Abstract
Anxiety disorders, namely generalized anxiety disorder, panic disorder, and phobias, are common, etiologically complex conditions with a partially genetic basis. Despite differing on diagnostic definitions based upon clinical presentation, anxiety disorders likely represent various expressions of an underlying common diathesis of abnormal regulation of basic threat-response systems. We conducted genome-wide association analyses in nine samples of European ancestry from seven large, independent studies. To identify genetic variants contributing to genetic susceptibility shared across interview-generated DSM-based anxiety disorders, we applied two phenotypic approaches: (1) comparisons between categorical anxiety disorder cases and super-normal controls, and (2) quantitative phenotypic factor scores derived from a multivariate analysis combining information across the clinical phenotypes. We used logistic and linear regression, respectively, to analyze the association between these phenotypes and genome-wide single nucleotide polymorphisms. Meta-analysis for each phenotype combined results across the nine samples for over 18 000 unrelated individuals. Each meta-analysis identified a different genome-wide significant region, with the following markers showing the strongest association: for case-control contrasts, rs1709393 located in an uncharacterized non-coding RNA locus on chromosomal band 3q12.3 (P=1.65×10−8); for factor scores, rs1067327 within CAMKMT encoding the calmodulin-lysine N-methyltransferase on chromosomal band 2p21 (P=2.86×10−9). Independent replication and further exploration of these findings are needed to more fully understand the role of these variants in risk and expression of anxiety disorders.
Keywords: anxiety disorder, anxiety, genetics, genome-wide association study, meta-analysis
Anxiety disorders (ADs), namely generalized anxiety disorder (GAD), panic disorder (PD), and phobias, are relatively common, often disabling conditions with lifetime prevalence of over 20% (Kessler et al, 2005). Family and twin studies suggest both genetic and environmental factors underlying their etiology, with moderate levels of familial aggregation (OR 3–6) and heritability (30–50%) (1). As with most complex genetic traits, many linkage and candidate gene association studies of ADs have been conducted, with little success in robustly identifying their susceptibility genes (2;3).
Genome-wide association studies (GWAS) have proven to be a successful method for the identification of common genetic variants that increase susceptibility to complex disease. Recently, GWAS of specific anxiety and related disorders such as PD (4;5), post-traumatic stress disorder (6–8), obsessive compulsive disorder (9;10), and phobias (11) have been published. However, these have been limited by small sample sizes and resulting low overall power to detect significant associations.
Despite differing on diagnostic definitions based upon clinical presentation, ADs likely represent various expressions of an underlying common diathesis of abnormal regulation of basic threat-response systems (12). ADs exhibit strong lifetime comorbidity with each other (13), with genetic epidemiologic studies pointing to shared genetic risk factors between them (14;15). Since clinical descriptions do not reflect underlying genetic architecture, traditional studies focused on individual ADs may not represent an effective study design for such phenotypes. A more informative approach would coordinate data from clusters of disorders with shared genetic risk factors (16). One such strategy is to model a latent anxiety liability factor indexing ADs with substantial genetic overlap. Also, for common disorders like ADs, disease states can be interpreted as extremes of continuous liability dimensions, as has been done for somatic illnesses like obesity and hypertension. Therefore, quantitative trait approaches, assuming a continuous liability distribution, can be used to construct informative latent psychiatric phenotypes (17). Analyzing AD phenotypes in a coordinated manner may represent a powerful approach for identifying susceptibility genes for ADs. This strategy has yielded some success, as demonstrated by prior reports from our group (18).
In the current study, we conducted genome-wide association analyses in nine large, independent samples. To identify genetic variants contributing to genetic susceptibility shared across the ADs, we applied two phenotypic approaches: (1) categorical case-control comparisons based upon having any AD diagnosis, and (2) quantitative phenotypic factor scores derived from a multivariate analysis combining information across the clinical phenotypes. We performed a meta-analysis for each phenotype across the nine samples for over 18 000 unrelated individuals using around 6.5 million imputed SNPs. This represents the largest genetic study to date of any of the ADs and the first of this magnitude to explicitly incorporate comorbidity structure directly into prediction of SNP effects.
Materials and Methods
Overview
We conducted parallel GWAS in nine samples of European ancestry and combined the results via meta-analysis. We applied two phenotypic strategies aimed at capturing common (pleiotropic) genetic effects shared across the five core ADs: GAD, PD, social phobia, agoraphobia, and specific phobias. We conducted two types of analyses in each sample based upon complementary approaches to modeling the comorbidity and common genetic risk across the ADs: (1) case-control (CC) comparisons, in which cases were designated as having “any AD” versus supernormal controls, and (2) quantitative factor scores (FS) estimated for every subject in the sample using confirmatory factor analysis.
Samples
Nine samples containing AD phenotypes from seven independent studies participating in the Anxiety NeuroGenetics STudy (ANGST) Consortium were included in the meta-analysis. Standardized assessment instruments were used to generate DSM-based AD diagnoses, with some exceptions. The samples were genotyped on various SNP arrays according to their original study designs. Genotype calling, quality control (QC), imputation, and association analyses were performed at each site under similar standard protocols. SNP imputation was conducted within each sample using IMPUTE2 (19) or MACH (20) software utilizing the full 1000 Genomes Project reference data (March 2012, release v3). Genomic locations were based on NCBI build 37/UCSC hg 19 data. After imputation, SNPs with MAF<0.01, poor imputation quality <0.30, and Hardy-Weinberg equilibrium p-value <10−6 were removed. See Supplement for study descriptions and Supplemental Table S1 for details of genotyping and QC procedures. Table 1 summarizes basic statistics by cohort.
Table 1.
Contributing Cohorts
| Cohort | Country | Mean age (s.d.) at interview | N Phenotyped | N Cases (ANX=2) | N Controls (ANX=0) | N Analyzed (FS/CC) |
|---|---|---|---|---|---|---|
| MGS Controls (43) | USA | 50.0 (16.4) | 2659 | 757 | 1059 | 2009/1336 |
| PsyCoLaus (44) | Switzerland | 50.9 (8.8) | 3575 | 1044 | 1351 | 2887/1955 |
| RS (45) | Netherlands | 66.5 (10.8) | 9686 | 1112 | 5459 | 7832/5379 |
| SHIP (46) | Germany | 55.4 (13.9) | 2279 | 581 | 890 | 2026/1379 |
| QIMR (47) | Australia | 41.3 (11.5) | 6147 | 1611 | 2544 | 2277/2156 |
| TRAILS (48) | Netherlands | 18.7 (0.7) | 1584 | 390 | 472 | 1155/614 |
| NESDA (49) / NTR (50;51) | Netherlands | 45.6 (14.2) /44.6 (12.7) | 4491 | 1521 | 2970 | NA/4491 |
| Total | 31 060 | 7016 | 14 745 | 18 186/17 310 |
Abbreviations: MGS, Molecular Genetics of Schizophrenia; RS, Rotterdam Study; SHIP, Study of Health in Pomerania; NESDA/NTR, The Netherlands Study of Depression and Anxiety / Netherlands Twin Registry; QIMR, Queensland Institute of Medical Research; TRAILS, Tracking Adolescents' Individual Lives Survey; FS, Factor Score; CC, Case-Control.
Genome-wide association analyses
To identify genetic variants contributing to genetic susceptibility shared across the ADs, we applied and compared two complementary phenotypic approaches: (1) categorical CC comparisons, and (2) quantitative phenotypic FS. For CC comparisons, AD cases were assigned to subjects meeting criteria for any lifetime AD (ANX=2) while control subjects were “super-normal”, i.e., having few or no clinical anxiety symptoms (ANX=0); those with subsyndromal ADs (ANX=1) were excluded from the CC analyses. For FS analyses, first exploratory factor analyses were conducted using Mplus (version 4) (39) separately in each sample, finding evidence for a single common factor model by scree plots. This was followed by confirmatory factor analyses that estimated a single FS for each subject from this common AD liability factor. (See Supplement for details of phenotype construction.) Association analyses were then performed in each study independently with imputed SNP dosages under an additive genetic model using logistic regression for CC phenotype and linear regression for quantitative FS phenotype. As covariates, we used sex and age at interview, as they were significant predictors of the phenotypes. Ancestry principal components were estimated for each sample and included on a sample-by-sample basis depending on their correlation with the outcome phenotypes. The quantile-quantile (Q-Q) plot was used to evaluate overall significance of the association test results and the genomic control factor λ.
Meta-analysis of GWAS
We performed an inverse-variance weighted, fixed-effects meta-analysis with all GWAS samples using METAL (21) (nine samples using CC phenotypes and eight using FS phenotypes). For each SNP, a pooled effect size, standard error, and p-value were computed. SNPs with low MAF (<0.05) were excluded, resulting in a final meta-analytic data set of around 6.5M SNPs. Cochran’s Q statistics and corresponding I2 statistics were used as heterogeneity metrics. Cochran’s Q statistic was computed by summing the squared deviations of each study’s estimate by weighting each study’s contribution in the same manner as in the meta-analyses. I2 measured the amount of heterogeneity that is not due to chance.
Q-Q and Manhattan plots were examined, and False Discovery Rate (FDR) q-values were calculated based on the p-values from the meta-analyses. Q-values provide a balance between type I and Type II errors and can be interpreted as the probability that a marker identified as significant is a false discovery (22).
Cross-validation
In order to examine overall consistency of association between datasets, we employed a leave-one-out procedure for internal cross-validation. At each step, we meta-analyzed eight of the nine CC GWAS samples as the “training” set (seven of the eight samples were used for FS), the results of which were then tested in the respective remaining target sample (“testing” set). The top associated SNPs in the training set (Ptraining<1×10−5, pruned to r2<0.4 within a 500-kb window) were used to test the replicability (Ptesting<0.05) and consistency of the direction of their effects with the top associated SNPs identified in each testing set. One thousand random permutations of phenotype allocation to an individual’s genome-wide genotypes were performed in each training-testing set pair, totaling 9,000 and 8,000 permutations in CC and FS, respectively. Across all sets, we compared the aggregate numbers of replicated SNPs and SNPs with the same direction of effect against the numbers expected by chance.
Gene-based tests
The SNP-based p-values derived from the meta-analyses were applied to gene-based association testing using KGG software (http://statgenpro.psychiatry.hku.hk/limx/kgg/) (23). No prioritization or preselection of genes was performed. Gene-based tests in KGG combine univariate association statistics to evaluate the cumulative evidence of association in a gene using extended Simes test (GATES) (24). SNPs were mapped onto 23 931 genes according to the gene coordinate information from NCBI, and SNPs within 10-kb of each gene were assigned to that gene. We considered genes with P<2×10−6 (=.05/23 931) as significant and those with q-value <0.1 as interesting (25).
Secondary Analyses
We conducted several secondary analyses, the details of which are described in the Supplement.
SNP-based Heritability. Genomic-relatedness-matrix restricted maximum likelihood (GREML), as implemented in the software program GCTA (26), was conducted in our largest cohort (RS) to estimate the total amount of variance explained by all analyzed SNPs. This was supplemented by a similar procedure in the full meta-analytic sample using LD score regression (27).
Polygenic Risk Profile Analyses. Given the observed high comorbidity between ADs and other psychiatric syndromes, genomic profile risk scores (GPRS) (28) were estimated to test the additive joint effects of multiple variants between our AD GWAS data as target samples and summary data from Psychiatric Genomics Consortium phase 1 (PGC1) schizophrenia (SCZ), bipolar disorder (BIP), and MDD as discovery samples.
RESULTS
GWAS Meta-analysis
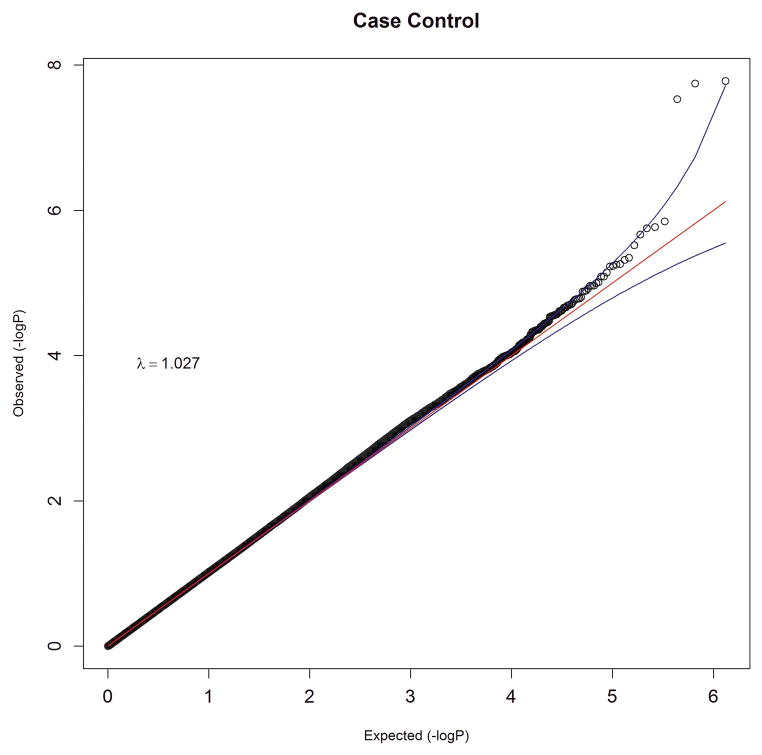
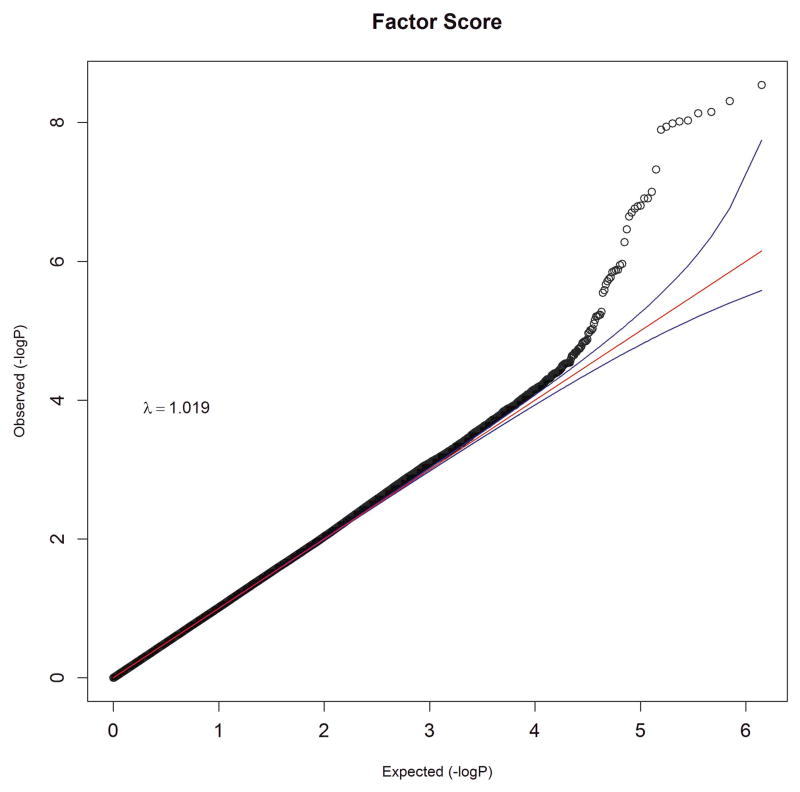
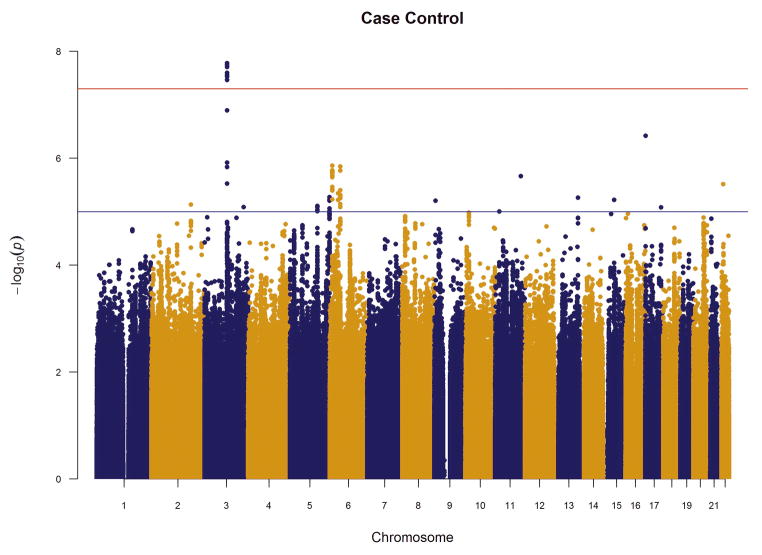
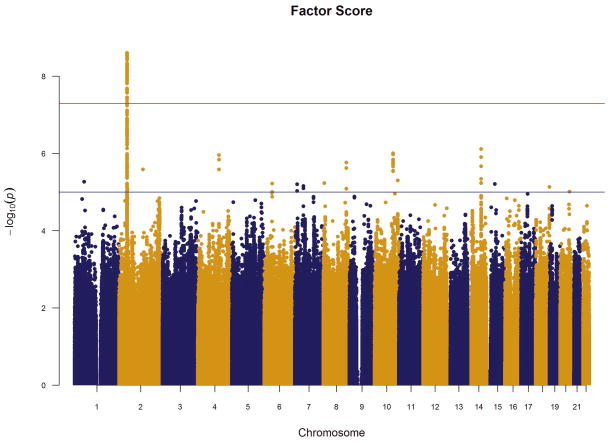
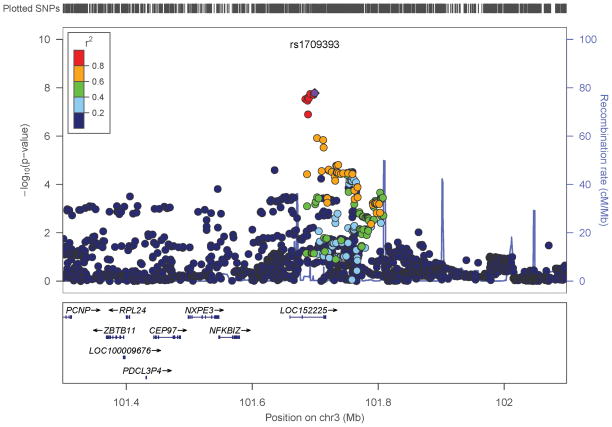
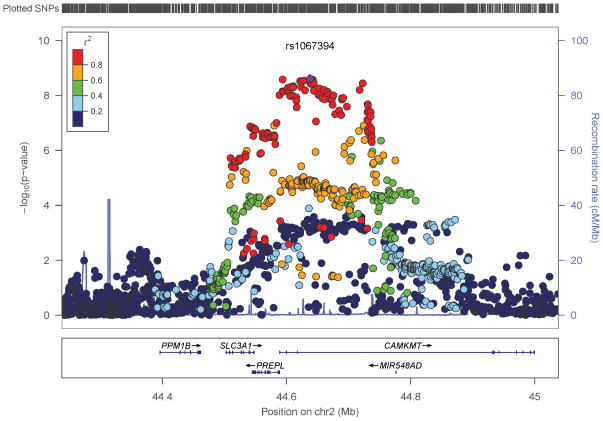
We performed an inverse-variance weighted, fixed-effects meta-analysis with all discovery GWAS data including approximately 6.5M common SNPs after applying post-imputation QC to each study. The genomic inflation factor λ ranged from 0.990 to 1.038 for all studies. The Q-Q plots of the meta-analyses for the CC and FS phenotypes are presented in Figure 1. Meta-analytic inflation factors were 1.03 and 1.02, suggesting little effect of population stratification. Manhattan plots are presented in Figure 2. Table 2 lists the LD-independent, genome-wide significant SNPs and associated regions. For the CC model, the strongest association was observed at rs1709393 located in an intron of an uncharacterized non-coding RNA locus LOC152225 on chromosome 3q12.3 (P=1.65×10−8; Q=0.027). Allelic frequencies were very similar across studies and ranged between 0.55 and 0.60. The most significant SNP in the FS model was rs1067327 on chromosome 2p21 within CAMKMT encoding the calmodulin-lysine N-methyltransferase (P=2.86×10−9; Q=0.0017) with LD extending into several adjacent genes. Allelic frequencies were consistent across studies, ranging from 0.32 to 0.36. Both of these SNPs were imputed with very high quality across studies (R2>0.93). As indicted in the forest plots (Supplementary Figure S1), no heterogeneity of effects was observed for either SNP. Figure 3 displays the regional SNP plots for these two genome-wide significant loci.
Figure 1.
Quantile-quantile plots of meta-analysis results for (a) case-control and (b) factor score phenotypes. Observed association results of −log10P , after LD-pruning at r2 of 0.4, are plotted against the expected distribution under the null hypothesis of no association.
Figure 2.
Manhattan plots of meta-analysis results for (a) case-control and (b) factor score phenotypes. Red horizontal line indicates the genome-wide significant p-value 5×10−8; blue line indicates the suggestive p-value=1×10−5.
Table 2.
Top association results for meta-analysis of SNP main effects for case-control and factor score phenotypes.
| Association with Original Phenotype
|
Cross Phenotype†
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNP | A12a | Frqmean | Chr | Positionb | Effectc (95% CI) | p- value | q- value | Directiond | p-value | Effectc |
| Case-Control | ||||||||||
| rs1709393 | TC | 0.58 | 3 | 101684480-101692234 | 0.860 (0.816-0.906) | 1.65e-8 | 0.027 | --------- | 0.0342 | −0.010 |
| Factor Score | ||||||||||
| rs1067327 | CG | 0.34 | 2 | 44588941-44678648 | 0.028 (0.019-0.038) | 2.86e–9 | 0.0017 | ++++++++ | 0.0002 | 1.123 |
Chr, chromosome; Frq, frequency of allele 1; CI, 95% confidence interval.
First allele is the reference allele, for which the effect is reported.
Position denotes the associated region surrounding the top SNP containing 1 or more genome-wide significant SNPs in LD (r2>0.4) with the top SNP.
Effect size represents odds ratio for case control and regression coefficient for factor score.
Direction of association is provided for each study in the following order: RS1, RS2, RS3, NTR/NESDA, MGS, PsyColaus, SHIP, QIMR, and TRAILS for case-control; RS1, RS2, RS3, MGS, PsyColaus, SHIP, QIMR, and TRAILS for factor score. Plus (+) indicates the association between the SNP and the corresponding anxiety phenotype is positive. Minus (−) indicates a negative direction of association.
Shown are the association results of the top SNPs in the other phenotype, i.e., factor score results of rs1709393 and case-control results of rs1067327.
Fig 3.
Regional plots around most significant SNPs in (a) case-control and (b) factor score model.
Given that the CC and FS phenotypic approaches provide conceptually different but otherwise complementary information, we estimated the overlap in their association signals. These phenotypes were highly correlated in the different cohorts (0.88–0.94). Overall rank-based correlations between the CC and FS association effects were 0.61. The degree of correlation increased with decreasing p-value threshold, ranging from 0.275 to 0.899 (Supplementary Table S3). The most significant SNPs all have the same direction of effect (top 1,000 SNPs in CC and top 1,500 SNPs in FS); indeed, among the approximately 30% of total SNPs with opposite sign, none had even suggestively significant association (P<1×10−5). However, among ~1.4M independent SNPS (pruned at r2=0.4), significantly more with P<10−5 were identified for the FS phenotype than for the CC phenotype: 42 verses 18 (test p-value=0.0034).
Cross-Validation
In the leave-one-out cross-validation analyses, the replication rate was significantly higher than expected by chance (Table 3). In CC, 18 of 173 tested SNPs across all leave-one out analyses replicated in the left out testing sets (permutation P=0.001) and the proportion of SNPs with the same direction of effect was 59.5% (sign test P=0.005). Of 315 tested SNPs in FS, 43 SNPs replicated (permutation P<0.001) and 77.8% had the same direction of effect (sign test P<0.001). Supplemental Figure S2 displays Manhattan plots of the training set meta-analyses conducted after leaving out each sample.
Table 3.
Results of leave-one-out cross-validation analyses
| Case-Control
|
Factor Score
|
|||||
|---|---|---|---|---|---|---|
| Testing Sample | NSNPa Ptraining<10−5 |
NSNPb Ptesting<0.05 |
NSNP (same direction) | NSNPa Ptraining<10−5 |
NSNPb Ptesting<0.05 |
NSNP (same direction) |
| MGS | 19 | 3 | 10 | 39 | 0 | 25 |
| GSK | 26 | 3 | 12 | 22 | 4 | 11 |
| RS1 | 24 | 0 | 13 | 42 | 6 | 36 |
| RS2 | 15 | 3 | 9 | 45 | 0 | 33 |
| RS3 | 22 | 0 | 10 | 46 | 21 | 35 |
| SHIP | 24 | 0 | 14 | 40 | 10 | 32 |
| NTR/NESDAc | 14 | 3 | 13 | - | - | - |
| QIMR | 13 | 3 | 9 | 35 | 0 | 32 |
| TRAILS | 16 | 3 | 13 | 46 | 2 | 41 |
| OVERALL | 173 | 18* (P=0.001) | 103* (P=0.005) | 315 | 43* (P<0.001) | 245* (P<0.001) |
P<0.05 thresholds applied for replication and sign tests; Replication and sign tests are conducted based on permutations with 9,000 iterations in case-control and 8,000 iterations in factor score phenotype under the null hypothesis of no association (1,000 iteration for each pair of training and testing sets).
The number of SNPs associated at Ptraining<1×10−5 with r2<0.4 in the leave-one-out meta-analysis using N-1 training samples after removing one testing sample at a time.
Of the SNPs with Ptraining<1×10−5 in the meta-analysis of the N-1 training samples, the number of replicated SNPs with one-sided Ptesting<0.05 in the left-out (testing) sample.
Only case-control phenotype is available for NTR/NESDA sample.
Gene-based tests
In the CC model, LOC152225 on 3q12.3 surpassed genome-wide significance (P=1.19×10−6; Q=0.028). In the FS model, three genes exceeded genome-wide significance: PREPL, CAMKMT, and SLC3A1 on chromosome 2 (Table 4). Supplementary Figure S3 depicts the Manhattan plots for these gene-based analyses.
Table 4.
Top associated genes (Q < 0.1) using gene-based tests
| Gene | p-valuea | q-value | SNPb with lowest P | Lowest Pc | Chr | Gene Feature |
|---|---|---|---|---|---|---|
| Case-Control | ||||||
| LOC152225 | 1.19E-06 | 0.028 | rs1709393 | 1.65E-08 | 3 | ncRNA |
| Factor Score | ||||||
| PREPL | 5.61E-08 | 0.001 | rs786618 | 6.99E-09 | 2 | intronic |
| CAMKMT | 3.15E-07 | 0.004 | rs1067327 | 2.86E-09 | 2 | intronic |
| SLC3A1 | 1.45E-06 | 0.012 | rs1142523 | 1.44E-07 | 2 | 3’UTR |
| LBX1 | 7.50E-06 | 0.045 | rs11190870 | 9.79E-07 | 10 | downstream |
| LBX1-AS1 | 1.33E-05 | 0.064 | rs594791 | 1.05E-06 | 10 | upstream |
We used LD pruning at r2 of 0.4 for gene-based tests. Boldface indicates genome-wide P < 2×10−6.
Gene-based p-value (bolded genome-wide significant P<2×10−6) .
Most significant SNP within the corresponding gene.
SNP-based p-value for the most significant SNP
Secondary Analyses (see Supplement for details)
SNP-based heritability. This was estimated by GREML using GCTA in the Rotterdam sample as 0.106 (SE=0.06, P=0.05) for FS phenotype and 0.138 (SE=0.18, P=0.2) for CC phenotype on the liability scale assuming 10% AD population prevalence. Within the margin of error, these were consistent with LD score regression using summary statistics in the full meta-analysis sample, with SNP heritability estimated as 0.072 (SE=0.028) for FS phenotype and 0.095 (SE =0.037) for CC phenotype.
Polygenic Risk Profile Analyses. GPRS from PGC-MDD explained a small but significant proportion of variance in CC ADs in QIMR (0.5%–0.7%), while SCZ and BIP each explained a somewhat smaller proportion of this variance varying by sample. These results were supported by LD score regression performed in the meta-analysis sample, estimating significant genetic correlation between ADs and MDD (r=0.68) but not between ADs and BIP or SCZ.
DISCUSSION
We conducted the largest and most comprehensive genetic study of the primary anxiety disorders (ADs) to date. Specifically, we integrated phenotypic information on GAD, PD, agoraphobia, social phobia, and specific phobias and combined this with genome-wide SNP data from nine large samples totaling over 18 000 subjects. We conducted parallel GWAS in these samples and statistically combined the results via meta-analysis, with the aim of detecting common variants that play a role in shared AD susceptibility.
While only an approximate representation of the underlying complexity of AD genetic mechanisms, our integrated phenotypic approaches successfully identified novel genetic variants that significantly associate with these composite AD phenotypes. The results were generally the same whether analyzing individual SNPs or genes. In the CC model, we identified a novel genome-wide association within an uncharacterized non-coding RNA locus LOC152225 on chromosome 3q12.3. We found no extant reports for this locus in PubMed or the NHGRI Catalogue of Published GWAS (www.genome.gov/gwastudies/). In the FS model, we detected genome-wide significant associations at SNPs in three genes within a large LD block on chromosome 2p21, each of which has reported expression in brain: (1) SLC3A1 encoding the large subunit of a heterodimeric dibasic /neutral amino acid transporter (solute carrier family 3 (amino acid transporter heavy chain), member 1); (2) PREPL encoding a putative prolyl endopeptidase belonging to the prolyl oligopeptidase family; and (3) CAMKMT encoding a calmodulin-lysine N-methyltransferase. This region is well-known for two contiguous gene-deletion syndromes, the hypotonia-cystinuria syndrome and the more severe 2p21 deletion syndrome (29). Deletion of SLC3A1 results in the autosomal-recessive form of cystinuria (30), while PREPL deletion causes hypotonia at birth, failure to thrive and growth hormone deficiency (31). The evolutionarily conserved class I protein methyltransferase encoded by CAMKMT acts in the formation of trimethyllysine in calmodulin which is involved in calcium-dependent signaling (32). Interestingly, GWAS of schizophrenia and bipolar disorder have highlighted other genes encoding proteins involved in calcium-dependent signaling (33). Although the most significant SNP, rs1067327, is located in an intron of CAMKMT , in silico analyses (Supplement) suggest rs698775 is the most likely functional candidate with a cis regulatory effect possibly specific to PREPL.
There is substantial phenotypic overlap between the CC and FS models used to capture the comorbidity and shared genetic risk among the ADs, and as expected, there was a high degree of concordance in the association signals genome-wide (Supplementary Table S3). The most significantly associated SNPs (p<.0.05) have very high correlation of association effects, suggesting they are tapping into strongly related AD risk factors. We note that, overall, the FS phenotype identified a larger number of associated SNPs than the CC model. This is likely due to several reasons: (1) this approach combines disorder information to capture individual differences on an underlying latent AD liability; (2) for high prevalence disorders, quantitative variables generally have greater power for genetic association than categorical variables (34;35); (3) the FS models generally involve larger sample sizes since they also include the subjects with subthreshold ADs (score=1); and (4) the FS model produces a phenotype that incorporates the observed relationship information (covariance) between the individual ADs. These findings support the use of quantitative phenotypic factors scores in future GWAS of comorbid psychiatric disorders assessed in the same individuals.
Several secondary analyses support our findings. First, we applied cross validation in the nine samples to examine the internal consistency of the results. We created sub-samples by iteratively removing the data of each of the individual samples and conducting meta-analysis with the remaining datasets. A highly significant proportion of the top results were consistently identified across these sub-analyses, suggesting the stability and validity of our findings. Next we estimated the genome-wide contribution via GREML and the complementary LD-score regression approach, producing generally consistent estimates of SNP heritability across samples included and methods applied. Similar to GWAS studies of many phenotypes (36), these estimates are substantially smaller than those predicted by twin studies of ADs. Finally, we tested the polygenic association between our results and those from other psychiatric disorders using GRPS, finding significant correlation of genetic risk between ADs and MDD but not between ADs and BIP or SCZ. The former result is consistent with large epidemiologic studies that report correlated genetic risk between ADs and MDD (see (15) for review) as well as a prior overlap seen for depression and anxiety scales (37).
A strength of this study is that we applied phenotypic strategies aimed at detecting genetic variants that play a central but non-specific role in AD susceptibility. This is counter to the approach taken in most psychiatric genetic studies which generally apply case-control comparisons for specific clinical diagnoses, sometimes followed by adjunct cross-disorder analyses. However, it has long been recognized that clinical nosology poorly reflects etiological mechanisms, with both genetic and environmental risk factors showing non-specific effects across disorders. ADs, despite their heterogeneous clinical presentations, likely represent various expressions of an underlying common diathesis of abnormal regulation of basic threat-response systems (12). Given the value of fear and anxiety for survival, there are likely sets of evolutionarily-conserved genes that regulate these basic biological responses. This is supported by twin studies that identify factors of common genetic risk across ADs in addition to disorder-specific genetic factors. With this in mind, we applied and compared two strategies for combining information across clinical phenotypes. The first is a simple CC approach, comparing cases defined as having “any AD” against supernormal controls. The second applied multivariate modeling of the covariation among the ADs using the common factor model to define a single continuous dimension of liability for which quantitative scores can be estimated for each subject. Our group has applied this approach in prior candidate gene association studies (18) and in a pilot GWAS in the MGS sample (38), but this is the first such application in a large GWAS meta-analysis. We note that this strategy is consistent with NIMH’s Research Domain Criteria (RDoC) initiative, which aims to serve as a framework for new approaches to research on mental disorders based on fundamental dimensions that cut across traditional disorder categories and more closely align with mechanisms that underlie psychopathology at various biological levels from genes to neural circuits (39). Also important to note is that ADs not only share genetic risk factors amongst themselves but also with other internalizing phenotypes like MDD (15), obsessive compulsive disorder (40), and personality traits like neuroticism and extroversion (41). It will be important for future studies to examine this broader pleiotropic spectrum either through cross-disorder GWAS as previously conducted for other psychiatric conditions (42) or by including these additional traits directly in the phenotypic construction with the ADs. It is possible that, by including AD cases with comorbid MDD, the genetic overlap between these conditions has influenced our results.
Several potential limitations of this study should be noted. First, although the total sample size far exceeds those from prior AD genetic studies, it is still relatively underpowered to detect common genetic variants of small effect expected for the genetic architecture of such complex phenotypes (37). Second, not all samples provided the same level of phenotypic coverage; in particular, some subjects in QIMR were missing diagnostic data for GAD or specific phobia. While this can produce bias, our forest plots, tests for heterogeneity, and internal validation analyses suggest that this likely did not bias our results. Third, consent agreements for some of the sites did not allow for sharing of subject data, so GWAS analyses had to be conducted separately using a standardized procedure and combined via meta-analysis. While this has been shown to approximate the power obtained when using raw data via mega-analysis (38), we were limited in our ability to conduct additional post-hoc analyses such as GPRS and GREML that require the use of raw GWAS data. Reassuringly, results obtained by applying LD score regression to summary statistics from the total meta-analysis sample were consistent with those using raw data from select individual samples. Fourth, the results apply only to subjects of European-ancestry and might not generalize to individuals of other genetic and cultural backgrounds. Finally, we combined all data available at the time of this study into a single meta-analysis rather than divide into discovery and replication samples. This was necessary due to the large sample sizes required to detect small effects of genes involved in complex traits like ADs. Internal cross-validation supported the robustness of our results but do not substitute for replication in well-powered, independent samples. At this time, we are unaware of other large data sets that could be used for replication of our results.
In summary, this study has identified several potentially novel susceptibility loci that increase shared risk across the primary ADs. Future studies are needed to (1) further confirm these findings via independent replication, (2) increase the total sample size to enhance power to detect additional loci, and (3) identify loci associated specifically with each particular AD not accounted for by the pleiotropic effects targeted in this study.
Supplementary Material
Acknowledgments
This overall project was supported by NIH grant R01MH87646 to JMH. TO was supported by a research fellowship from the Japan Society for the Promotion of Science (no. 21-8373).
MGS
Samples and associated phenotype data for the MGS study were collected under the following grants: NIMH Schizophrenia Genetics Initiative U01s: MH046276 (CR Cloninger), MH46289 (C Kaufmann), and MH46318 (MT Tsuang); and MGS Part 1 (MGS1) and Part 2 (MGS2) R01s: MH67257 (NG Buccola), MH59588 (BJ Mowry), MH59571 (PV Gejman), MH59565(Robert Freedman), MH59587 (F Amin), MH60870 (WF Byerley), MH59566 (DW Black), MH59586 (JM Silverman), MH61675 (DF Levinson), and MH60879 (CR Cloninger).
Rotterdam Study (RS)
This work is supported by the Research Institute for Diseases in the Elderly (014-93-015; RIDE2), the Netherlands Genomics Initiative (NGI)/Netherlands Consortium for Healthy Ageing (NCHA) project No. 050-060-810. The work of Henning Tiemeier is supported by Vidi (grant 017.106.370). The Rotterdam Study is funded by Erasmus Medical Center, Rotterdam, the Netherlands Organization for the Health Research and Development (ZonMw), the Ministry of Education, Culture and Science, and the Ministry for Health, Welfare and Sports.
SHIP
SHIP is part of the Community Medicine Research net of the University of Greifswald, Germany, which is funded by the Federal Ministry of Education and Research (grants no. 01ZZ9603, 01ZZ0103, and 01ZZ0403), the Ministry of Cultural Affairs and the Social Ministry of the Federal State of Mecklenburg-West Pomerania. Genome-wide data have been supported by the Federal Ministry of Education and Research (grant no. 03ZIK012) and a joint grant from Siemens Healthcare, Erlangen, Germany and the Federal State of Mecklenburg-West Pomerania. This work was also funded by the German Research Foundation (DFG: GR 1912/5-1). The University of Greifswald is a member of the Caché Campus program of the InterSystems GmbH.
QIMR
The QIMR samples are made available through the generous and willing participation of twins and their families registered at the Australian Twin Registry and through grant funding awarded from many grant funding bodies including the Australian National Health and Medical Research Council (241944, 339462, 389927, 389875, 389891, 389892, 389938, 442915, 442981, 496675, 496739, 552485, 552498, 613608), the FP-5 GenomEUtwin Project (QLG2-CT- 2002-01254), the US National Institutes of Health (NIH grants AA07535, AA10248, AA13320, AA13321, AA13326, AA14041, MH66206, DA12854, DA019951), and the Center for Inherited Disease Research, Baltimore. We thank Dixie Statham (sample collection); Leanne Wallace, Anthony Caracella and staff of the Molecular Epidemiology Laboratory (DNA processing); David Smyth, Harry Beeby, and Daniel Park (IT support). Statistical analyses were partly conducted at the Genetic Cluster Computer (http://www.geneticcluster.org), which is financially supported by the Netherlands Scientific Organization (NWO 480-05-003). EBM (1053639), SEM, DN, GWM, NRW (613602, 1078901) are supported by the National Health and Medical Research Council Fellowship Scheme.
PsyCoLaus
The CoLaus|PsyCoLaus study was and is supported by research grants from GlaxoSmithKline, the Faculty of Biology and Medicine of Lausanne, and the Swiss National Science Foundation (grants 3200B0–105993, 3200B0-118308, 33CSCO-122661, 33CS30-139468 and 33CS30-148401).
TRAILS
Participating centers of TRAILS include various departments of the University Medical Center and University of Groningen, the Erasmus University Medical Center Rotterdam, the University of Utrecht, the Radboud Medical Center Nijmegen, and the Parnassia Bavo group, all in the Netherlands. TRAILS has been financially supported by various grants from the Netherlands Organization for Scientific Research NWO (Medical Research Council program grant GB-MW 940-38-011; ZonMW Brainpower grant 100-001-004; ZonMw Risk Behavior and Dependence grants 60-60600-97-118; ZonMw Culture and Health grant 261-98-710; Social Sciences Council medium-sized investment grants GB-MaGW 480-01-006 and GB-MaGW 480-07-001; Social Sciences Council project grants GB-MaGW 452-04-314 and GB-MaGW 452-06-004; NWO large-sized investment grant 175.010.2003.005; NWO Longitudinal Survey and Panel Funding 481-08-013), the Dutch Ministry of Justice (WODC), the European Science Foundation (EuroSTRESS project FP-006), Biobanking and Biomolecular Resources Research Infrastructure BBMRI-NL (CP 32), the participating universities, and Accare Center for Child and Adolescent Psychiatry. Statistical analyses were carried out on the Genetic Cluster Computer (http://www.geneticcluster.org), which is financially supported by the Netherlands Scientific Organization (NWO 480-05-003) along with a supplement from the Dutch Brain Foundation.
NESDA/NTR
Funding was obtained from the Netherlands Organization for Scientific Research (Geestkracht program grant 10-000-1002; 904-61-090, 985-10- 002, 904-61-193, 480-04-004, 400-05-717, 912-100-20; Spinozapremie 56-464-14192); the Center for Medical Systems Biology (CSMB, NWO Genomics), Biobanking and Biomolecular Resources Research Infrastructure (BBMRI-NL), VU University’s Institutes for Health and Care Research (EMGO+) and Neuroscience Campus Amsterdam, European Research Council (ERC, 230374),National Institutes of Health (NIH, R01D0042157-01A, MH081802, Grand Opportunity grants 1RC2 MH089951 and 1RC2 MH089995). Part of the genotyping and analyses were funded by the Genetic Association Information Network (GAIN) of the Foundation for the National Institutes of Health. Computing was supported by BiG Grid, the Dutch e-Science Grid, which is financially supported by NWO.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
Supplementary information is available at Molecular Psychiatry’s website.
Summary statistics for the two meta-analyses will be made available at the Psychiatric Genomics Consortium downloads website: http://www.med.unc.edu/pgc/downloads.
Reference List
- 1.Hettema JM, Neale MC, Kendler KS. A review and meta-analysis of the genetic epidemiology of anxiety disorders. Am J Psychiatry. 2001 Oct;158(10):1568–78. doi: 10.1176/appi.ajp.158.10.1568. [DOI] [PubMed] [Google Scholar]
- 2.Smoller JW, Block SR, Young MM. Genetics of anxiety disorders: the complex road from DSM to DNA. Depress Anxiety. 2009;26(11):965–75. doi: 10.1002/da.20623. [DOI] [PubMed] [Google Scholar]
- 3.Maron E, Hettema JM, Shlik J. Advances in molecular genetics of panic disorder. Mol Psychiatry. 2010 Jul;15(7):681–701. doi: 10.1038/mp.2009.145. [DOI] [PubMed] [Google Scholar]
- 4.Erhardt A, Czibere L, Roeske D, Lucae S, Unschuld PG, Ripke S, et al. TMEM132D, a new candidate for anxiety phenotypes: evidence from human and mouse studies. Mol Psychiatry. 2011 Jun;16(6):647–63. doi: 10.1038/mp.2010.41. [DOI] [PubMed] [Google Scholar]
- 5.Otowa T, Kawamura Y, Nishida N, Sugaya N, Koike A, Yoshida E, et al. Meta-analysis of genome-wide association studies for panic disorder in the Japanese population. Transl Psychiatry. 2012;2:e186. doi: 10.1038/tp.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Logue MW, Baldwin C, Guffanti G, Melista E, Wolf EJ, Reardon AF, et al. A genome-wide association study of post-traumatic stress disorder identifies the retinoid-related orphan receptor alpha (RORA) gene as a significant risk locus. Mol Psychiatry. 2013 Aug;18(8):937–42. doi: 10.1038/mp.2012.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie P, Kranzler HR, Yang C, Zhao H, Farrer LA, Gelernter J. Genome-wide association study identifies new susceptibility loci for posttraumatic stress disorder. Biol Psychiatry. 2013 Nov 1;74(9):656–63. doi: 10.1016/j.biopsych.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guffanti G, Galea S, Yan L, Roberts AL, Solovieff N, Aiello AE, et al. Genome-wide association study implicates a novel RNA gene, the lincRNA AC068718.1, as a risk factor for post-traumatic stress disorder in women. Psychoneuroendocrinology. 2013 Dec;38(12):3029–38. doi: 10.1016/j.psyneuen.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stewart SE, Yu D, Scharf JM, Neale BM, Fagerness JA, Mathews CA, et al. Genome-wide association study of obsessive-compulsive disorder. Mol Psychiatry. 2013 Jul;18(7):788–98. doi: 10.1038/mp.2012.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mattheisen M, Samuels JF, Wang Y, Greenberg BD, Fyer AJ, McCracken JT, et al. Genome-wide association study in obsessive-compulsive disorder: results from the OCGAS. Mol Psychiatry. 2014 May 13; doi: 10.1038/mp.2014.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walter S, Glymour MM, Koenen K, Liang L, Tchetgen Tchetgen EJ, Cornelis M, et al. Performance of polygenic scores for predicting phobic anxiety. PLoS One. 2013;8(11):e80326. doi: 10.1371/journal.pone.0080326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Craske MG, Rauch SL, Ursano R, Prenoveau J, Pine DS, Zinbarg RE. What is an anxiety disorder? Depress Anxiety. 2009;26(12):1066–85. doi: 10.1002/da.20633. [DOI] [PubMed] [Google Scholar]
- 13.Comorbidity of Mood and Anxiety Disorders. Washington, DC: American Psychiatric Press; 1990. [Google Scholar]
- 14.Hettema JM, Prescott CA, Myers JM, Neale MC, Kendler KS. The structure of genetic and environmental risk factors for anxiety disorders in men and women. Arch Gen Psychiatry. 2005 Feb;62(2):182–9. doi: 10.1001/archpsyc.62.2.182. [DOI] [PubMed] [Google Scholar]
- 15.Middeldorp CM, Cath DC, van Dyck R, Boomsma D. The co-morbidity of anxiety and depression in the perspective of genetic epidemiology. A review of twin and family studies. Psychol Med. 2005;35(May):611–24. doi: 10.1017/s003329170400412x. [DOI] [PubMed] [Google Scholar]
- 16.Kendler KS, Prescott CA, Myers J, Neale MC. The structure of genetic and environmental risk factors for common psychiatric and substance use disorders in men and women. Arch Gen Psychiatry. 2003 Sep;60(9):929–37. doi: 10.1001/archpsyc.60.9.929. [DOI] [PubMed] [Google Scholar]
- 17.Plomin R, Haworth CM, Davis OS. Common disorders are quantitative traits. Nat Rev Genet. 2009 Dec;10(12):872–8. doi: 10.1038/nrg2670. [DOI] [PubMed] [Google Scholar]
- 18.Hettema JM, An SS, Neale MC, Bukszar J, van den Oord EJ, Kendler KS, et al. Association between glutamic acid decarboxylase genes and anxiety disorders, major depression, and neuroticism. Molecular Psychiatry. 2006 May 23; doi: 10.1038/sj.mp.4001845. [DOI] [PubMed] [Google Scholar]
- 19.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012 Aug;44(8):955–9. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010 Dec;34(8):816–34. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010 Sep 1;26(17):2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003 Aug 5;100(16):9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li MX, Sham PC, Cherny SS, Song YQ. A knowledge-based weighting framework to boost the power of genome-wide association studies. PLoS One. 2010;5(12):e14480. doi: 10.1371/journal.pone.0014480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li MX, Gui HS, Kwan JS, Sham PC. GATES: a rapid and powerful gene-based association test using extended Simes procedure. Am J Hum Genet. 2011 Mar 11;88(3):283–93. doi: 10.1016/j.ajhg.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van den Oord EJ, Sullivan PF. False discoveries and models for gene discovery. Trends Genet. 2003 Oct;19(10):537–42. doi: 10.1016/j.tig.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 26.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011 Jan 7;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Patterson N, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015 Mar;47(3):291–5. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wray NR, Lee SH, Mehta D, Vinkhuyzen AA, Dudbridge F, Middeldorp CM. Research review: Polygenic methods and their application to psychiatric traits. J Child Psychol Psychiatry. 2014 Oct;55(10):1068–87. doi: 10.1111/jcpp.12295. [DOI] [PubMed] [Google Scholar]
- 29.Martens K, Jaeken J, Matthijs G, Creemers JW. Multi-system disorder syndromes associated with cystinuria type I. Curr Mol Med. 2008 Sep;8(6):544–50. doi: 10.2174/156652408785747997. [DOI] [PubMed] [Google Scholar]
- 30.Saravakos P, Kokkinou V, Giannatos E. Cystinuria: current diagnosis and management. Urology. 2014 Apr;83(4):693–9. doi: 10.1016/j.urology.2013.10.013. [DOI] [PubMed] [Google Scholar]
- 31.Martens K, Derua R, Meulemans S, Waelkens E, Jaeken J, Matthijs G, et al. PREPL: a putative novel oligopeptidase propelled into the limelight. Biol Chem. 2006 Jul;387(7):879–83. doi: 10.1515/BC.2006.111. [DOI] [PubMed] [Google Scholar]
- 32.Magnani R, Dirk LM, Trievel RC, Houtz RL. Calmodulin methyltransferase is an evolutionarily conserved enzyme that trimethylates Lys-115 in calmodulin. Nat Commun. 2010;1:43. doi: 10.1038/ncomms1044. [DOI] [PubMed] [Google Scholar]
- 33.Neale BM, Sklar P. Genetic analysis of schizophrenia and bipolar disorder reveals polygenicity but also suggests new directions for molecular interrogation. Curr Opin Neurobiol. 2015 Feb;30C:131–8. doi: 10.1016/j.conb.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 34.Yang J, Wray NR, Visscher PM. Comparing apples and oranges: equating the power of case-control and quantitative trait association studies. Genet Epidemiol. 2010 Apr;34(3):254–7. doi: 10.1002/gepi.20456. [DOI] [PubMed] [Google Scholar]
- 35.van der Sluis S, Posthuma D, Nivard MG, Verhage M, Dolan CV. Power in GWAS: lifting the curse of the clinical cut-off. Mol Psychiatry. 2013 Jan;18(1):2–3. doi: 10.1038/mp.2012.65. [DOI] [PubMed] [Google Scholar]
- 36.Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet. 2012 Jan 13;90(1):7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Demirkan A, Penninx BW, Hek K, Wray NR, Amin N, Aulchenko YS, et al. Genetic risk profiles for depression and anxiety in adult and elderly cohorts. Mol Psychiatry. 2011 Jul;16(7):773–83. doi: 10.1038/mp.2010.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Otowa T, Maher BS, Aggen SH, McClay JL, van den Oord EJ, Hettema JM. Genome-wide and gene-based association studies of anxiety disorders in European and African American samples. PLoS One. 2014;9(11):e112559. doi: 10.1371/journal.pone.0112559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Insel T, Cuthbert B, Garvey M, Heinssen R, Pine DS, Quinn K, et al. Research domain criteria (RDoC): toward a new classification framework for research on mental disorders. Am J Psychiatry. 2010 Jul;167(7):748–51. doi: 10.1176/appi.ajp.2010.09091379. [DOI] [PubMed] [Google Scholar]
- 40.Bienvenu OJ, Samuels JF, Wuyek LA, Liang KY, Wang Y, Grados MA, et al. Is obsessive-compulsive disorder an anxiety disorder, and what, if any, are spectrum conditions? A family study perspective. Psychol Med. 2012 Jan;42(1):1–13. doi: 10.1017/S0033291711000742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bienvenu OJ, Hettema JM, Neale MC, Prescott CA, Kendler KS. Low extraversion and high neuroticism as indices of genetic and environmental risk for social phobia, agoraphobia, and animal phobia. Am J Psychiatry. 2007 Nov;164(11):1714–21. doi: 10.1176/appi.ajp.2007.06101667. [DOI] [PubMed] [Google Scholar]
- 42.Cross-Disorder Group of the Psychiatric GWAS Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013 Apr 20;381(9875):1371–9. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanders AR, Levinson DF, Duan J, Dennis JM, Li R, Kendler KS, et al. The Internet-based MGS2 control sample: self report of mental illness. Am J Psychiatry. 2010 Jul;167(7):854–65. doi: 10.1176/appi.ajp.2010.09071050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Preisig M, Waeber G, Vollenweider P, Bovet P, Rothen S, Vandeleur C, et al. The PsyCoLaus study: methodology and characteristics of the sample of a population-based survey on psychiatric disorders and their association with genetic and cardiovascular risk factors. BMC Psychiatry. 2009;9:9. doi: 10.1186/1471-244X-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hofman A, Breteler MM, van Duijn CM, Krestin GP, Pols HA, Stricker BH, et al. The Rotterdam Study: objectives and design update. Eur J Epidemiol. 2007;22(11):819–29. doi: 10.1007/s10654-007-9199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmidt CO, Watzke AB, Schulz A, Baumeister SE, Freyberger HJ, Grabe HJ. The lifetime prevalence of mental disorders in north-eastern Germany. What is the influence of earlier mental morbidity on survey participation and prevalence estimates? Results from the SHIP-study. Psychiatr Prax. 2013 May;40(4):192–9. doi: 10.1055/s-0033-1343100. [DOI] [PubMed] [Google Scholar]
- 47.Jardine R, Martin NG, Henderson AS. Genetic covariation between neuroticism and the symptoms of anxiety and depression. Genet Epidemiol. 1984;1(2):89–107. doi: 10.1002/gepi.1370010202. [DOI] [PubMed] [Google Scholar]
- 48.Oldehinkel AJ, Rosmalen JG, Buitelaar JK, Hoek HW, Ormel J, Raven D, et al. Cohort Profile Update: the TRacking Adolescents' Individual Lives Survey (TRAILS) Int J Epidemiol. 2015 Feb;44(1):76–76n. doi: 10.1093/ije/dyu225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Penninx BW, Beekman AT, Smit JH, Zitman FG, Nolen WA, Spinhoven P, et al. The Netherlands Study of Depression and Anxiety (NESDA): rationale, objectives and methods. Int J Methods Psychiatr Res. 2008;17(3):121–40. doi: 10.1002/mpr.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boomsma DI, Vink JM, Van Beijsterveldt TC, de Geus EJ, Beem AL, Mulder EJ, et al. Netherlands Twin Register: a focus on longitudinal research. Twin Research. 2002 Oct;5(5):401–6. doi: 10.1375/136905202320906174. [DOI] [PubMed] [Google Scholar]
- 51.Boomsma DI, de Geus EJ, Vink JM, Stubbe JH, Distel MA, Hottenga JJ, et al. Netherlands Twin Register: from twins to twin families. Twin Research and Human Genetics. 2006 Dec;9(6):849–57. doi: 10.1375/183242706779462426. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.