Abstract
Protein target structures for the Critical Assessment of Structure Prediction round 11 (CASP11) and CASP ROLL were split into domains and classified into categories suitable for assessment of template-based modeling (TBM) and free modeling (FM) based on their evolutionary relatedness to existing structures classified by the Evolutionary Classification of Protein Domains (ECOD) database. First, target structures were divided into domain-based evaluation units. Target splits were based on the domain organization of available templates as well as the performance of servers on whole targets compared to split target domains. Second, evaluation units were classified into TBM and FM categories using a combination of measures that evaluate prediction quality and template detectability. Generally, target domains with sequence-related templates and good server prediction performance were classified as TBM, whereas targets without sequence-identifiable templates and low server performance were classified as FM. As in previous CASP experiments, the boundaries for classification were blurred due to the presence of significant insertions and deteriorations in the targets with respect to homologous templates, as well as the presence of templates with partial coverage of new folds. The FM category included 45 target domains, which represents an unprecedented number of difficult CASP targets provided for modeling.
Keywords: protein structure, CASP11, classification, fold space, sequence homologs, structure analogs, free modeling, template based modeling, structure prediction
Introduction
The Critical Assessment of Structure Prediction, round 11 (CASP11) aimed to provide an objective evaluation of current state-of-the-art methodologies in protein structure prediction. Participants submitted models for targets whose structures were unknown or withheld from public release during the CASP11 timeframe. In addition to the traditional CASP objective, CASP ROLL released difficult targets for prediction year-round. Models were assessed for their similarity to the target structure to reveal the performance of both automated and manual structure prediction methods. In addition to evaluation of tertiary structure prediction, CASP11 included new initiatives to address the biological relevance of structure models and to assess modeling of oligomeric interactions in collaboration with Critical Assessment of Protein Interactions (CAPRI). Addressing all of these objectives requires in-depth knowledge of target protein sequence-structure-function relationships revealed through evolutionary classification. These relationships were assigned by taking advantage of a pre-existing evolutionary-based hierarchal classification of existing fold space: the Evolutionary Classification of Protein Domains (ECOD) database 1.
The experimental protein structure community contributed 100 target structures (designated T0759-T0858) to CASP11. CASP organizers divided these targets into two prediction tracts based on sequence relationship to known structures [see PMID: 2403855 for details of the procedure]. Those lacking strong sequence similarity were released to all prediction groups (55 Targets) and those with apparent sequence similarity were for servers only (45 Targets). Several structural genomics centers contributed targets to CASP11, including 32 from the Joint Center for Structural Genomics (JCSG), 4 from the Structural Genomics Consortium (SGC), 8 from the Midwest Center for Structural Genomics (MCSG), 5 from the Northeast Structural Genomics Consortium (NESG), 6 from the New York Structural Genomics Research Center (NYSGRC), 4 from the Seattle Structural Genomics Center for Infectious Disease (SSGCID) and 1 from the Natural Product Biosynthesis Protein Structure Initiative (NatPro). Non-SGI research Centers and other research groups submitted the remaining 40 targets. Seven targets were designated for interaction prediction only (T0787/788, T0797/798, T0840/841 and T0825). Five targets (T0779, T0842, T0844, T0846 and T0850) were canceled as not having structure solved in time for the CASP meeting, and two more targets (T0778 and T0809) were canceled for details of their structure being prematurely released by another experimental group. An additional three structures (T0789-T0791), which were originally released for all-group prediction, became available before the target expired for manual prediction and were redefined as “server only”.
CASP ROLL included 30 target structures (designated R0002 and R0019-R0047). ROLL targets were contributed by JCSG (18 Targets) as well as non-SGI research Centers and others (12 Targets). Two of the targets (R0039 and R0041) were cancelled. To augment the number of FM targets for evaluation, nine of the ROLL targets were also released for the traditional CASP11 evaluation (R0024/T0775, R0026/T0785, R0030/T0806, R0042/T0794, R0043/T0763, R0044/T0771, R0045/T0777, R0046/T0765, and R0047/T0767).
Table 1 outlines CASP11 target proteins in the context of evolutionary relationships to known folds, which provided the basis for classification. Information from this table contributed to the main goals of this report: 1) defining evolutionary domains within targets and splitting them into reasonable evaluation units, 2) providing a basis for attributing evaluation units to template-based modeling (TBM) and template free modeling (FM) categories, and 3) providing structure-function relationships to evaluate the biological relevance of models in the accompanying evaluation papers. Several challenging examples of target categorization are discussed with respect to these goals.
Table 1.
Overview of CASP11 Targets
| Domain | Class | Temp | Level | Architecture | H/XGroup:Name |
|---|---|---|---|---|---|
| T0759-D1 | TBM | 1lm5B | Family | a+b duplicates or obligate multimers | H:beta-hairpin-alpha-hairpin repeat |
| T0759-D2 | TBM | 1lm5B | Family | a+b duplicates or obligate multimers | H:beta-hairpin-alpha-hairpin repeat |
| T0760-D1* | TBM | 3k0yA | Family Family |
beta barrels beta sandwiches |
H:OB domain in putative lipoprotein BF3042-related proteins H:Ig-like domain in putative lipoprotein BF3042-related proteins |
| T0761-D1 | FM | 3kiiB | X-group | a+b two layers | X:Cystatin-like |
| T0761-D2 | FM | 3kztA | X-group | a+b two layers | X:Cystatin-like |
| T0762-D1 | TBM | 4ib2A | Family | a/b three-layered sandwiches | H:Periplasmic binding protein-like II |
| T0763-D1 | FM | 2mctA | X-group | a+b two layers | X:Uncharacterized protein ZP 02042476.1 |
| T0764-D1 | TBM | 1qlwA | Family | a/b three-layered sandwiches | H:alpha/beta-Hydrolases |
| T0765-D1 | TBM | 2dt9A | X-group | a+b two layers | X:Alpha-beta plaits |
| T0766-D1 | TBM | 4orlA | Family | a+b two layers | H:NTF2-like |
| T0767-D1 | TBM | 4gl6A | H-group | a+b two layers | H:hypothetical protein (RUMGNA 01148) |
| T0767-D2 | FM | 4gl6A | H-group | combined into above | |
| T0768-D1 | TBM | 4gt6A | Family | beta duplicates or obligate multimers | H:Leucine-rich repeats |
| T0769-D1 | TBM | 2y4eB | X-group | a+b two layers | X:Alpha-beta plaits |
| T0770-D1 | TBM | 3p1uB | H-group | alpha superhelices | H:ARM repeat |
| T0771-D1 | FM | 1xx3A | X-group | a+b two layers | X:TolA/TonB C-terminal domain |
| T0772-D1 | TBM | 3hbkA | Family | beta sandwiches | H:Concanavalin A-like |
| T0773-D1 | TBM | 3opkA | X-group | a+b two layers | X:Alpha-beta plaits |
| T0774-D1* | TBM | 4k4kB | Family Family |
beta sandwiches beta sandwiches |
H:Immunoglobulin-related H:Immunoglobulin-related |
| T0775-D1 | FM | 2yvvA | H-group | beta duplicates or obligate multimers | H:Phage tail fiber protein trimerization domain |
| T0775-D2 | FM | 1qa2A | H-group | beta duplicates or obligate multimers | H:Phage tail fiber protein trimerization domain |
| T0775-D3 | FM | 3vtoP | H-group | beta duplicates or obligate multimers | H:Phage tail fiber protein trimerization domain |
| T0775-D4 | FM | 2xc1C | H-group | beta duplicates or obligate multimers | H:Phage tail fiber protein trimerization domain |
| T0775-D5 | FM | 4a0tA | H-group | beta duplicates or obligate multimers | H:Phage tail fiber protein trimerization domain |
| T0775-D6 | FM | 2xc1B | H-group | beta duplicates or obligate multimers | H:Phage tail fiber protein trimerization domain |
| T0776-D1 | TBM | 4jj4B | Family | a/b three-layered sandwiches | H:SGNH hydrolase |
| T0777-D1 | FM | 1fftF | New | alpha complex topology | new to ecod |
| T0780-D1 | TBM | 2kq1A | Family | a+b complex topology | H:Insert subdomain of RNA polymerase alpha subunit |
| T0780-D2 | TBM | 2kxyA | Family | a+b complex topology | H:Insert subdomain of RNA polymerase alpha subunit |
| T0781-D1 | FM | 3gwrB | H-group | a+b two layers | H:NTF2-like |
| T0781-D2 | TBM | 3gwrB | H-group | a+b two layers | H:NTF2-like |
| T0782-D1 | TBM | 3ebkA | X-group | beta barrels | X:Lipocalins/Streptavidin |
| T0783-D1 | TBM | 1vpaA | Family | a/b three-layered sandwiches | H:Nucleotide-diphospho-sugar transferases |
| T0783-D2 | TBM | 2ptgA | X-group | a/b three-layered sandwiches | X:Rossmann-like |
| T0784-D1 | TBM | 3u6gA | Family | beta sandwiches | H:DUF4425 |
| T0785-D1 | FM | 1zruA | H-group | beta sandwiches | H:Domain in virus attachment proteins |
| T0786-D1* | TBM | 2q4hA | H-group H-group |
a+b three layers a+b three layers |
H:HIT-like H:HIT-like |
| T0789-D1 | FM | 3sb1A | X-group | a+b two layers | X:hydrogenase expression protein-like |
| T0789-D2 | FM | 3sb1A | X-group | combined into above | |
| T0790-D1 | FM | 3sb1A | X-group | a+b two layers | X:hydrogenase expression protein-like |
| T0790-D2 | FM | 3sb1A | X-group | combined into above | |
| T0791-D1 | FM | 3sb1B | X-group | a+b two layers | X:hydrogenase expression protein-like |
| T0791-D2 | FM | 3sb1B | X-group | combined into above | |
| T0792-D1 | TBM | 2lh9A | Family | alpha arrays | H:HTH |
| T0793-D1 | FM | 1g6zA | X-group | beta barrels | X:SH3 |
| T0793-D2 | FM | 2ozxA | New | few SS elements | new to ecod |
| T0793-D3 | TBM | 4fdgC | H-group | a/b three-layered sandwiches | H:P-loop domains-related |
| T0793-D4 | TBM | 4fdgC | H-group | combined into above | |
| T0793-D5 | FM | 1v3fA | H-group | alpha arrays | H:HTH |
| T0794-D1 | TBM | 1j31C | Family | a+b four layers | H:Carbon-nitrogen hydrolase |
| T0794-D2 | FM | 2wmfA | X-group | beta sandwiches | X:Glycosyl hydrolase domain-like |
| T0795-D1 | TBM | 3zpfA | H-group | beta sandwiches | H:Domain in virus attachment proteins |
| T0796-D1 | TBM | 2d42A | Family | beta complex topology | H:Aerolisin/ETX pore-forming domain-related |
| T0799-D1 | FM | 2xc1B | H-group | beta duplicates or obligate multimers | H:Phage tail fiber protein trimerization domain |
| T0799-D2 | FM | 1v0eA | H-group | beta duplicates or obligate multimers | H:Phage tail fiber protein trimerization domain |
| T0799-D3 | TBM | 2xc1B | H-group | beta duplicates or obligate multimers | H:Phage tail fiber protein trimerization domain |
| T0799-D4 | TBM | 3gw6D | H-group | alpha arrays | H:Intramolecular chaperone domain in virus tail spike protein |
| T0800-D1 | TBM | 3lycL | H-group | beta duplicates or obligate multimers | H:Pectin lyase-like |
| T0801-D1 | TBM | 3dr4C | Family | a/b three-layered sandwiches | H:PLP-dependent transferases |
| T0802-D1 | FM | 2wstC | H-group | beta sandwiches | H:Domain in virus attachment proteins |
| T0803-D1 | TBM | 1t92A | H-group | a+b two layers | H:Pili subunits |
| T0804-D1 | FM | 4gu3C | H-group | beta duplicates or obligate multimers | H:Triple beta-spiral |
| T0804-D2 | FM | 2j1kF | H-group | beta sandwiches | H:Domain in virus attachment proteins |
| T0805-D1 | TBM | 2iskB | Family | a+b two layers | H:FMN-dependent nitroreductase-like |
| T0806-D1 | FM | 2q07A | X-group | a/b three-layered sandwiches | X:Other Rossmann-like structures with the crossover |
| T0807-D1 | TBM | 3lutA | Family | a/b barrels | H:TIM barrels |
| T0808-D1 | TBM | 3zm8A | H-group | beta sandwiches | H:Concanavalin A-like |
| T0808-D2 | FM | 4hfsB | H-group | beta sandwiches | H:Concanavalin A-like |
| T0810-D1 | FM | 4c0eA | H-group | alpha superhelices | H:ARM repeat |
| T0810-D2 | TBM | 2c1gA | Family | a/b barrels | H:Glycoside hydrolase/deacetylase |
| T0811-D1 | TBM | 1b9bA | Family | a/b barrels | H:TIM barrels |
| T0812-D1 | TBM | 1v0aA | H-group | beta sandwiches | H:Concanavalin A-like |
| T0813-D1* | TBM | 3gggC | Family Family |
a/b three-layered sandwiches alpha arrays |
H:Rossmann-related H:6-phosphogluconate dehydrogenase C-terminal domain-like |
| T0814-D1 | FM | 3ac0A | H-group | beta sandwiches | H:Immunoglobulin-related |
| T0814-D2 | FM | 2je8B | H-group | beta sandwiches | H:Immunoglobulin-related |
| T0814-D3 | TBM | 1eo5A | H-group | beta sandwiches | H:Immunoglobulin-related |
| T0815-D1 | TBM | 3gzrB | Family | a+b two layers | H:NTF2-like |
| T0816-D1 | TBM | 1bqbA | X-group | a/b three-layered sandwiches | X:Zincin-like |
| T0817-D1* | TBM | 4dcxB | Family Family |
a/b three-layered sandwiches a+b two layers |
H:Periplasmic binding protein-like II H:Oligo-peptide binding protein (OPPA) insertion domain |
| T0817-D2 | TBM | 2nooA | Family | a/b three-layered sandwiches | H:Periplasmic binding protein-like II |
| T0818-D1 | TBM | 2mc8A | H-group | a+b two layers | H:NTF2-like |
| T0819-D1 | TBM | 3getA | Family | a+b two layers | H:C-terminal domain in some PLP-dependent transferases |
| T0820-D1 | FM | 2q1hA | New | alpha obligate multimers | new to ecod |
| T0820-D2 | TBM | 2f23B | X-group | a+b duplicates or obligate multimers | X:FKBP-like |
| T0821-D1 | TBM | 1w3bA | Family | alpha superhelices | H:ARM repeat |
| T0822-D1 | TBM | 2zxqA | H-group | beta sandwiches | H:Concanavalin A-like |
| T0823-D1 | TBM | 1lqaB | H-group | a/b barrels | H:TIM barrels |
| T0824-D1 | FM | 1g8tB | H-group | few SS elements | H:His-Me finger endonucleases |
| T0826-D1 | FM | 4od4A | New | alpha bundles | new to ecod |
| T0826-D2 | TBM | 4kayB | Family | a/b three-layered sandwiches | H:Alkaline phosphatase-like |
| T0827-D1 | TBM | 4k92A | H-group | alpha superhelices | H:ARM repeat |
| T0827-D2 | FM | 1iqrA | New | alpha complex topology | new to ecod |
| T0828-D1 | TBM | 1wk0A | H-group | beta sandwiches | H:Immunoglobulin-related |
| T0828-D2 | TBM | 3vtyC | H-group | alpha superhelices | H:ARM repeat |
| T0829-D1 | TBM | 3k8rA | X-group | a+b two layers | X:NE0471 N-terminal domain-like |
| T0830-D1 | TBM | 3wakA | Family | alpha bundles | H:STT3/PglB/AglB transmembrane domain |
| T0830-D2 | TBM | 3wakA | H-group | a/b three-layered sandwiches | H:STT3/PglB/AglB core domain |
| T0831-D1 | TBM | 3qkyA | X-group | alpha superhelices | X:Repetitive alpha hairpins |
| T0831-D2 | FM | 3zcjD | X-group | alpha bundles | X:Spectrin repeat-like |
| T0832-D1 | FM | 1dmwA | H-group | a+b complex topology | H:Aromatic aminoacid monoxygenases, catalytic and oligomeric |
| T0833-D1 | TBM | 3mswA | Family | beta meanders | H:Uncharacterized protein BF3112 |
| T0834-D1 | FM | 2grgA | X-group | a+b three layers | X:Profilin-like |
| T0834-D2 | FM | 4gyoB | X-group | alpha bundles | X:Repetitive alpha hairpins |
| T0835-D1 | TBM | 3wkfA | Family | alpha superhelices | H:alpha/alpha toroid |
| T0836-D1 | FM | 2fynM | H-group | alpha bundles | H:Transmembrane heme-binding four-helical bundle |
| T0837-D1 | FM | 2af7D | X-group | alpha arrays | X:AhpD-like |
| T0838-D1 | TBM | 2lnjA | H-group | a+b three layers | H:Mog1p/PsbP-like |
| T0839-D1* | TBM | 1hf8A | Family Family |
alpha bundles alpha superhelices |
H:GAT-like domain H:ARM repeat |
| T0843-D1* | TBM | 2ogeD | Family Family |
a/b three-layered sandwiches a+b two layers |
H:PLP-dependent transferases H:C-terminal domain in some PLP-dependent transferases |
| T0845-D1 | TBM | 2cqvA | H-group | beta sandwiches | H:Immunoglobulin-related |
| T0845-D2 | TBM | 3dsmA | H-group | beta duplicates or obligate multimers | H:beta-propeller |
| T0847-D1 | TBM | 1bysA | H-group | a/b three-layered sandwiches | H:Phospholipase D/nuclease |
| T0848-D1 | TBM | 3cfuA | Family | beta sandwiches | H:Antigen MPT63/MPB63 (immunoprotective extracellular protein) |
| T0848-D2 | TBM | 3d9rD | H-group | a+b two layers | H:NTF2-like |
| T0849-D1* | TBM | 1tu8A | Family Family |
alpha superhelices mixed a+b and a/b |
H:Glutathione S-transferase (GST)-C H:Thioredoxin-like |
| T0851-D1* | TBM | 3gg2D | Family Family |
a/b three-layered sandwiches alpha arrays |
H:Rossmann-related H:6-phosphogluconate dehydrogenase C-terminal domain-like |
| T0852-D1 | TBM | 3gffA | Family | a/b three-layered sandwiches | H:alpha/beta-Hydrolases |
| T0852-D2 | TBM | 2oevA | H-group | alpha superhelices | H:ARM repeat |
| T0853-D1 | TBM | 1yn4A | X-group | a+b two layers | X:beta-Grasp |
| T0853-D2 | TBM | 1yn4A | X-group | a+b two layers | X:beta-Grasp |
| T0854-D1 | TBM | 2hszA | Family | a/b three-layered sandwiches | H:HAD domain-related |
| T0854-D2 | TBM | 2ah5A | Family | a/b three-layered sandwiches | H:HAD domain-related |
| T0855-D1 | FM | 1ikjA | New | a+b two layers | new to ecod |
| T0856-D1 | TBM | 3tojB | Family | beta sandwiches | H:Concanavalin A-like |
| T0857-D1 | TBM | 3lxuX | H-group | beta sandwiches | H:Immunoglobulin-related |
| T0858-D1* | TBM | 2v3eA | Family | a/b barrels | H:TIM barrels |
Defining Evaluation Units for CASP11 Targets
Domains represent structurally compact evolutionary modules in proteins and serve as the basic units of folding 2–4. Domains can be mobile, and their relative placement can depend on factors such as the presence of a ligand or crystal packing for X-ray structures. As such, evaluation of CASP targets is traditionally domain-based. We considered several criteria for parsing CASP11 targets into domains: 1) compactness of secondary structure elements and the presence of a hydrophobic core, 2) self-similarity or internal duplications, 3) sequence continuity, and 4) similarity to other protein sequences and structures measured by methods such as PSI-BLAST5 or HHPRED6 for sequence or Local-Global Alignment (LGA)7 for structure. Side-chain orientations and interactions of residues that border the potential split areas were inspected to define precise domain boundaries. For a few difficult domain splits, we generated test splits and considered server performance to determine boundaries.
For each multidomain target, the decision to split identified domains into evaluation units was ultimately based on server performance. This strategy was implemented because predictions for individual domains were sometimes better than for their assembly, and we wanted to minimize scoring penalties that arose from differences in the relative packing or the prediction difficulty of the individual units. On the other hand, performance on individual domains could approach that of the assembled domains. For such cases splitting did not tend to reveal interesting prediction features, and the targets were better treated as single evaluation units to promote development of methods that find correct domain assembly. To select targets that required splitting, we consulted ‘Grishin Plots’ developed for CASP88 and first implemented for official classification in CASP99 that plot the weighted sum of GDT_TS scores for individual domains versus the GDT_TS scores of the combined domains. Generally a slope of the zero-intercept best fit line above 1.3 required splitting. Comparison of domain-based server predictions with whole chain server predictions revealed that 30 targets require domain-based evaluation and 13 targets with defined domains were kept as single evaluation units. For non-overlapping CASP ROLL targets, nine were split into domains for evaluation. To exemplify the procedure for defining evaluation units in CASP11, we highlight two examples below.
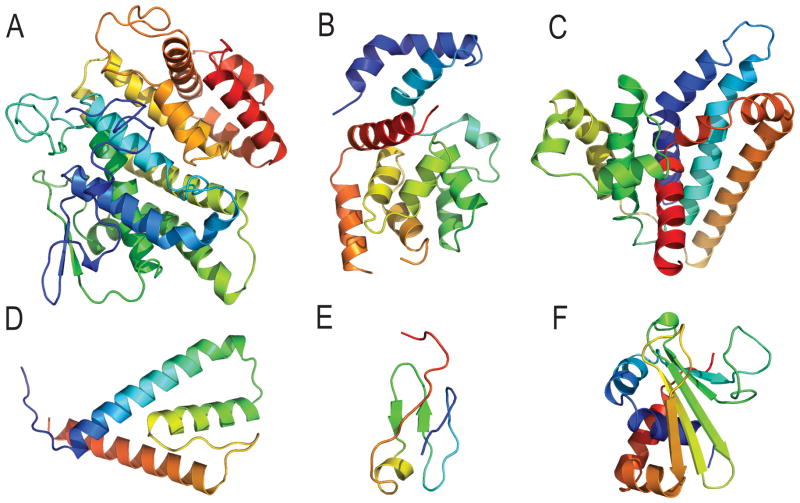
Based on visual inspection, targets T0759 and T0786 were both initially considered as two-domain targets: T0759 exhibited independent hydrophobic cores (D1: 12–45 and D2: 46–107, Figure 1A), while T0786 - internal fold duplication (D1:37–136 and D2:137–253, Figure 1D). The Grishin plots, though, suggested different split scenarios for their evaluation, supporting the detailed inspection of the targets (below).
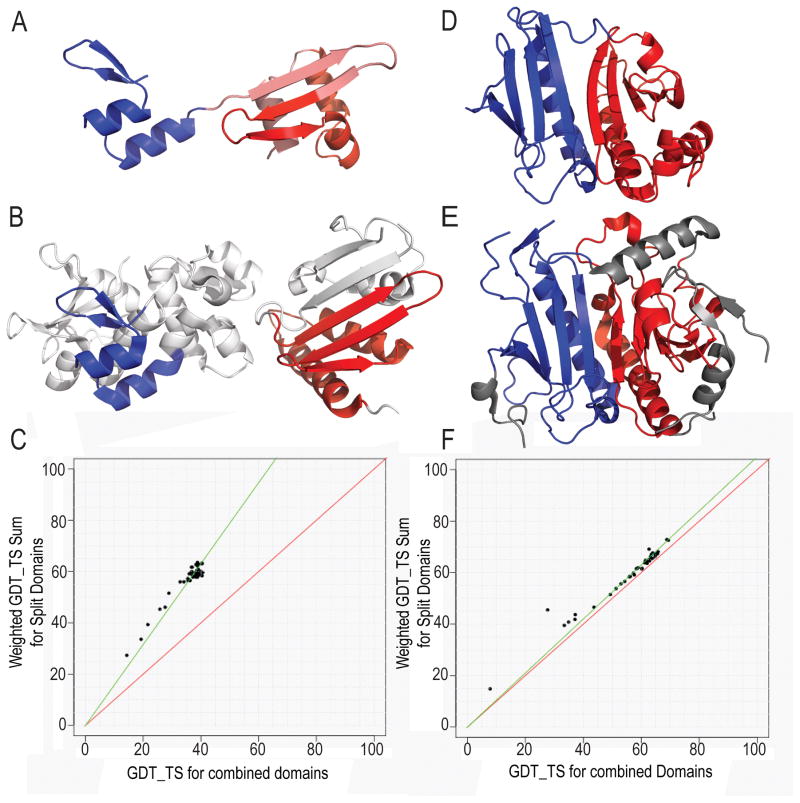
Figure 1. CASP11 Target domain splits.
The procedure for splitting targets into domains for evaluation is illustrated using examples. A) Target T0759 was split into an N-terminal (blue) and a C-terminal (salmon/red) domain, based on the presence of separate hydrophobic cores. The domains are sequence-detected repeating units, with the C-terminal core of the repeat (red) being elaborated by additional secondary structures (salmon). B) The closest homologous template to the T0759 core N-terminal domain (1lm5, blue)differs from the closest template analog to the T0759 C-terminal elaborated domain duplication (3cwx, red). C) The Grishin plot performance comparison for T0759 suggests splitting the domains into two evaluation units based on the increased scores of split domains. D) Target T0786 was split into an N-terminal (blue) domain and a C-terminal (red) domain based on an internal fold duplication. E) The closest template has the same domain duplication (2q4h, blue and red) arranged similarly as target T0786. F) The Grishin plot slope close to 1 suggests the split is not necessary for target T0786 evaluation.
Target T0759 contained a duplicated plectin superfamily motif in the target sequence annotated by Conserved Domain Database (CDD)10. However, the closest LGA templates to each domain exhibit a different fold (Figure 1B). The D1 template (1lm5) contains several plectin superfamily domains classified in ECOD as homologous β-hairpinc-α-hairpin repeats (Figure 1B left panel). The plectin superfamily-like repeat in D2 is elaborated with additional secondary structure elements that almost double the size of the core fold (62 residues for D2 vs. 34 for D1). The additional secondary structural elements mask the evolutionary core, making D2 most closely resemble an analogous template fold (3cwx, type III secretory system chaperone-like X-group in ECOD) and increasing its prediction difficulty (Figure 1B right panel). Accordingly, the Grishin plot for this target suggested splitting the domains for evaluation (Figure 1C).
The sequence of T0786 included a CDD-annotated N-terminal Histidine triad (HIT)-like superfamily motif that covered D1, but the duplicated C-terminal domain was not recognized by CDD. The closest LGA template (2q4h) to the full-length Target T0786 structure includes the duplication, with each domain classified by ECOD as HIT-related. The HIT-related domains of both the target and template assemble similarly (Figure 1E), and the Grishin plot suggested that the target structure does not require splitting (Figure 1F).
Difficult splits: domain swaps and crystal packing
The CASP11 targets included a number of difficult domain split cases. For example, certain regions protruded away from the structures, lacking interactions with the rest of the domain. The protrusions resulted from 1) domain swaps, where an exchange of secondary structural elements occurred between protein chains or domains; 2) crystal packing, where an extension adopted a potentially non-physiological conformation through interactions with other chains in the crystal; or 3) protein oligomerization, where a partial non-globular domain or set of secondary structures interacted with other chains to form a compact globular unit. These protruding segments remain difficult to predict in absence of having a similar swap or assembly present in existing structure templates. For example, a short C-terminal segment of the server-only target T0805 swaps with another chain. The same swapped C-terminal segment is present in the top BLAST hit (3ge6), and the predictions followed the swapped extension with good server performance (top GDT_TS 76.27). Thus, we kept the protruding extension for this target as part of the domain.
Other more difficult swap examples that lacked reliably detected templates required generating a test case of an artificially swapped domain for performance evaluation. For example, a C-terminal helix from a domain (D1) in Target T0831 swaps with a neighboring chain. Performance on an artificially swapped domain containing portions of both chains (1–108 combined with 353–417) was not improved (top server scores are identical at GDT_TS 41.13), so the helical extension was kept as part of D1. Two additional example targets (T0761 and T0767) had relatively short N-terminal extensions that formed peripheral interactions with the C-terminal domains. For such extensions that lack similar templates, we chose to eliminate the extension rather than create domains with either discontinuous sequence (adding the extension to the C-terminal domain) or non-globular structure (keeping the extension on the N-terminal domain). Domains that adopt alternate conformations in each chain (i.e. T0771) were considered mobile and removed. Other protruding regions appeared to be involved in crystal packing. For example, T0772 has an N-terminal extension that interacts with other chains in the crystal, is not present in templates, and was therefore excluded from analysis.
Difficult splits: obligate oligomers
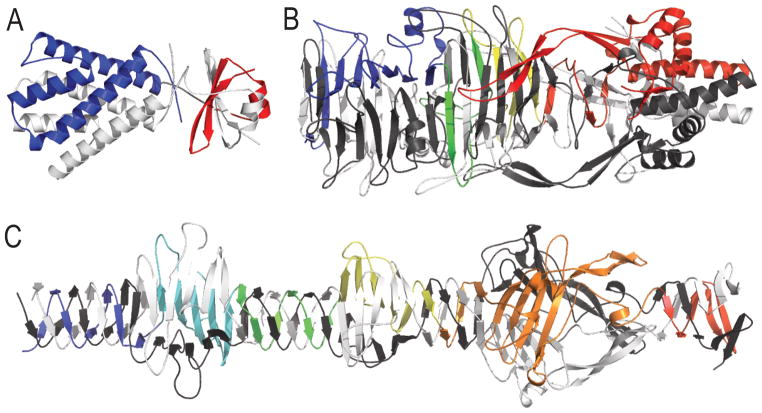
Target T0820 forms an obligate dimer with two domains. The N-terminal domain T0820-D1 adopts an almost linear array of α-helices that pack together with the corresponding array from the second chain to form a globular unit. This domain is linked to the C-terminal domain T0820-D2 through an apparently flexible linker that adopts different conformations in each chain. T0820-D2 adopts a β-α-β-hairpin that swaps with itself through an interleaving of the β-strands (Figure 2A). However, the T0820-D2 N-terminus falls on the opposite side of the domain as the C-terminus, requiring an artificial assembly of the swap to exclude the N-terminal β-strand to form a β-hairpin-α-helix. The sequence from marine bacteriophage metagenomic data detected no similar sequences among any of the NCBI databases, and existing structures did not serve as good topological templates. Nonetheless, the top HHPRED-detected template (2f23) covered a portion of T0820-D2 that correctly aligned to the β-hairpin. Template similarity extended to include the upstream α-helix (see template similarity in Fig 3 below). Given the correct orientation of the template hit (α-β-hairpin) and the fact that the swap would cause a permutation of the fold (β-hairpin-α), we chose to keep the two domains without the swap, excluding the flexible linker.
Figure 2. Difficult domain splits in obligate oligomers.
A) The N-terminal (blue) and C-terminal (red) domains in Target T0820 adopt an obligate dimer with a second chain (white) through a C-terminal domain swap. The phage tail proteins form obligate trimers through alternating integrated b-strands and meandering b-strands B) in target T0799, with three alternating trimerization domains colored inblue, green and yellow, followed by a chaperone domain in red; and C) in target T0775, with six defined alternating domains in rainbow.
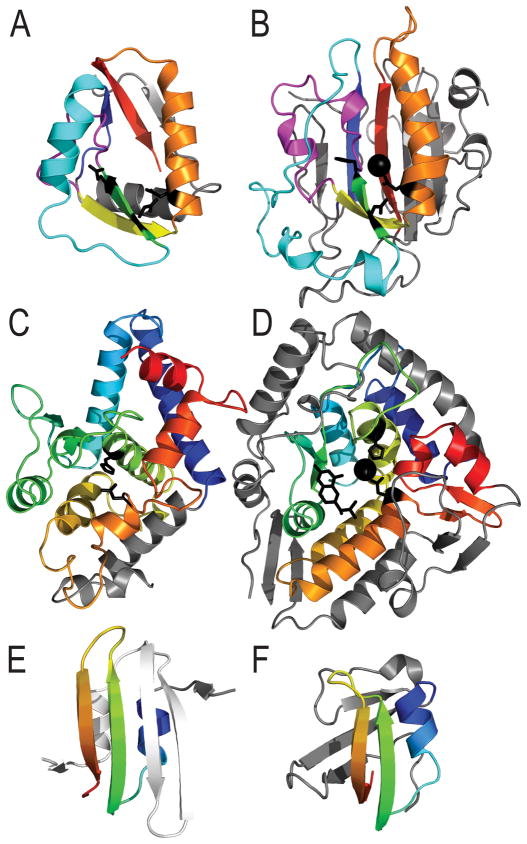
Figure 3. Difficult evolutionary target classifications.
Similar structural elements between target and template are colored in rainbow, with insertions in gray. A) The target T0824-D1 retains a similar placement of the active site (black sidechains) and unusual structure feature (magenta), yet has a significant deterioration of the fold present in B) the closest template (1g8t), active site marked by black sphere. C) The target T0832 retains a similar placement of the active site (black sidechains), yet has a significant deterioration of the fold present in D) the closest template (1dmw). E) A relatively well-predicted new fold of the swapped domain in target T0820-D2 (rainbow) can be modeled over a significant portion of the fold by F) a partially detected domain template (2f23).
Two CASP11 targets represent bacteriophage tail fibre proteins (T0775 and T0799). Both of these targets adopt long and extended conformations through trimerization of alternating β-meanders and interleaved β-strands. While several templates exist that represent tail fibre trimerization domains, their structural diversity, combined with rapidly diverging phage sequence, tend to prevent sequence detection. One of the targets (T0779), however, includes a C-terminal chaperone domain that is artificially attached to the tail fibre due to mutation of a protease cleavage site. This C-terminal chaperone has a detectable template (3gw6) and was split based on the presence of similar structural elements in the template (Figure 2B). Since the tail fibre trimerization domains have repeating structural units of interleaved β-strands followed by β-meanders, we initially made splits according to the repeating interleaved β-strand/β-meander units. However, some structure templates exist with repeating units that begin from the β-meander. To allow for all possible template-based predictions, we chose to split each independent interleaved unit from the meander unit (Figure 2 B and C), extending the boundary for T0775-D5 to include a partially detected HHPRED template. Those domains that were shorter than the cutoff for running various structure evaluation metrics were assigned to domains on either side.
Evolutionary relationship of targets to fold space
CASP predictions have been traditionally assessed according to categories, which are currently designated as template-based modeling (TBM) and free modeling (FM). However, classifying evaluation units by these categories has become increasingly difficult over the course of the CASP rounds. These categories are typically defined based on the presence of existing detectable templates in the PDB that can be used for modeling. However, such a distinction requires a priori knowledge of each of the participating prediction methods’ abilities to find templates. To help overcome this problem and determine if a template could exist, we attempted to classify each target based on their evolutionary relatedness to existing fold space. Those targets that are homologous to templates are related by sequence (i.e. detected using PSI-BLAST or HHPRED) and were categorized as confident TBM, whereas those targets that adopt new topological folds should not have templates and fell within confident FM. However, many of the templates did not possess such clear-cut evolutionary relationships to existing structure space and required additional considerations for homology designations. Our eventual classification scheme was developed based on knowledge of evolutionary relationships (i.e. using confident TBM and FM targets), but included more objective scores to provide the ultimate distinction of all targets between categories (see Assigning Evaluation Units to TBM and FM Categories section below).
Clear-cut cases of sequence-related TBM templates existed for many CASP11 targets such as T0833-D1, which belongs to the Pfam-designated11 domain of unknown function DUF3836. The closest LGA template (3msw, LGA_S 76.78) also belongs to DUF3836, although the core 7-stranded β-meander fold present in both structures is elaborated by an additional β-hairpin in the template. Such close sequence-related targets also tended to be predictable, as exemplified by relatively good server performance on T0833-D1 (top server GDT_TS 77.55). However, two of the sequence-related targets that were designated in the same Pfam family as their templates were quite distant structurally. One of these structures was previously mentioned as a domain split example, T0759-D2, and includes elaborated secondary structure elements that almost double the core fold of the template, resulting in a relatively low structure similarity score (LGA_S 41.77). This divergence of duplicated domains might reflect the ability of duplications to evolve quickly12. In fact, CASP11 includes 10 targets with potentially fast evolving domain duplications (T0761, T0781, T0789, T0790, T0791, T0808, T0814, T0817, T053, and T0854). The other target T0774-D1 and its template both belong to DUF3988, yet their structures are also distant with a relatively low structure similarity score (LGA_S 39.37). Performance on these sequence-related targets tended to correlate with the structure distance between the target and template (i.e. top server GDT_TS 46.37 for T0759-D2 and 44.90 for T0774-D1).
Many of the targets that lacked clear sequence relationships to existing templates (i.e. same PFAM family), yet maintained similar folds as existing structure templates, possessed significant deletions or insertions to the core topology. Accordingly, these examples caused prediction quality to decrease and posed more of a challenge for classification. The NucB DNase target T0824-D1 (Figure 3A) topology represents a significant deterioration that lacks a β-sheet of the top SM2 nuclease template (1g8t) fold (Figure 3B). The template is classified in ECOD as belonging to the His-Me finger endonucleases H-group. Despite the deterioration and the structure distance (low LGA_S score 45.81), as pointed out in the Structural Classification of Proteins (SCOP) database13 for this fold, both retain an unusual omega loop structure feature in the core α-β-omega loop-β-α topology that forms the nuclease active site, causing us to classify the two as homologs. A similar example of significant domain deterioration was noted in the mainly α-helical structure of target T0832-D1 (Figure 3C). The phenylalanine dehydroxylase template (Figure 3D, 1dmw) includes numerous elaborations to the fold present in the target and is structurally distant (low LGA_S score 29.74). However, both retain conserved residues and an unusual β-α-β structure feature that forms the active site, and we classified them as homologs.
An additional source of CASP11 classification difficulty occurred when detected partial templates covered significant portions of new folds. One of the aforementioned swapped target domains, T0820-D2, falls into this category (Figure 3E). While not the top LGA template, a FKBB-like structure (2f23) retains the β-hairpin-α-helix present in the swapped domain (Figure 3F) and was even detected by HHPRED as the top hit (82.5% probability). Due to the relatively high probability of sequence detection, as well as the presence of a similar duplication in the template, we chose to classify the target at the ECOD X-group level as opposed to being a new fold. The presence of these more difficult cases among the CASP11 targets tended to blur the lines of categorization. Additionally, many of today’s prediction methods are hybrid approaches designed to find and combine weakly similar partial templates (i.e. fragment-based methods). The same methods are incorporated into automated servers that are trained to perform on the entire spectrum of CASP targets, making the category distinction somewhat obsolete for methods evaluation. Nevertheless, the categories provided an important means for applying different types of evaluation methodology to different types of targets.
We attempted to classify all CASP11 target structures based on evolutionary relatedness to existing templates. The ECOD database1 provided the basis for defining template fold space. Each new target was assigned to the hierarchical ECOD categories: including a close family homology level (F-group), a more distant homology level (H-group), and a level of similar overall topology that lacks evidence for homology (X-group). Many of the targets with significant sequence/structure similarity were classified automatically by the ECOD pipeline or were easily distinguished by manual inspection (55 domains in 50 targets). These target domains were considered confidently assigned as TBM. However for many of the target domains, the distinction between H-group and X-group required expert manual analysis of various scores provided by ECOD, of potentially similar functional sites, or of unusual structural features that provide additional justification for homology. Some of the target domains with more questionable homology assignment were initially noted as unknown, while others lacking any similarity to existing structure topologies or with undetectable or distant templates were noted as FM (33 domains in 26 targets). The pie chart in Figure 4A summarizes the classification of CASP11 templates into the ECOD hierarchy.
Figure 4. Evolutionary classification of CASP11 targets using ECOD.
A) Targets are distributed into ECOD hierarchy, with 42 classified at the family level with closely related structures, 50 classified at the H-group level with more distantly related structure homologs, 28 classified at the X-group level with structures having similar topology, but questionable homology, and 6 targets classified as new folds. B) The distribution of targets into ECOD architectures shows relatively equal distribution among traditionally categorized classes of all-α (highlighted pink), all-β (highlighted lavender), α/β (highlighted light blue) and α+β (highlighted light green). C). Some ECOD architectures are overrepresented in CASP11 targets (blue bars) and some are underrepresented in CASP11 targets (red bars) as compared to all ECOD classified PDB structures.
The evolutionary classified target domains fall into 19 ECOD-defined architectures (Figure 4B), excluding the extended segments and special-cases architectures present in the database. The targets distribute among these architectures into roughly equivalent classes of beta, alpha, α/β, and α+β; suggesting that they are not skewed towards any one class. To see how the architecture distributions of CASP11 targets compare to those present in the current PDB, we calculated two ratios. The first ratio represents the CASP distribution among architectures (CASPr: number of targets in the architecture/total number of targets) and the second represents the PDB distribution among architectures (PDBr:number of chains classified in the architecture/total number of chains). To visualize the under or over representation of the architectures, we calculated the ratio difference as (CASPr-PDBr)/(CASPr+PDBr), where overrepresented CASP targets will be positive and underrepresented CASP targets will be negative (Figure 4B). Notable overrepresented categories include obligate multimers in the beta, alpha, and α+β classes, which might complicate predictions in the absence of good templates. Similarly, the overrepresented alpha superhelices category often contains repetitive units that are difficult to align or have folds that have diverged significantly, and need to be treated as special cases for structure modeling14,15.
Assigning Evaluation Units to TBM and FM Categories
Due to the subjective nature of imposing homology distinctions on the CASP11 targets that rely on manual considerations of multiple factors, we tested a number of numerical criteria for their ability to separate TBM from FM evaluation units. Such a strategy was applied to CASP9 targets9, which attempted to categorize domains based an automated score that reflected prediction difficulty 8. As revealed in previous CASP rounds, structure modeling has not worked reliably in the absence of templates, and poor prediction quality remained a good indicator of an FM target. Unfortunately, this definition might exclude the possibility of measured progress, as it tends to assign unusually high quality FM predictions to TBM. We hoped to reduce the influences of such predictions by using score averages to reflect prediction quality, assuming the unusually high quality prediction scores should be normalized by the majority of the remaining scores. Using performance-based measures for a classification scheme that will eventually evaluate performance also has conceptual drawbacks. However, we hoped to limit this problem by including only performance-based measures of the server results that were provided to the entire prediction community during the CASP experiment. The first two chosen scores, average GDT_TS of server models above random and number of server models above random, were the same measures used for target domain classification in CASP99. We combined these two scores with the average GDT_TS of all server models to reflect the prediction difficulty of CASP11 targets.
Previously, CASP9 target sequences were carefully evaluated by team members prior to structure release to decide if templates could be detected9. For CASP11, we wanted to emulate the ability to detect a template through more objective scores. A method that reflects sensitive sequence-based template detection (HHPRED) has been used as a benchmark of template detection in past CASP rounds, and the prediction center provided HHPRED results as sequences were released for prediction. Therefore, we chose to use one of the HHPRED scores (HHPRED Probability) as a measure of template detection ability. Where possible, we selected template homologs determined by our evolutionary classification among the HHPRED results, or we selected analogs that provided reasonable partial templates. For those targets without detected templates, a zero probability was assigned. To further evaluate the target distance to the closest template, we included the LGA_S score between the target and chosen template as a second measure.
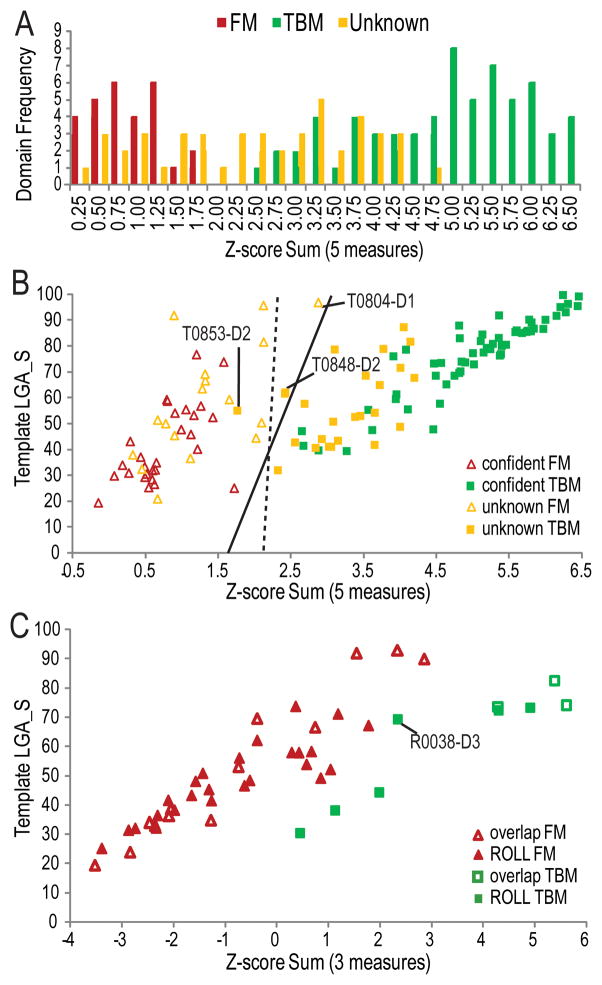
In order to combine the chosen measures, each was converted to a Z-score, and the Z-scores were summed. The distribution of Z-score sums for domains that were confidently assigned to FM and TBM using evolutionary considerations suggests that the two categories split around a Z-score sum of 2.25 (Figure 5A, between red and green bars). However, several of the unknown domains also tended to cluster around this value. To further visualize the score distributions, we plotted the Z-score sum against one of the measures of template distance (LGA_S between target and chosen template). Categorization of these scores was performed automatically using two methodologies: 1) Support Vector Machine (SVM) with a linear kernel (using python scikit-learn package with penalty of the error term as 0.5) and 2) logistic regression with lasso regularization (using matlab lassoglm module) (Figure 5B, dotted line for method 1 and line for method 2). Questionable target domains near the automatically computed bounds were carefully inspected and assigned to either the FM (Figure 5B, triangles) or TBM (Figure 5B, squares) categories and are discussed in the following section.
Figure 5. CASP11 target domain score distributions.
Five scores reflecting prediction quality (average GDT_TS scores of server models, average GDT_TS score of first server models above random, and number of first server models above random) and template distance (LGA_S to chosen template and HHPRED probability to homologous template) were combined as Z-score sums. A) A distribution of Z-score sum frequencies highlights the distinction (around 2.25) between confidently assigned FM (red bars) and TBM domains (green bars), with unknown domains distributed in the middle (yellow bars). B) A scatter plot of the Z-score sum vs. the template LGA_S is colored as above and highlights the final categorization into FM (empty triangles) and TBM (filled squares). An automatically defined categorization boundary using SVM with linear kernel (dashed line) differs slightly from that defined using logic regression (solid line). Target domains that blur the boundaries of categorization are labeled. C) A scatter plot of CASP ROLL targets overlapping with CASP11 FM (open markers) and targets unique to CASP ROLL (filled markers) illustrates categorization into FM (red triangles) and TBM (green squares) based on Z-score combination of measures (top first model GDT_TS, LGA_S to template, and HHPred Probability).
The majority of CASP11 target domains classified as FM had either reasonable structure templates that were undetectable (LGA_S above 50, 20 domains) or distantly related structures that did not serve as adequate templates (LGA_S below 50, 19 domains), with only 6 domains being potential new folds. Many of the unknown FM targets (Figure 5, yellow) fall into the first category, having relatively good structure templates that were not detected by prediction methods. For example, all but one domain (T0775-D5) of the split T0775 phage tail trimerization units had reasonable template homologs that were not used for modeling (LGA_S range 59.29 to 95.8). These target domains exhibited a significant shift upwards toward higher LGA_S scores and caused a large spread in the Fig. 5B performance plot for FM domains. The plot could resolve several of the targets into categories that would have otherwise been masked.
The total number of FM target domains (45) represents an unprecedented number of difficult domains provided for CASP modeling. The reasons for the observed difficulty include the previously discussed presence of significant insertions and deletions with respect to templates, rapidly diverging domain duplications in targets, overrepresentation of obligate multimers in the beta, alpha, and α+β classes, and the overrepresentation of repetitive elements like α-superhelices. The CASP11 targets also included the following difficult cases: 1) rapidly diverging viral or phage sequences (9 targets), 2) effective singleton sequences (9 targets) including two engineered and one environmental metagenome sequence, and 3) special sequence features such as signal sequences (34 targets), His-tags (6 targets) that interfere with prediction and transmembrane helices (3 targets) that have relatively low sequence complexity compared to soluble folds and less template examples in the PDB.
The unprecedented number of difficult FM target domains was due in part to the inclusion of 9 CASP ROLL targets in CASP11. These targets were split into 16 evaluation units, which were classified into 13 FM and 3 TBM by the procedure outlined above. Knowing this target distribution, we chose to use for categorization three measures that take into account the fewer number of ROLL predictions and do not require calculation of random models (GDT_TS mode, LGA_S to template, and HHPred Probability). A plot of the template LGA_S against the Z-score Sum of these measures (Figure 5C) separates the previously categorized CASP ROLL FM domains (open red diamonds) from the TBM domains (open green squares), and suggests a boundary for categorization of the remaining CASP Roll domains into FM (red diamonds) and TBM (green squares). The ROLL target domains classified as TBM had confident HHPred probability (>90%) for correct templates, including one of the ROLL target domains (R0038-D3) that approaches the separation boundary. The non-overlapping CASP ROLL category excluded 6 domains that were considered as TBM, resulting in 25 additional FM domains (38 total ROLL domains for evaluation).
Difficult classification examples and the new folds
Several targets blurred the boundaries of CASP11 target categorization into TBM and FM. For example, target T0804-D1 adopts an obligate oligomer triple beta spiral repeating unit. It has a homologous template in the PDB, yet the model quality was poor (top GDT_TS 50.52). The top template (LGA_S 96.97) is also a triple beta spiral (4gu3C), and only a single prediction declared a triple beta spiral template homolog (1qiu) as a parent. Otherwise, the poor performance for this target resulted from a logistic problem of not being able to split the two short repeating β-hairpin units. The β-hairpins were modeled by many of the top-performing groups using analogous β-trefoil templates (i.e. 24 groups declared 3iir as a parent), which adopt an alternate relative orientation of β-hairpin repeats. Thus, these top server scores predicted a single repeat correct (GDT_TS scores approach 50). The resulting poor performance and choice of analogous template for modeling led us to classify this target as FM.
Two different CASP11 target domains were included as TBM that tended to cross the boundary towards FM: T0848-D2 and T0853-D2. T0848-D2 was classified in the NTF2-like H-group, together with the top LGA template (3d9r) and the top HHPRED identified sequence (3k7c). Despite the confident assignment made by sequence (HHPRED probability 95.5% over the domain), the top structure-related template was quite distant (LGA_S 30.48) and proved to be a poor template (TOP server GDT_TS 38.12). Since the T0848-D2 templates were identified by sequence, and the prediction quality approached that of the top template, we chose to classify T0848-D2 as TBM. T0853-D2 is a duplication of the ubiquitin-related β-grasp fold from the N-terminal domain of the same target. While the N-terminal domain found a ubiquitin-related template as the top hit (LGA_S 57.71), T0853-D2 found an analogous diaminopimelate epimerase-like fold as the top template (4k7g, LGA_S 62.66). Additionally, the N-terminal domain also identified a ubiquitin-related sequence with almost complete coverage using HHPRED, albeit with a relatively low score (probability 41.1), while T0853-D2 did not. Given this divergence of the duplicated C-terminal domain from an N-terminal TBM domain, we also classified T0853-D2 as TBM.
After classification of all CASP11 target domains, six were considered as new folds that lacked significant overlap of topology with existing structures classified in ECOD. Two of the new folds adopt alpha complex topology architectures: T0777-D1 (Figure 6A) and T0827-D2 (Figure 6B). T0820-D1 adopts an alpha obligate multimers architecture (Figure 6C). T0826-D1 adopts an alpha bundles architecture (Figure 6D). T0793-D2 adopts a few secondary structure elements architecture (Figure 6E), and T0855-D1 adopts an α+β two layers architecture (Figure 6F).
Figure 6. New Folds.
New folds are depicted in cartoon and colored in rainbow from the N-terminus to the C-terminus: A) complex alpha T0777-D1, and B) T0827D2, C) alpha obligate multimer T0820-D1, D) alpha bundle T0826-D1, E) few SSEs T0793-D2, and F)α+β two layer T0855-D1.
Oligomerization Targets
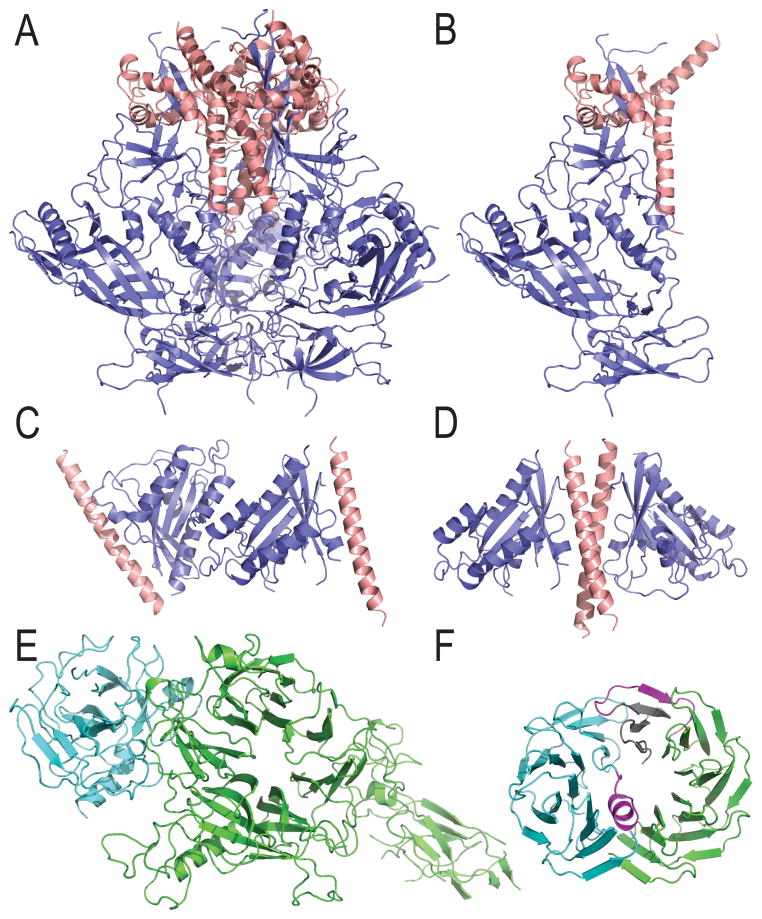
A new initiative in CASP11 included a CAPRI-style evaluation of oligomeric interaction. Seven CASP11 targets qualified for this type of prediction: T0787 with T0788, T0797 with T0798, T0840 with T0841, and T0825 with itself. The HIV-1 envelope spike is formed by a hexameric complex of three GP41 (T0787) and three GP120 (T0788) protein subunits. The functional oligomeric hexamer adopts a trimer (Figure 7A) of GP41/GP120 heterodimers (Figure 7B). The second example represents a complex between a leucine zipper of cGMP-Dependent Protein Kinase II (PGKII) (T0797) and Rab11b (T0798). The two targets form a dimer of heterodimers in the crystal unit. However, the dimer in the crystal unit is formed by Rab11b interactions (Figure 7C), placing the PGKII α-helices on opposite ends. In order to form the appropriate leucine zipper, crystal contacts must be considered (Figure 7D). The RON receptor tyrosine kinase extracellular domains (T0840) form a one-to- one complex (Figure 7E) with macrophage stimulating protein (T0841). Target T0825 is a synthetic β-propeller structure with two chains of identical sequence adopting alternate conformations to oligomerize (Figure 7F).
Figure 7. Oligomeric interactions.
A) Hexameric complex contains three T0787 (salmon cartoon) and three T0788 (slate cartoon) protein subunits, formed by a trimer of B) T0787/T0788 heterodimers. C) T0797 (salmon cartoon) and T0798 (slate cartoon) form a dimer of heterodimers in the crystal unit, D) forming the functional T0797 leucine zipper requires considering crystal contacts. E) T0840 (green cartoon) forms a one to one complex with T0841 (cyan cartoon). F) T0825 has two identical chains (cyan and green cartoon) that adopt alternate conformations (magenta) to dimerize into a complete β-propeller.
Acknowledgments
We thank Hua Cheng for critical contributions to the ECOD classification and helpful discussions, and the CASP organizers for their invitation to participate in CASP11. This research was supported by the National Institutes of Health (GM094575 to NVG), the Welch Foundation (I-1505 to NVG), and R01 (GM084453 to RLD).
References
- 1.Cheng H, Schaeffer RD, Liao Y, Kinch LN, Pei J, Shi S, Kim BH, Grishin NV. ECOD: an evolutionary classification of protein domains. PLoS Comput Biol. 2014;10(12):e1003926. doi: 10.1371/journal.pcbi.1003926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bork P. Shuffled domains in extracellular proteins. FEBS letters. 1991;286(1–2):47–54. doi: 10.1016/0014-5793(91)80937-x. [DOI] [PubMed] [Google Scholar]
- 3.Richardson JS. The anatomy and taxonomy of protein structure. Advances in protein chemistry. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- 4.Wetlaufer DB. Nucleation, rapid folding, and globular intrachain regions in proteins. Proceedings of the National Academy of Sciences of the United States of America. 1973;70(3):697–701. doi: 10.1073/pnas.70.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic acids research. 2005;33(Web Server issue):W244–248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zemla A. LGA: A method for finding 3D similarities in protein structures. Nucleic Acids Res. 2003;31(13):3370–3374. doi: 10.1093/nar/gkg571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raman S, Vernon R, Thompson J, Tyka M, Sadreyev R, Pei J, Kim D, Kellogg E, DiMaio F, Lange O, Kinch L, Sheffler W, Kim BH, Das R, Grishin NV, Baker D. Structure prediction for CASP8 with all-atom refinement using Rosetta. Proteins. 2009;77(Suppl 9):89–99. doi: 10.1002/prot.22540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kinch LN, Shi S, Cheng H, Cong Q, Pei J, Mariani V, Schwede T, Grishin NV. CASP9 target classification. Proteins. 2011;79(Suppl 10):21–36. doi: 10.1002/prot.23190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Bryant SH. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015;43(Database issue):D222–226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M. Pfam: the protein families database. Nucleic acids research. 2014;42(Database issue):D222–230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohno S. Evolution by gene duplication. London, New York: Allen & Unwin; Springer-Verlag; 1970. p. xv.p. 160. [Google Scholar]
- 13.Lo Conte L, Ailey B, Hubbard TJ, Brenner SE, Murzin AG, Chothia C. SCOP: a structural classification of proteins database. Nucleic acids research. 2000;28(1):257–259. doi: 10.1093/nar/28.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andrade MA, Petosa C, O’Donoghue SI, Muller CW, Bork P. Comparison of ARM and HEAT protein repeats. Journal of molecular biology. 2001;309(1):1–18. doi: 10.1006/jmbi.2001.4624. [DOI] [PubMed] [Google Scholar]
- 15.Kajava AV. Review: proteins with repeated sequence--structural prediction and modeling. Journal of structural biology. 2001;134(2–3):132–144. doi: 10.1006/jsbi.2000.4328. [DOI] [PubMed] [Google Scholar]