10.1016/0167-5699(84)90137-3. [DOI] [PubMed] [Google Scholar]
- 88.Deacock S, Schwarer A, Batchelor R, Goldman J, Lechler R. A Rapid Limiting Dilution Assay for Measuring Frequencies of Alloreactive, Interleukin-2-Producing T Cells in Humans. J Immunol Methods. 1992 Feb 14;147(1):83–92. doi: 10.1016/s0022-1759(12)80032-9. [DOI] [PubMed] [Google Scholar]
- 89.Hornick PI, Brookes PA, Mason PD, et al. Optimizing a Limiting Dilution Culture System for Quantifying the Frequency of Interleukin-2-Producing Alloreactive T Helper Lymphocytes. Transplantation. 1997 Aug 15;64(3):472–479. doi: 10.1097/00007890-199708150-00017. [DOI] [PubMed] [Google Scholar]
- 90.Schwarer AP, Jiang YZ, Brookes PA, et al. Frequency of Anti-Recipient Alloreactive Helper T-Cell Precursors in Donor Blood and Graft-Versus-Host Disease after Hla-Identical Sibling Bone-Marrow Transplantation. Lancet. 1993 Jan 23;341(8839):203–205. doi: 10.1016/0140-6736(93)90067-q. [DOI] [PubMed] [Google Scholar]
- 91.Mestre M, Massip E, Bas J, et al. Longitudinal Study of the Frequency of Cytotoxic T Cell Precursors in Kidney Allograft Recipients. Clin Exp Immunol. 1996 Apr;104(1):108–114. doi: 10.1046/j.1365-2249.1996.d01-657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hu H, Robertus M, de Jonge N, et al. Reduction of Donor-Specific Cytotoxic T Lymphocyte Precursors in Peripheral Blood of Allografted Heart Recipients. Transplantation. 1994 Dec 15;58(11):1263–1268. [PubMed] [Google Scholar]
- 93.Hornick PI, Mason PD, Baker RJ, et al. Significant Frequencies of T Cells with Indirect Anti-Donor Specificity in Heart Graft Recipients with Chronic Rejection. Circulation. 2000 May 23;101(20):2405–2410. doi: 10.1161/01.cir.101.20.2405. [DOI] [PubMed] [Google Scholar]
- 94.Heeger PS, Greenspan NS, Kuhlenschmidt S, et al. Pretransplant Frequency of Donor-Specific, Ifn-Gamma-Producing Lymphocytes Is a Manifestation of Immunologic Memory and Correlates with the Risk of Posttransplant Rejection Episodes. J Immunol. 1999 Aug 15;163(4):2267–2275. [PubMed] [Google Scholar]
- 95.Ashoor I, Najafian N, Korin Y, et al. Standardization and Cross Validation of Alloreactive Ifngamma Elispot Assays within the Clinical Trials in Organ Transplantation Consortium. Am J Transplant. 2013 Jul;13(7):1871–1879. doi: 10.1111/ajt.12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nickel P, Presber F, Bold G, et al. Enzyme-Linked Immunosorbent Spot Assay for Donor-Reactive Interferon-Gamma-Producing Cells Identifies T-Cell Presensitization and Correlates with Graft Function at 6 and 12 Months in Renal-Transplant Recipients. Transplantation. 2004 Dec 15;78(11):1640–1646. doi: 10.1097/01.tp.0000144057.31799.6a. [DOI] [PubMed] [Google Scholar]
- 97.Augustine JJ, Siu DS, Clemente MJ, Schulak JA, Heeger PS, Hricik DE. Pre-Transplant Ifn-Gamma Elispots Are Associated with Post-Transplant Renal Function in African American Renal Transplant Recipients. Am J Transplant. 2005 Aug;5(8):1971–1975. doi: 10.1111/j.1600-6143.2005.00958.x. [DOI] [PubMed] [Google Scholar]
- 98.Hricik DE, Rodriguez V, Riley J, et al. Enzyme Linked Immunosorbent Spot (Elispot) Assay for Interferon-Gamma Independently Predicts Renal Function in Kidney Transplant Recipients. Am J Transplant. 2003 Jul;3(7):878–884. doi: 10.1034/j.1600-6143.2003.00132.x. [DOI] [PubMed] [Google Scholar]
- 99.Hricik DE, Augustine J, Nickerson P, et al. Interferon Gamma Elispot Testing as a Risk-Stratifying Biomarker for Kidney Transplant Injury: Results from the Ctot-01 Multicenter Study. Am J Transplant. 2015 Jul 30; doi: 10.1111/ajt.13401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Augustine JJ, Poggio ED, Heeger PS, Hricik DE. Preferential Benefit of Antibody Induction Therapy in Kidney Recipients with High Pretransplant Frequencies of Donor-Reactive Interferon-Gamma Enzyme-Linked Immunosorbent Spots. Transplantation. 2008 Aug 27;86(4):529–534. doi: 10.1097/TP.0b013e31818046db. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.VanBuskirk AM, Burlingham WJ, Jankowska-Gan E, et al. Human Allograft Acceptance Is Associated with Immune Regulation. J Clin Invest. 2000 Jul;106(1):145–155. doi: 10.1172/JCI9171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rodriguez DS, Jankowska-Gan E, Haynes LD, et al. Immune Regulation and Graft Survival in Kidney Transplant Recipients Are Both Enhanced by Human Leukocyte Antigen Matching. Am J Transplant. 2004 Apr;4(4):537–543. doi: 10.1111/j.1600-6143.2004.00385.x. [DOI] [PubMed] [Google Scholar]
- 103.Burlingham WJ, Jankowska-Gan E. Mouse Strain and Injection Site Are Crucial for Detecting Linked Suppression in Transplant Recipients by Trans-Vivo Dth Assay. Am J Transplant. 2007 Feb;7(2):466–470. doi: 10.1111/j.1600-6143.2006.01627.x. [DOI] [PubMed] [Google Scholar]
- 104.Haynes LD, Jankowska-Gan E, Sheka A, et al. Donor-Specific Indirect Pathway Analysis Reveals a B-Cell-Independent Signature Which Reflects Outcomes in Kidney Transplant Recipients. Am J Transplant. 2012 Mar;12(3):640–648. doi: 10.1111/j.1600-6143.2011.03869.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Frentsch M, Arbach O, Kirchhoff D, et al. Direct Access to Cd4+ T Cells Specific for Defined Antigens According to Cd154 Expression. Nat Med. 2005 Oct;11(10):1118–1124. doi: 10.1038/nm1292. [DOI] [PubMed] [Google Scholar]
- 106.Ashokkumar C, Talukdar A, Sun Q, et al. Allospecific Cd154+ T Cells Associate with Rejection Risk after Pediatric Liver Transplantation. Am J Transplant. 2009 Jan;9(1):179–191. doi: 10.1111/j.1600-6143.2008.02459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cortesini R, Renna-Molajoni E, Cinti P, et al. Tailoring of Immunosuppression in Renal and Liver Allograft Recipients Displaying Donor Specific T-Suppressor Cells. Hum Immunol. 2002 Nov;63(11):1010–1018. doi: 10.1016/s0198-8859(02)00442-1. [DOI] [PubMed] [Google Scholar]
- 108.Li Y, Koshiba T, Yoshizawa A, et al. Analyses of Peripheral Blood Mononuclear Cells in Operational Tolerance after Pediatric Living Donor Liver Transplantation. Am J Transplant. 2004 Dec;4(12):2118–2125. doi: 10.1111/j.1600-6143.2004.00611.x. [DOI] [PubMed] [Google Scholar]
- 109.Mazariegos GV, Zahorchak AF, Reyes J, et al. Dendritic Cell Subset Ratio in Peripheral Blood Correlates with Successful Withdrawal of Immunosuppression in Liver Transplant Patients. Am J Transplant. 2003 Jun;3(6):689–696. doi: 10.1034/j.1600-6143.2003.00109.x. [DOI] [PubMed] [Google Scholar]
- 110.Codarri L, Vallotton L, Ciuffreda D, et al. Expansion and Tissue Infiltration of an Allospecific Cd4+Cd25+Cd45ro+Il-7ralphahigh Cell Population in Solid Organ Transplant Recipients. J Exp Med. 2007 Jul 9;204(7):1533–1541. doi: 10.1084/jem.20062120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Israeli M, Yussim A, Mor E, Sredni B, Klein T. Preceeding the Rejection: In Search for a Comprehensive Post-Transplant Immune Monitoring Platform. Transpl Immunol. 2007 Jul;18(1):7–12. doi: 10.1016/j.trim.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 112.Kowalski R, Post D, Schneider MC, et al. Immune Cell Function Testing: An Adjunct to Therapeutic Drug Monitoring in Transplant Patient Management. Clin Transplant. 2003 Apr;17(2):77–88. doi: 10.1034/j.1399-0012.2003.00013.x. [DOI] [PubMed] [Google Scholar]
- 113.Li B, Hartono C, Ding R, et al. Noninvasive Diagnosis of Renal-Allograft Rejection by Measurement of Messenger Rna for Perforin and Granzyme B in Urine. N Engl J Med. 2001 Mar 29;344(13):947–954. doi: 10.1056/NEJM200103293441301. [DOI] [PubMed] [Google Scholar]
- 114.Afaneh C, Muthukumar T, Lubetzky M, et al. Urinary Cell Levels of Mrna for Ox40, Ox40l, Pd-1, Pd-L1, or Pd-L2 and Acute Rejection of Human Renal Allografts. Transplantation. 2010 Dec 27;90(12):1381–1387. doi: 10.1097/TP.0b013e3181ffbadd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Muthukumar T, Dadhania D, Ding R, et al. Messenger Rna for Foxp3 in the Urine of Renal-Allograft Recipients. N Engl J Med. 2005 Dec 1;353(22):2342–2351. doi: 10.1056/NEJMoa051907. [DOI] [PubMed] [Google Scholar]
- 116.Flechner SM, Kurian SM, Head SR, et al. Kidney Transplant Rejection and Tissue Injury by Gene Profiling of Biopsies and Peripheral Blood Lymphocytes. Am J Transplant. 2004 Sep;4(9):1475–1489. doi: 10.1111/j.1600-6143.2004.00526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ho J, Rush DN, Gibson IW, et al. Early Urinary Ccl2 Is Associated with the Later Development of Interstitial Fibrosis and Tubular Atrophy in Renal Allografts. Transplantation. 2010 Aug 27;90(4):394–400. doi: 10.1097/TP.0b013e3181e6424d. [DOI] [PubMed] [Google Scholar]
- 118.Hricik DE, Nickerson P, Formica RN, et al. Multicenter Validation of Urinary Cxcl9 as a Risk-Stratifying Biomarker for Kidney Transplant Injury. Am J Transplant. 2013 Oct;13(10):2634–2644. doi: 10.1111/ajt.12426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ling XB, Sigdel TK, Lau K, et al. Integrative Urinary Peptidomics in Renal Transplantation Identifies Biomarkers for Acute Rejection. J Am Soc Nephrol. 2010 Apr;21(4):646–653. doi: 10.1681/ASN.2009080876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Newell KA, Asare A, Kirk AD, et al. Identification of a B Cell Signature Associated with Renal Transplant Tolerance in Humans. J Clin Invest. 2010 Jun;120(6):1836–1847. doi: 10.1172/JCI39933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sagoo P, Perucha E, Sawitzki B, et al. Development of a Cross-Platform Biomarker Signature to Detect Renal Transplant Tolerance in Humans. J Clin Invest. 2010 Jun;120(6):1848–1861. doi: 10.1172/JCI39922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dugast E, Chesneau M, Soulillou JP, Brouard S. Biomarkers and Possible Mechanisms of Operational Tolerance in Kidney Transplant Patients. Immunol Rev. 2014 Mar;258(1):208–217. doi: 10.1111/imr.12156. [DOI] [PubMed] [Google Scholar]
- 123.Sanchez-Fueyo A. Tolerance Profiles and Immunosuppression. Liver Transpl. 2013 Nov;19(Suppl 2):S44–48. doi: 10.1002/lt.23749. [DOI] [PubMed] [Google Scholar]
- 124.Bohne F, Martinez-Llordella M, Lozano JJ, et al. Intra-Graft Expression of Genes Involved in Iron Homeostasis Predicts the Development of Operational Tolerance in Human Liver Transplantation. J Clin Invest. 2012 Jan;122(1):368–382. doi: 10.1172/JCI59411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Suthanthiran M, Schwartz JE, Ding R, et al. Urinary-Cell Mrna Profile and Acute Cellular Rejection in Kidney Allografts. N Engl J Med. 2013 Jul 4;369(1):20–31. doi: 10.1056/NEJMoa1215555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Scheinberg P, Price DA, Ambrozak DR, Barrett AJ, Douek DC. Alloreactive T Cell Clonotype Recruitment in a Mixed Lymphocyte Reaction: Implications for Graft Engineering. Exp Hematol. 2006 Jun;34(6):788–795. doi: 10.1016/j.exphem.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 127.Michalek J, Collins RH, Hill BJ, Brenchley JM, Douek DC. Identification and Monitoring of Graft-Versus-Host Specific T-Cell Clone in Stem Cell Transplantation. Lancet. 2003 Apr 5;361(9364):1183–1185. doi: 10.1016/S0140-6736(03)12917-0. [DOI] [PubMed] [Google Scholar]
- 128.Watson D, Zhang GY, Sartor M, Alexander SI. “Pruning” of Alloreactive Cd4+ T Cells Using 5- (and 6-)Carboxyfluorescein Diacetate Succinimidyl Ester Prolongs Skin Allograft Survival. J Immunol. 2004 Dec 1;173(11):6574–6582. doi: 10.4049/jimmunol.173.11.6574. [DOI] [PubMed] [Google Scholar]
- 129.Hu M, Watson D, Zhang GY, et al. Long-Term Cardiac Allograft Survival across an Mhc Mismatch after “Pruning” of Alloreactive Cd4 T Cells. J Immunol. 2008 May 15;180(10):6593–6603. doi: 10.4049/jimmunol.180.10.6593. [DOI] [PubMed] [Google Scholar]
- 130.Kawai T, Cosimi AB, Spitzer TR, et al. Hla-Mismatched Renal Transplantation without Maintenance Immunosuppression. New England Journal of Medicine. 2008;358(4):353–361. doi: 10.1056/NEJMoa071074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kawai T, Sachs DH, Sykes M, Cosimi AB. Hla-Mismatched Renal Transplantation without Maintenance Immunosuppression. N Engl J Med. 2013 May 9;368(19):1850–1852. doi: 10.1056/NEJMc1213779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kawai T, Sachs DH, Sprangers B, et al. Long-Term Results in Recipients of Combined Hla-Mismatched Kidney and Bone Marrow Transplantation without Maintenance Immunosuppression. Am J Transplant. 2014 Jul;14(7):1599–1611. doi: 10.1111/ajt.12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.LoCascio SA, Morokata T, Chittenden M, et al. Mixed Chimerism, Lymphocyte Recovery, and Evidence for Early Donor-Specific Unresponsiveness in Patients Receiving Combined Kidney and Bone Marrow Transplantation to Induce Tolerance. Transplantation. 2010 Dec 27;90(12):1607–1615. doi: 10.1097/TP.0b013e3181ffbaff. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Andreola G, Chittenden M, Shaffer J, et al. Mechanisms of Donor-Specific Tolerance in Recipients of Haploidentical Combined Bone Marrow/Kidney Transplantation. Am J Transplant. 2011 Jun;11(6):1236–1247. doi: 10.1111/j.1600-6143.2011.03566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Khan A, Tomita Y, Sykes M. Thymic Dependence of Loss of Tolerance in Mixed Allogeneic Bone Marrow Chimeras after Depletion of Donor Antigen. Peripheral Mechanisms Do Not Contribute to Maintenance of Tolerance. Transplantation. 1996 Aug 15;62(3):380–387. doi: 10.1097/00007890-199608150-00014. [DOI] [PubMed] [Google Scholar]
- 136.Muraro PA, Robins H, Malhotra S, et al. T Cell Repertoire Following Autologous Stem Cell Transplantation for Multiple Sclerosis. J Clin Invest. 2014 Mar;124(3):1168–1172. doi: 10.1172/JCI71691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Miqueu P, Degauque N, Guillet M, et al. Analysis of the Peripheral T-Cell Repertoire in Kidney Transplant Patients. Eur J Immunol. 2010 Nov;40(11):3280–3290. doi: 10.1002/eji.201040301. [DOI] [PubMed] [Google Scholar]
- 138.Robins H, Desmarais C, Matthis J, et al. Ultra-Sensitive Detection of Rare T Cell Clones. J Immunol Methods. 2012 Jan 31;375(1–2):14–19. doi: 10.1016/j.jim.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Emerson RO, Mathew JM, Konieczna IM, Robins HS, Leventhal JR. Defining the Alloreactive T Cell Repertoire Using High-Throughput Sequencing of Mixed Lymphocyte Reaction Culture. PLoS One. 2014;9(11):e111943. doi: 10.1371/journal.pone.0111943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Dziubianau M, Hecht J, Kuchenbecker L, et al. Tcr Repertoire Analysis by Next Generation Sequencing Allows Complex Differential Diagnosis of T Cell-Related Pathology. Am J Transplant. 2013 Nov;13(11):2842–2854. doi: 10.1111/ajt.12431. [DOI] [PubMed] [Google Scholar]
- 141.Gorochov G, Larsen M, Parizot C, et al. Comment on “Tracking Donor-Reactive T Cells: Evidence for Clonal Deletion in Tolerant Kidney Transplant Patients”. Sci Transl Med. 2015 Jul 22;7(297):297l, e291. doi: 10.1126/scitranslmed.aab1994. [DOI] [PubMed] [Google Scholar]
- 142.DeWolf S, Morris H, Shen Y, Sykes M. Author Response to Comment on “Tracking Donor-Reactive T Cells: Evidence for Clonal Deletion in Tolerant Kidney Transplant Patients”. Sci Transl Med. 2015 Jul 22;7(297):297lr291. doi: 10.1126/scitranslmed.aac9461. [DOI] [PubMed] [Google Scholar]
- 143.Thome JJ, Yudanin N, Ohmura Y, et al. Spatial Map of Human T Cell Compartmentalization and Maintenance over Decades of Life. Cell. 2014 Nov 6;159(4):814–828. doi: 10.1016/j.cell.2014.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lerut J, Sanchez-Fueyo A. An Appraisal of Tolerance in Liver Transplantation. Am J Transplant. 2006 Aug;6(8):1774–1780. doi: 10.1111/j.1600-6143.2006.01396.x. [DOI] [PubMed] [Google Scholar]
- 145.Whitehouse GP, Sanchez-Fueyo A. Immunosuppression Withdrawal Following Liver Transplantation. Clin Res Hepatol Gastroenterol. 2014 Dec;38(6):676–680. doi: 10.1016/j.clinre.2014.06.011. [DOI] [PubMed] [Google Scholar]