Abstract
Human homolog double minute 2 (hdm2), an oncoprotein, which binds to tumor suppressor p53 to facilitate its degradation, has been known to contribute to tumorigenesis. Its splicing variants are reported to be highly expressed in many cancers and can be induced by ultraviolet B light (UVB). However, the mechanisms of how UVB radiation induces hdm2 alternative splicing still remain unclear. In this study, we investigated the roles of two common splicing factors, heterogeneous nuclear ribonucleoproteins (hnRNP) A1 and serine/arginine-rich splicing factor 1 (SRSF1), in regulating UVB-induced hdm2 splicing. Our study indicated that while the expression of both hnRNP A1 and SRSF1 are induced, only hnRNP A1 is involved in hdm2 alternative splicing upon UVB irradiation. Overexpression of hnRNP A1 resulted in decrease of full-length hdm2 (hdm2-FL) and increase of hdm2B, one of hdm2 alternate-splicing forms; while down-regulated hnRNP A1 expression led to the decrease of the hdm2-FL and hdm2B in HaCaT cells. Protein-mRNA binding assay confirmed that UVB irradiation could increase the binding of hnRNP A1 to hdm2 pre-mRNA. In conclusion, we elucidated that UVB induces alternative splicing of hdm2 via increasing the expression and the binding of hnRNP A1 to hdm2 full-length mRNA.
INTRODUCTION
DNA damage is a major cellular consequence upon exposure to ultraviolet light (UV) and ionizing radiation resulting in gene alternative splicing (1), which is involved in many cell processes and diseases including cancers (2–5). One of UV-induced alternative spliced genes is mouse double minute 2 (mdm2) (6–10). The full-length mdm2 (mdm2-FL), as an ubiquitin ligase, can bind to tumor suppressor p53 and facilitate its degradation (11). Therefore, mdm2-FL is recognized as an oncoprotein to down-regulate the expression and activity of p53 (12). However, mdm2B (also known as mdm2alt1), an UV-induced common splicing variant of mdm2, has been shown to promote mutant p53 accumulation and gain-of-function in tumorigenesis (13). Our previous report indicated that UVB induces alternative splicing of hdm2, a human homology of mdm2, in a reactive oxygen species (ROS) and p53 dependent manner (10). However it remains unclear regarding what splicing factors are involved in the alternative splicing of hdm2 post UVB irradiation.
Splicing factors including heterogeneous nuclear ribonucleoproteins (hnRNP) and serine/arginine-rich splicing factor 1 [SRSF1; also known as alternative splicing factor 1 (ASF1) or pre-mRNA-splicing factor SF2 (SF2)] can effectively regulate pre-mRNA splicing, which is regarded as a mechanism to increase diversity of the proteome (14,15). Previous report indicated that an hnRNP family member hnRNP A1 and SRSF1 could efficiently regulate gene alternative splicing (16–18). In addition, the expression level and activity of hnRNP A1 and SRSF1 are up-regulated in multiple human cancers, which is believed to be related to motility and proliferation of cancer cells (16,19,20). In this study, we determine the involvement of hnRNP A1 and SRSF1 in UVB-induced alternative splicing of hdm2. Our results demonstrated that hnRNP A1, but not SRSF1, regulates alternative splicing of hdm2 in HaCaT cells exposed to UVB irradiation.
MATERIALS AND METHODS
Cell culture
HaCaT cells (a generous gift from Dr. Li Zhong, Chongqing University) were grown in DMEM (Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco) and 1% Penicillin-Streptomycin (P/S, Hyclone). The cells were incubated at 37 °C in 5% CO2.
UVB irradiation
Two 8-watts UVB Lamps (SANKYO, Japan) were used in the experiments. The intensity of UVB was standardized by UV-meter (Photoelectric Instrument Factory of Beijing Normal University, China). The culture media was removed from the cells during the irradiation and fresh medium was added to the cells after UVB irradiation. 50 mJ/cm2 of UVB was used for all the experiments. HaCaT cells are harvested or tested at 6 h post UVB irradiation. For N-acetyl-L-cysteine (L-NAC) (Sigma) treatment, the HaCaT cells were pretreated with 10 mM L-NAC for 1 h before UVB irradiation. The cells were continuously incubated with L-NAC after UVB irradiation until harvesting.
Semi-quantitative PCR
Total RNA was extracted using the TRIzol reagent (Invitrogen, USA) according to the manufacturer’s protocol. First strand cDNA was reverse-transcribed from the RNA using the PrimeScript™ RT reagent Kit with gDNA Eraser (TaKaRa, Japan).
The primers for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were:
5′-GGAGCGAGATCCCTCCAAAAT-3′(sense)
5′-GGCTGTTGTCATACTTCTCATGG-3′(antisense)
The primers detecting hdm2-FL and hdm2B were:
5′-GAATCATCGGACTCAGGTACATC-3′(hdm2-FL-F)
5′-TCTGTCTCACTAATTGCTCTCCT-3′(hdm2-FL-R)
5′-TGAGGAGCAGGCAAATGTG-3′(hdm2B-F)
5′-CTTTGTCTTGGGTTTCTTCC-3′(hdm2B-R)
The primers detecting hdm2-FL and all the variants were:
5′-TACCTACTGATGGTGCTG-3′ (hdm2-F)
5′-CTTTGTCTTGGGTTTCTT-3′ (hdm2-R)
The results were normalized against GAPDH as an endogenous control. The DNA polymerase was purchased from Tiangen (China). The PCR parameters were set as 95 °C for 5 min, followed by 40 cycles of 95 °C for 30 s, 53.5 °C for 30 s and 72 °C for 2 min.
Western blot
Cells were lysed in RIPA lysis buffer (50 mM Tris-HCl pH7.5, 1% NP-40, 0.5 mM EDTA, 0.1% SDS, 150 mM NaCl, 0.5% Sodium deoxycholate) with PMSF (Beyotime, China). Cell lysate was centrifuged at 14,000 rpm for 10 min at 4°C. The protein concentration was measured by BCA assay (Beyotime, China). The protein samples were separated by 15% SDS-PAGE, and then transferred to polyvinylidene fluoride (PVDF) membranes (PALL, USA). Membranes were blocked in TBST with 5% nonfat milk and then incubated with primary antibodies at 4 °C overnight. After washing with TBST, the membranes were incubated with corresponding secondary antibodies for 1h at room temperature. Following a triple washing step with TBST, the bands were visualized with the chemiluminescence method (Millipore, USA). Antibodies including β-actin, GAPDH, hnRNP A1, SRSF1 and HRP-conjugated anti-rabbit or anti- mouse secondary antibodies were purchased from Santa Cruz (USA).
ROS measurement
CM-H2DCFDA (Invitrogen) was used to measure the total ROS level in cells. CM-H2DCFDA was dissolved in DMSO to a stock solution of 500 μM and diluted in PBS to final concentration of 5 μM. CM-H2DCFDA was added into cells 1 h prior to UVB exposure and the reading of fluorescence dye was recorded at 6 h post UVB irradiation using luminometer (Molecular devices Spectra Max M2).
Construction and transfection of the protein expression vectors and siRNAs
Mammalian expression vector pcDNA3.1(+) (Addgene) was used to express hnRNP A1, SRSF1, Hdm2-FL and Hdm2B. All the inserted sequences were confirmed by DNA sequencing (Sangon, China). Total cDNA prepared from HaCaT cells was used as template for PCR using the following primers. The amplify hnRNP A1 and SRSF1 gene fragments were subcloned into Hind III and EcoR I or Hind III and Xho I MCS sites of pcDNA3.1(+) respectively.
| hnRNP A1: | Forward: 5′-CCCAAGCTTGGGATGTCTAAGTCAGAGTCCCCCA-3′ |
| Reverse: 5′-CCGGAATTCCGGTTAGAACCTCCTGCCACTGCC-3′ | |
| SRSF1: | Forward: 5′-CCCAAGCTTGGGATGTCAGGAGGTGGTGTGATTCG-3′ |
| Reverse 5′-CCGCTCGAGCGGTTATGTACGAGAGCGAGA-3′. |
Total cDNA prepared from human mesenchymal stem cells (MSC) cells was used as template for PCR using the following primers. The amplify hdm2-FL gene fragment was subcloned into BamH I and Xho I MCS sites of pcDNA3.1(+).
| Hdm2-FL: | Forward: 5′-CGCGGATCCATGGTGAGGAGCAGGCAAAT-3′ |
| Reverse: 5′-CCGCTCGAGCTAGGGGAAATAAGTTAGCACAATC-3′. |
The cloned hdm2-FL gene was then used as the template to amplify hdm2B fragment through an overlapping PCR method. Two gene fragments of hdm2B were amplified separately using the following two pairs of primers, and then joined into one complete hdm2B, which was subcloned into BamH I and Xho I MCS sites of pcDNA3.1(+).
| Hdm2B: | Forward-1: 5′-CGCGGATCCATGGTGAGGAGCAGGCAAAT-3′ |
| Reverse-1: 5′-TTTCCAATAGTCAGCCAGGGTCTCTTGTTCCGAA-3′ | |
| Forward-2: 5′-GCTGACTATTGGAAATGCACTT-3′ | |
| Reverse-2: 5′-CCGCTCGAGCTAGGGGAAATAAGTTAGCACAATC-3′. |
siRNAs targeting hnRNP A1 and SRSF1 were synthesized at GenePharma (China).
The sequences of siRNAs were as following.
| hnRNP A1: | 5′-CAGCUGAGGAAGCUCUUCA-3′ |
| SRSF1: | 5′-ACGAUUGCCGCAUCUACGU-3′ (21). |
The expression vectors were transfected into HaCaT cells using Effectene Transfection Reagent (Qiagen, Germany) and siRNAs were transfected into HaCaT cells using X-tremeGENE siRNA Transfection Reagent (Roche, USA) following the manufacturer’s instructions. The proteins expression was determined by western blot or semi-quantitative RT-PCR at 48 h post transfection.
Cell viability
HaCaT cells were seeded into the 96-well plate at 2000 cells/well and cultured overnight. pcDNA3.1(+)-hdm2-FL and pcDNA3.1(+)-hdm2B were transfected into HaCaT cells. The cells were radiated by UVB at 48 h post transfection and the cell viability was measured at 6 and 24 h post UVB using a CCK-8 assay (Beyotime, China). Each experiment was repeated for at least three times. Data were analyzed statistically using the t-test and ANOVA. P value (p) <0.05 was set as the level of statistically significant difference.
RNA Immunoprecipitation (RIP)
RIP assay was carried out as previously described (22,23). In brief, HaCaT cells were seeded into a 100 mm cell culture dish. At 70–80% confluent, the cells were harvested and resuspended in 2 mL PBS, 2 mL nuclear isolation buffer (1.28 M sucrose, 40 mM Tris-HCl pH 7.5, 20 mM MgCl2, 4% Triton X-100) and 6 ml water, and incubated on ice for 20 min. Nuclei was isolated by centrifugation at 2,500 g for 15 min, and nuclear pellet was resuspended in 1 mL RIP buffer (150 mM KCl, 25 mM Tris pH 7.4, 5 mM EDTA, 0.5 mM DTT, 0.5% NP40, 100 U/ml RNAase, protease inhibitor cocktail). Then nuclei were mechanically sheared by a Dounce homogenizer, and nuclear membrane and debris were pelleted and discarded by centrifugation. hnRNP A1 antibody was added to supernatant and incubated for 2 h at 4 °C. Then, 40 μL Protein-A/G beads (Thermo Scientific, USA) were added and incubated for 1 h at 4 °C with gentle rotation. Unbound material was washed off beads by 3 times RIP buffer wash and one time PBS wash. Beads were then resuspended in 1 mL Trizol reagent. RNA was extracted and transcribed into cDNA. PCR was performed to detect hdm2 and its variants.
RESULTS
hdm2 is spliced to hdm2B in keratinocyte cells upon UVB irradiation
We first investigate that if UVB induces hdm2 alternative splicing in HaCaT cells, and the specific form of splicing variant hdm2B, one of the common hdm2 splicing variants which lacks the p53-binding domain (Fig. 1A). RT-PCR using hdm2B specific primers was performed to detect the full-length hdm2 (hdm2-FL) and hdm2B. Our data indicated that a decrease of hdm2-FL mRNA was correlated with an increase of hdm2B mRNA at 6 h post UVB (Fig. 1B). The universal primers spanning Exon 2 to Exon 11 were also used to detect hdm2 alternative splicing after UVB irradiation, and the results were consistent to the data using specific primers (Fig. 1C). The hdm2B band was confirmed by DNA sequencing. Our result indicated that UVB induces an alternative splicing of hdm2 mRNA resulting in the production of hdm2B.
Figure 1.
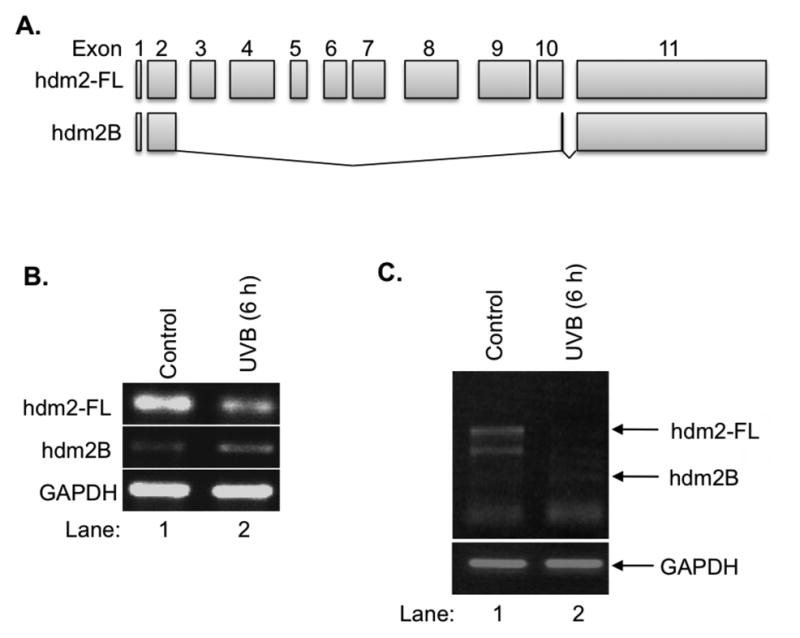
UVB-induced hdm2 splicing. A: UVB-induced hdm2 splicing led to the decrease of hdm2-FL and increase of hdm2B. B: The protein schematics of hdm2 and hdm2B. C: UVB-induced alternative splicing of hdm2 determined by RT-PCR with universal primers.
UVB radiation regulating the expression of hnRNP A1 and SRSF1
We next determine the involvement of hnRNP A1 and SRSF1, two common splicing factors, in regulation of UVB-induced alternative splicing of hdm2. The protein level of hnRNP A1 and SRSF1 was determined by western blot at 6 h after UVB irradiation. Our results showed that the protein level of both hnRNP A1 and SRSF1 were induced by approximately 20% upon UVB irradiation (Fig. 2A, 2B), which suggested that these splicing factors might be involved in the alternative splicing process of hdm2.
Figure 2.
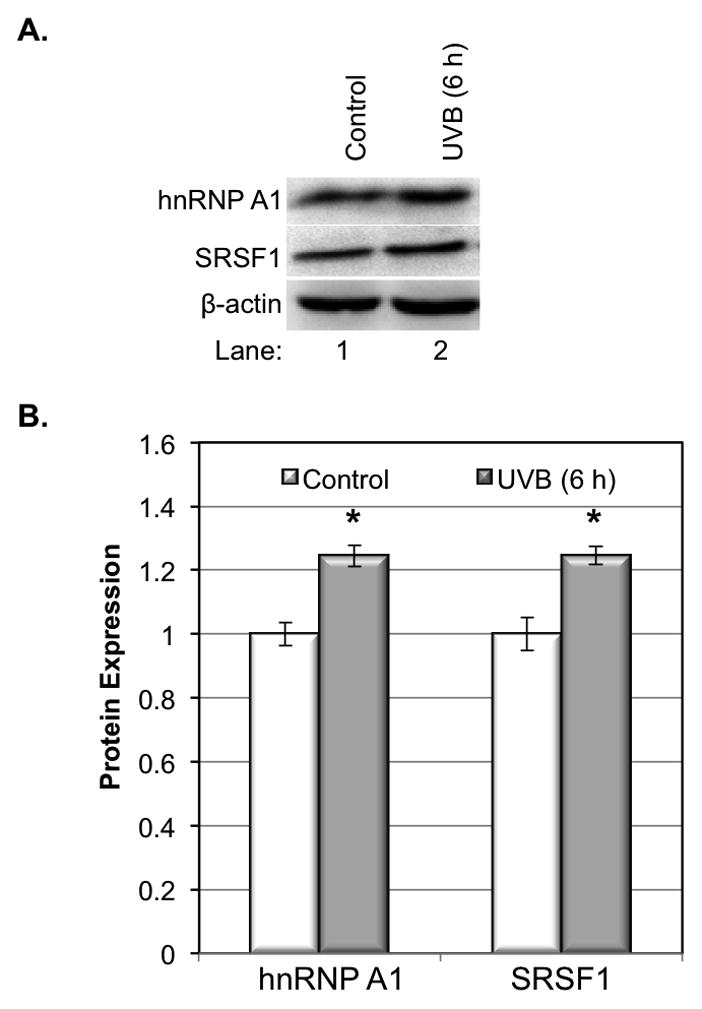
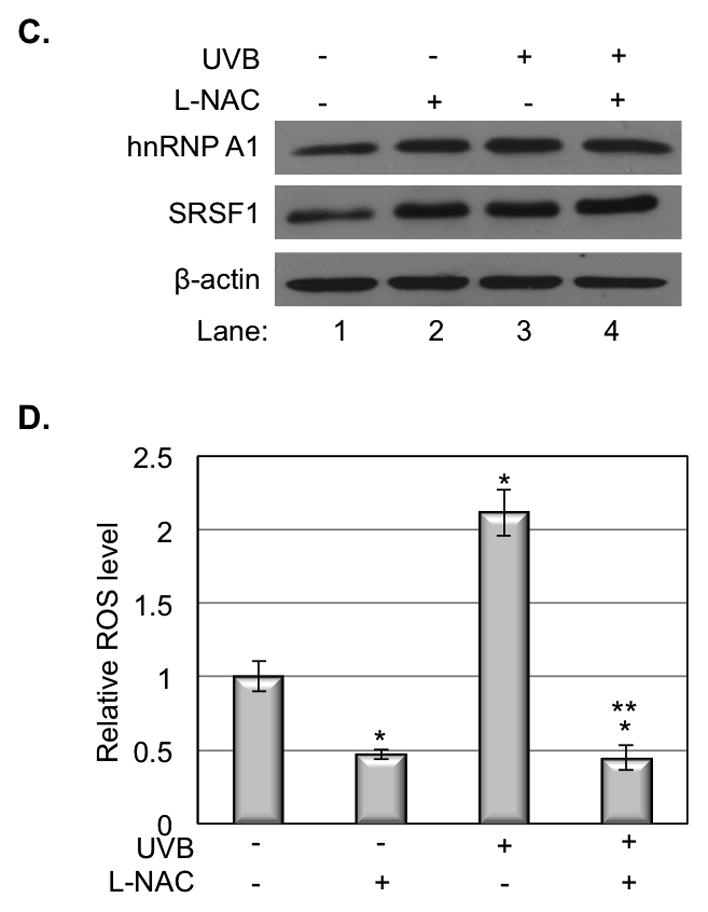
hnRNP A1 and SRSF1 expression in response to UVB. A. The expression of hnRNP A1 and SRSF1 increased at 6 h but decreased at 24 h post UVB irradiation. B. The quantity of protein was estimated by image density analysis software (Image J) and the data were shown as the means±S.D. *<0.05. C. Inhibition of the ROS by adding L-NAC could promote the expression of hnRNP A1 and SRSF1 in normal cells or UVB radiated cells. D. ROS in HaCaT cells was measured at 6 h post UVB irradiation. The data represents three sets of independent experiments. *p<0.05 versus corresponded control group, **p<0.05 versus UV alone.
Our previous study indicated that ROS is involved in UVB-induced alternative splicing of hdm2 (10). To determine whether ROS plays a role in regulation of the expression of splicing factors, we analyzed the effect of L-NAC on hnRNP A1 and SRSF1 protein expression after UVB irradiation. Our results showed that either L-NAC treatment or UVB irradiation induced the expression of both proteins (Fig. 2C, Lane 2, 3 vs. 1). However, the double treatment of L-NAC and UVB did not further increase the expression of hnRNP A1 and SRSF1 proteins (Fig. 2C, Lane 4 vs. 2. 3), indicating the expression of these two proteins are not linearly correlated to the ROS level in the cells (Fig. 2D).
hnRNP A1 is involved in hdm2 alternative splicing process
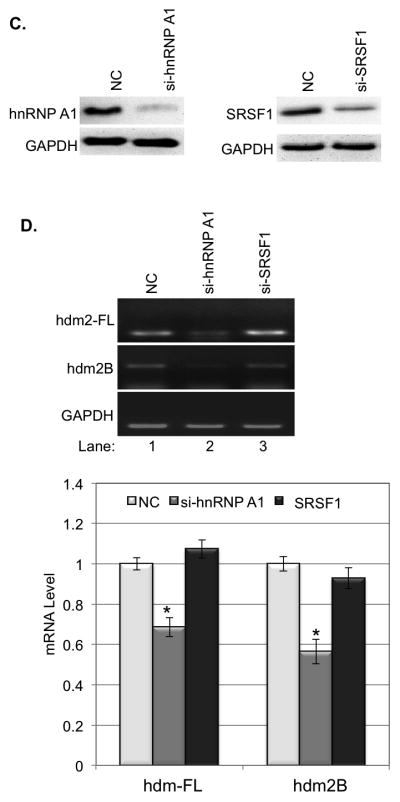
To determine whether a change in expression of these splicing factors will affect the alternative splicing of hdm2, we first transiently transfected pcDNA3.1(+)-hnRNP A1 or pcDNA3.1(+)-SRSF1 into HaCaT cells and the expression of hnRNP A1 and SRSF1 in the transfected cells were confirmed by western blot (Fig. 3A). Our data showed that overexpression of hnRNP A1, but not SRSF1, in HaCaT cells led to a 17% increase of hdm2B mRNA and a 30% decrease of hdm2-FL mRNA (Fig. 3B).
Figure 3.
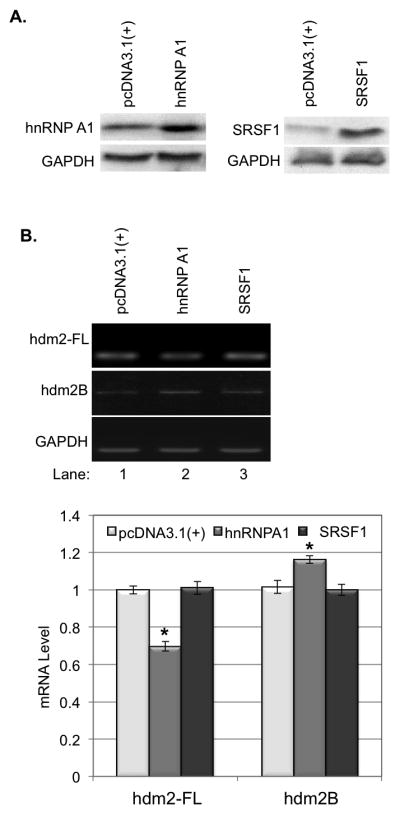
The influences of hnRNP A1 and SRSF1 on the expression of hdm2-FL and hdm2B. A. hnRNP A1 and SRSF1 were overexpressed in HaCaT cells by transient gene expression. B. Overexpression hnRNP A1 resulted in decrease of hdm2-FL and increase of hdm2B, while Overexpression of SRSF1 showed no significantly effect on the expression of hdm2-FL and hdm2B. The quantity of mRNA was estimated by image density analysis software (Image J) and the data were shown as the means±S.D. *<0.05. C. hnRNP A1 and SRSF1 were knockdown in HaCaT cells by siRNAs. D. Kockdown hnRNP A1 expression in HaCaT cells led to the decrease of hdm2-FL and hdm2B. down-regulation of SRSF1 showed no significantly effect on the expression of hdm2-FL and hdm2B. The quantity of mRNA was also estimated by image density analysis software (Image J) and the data were shown as the means±S.D. *<0.05.
We then transiently transfected hnRNP A1-targeting or SRSF1-targeting siRNA into HaCaT cells and the knockdown of hnRNP A1 and SRSF1 expression was determined by western blot (Fig. 3C). Our data showed that down-regulation of showed knockdown of hnRNP A1, but not SRSF1, expression in the cells led to a decrease of both hdm2-FL and hdm2B (Fig. 3D). These results indicated that splicing factor hnRNP A1, but not SRSF1, is involved in the regulation of alternative splicing of hmd2.
hnRNP A1 is involved in hdm2 splicing upon UVB irradiation
To determine whether hnRNP A1 is involved in the regulation of UVB-induced alternative splicing of hdm2, we examined the splicing of hdm2 in parent and hnRNP A1 overexpressing HaCaT cells that were treated or not treated with UVB radiation. Our data showed that while hnRNP A1 overexpression or UVB treatment alone could slightly effect on the alternative splicing (Fig. 4A, Lanes 2, 3 vs. 1), the combined treatment significantly reduced hdm2-FL and increase hdm2B expression (Fig. 4A, Lane 4 vs. 1).
Figure 4.
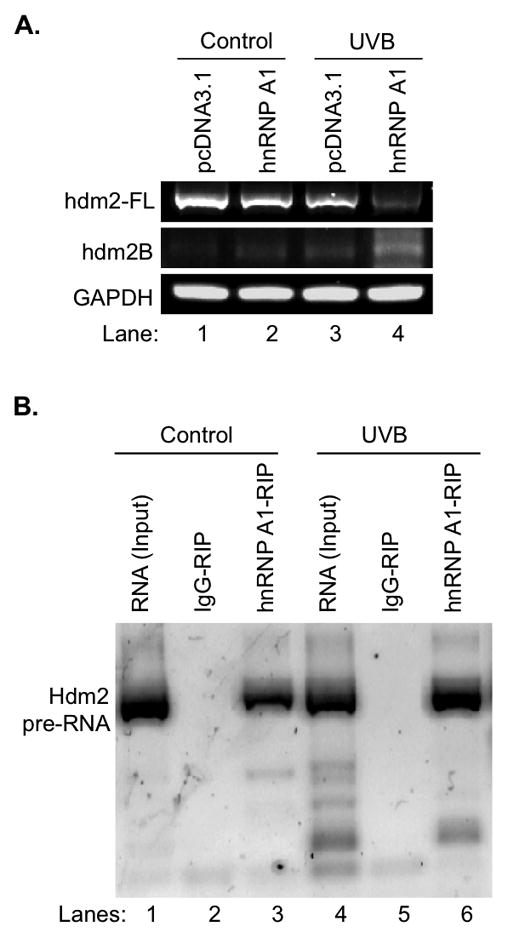
hnRNP A1 regulates hdm2 splicing upon UVB irradiation. A. Overexpression of hnRNP A1 further decreased hdm2-FL and increased hdm2B in HaCaT cells after UVB irradiation. B. RIP-RT-PCR showed UVB irradiation enhanced the binding of hnRNP A1 with hdm2 mRNA in vivo.
To determine whether an increased alternative splicing is associated with an increased binding of hnRNP A1 to hdm2 pre-RNA post UVB, a RNA immunoprecipitation (RIP) assay was performed. Our data showed that while the negative control IgG-RIP gave no detectable signal with or without UVB irradiation (Fig. 4B, Lanes 2 and 5), hdm2 pre-mRNA was co-immunoprecipitated with hnRNP A1 (Fig. 4B, Lanes 3 and 6); and the binding of hnRNA A1 and hdm2 pre-mRNA was increased upon UVB irradiation (Fig. 4B, Lane 6 vs. 3).
The splicing of hdm2-FL affects cell viability upon UVB irradiation
Finally, we determine whether an overexpression of hdm2-FL or hdm2B affects the viability of HaCaT cells after UVB irradiation. Our data showed that while overexpression of either proteins has no statistically significant effect on HaCaT cell death at 6 h post UVB irradiation, overexpression of hdm2-FL increased the cell viability by 13% increase and overexpression of hdm2B decrease cell viability by 5% at 24 h post UVB irradiation (Fig. 5). These results indicated that UVB-induced alternative splicing of hdm2, which reduces hdm2-FL but increases hdm2B expression.
Figure 5.
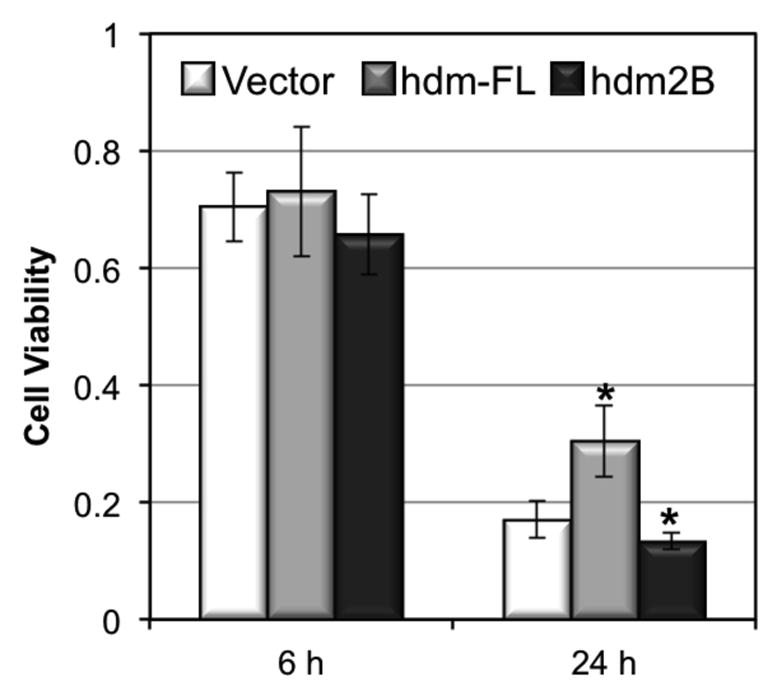
The effect of overexpression of hdm2-FL and hdm2B on cell viability upon UVB irradiation. Cells were transiently transfected with pcDNA3.1(+) vector, and pcDNA3.1(+) inserted with hdm2-FL and hdm2B expression genes. 48 h after transfection, cells were exposed to UVB radiation and cell viability was determined at 6 and 24 h post UVB irradiation.
DISCUSSION
We previously showed that the alternative splicing of hdm2 is induced by UVB (10). However, the splicing factor(s) that are involved in regulation of hdm2 alternative splicing post UVB irradiation have never been identified. In this study, we examined the involvement of two common splicing factors, hnRNP A1 and SRSF1, in UVB-induced alternative splicing of hdm2. Our results indicated that while UVB induces the expression of both hnRNP A1 and SRSF1, only hnRNP A1 is involved in the regulation of alternative splicing of hdm2 (Figs. 2, 3). More specifically, the expression level of hnRNP A1 is positively correlated to the formation of hnRNP A1-hdm2 pre-mRNA complex and hdm2B expression (Figs. 2–4) suggesting UVB-induced expression of hnRNP A1 upregulates the alternative splicing of hdm2 and the expression of hdm2B. The relationship between the expression of hnRNP A1 and hdm2-FL is more complicated because either overexpression or knockdown of hnRNP A1 reduces the expression of hdm2-FL (Figs. 3B, 3C). The reduction of hdm2-FL expression in hnRNP A1 overexpression cells is expected. However it is a surprise that reduced hnRNP A1 expression in HaCaT cells led to the decrease of both mdm2-FL and mdm2B (Fig. 3D). One potential explanation could be that hnRNP A1 positively regulates NF-κB activity (24,25), which has been shown to upregulate mdm2 expression via both p53 dependent and independent pathways (26,27). Therefore, the decreased hdm2-FL expression could be due to a reduced background activity of NF-κB upon hnRNP A1 knockdown.
Furthermore, we demonstrated that ROS is involved in regulation of the expression of both hnRNP A1 and SRSF1 (Fig. 2C). While the mechanism is not clear, this is the first time it has been shown that the expressions of hnRNP A1 and SRSF1 can be upregulated. Interestingly, the expressions of the two splicing factors are induced by the treatment of either free radical scavenger L-NAC or ROS-inducer UVB, which suggests that a proper level of ROS needs to be maintained for keeping a sturdy level of the splicing factors and possibly also normal splicing of hdm2. When ROS level is too high or too low, the expression of the splicing factors will be induced in HaCaT cells. This interpretation could also explain why the double treatment of L-NAC and UVB could not further induce the expression of the factors because the ROS levels in cells treated with L-NAC alone or L-NAC/UVB in HaCaT cells were at the same level, which was beyond the background ROS level (Fig. 2D). However, this result does not totally agree with our previous report indicating that UVB-induced ROS production inhibits the alternative splicing of hdm2 in H1299 human lung cancer cells (10). The disagreement could be due to the fact that the cancer cells are under higher oxidative-stress and they have adapted to this environment. Thus the further elevation of ROS induced by UVB could not further induce the alternative splicing of hdm2 as a lowered ROS caused by L-NAC could do.
In addition to the mechanism in regulation of hdm2 splicing after UVB irradiation, our data showed that the alternative splicing of hdm2 might play a role in regulation of UVB-induced HaCaT cell death (Fig. 5). It was known that hdm2B can bind to hdm2-FL and inhibit the interaction of hdm2-FL with p53 and thus enhances the transcriptional activity of p53 (28), which regulates UVB-induced apoptosis (29,30). HaCaT cells carry a mutated p53 (31) that sensitize the cells to UVB irradiation (30). Our results suggest that the overexpressed hdm2-FL is able to bind to the mutated p53 and protect the cells from UVB-induced apoptosis. In summary, our results indicated that UVB induces hnRNP A1 expression via a ROS-dependent mechanism; and hnRNP A1 plays a role in the alternative splicing of hdm2 and HaCaT apoptosis upon UVB irradiation.
Acknowledgments
This work was supported by the Natural Science Foundation of China (No.81261120383, 31170890, to Tang L), the Fundamental Research Funds for the Central Universities (No.106112015CDJZR158804, to Tang L) and National Institutes of Health (RO1 CA86926S1, to Wu S).
References
- 1.Sprung CN, Li J, Hovan D, McKay MJ, Forrester HB. Alternative transcript initiation and splicing as a response to DNA damage. PLoS One. 2011;6:e25758. doi: 10.1371/journal.pone.0025758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garcia-Blanco MA, Baraniak AP, Lasda EL. Alternative splicing in disease and therapy. Nat Biotechnol. 2004;22:535–46. doi: 10.1038/nbt964. [DOI] [PubMed] [Google Scholar]
- 3.Fan X, Tang L. Aberrant and alternative splicing in skeletal system disease. Gene. 2013;528:21–6. doi: 10.1016/j.gene.2013.06.027. [DOI] [PubMed] [Google Scholar]
- 4.He L, Bai Q, Tang L. Alternative Splicing Regulates Pluripotent State in Pluripotent Stem Cells. Curr Stem Cell Res Ther. 2014 doi: 10.2174/1574888x09666141112115525. [DOI] [PubMed] [Google Scholar]
- 5.Rajan P, Elliott DJ, Robson CN, Leung HY. Alternative splicing and biological heterogeneity in prostate cancer. Nat Rev Urol. 2009;6:454–60. doi: 10.1038/nrurol.2009.125. [DOI] [PubMed] [Google Scholar]
- 6.Dias CS, Liu Y, Yau A, Westrick L, Evans SC. Regulation of hdm2 by stress-induced hdm2alt1 in tumor and nontumorigenic cell lines correlating with p53 stability. Cancer Res. 2006;66:9467–73. doi: 10.1158/0008-5472.CAN-05-3013. [DOI] [PubMed] [Google Scholar]
- 7.Chandler DS, Singh RK, Caldwell LC, Bitler JL, Lozano G. Genotoxic stress induces coordinately regulated alternative splicing of the p53 modulators MDM2 and MDM4. Cancer research. 2006;66:9502–8. doi: 10.1158/0008-5472.CAN-05-4271. [DOI] [PubMed] [Google Scholar]
- 8.Singh RK, Tapia-Santos A, Bebee TW, Chandler DS. Conserved sequences in the final intron of MDM2 are essential for the regulation of alternative splicing of MDM2 in response to stress. Experimental cell research. 2009;315:3419–32. doi: 10.1016/j.yexcr.2009.07.017. [DOI] [PubMed] [Google Scholar]
- 9.Wong A, Zhang S, Mordue D, Wu JM, Zhang Z, Darzynkiewicz Z, Lee EY, Lee MY. PDIP38 is translocated to the spliceosomes/nuclear speckles in response to UV-induced DNA damage and is required for UV-induced alternative splicing of MDM2. Cell Cycle. 2013;12:3184–93. doi: 10.4161/cc.26221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tong L, Wu S. ROS and p53 in regulation of UVB-induced HDM2 alternative splicing. Photochem Photobiol. 2015;91:221–4. doi: 10.1111/php.12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Honda R, Yasuda H. Activity of MDM2, a ubiquitin ligase, toward p53 or itself is dependent on the RING finger domain of the ligase. Oncogene. 2000;19:1473–6. doi: 10.1038/sj.onc.1203464. [DOI] [PubMed] [Google Scholar]
- 12.Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–53. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 13.Zheng T, Wang J, Zhao Y, Zhang C, Lin M, Wang X, Yu H, Liu L, Feng Z, Hu W. Spliced MDM2 isoforms promote mutant p53 accumulation and gain-of-function in tumorigenesis. Nat Commun. 2013;4:2996. doi: 10.1038/ncomms3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Z, Fu XD. Regulation of splicing by SR proteins and SR protein-specific kinases. Chromosoma. 2013;122:191–207. doi: 10.1007/s00412-013-0407-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez-Contreras R, Cloutier P, Shkreta L, Fisette JF, Revil T, Chabot B. hnRNP proteins and splicing control. Adv Exp Med Biol. 2007;623:123–47. doi: 10.1007/978-0-387-77374-2_8. [DOI] [PubMed] [Google Scholar]
- 16.Bonomi S, di Matteo A, Buratti E, Cabianca DS, Baralle FE, Ghigna C, Biamonti G. HnRNP A1 controls a splicing regulatory circuit promoting mesenchymal-to-epithelial transition. Nucleic Acids Res. 2013;41:8665–79. doi: 10.1093/nar/gkt579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anczukow O, Rosenberg AZ, Akerman M, Das S, Zhan L, Karni R, Muthuswamy SK, Krainer AR. The splicing factor SRSF1 regulates apoptosis and proliferation to promote mammary epithelial cell transformation. Nat Struct Mol Biol. 2012;19:220–8. doi: 10.1038/nsmb.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghigna C, Giordano S, Shen H, Benvenuto F, Castiglioni F, Comoglio PM, Green MR, Riva S, Biamonti G. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol Cell. 2005;20:881–90. doi: 10.1016/j.molcel.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 19.Guo R, Li Y, Ning J, Sun D, Lin L, Liu X. HnRNP A1/A2 and SF2/ASF regulate alternative splicing of interferon regulatory factor-3 and affect immunomodulatory functions in human non-small cell lung cancer cells. PLoS One. 2013;8:e62729. doi: 10.1371/journal.pone.0062729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.David CJ, Manley JL. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 2010;24:2343–64. doi: 10.1101/gad.1973010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cartegni L, Hastings ML, Calarco JA, de Stanchina E, Krainer AR. Determinants of exon 7 splicing in the spinal muscular atrophy genes, SMN1 and SMN2. Am J Hum Genet. 2006;78:63–77. doi: 10.1086/498853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao TT, Graber TE, Jordan LE, Cloutier M, Lewis SM, Goulet I, Cote J, Holcik M. hnRNP A1 regulates UV-induced NF-kappaB signalling through destabilization of cIAP1 mRNA. Cell Death Differ. 2009;16:244–52. doi: 10.1038/cdd.2008.146. [DOI] [PubMed] [Google Scholar]
- 25.Sahu I, Sangith N, Ramteke M, Gadre R, Venkatraman P. A novel role for the proteasomal chaperone PSMD9 and hnRNPA1 in enhancing IkappaBalpha degradation and NF-kappaB activation - functional relevance of predicted PDZ domain-motif interaction. FEBS J. 2014;281:2688–709. doi: 10.1111/febs.12814. [DOI] [PubMed] [Google Scholar]
- 26.Gudkov AV, Gurova KV, Komarova EA. Inflammation and p53: A Tale of Two Stresses. Genes Cancer. 2011;2:503–16. doi: 10.1177/1947601911409747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Busuttil V, Droin N, McCormick L, Bernassola F, Candi E, Melino G, Green DR. NF-kappaB inhibits T-cell activation-induced, p73-dependent cell death by induction of MDM2. Proc Natl Acad Sci U S A. 2010;107:18061–6. doi: 10.1073/pnas.1006163107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evans SC, Viswanathan M, Grier JD, Narayana M, El-Naggar AK, Lozano G. An alternatively spliced HDM2 product increases p53 activity by inhibiting HDM2. Oncogene. 2001;20:4041–9. doi: 10.1038/sj.onc.1204533. [DOI] [PubMed] [Google Scholar]
- 29.Ouhtit A, Muller HK, Gorny A, Ananthaswamy HN. UVB-induced experimental carcinogenesis: dysregulation of apoptosis and p53 signalling pathway. Redox Rep. 2000;5:128–9. doi: 10.1179/135100000101535447. [DOI] [PubMed] [Google Scholar]
- 30.Henseleit U, Zhang J, Wanner R, Haase I, Kolde G, Rosenbach T. Role of p53 in UVB-induced apoptosis in human HaCaT keratinocytes. J Invest Dermatol. 1997;109:722–7. doi: 10.1111/1523-1747.ep12340708. [DOI] [PubMed] [Google Scholar]
- 31.Lehman TA, Modali R, Boukamp P, Stanek J, Bennett WP, Welsh JA, Metcalf RA, Stampfer MR, Fusenig N, Rogan EM, et al. p53 mutations in human immortalized epithelial cell lines. Carcinogenesis. 1993;14:833–9. doi: 10.1093/carcin/14.5.833. [DOI] [PubMed] [Google Scholar]