Abstract
Purpose
Using optimized fast volumic echo planar spectroscopic imaging (3D-EPSI), we aimed to detect local metabolic abnormalities over the complete human brain in multiple sclerosis patients.
Materials and methods
Weighted mean combination of two 3D-EPSI covering the whole brain acquired at 3T in AC-PC and AC-PC+15° axial planes was performed to obtain high quality metabolite maps for five metabolites: N-acetyl aspartate (NAA), glutamate+glutamine (Glx), choline (Cho), myo-inositol (m-Ins) and creatine+phosphocreatine (tCr). After spatial normalisation, maps from 19 patients suffering from relapsing-remitting multiple sclerosis were compared to 19 matched controls using statistical mapping analyses to determine the topography of metabolic abnormalities. Probabilistic white matter (WM) T2 lesion maps and grey matter (GM) atrophy maps were also generated.
Results
Two-group ANOVA (SPM8, p<0.005, FDR corrected p<0.05 at the cluster level with age and sex as confounding covariates) comparing Patients and controls matched for age and sex showed clusters of abnormal metabolite levels with i) decreased NAA (around −15%) and Glx (around 20%) predominantly in GM within prefrontal cortices, motor cortices, bilateral thalami and mesial temporal cortices in line with neuronal/neuro-astrocytic dysfunction, ii) increased m-Ins (around +20%) inside WM T2 lesions and in the normal appearing WM of temporal-occipital lobes suggesting glial activation.
Conclusion
We demonstrated the ability to map non-invasively over the complete brain - from vertex to cerebellum – with a validated sequence, the metabolic abnormalities associated with MS, for characterizing the topography of pathological processes affecting widespread areas of WM and GM and its functional impact.
Keywords: Proton Magnetic resonance spectroscopic imaging, Multiple Sclerosis, whole brain, statistical mapping analysis, neurodegeneration, inflammation
INTRODUCTION
Magnetic resonance spectroscopic imaging (MRSI) is a way to study in vivo brain metabolism in a non-invasive manner (1). Nevertheless, MRSI suffers from several limitations including poor signal to noise ratio (SNR), strong susceptibility to B0 inhomogeneity, limited spatial coverage and long acquisition times. For the metabolic characterization of diffuse neurological pathologies such as multiple sclerosis (MS), whole brain coverage is useful to obtain a good overview of the topography of the various pathophysiological processes. Global approaches have also been proposed to obtain whole brain NAA with non-localized magnetic resonance spectroscopy (MRS) techniques with the major advantages of good SNR and good reproducibility (2), but because of RF field inhomogeneity over the whole brain and regional B0 inhomogeneities at tissue interfaces, up to 30% of brain tissue signal were not covered (3).
Methodological improvements have been proposed based on fast volumic echo planar spectroscopic imaging (3D-EPSI), interleaved water and metabolite acquisitions and automatic spectral fitting (4, 5). While these methods provide improved spatial coverage there remain limitations for sampling of the entire brain (6). Nevertheless, partial brain sampling has been reported to be sufficient to conduct statistical mapping analyses comparing metabolic maps between different group of healthy subjects (7). Concurrently, a combination of two fast short TE MRSI acquired in two different orientations (AC-PC and AC-PC +15°) has been used to obtain high quality cerebral metabolite maps covering the whole brain of healthy controls in about 45min (8). These complete brain metabolite maps may be of value to determine the brain topography of metabolic abnormalities associated with various brain diseases such as MS. Indeed, MRS and MRSI have evidenced abnormalities in brain metabolism associated with MS within white matter (WM) T2 lesions (9), Normal Appearing White Matter (NAWM), cortical Grey Matter (GM) (10), hippocampus and thalamus (11). MRS/MRSI has also helped to characterize the metabolic profiles of different stages and/or forms of MS, from the earliest stage of the disease in radiological isolated syndromes (12) and clinically isolated syndromes (10), classical forms (13) or benign forms of relapsing remitting MS (RRMS) (14), to the progressive forms of MS (13). Nevertheless, so far, the topographical dimension has either not been or only partially been assessed using single voxel spectroscopy (SVS) or 2D/3D-MRSI respectively (15). Indeed so far, SVS (16), 2D - MRSI (17) or 3D-MRSI (15) have permitted the exploration of only one limited predefined brain region, one brain slice or one brain volume respectively, limited to 60% of the entire brain (18).
Using optimized fast volumic echo planar spectroscopic imaging (3D-EPSI), we aimed to detect local metabolic abnormalities over the complete human brain in multiple sclerosis patients.
MATERIALS AND METHODS
Subjects
Nineteen healthy volunteers (mean age = 29.8 years± 9.5, range = 20-60, 10 women, 9 men) and nineteen RRMS patients (mean age = 36 years± 10.9, range = 23-59, 12 women, 7 men) were included in this study. The two groups were not different for sex (p > 0.05) or age (p > 0.05). All subjects gave their written consent to participate to the study, which was approved by the local ethics committee.
MRI
Imaging was performed on a 3T Magnetom Verio system (Siemens, Erlangen, Germany) using a standard 32-channel receiver head coil. The imaging protocol included localizer images (3 planes, 5 slices), sagittal T1-weighted Flash, and high resolution axial 3D T1-weighted Magnetization Prepared RApid Gradient Echo (MPRAGE) sequence (TE/TR/TI = 3.44ms/2150ms/1100ms; flip angle = 8°; voxel size = 1×1×1mm3, FOV = 256mm; matrix = 2562; partitions=176, TA=5min). Axial T2-weighted (TE1/TE2/TR = 11ms/90ms/8500ms; 49 contiguous slices; thickness = 3mm; flip angle = 150°; FOV = 250 mm; matrix = 2562; TA = 4:08min) and sagittal FLuid Attenuated Inversion Recovery (FLAIR) (TE/TR = 395ms/5000ms; flip angle = 70°; voxel size = 1×1×1mm3; FOV = 250 mm; matrix = 2562; partitions = 160; TA = 5:52min) sequences were additionally performed in the patient cohort to evaluate T2 lesion load. After the MRSI acquisitions, T1-weighted images were acquired pre- and post-Gadolinium injection (TE/TR = 8.4ms/500ms; 45 contiguous slices; thickness = 3mm; flip angle = 70°; FOV = 250 mm; matrix = 2562; TA = 4:22min) to identify active lesions (blood brain barrier disruption).
MRSI
Two axial fast 3D-EPSI (4) were included with two different orientations, first in the AC-PC plane and secondly in the AC-PC +15° plane (Figure A) (8). RF power settings and head position were identical to the MRI protocol, with the addition of CHESS water suppression (40 Hz), lipid inversion nulling with TI = 198 ms, TE/TR = 20ms/1710ms, flip angle = 73°, 50*50*18 k-space points, voxel size = 1×1×1cm3, FOV = 280*280*180mm3, acquisition time = 17:48min.
Figure A.
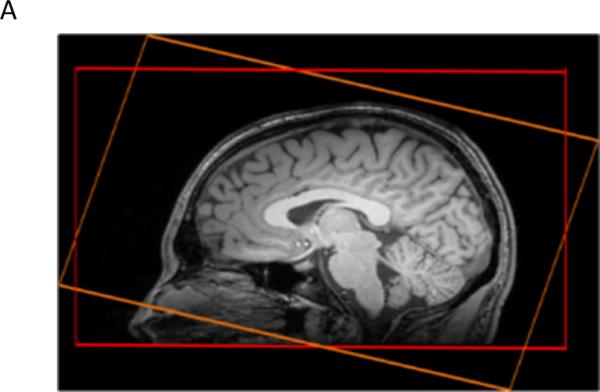
Positions of the two whole brain short TE 3D-EPSI sequences acquired in the AC-PC (red) and AC-PC+15° (orange) axial planes.
The shim was adjusted (first and second order) using the automatic 3D mapshim procedure (performed at least twice to get a stable phase map) for each orientation. Then, manual shimming (procedure duration about 5min) was performed until obtaining a water line width below 30 Hz in the acquisition volume. Frequency was adjusted before each acquisition.
Data Processing
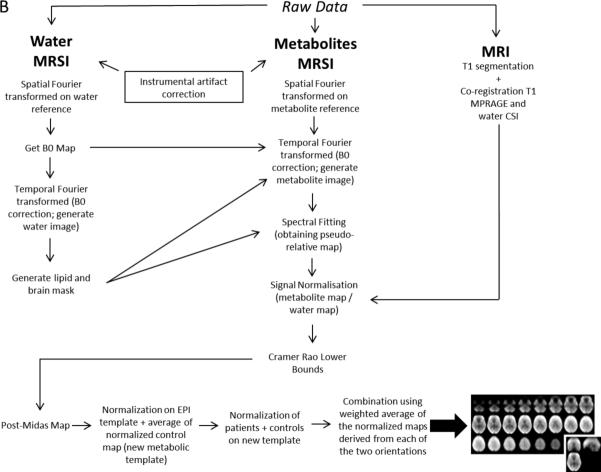
Metabolite maps were obtained for each subject (for both patients and controls), each orientation (AC-PC and AC-PC +15°) and each metabolite (NAA, Glx, Cho, m-Ins, tCr). As explained in details by Lecocq and coworkers (8), the two angles of MRSI orientations were chosen in order to get sufficient differences in acquisition planning between the two orientations to obtain good quality spectra on as large a brain area as possible (optimally whole brain) on at least one acquisition, but also keeping a reasonable angulation to allow accurate automatic normalization procedure for the two orientations (SPM8, Welcome Trust center for Neuroimaging at University College London). The post-processing pipeline described in Figure B, used the Metabolite Imaging and Data Analyses System (MIDAS) software (19) including B0 map correction, lipid suppression, tissue volume fraction through T1 segmentation, spectral fitting, exclusion of outlier voxels based on Cramer Rao bounds (CRB) and signal normalisation with the interleaved water signal acquired (5).
Figure B.
Pipeline used to obtain the optimized metabolite maps (MRI = Magnetic Resonance Imaging; MRSI = Magnetic Resonance Spectroscopic Imaging; MPRAGE = Magnetization Prepared RApid Gradient Echo; CSI = Chemical Shift Imaging)
In order to give an estimate of the absolute concentrations of metabolites, we used as reference the GM concentration of NAA of 10mM obtained in (8) using the same 3D-EPSI sequence. The correction factor from the arbitrary unit (AU) to mM was then derived from mean GM NAA value of controls and applied to all metabolite maps first expressed in AU, a unit already normalized by the water signal and the number of protons.
All the following steps were performed with SPM 8. First, metabolic maps of controls were spatially normalized using the EPI template of SPM8. Second, a new metabolic template was built based on the average of normalized maps of the entire control group. Maps of coefficient of variation (standard deviation divided by average, CV=(SD/AV)) were also created from all control data. Third, all subjects’ maps (patients and controls) were spatially normalized onto the new metabolic templates (one for each metabolite). Finally, a combination image was created using the weighted average of the normalized maps derived from each of the two orientations, calculated using:
With SD and AV the standard deviation and the average over the control group of the voxel value respectively for the AC-PC or the AC-PC+15° orientations.
T2-weigthed axial imaging performed on RRMS patients allowed localization and manual delineation of white matter lesions by an experimented neurologist (BA) with MRIcro software (Center for Brain Imaging, Columbia, USA). After normalisation on the T2 template in SPM8, lesions masks were averaged in order to obtain probabilistic maps thresholded at 30 %.
GM/WM/CSF segmentation was generated in all subjects using the VBM8 (SPM 8 toolbox to provide voxel based morphometry). GM masks of patients and controls were compared using voxel-wise two-group ANOVA (SPM8, p<0.005, FDR corrected p<0.05 at the cluster level with age and sex as confounding covariates) in order to create GM atrophy maps.
Clinical score assessment
The Multiple Sclerosis Functional Composite (MSFC), including Time 25-Foot Walk (TFW), 9-Hole Peg Test (9HPT) and Paced Auditory Serial Addition Test (PASAT) was determined for the patient group the day of the MR exam.
Statistical analyses
Metabolite concentrations between RRMS patients and controls were compared using voxel-wise two-group ANOVA (SPM8, p<0.005, FDR corrected p<0.05 at the cluster level with age and sex as confounding covariates). Identical statistical mapping analyses were performed for each metabolite: N-acetyl aspartate (NAA), glutamate + glutamine (Glx), choline (Cho), myo-inositol (m-Ins) and creatine (tCr) as independent tests. For each metabolite, an inclusive brain mask was used to exclude all the voxels outside the brain. Labeling of significant clusters was performed using the WFU PickAtlas toolkit (SPM 8).
To determine the potential relationship between metabolite abnormalities and clinical deficits assessed by MSFC in RRMS patients, voxel-wise multiple regression analyses were performed (SPM8, p <0.005, age and sex as confounding covariates) using masks of metabolite abnormalities.
RESULTS
Clinical features of RRMS patients
Median disease duration was 4.8 years ranging from 2.8 to 19 years. Median EDSS was 1 ranging from 0 to 4. Median MSFC was −0.12 ranging from −5.82 to 1.58. Fifteen out of the 19 patients (79%) were under immunosuppressive or modulator treatments but all subjects were in a stable stage of the disease (no relapse and no Gadolinium-enhancing lesions).
Metabolic templates, macroscopic WM T2 lesions and GM atrophy
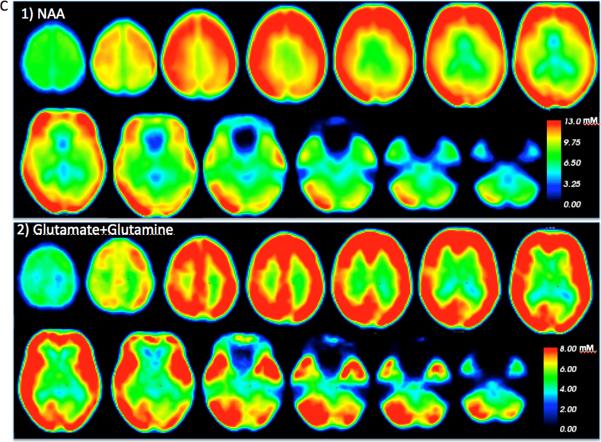
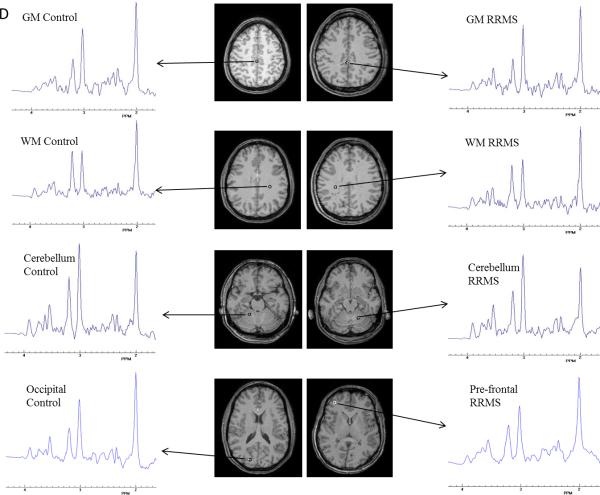
According to the pipeline described in Figure B, templates of each metabolite (NAA, Glx, Cho, m-Ins, and tCr) were built with MRSI data from all controls for each metabolite. Example of the qualities of the templates is shown in Figure C for NAA and Glx. Examples of spectra obtained from healthy control and RRMS patient for WM, cortical GM and cerebellum are shown in Figure D.
Figure C.
Whole brain quantitative 1) NAA and 2) Glx maps obtained from controls
Figure D.
Sample spectra from a healthy control and a RR patient from central WM, cortical GM, cerebellum, occipital and pre-frontal areas for highlight the spectra quality in the whole brain
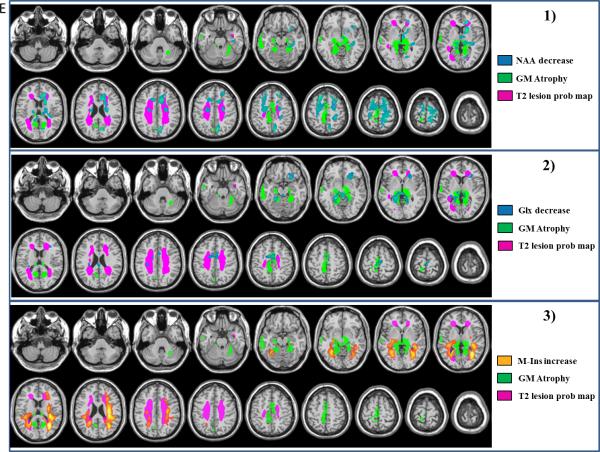
The probabilistic map of WM T2 lesions (yellow map in Figure E.1) showed macroscopic demyelinated areas in bilateral peri-ventricular regions, anterior and posterior corpus callosum, optic radiations and centrum semiovale areas in the group of patients with RRMS.
Figure E.
Topography of metabolic abnormalities, macroscopic WM lesions on T2 and GM atrophy of RRMS patients relative to controls. Maps of GM atrophy (green) and WM T2 lesion probability map (purple) are overlaid with maps of 1) NAA decreases, 2) Glx decreases (decreases represented in blue) and 3) m-Ins increases (increases represented inorange). (ANOVA p<0.005, FDR corrected at the cluster level p<0.05 with age and sex as confounding covariates).
GM atrophy (green map Figure E.1) was observed in RRMS patients relative to controls (ANOVA, p<0.005, FDR corrected at the cluster level p<0.05, age and sex as confounding covariates) within the median and superior temporal gyri, the pre and post central gyri, the anterior and posterior cingulate gyri and the median frontal gyrus. GM atrophy was also observed within bilateral thalami, hippocampal and parahippocampal gyri as well as in the anterior and medial cerebellum (Supl Table 1).
Topography of metabolic abnormalities in the group of RRMS patients
Statistical mapping of NAA decreases in RRMS showed significant clusters (p<0.005, FDR corrected p<0.05 at the cluster level) within the superior and medial frontal gyri, the pre- and post-central gyri, the anterior cingulate, the hippocampus, the putamen and the thalamus (Figure E.1 and Supl Table 2). NAA was also decreased within WM close to the frontal, paracentral and cingulate gyri, and inside the corpus callosum (Supl Table 2). From a quantitative point of view, in the mask defined by the significant clusters of NAA decreases in RRMS patients, NAA concentrations were 8.50±1.00mM for MS patients compared to 10.00±0.85mM for controls (−15%).
Statistical mapping of Glx decreases in RRMS (Figure E.2) showed significant clusters (p<0.005, FDR corrected p<0.05 at the cluster level) within the medial and inferior frontal gyri, the supplementary motor area, the medial cingulate, the caudate nucleus, the accumbens nuclei, the putamen, the thalamus and the parahippocampal gyrus. Decreases in Glx were also observed within the WM inside the frontal, the cingulate and the post-central brain areas and in the corpus callosum (Supl Table 3). In the mask defined by the significant clusters of Glx decreases in RRMS patients, Glx concentrations were 5.20±0.70mM for MS patients compared to 6.5±1.1mM for controls (−20%).
Statistical mapping of m-Ins increases showed significant clusters in RRMS (Figure E.3) within the WM in bilateral temporal, post-central, cingulate and frontal brain areas, inside the corpus callosum as well as within the left thalamus and the superior temporal gyrus (Supl Table 4).
At the same statistical threshold (p<0.005, FDR corrected p<0.05 at the cluster level) we did not find any significant increases or decreases for Cho and tCr between patients and controls. In the mask defined by the significant clusters of myo-Ins increases in RRMS patients, myo-Ins concentrations were 5.30±0.41mM for MS patients compared to 4.30±0.60mM for controls (+22%).
Co-localization of metabolic abnormalities, WM T2 lesions and GM atrophy
Decreases in NAA and Glx levels were observed in regions spared by atrophy except for thalamus, and pre- and post- central gyri where metabolite decreases were partially co-localized with GM atrophy maps (Figure E.1-2).
Increases of m-Ins were observed within WM T2 lesions especially in peri-ventricular lesional areas but were also present in the NAWM and GM areas (Figure E.3).
Correlations between topography of metabolic abnormalities and MSFC in RRMS
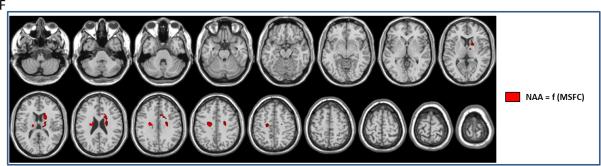
Statistical mapping analysis (multiple regression p<0.005, with age and sex as confounding covariates, masked with the metabolites patients abnormalities (Figure E.1-3)) looking at significant correlations between local NAA levels and MSFC showed significant clusters of positive correlation within bilateral thalamus, bilateral caudate nucleus, the right medial part of the corpus callosum and bilateral WM close to pre-central gyrus (Figure F and Supl Table 5).
Figure F.
Clusters of significant correlations between NAA levels and MSFC scores (SPM8, multiple regression p<0.005, k=5).
We did not found significant cluster (multiple regression p<0.005) between MSFC and the others metabolite abnormalities (Glx and m-Ins).
DISCUSSION
The present MR Spectroscopic imaging study reports the topography over the brain, from vertex to cerebellum, of the alteration of five metabolites (NAA, Glx, Cho, m-Ins and tCr) associated with a diffuse brain disease, namely RRMS. It also showed the predominant impact of NAA decrease within the motor and the prefrontal systems onto the clinical deficit assessed by the MSFC score in RRMS patients.
Using the technique presented above, we succeeded in providing a voxel by voxel comparison between patients and controls, permitting the assessment of the whole brain topography of metabolic disorders associated with MS processes. To this end, we have used a combination of an optimized acquisition protocol with a fast MRSI technique (3D-EPSI) (4), combining EPI and spectroscopy with an automated post-processing pipeline (MIDAS + SPM8) allowing us to obtain high quality whole brain 3D metabolite maps (5). Quality assurance of metabolite maps obtained in each orientation was assessed by Cramer Rao Bounds included in the MIDAS software (19). The first step used in order to improve reliability of large brain coverage with 3D-MRSI at high field was to correct for radiofrequency field inhomogeneities and B0 inhomogeneities responsible for spatial variations in contrast and sensitivity. To optimize the sampling of distal regions, a combination of two 3D-MRSI acquisitions acquired in two different orientation planes (i.e. anterior commissure-posterior commissure (AC-PC) and AC-PC +15°) benefited from the best spectrum quality of each acquisition (8). The choice of these orientations was dominated by the need to get sufficiently different brain coverage with the two MRSI volumes provided by a large difference in the orientation tilt but also to enable the coregistration step to be accurately performed to get the two volumes aligned according to the MNI template, required for statistical mapping analysis.
Increase in m-Ins has been related to glia proliferation (1, 20) present in WM T2 lesions, both in NAWM (21) and in cortical GM (22) as well as in the thalamus (11). In line with these previous reports derived from restricted sampling of predetermined brain regions, the whole brain approach using statistical mapping analysis without any a priori assumptions such as the location of metabolic abnormalities, showed in patients concordant significant increases in m-Ins mainly located in the white matter within the periventricular NAWM (10, 21), the demyelinated periventricular WM regions within bilateral temporal and frontal lobes (23), and within the left thalamus and the superior temporal gyrus (11).
These observations support the potential of whole brain 3D-EPSI to depict and locate encapsulated inflammatory processes accompanying MS and which are difficult to observe in SVS or 2D/3D MRSI.
Biochemical and histological studies performed on animal models of MS and/or human brain slices (24) have highlighted the large extent of neuronal damage driven by mitochondrial dysfunction leading to energy deficiency (25). These neurodegenerative processes have been observed in WM T2 lesions (26), NAWM (27), cortical GM (27), thalamus (28), hypothalamus, caudate nucleus (29) and cerebellum (30). MRS/MRSI enables characterization of the extent of mitochondrial neuronal dysfunction occurring as a result of energy depletion through the in vivo quantification of NAA levels, a metabolite synthetized specifically in neuronal mitochondria (25). In MS, decrease in NAA has been reported in WM T2 lesions (17) but also in the NAWM (31) and the subcortical GM (32). Likewise, we also observed decreased levels of Glutamate+Glutamine (Glx) that have been shown to play a role in neuro-astrocyte exchanges, in excitotoxicity (Glutamate) and in NH3 metabolism regulation (Glutamine) (33) in agreement with animal models (24) and human biochemical and histological studies (34).
The present statistical mapping analyses evidenced the preferential location of NAA and Glx alterations throughout the entire brain. Such metabolic changes were mainly located in the GM, more precisely within the limbic system (cingulate gyrus, hippocampus, parahippocampal gyrus), the motor system (central and pre-central gyri, putamen, supplementary motor area) and in the prefrontal areas (inferior, medial and superior frontal gyri). These results are in line with previous MR studies evidencing structural, functional or metabolic damage i) within the hippocampus, the entorhinal cortex and the cingulate gyrus (35), areas involved in in working memory, attention and speed of information processing and ii) within the pre-central gyrus and the prefrontal areas involved in motor control and executive functions (36). Interestingly, in most of these regions, decreases in NAA and Glx occurred without any regional GM atrophy or WM T2 lesions, suggesting early neuronal dysfunctions before appearance of any macroscopic lesions. Therefore as noted before, MRSI appears to be a powerful non-invasive method to evidence and monitor neuroaxonal dysfunction over the entire brain.
Unlike some previous literature (37), we did not observe, with short TE 3D-EPSI, Cho or tCr abnormalities in patients compared voxel by voxel to controls. This lack of Cho increase can be linked to the random distribution of Cho increase undetectable in this voxelwise between group analysis.
Using as a reference, the GM NAA concentrations (10mM) observed by Lecocq and coworkers in controls using the same 3D-EPSI sequence with B1 correction and absolute water concentration correction (8), we observed a drop of −15% in NAA concentrations and a drop −20% in Glx concentrations of GM of RRMS patients, as well as a 22% increase of myo-Ins within the WM of patients relative to controls. These variations were in the same range as those observed by Vingara and coworkers comparing controls and RRMS patients (31).
The MSFC score is a composite score testing both motor capacities of leg and arm, and cognitive performances. MSFC provides continuous values sensitive to low clinical deficits that could affect patients even at the early stage of MS. The voxel-wise multiple regression analysis looking at positive correlations between local NAA levels and MSFC, but not with Glx and m-Ins, demonstrated in patients significant clusters of positive correlation within pre-frontal WM regions likely to correspond to tracts linking frontal, parietal and occipital lobes and in thalamus. These regions are known to be involved in the working memory network, affected early in MS (38). Correlations between NAA levels and MSFC were also present in bilateral caudate nucleus and in corpus callosum, areas engaged in movement control and already described as being affected in MS (39, 40). Striking is the fact that, among the different pathophysiological processes characterized by this multiple metabolic mapping technique, the mitochondrial neuronal dysfunction within areas belonging to the working memory and the motor systems reflected by NAA decrease appeared as the predominant factor associated with clinical deficit. Glial proliferation observed within NAWM and demyelinated lesions (increased m-Ins), and neuro-axonal exchange dysfunction (decreased Glx) were showed to play here a limited role onto clinical deficit in this group of RRMS patients.
The main limitation of this study is the relatively limited sample size. Nevertheless all patients were in RR phase, with a low clinical deficit (median EDSS = 1 [0-4]), and in a stable stage of the disease distant from inflammatory episodes, and well-matched with the control group in terms of age and sex.
Although metabolite maps were not expressed in absolute concentrations, due to the requirement that this would greatly increase acquisition times, our approach provides normalized maps of individual metabolites using the interleaved water signal acquired during the 3D-EPSI sequence (8) as a reference that can be implemented across multiple sites (6). As a proof of concept, this study has validated statistical mapping analysis as applied to whole brain 3D-EPSI as an in vivo and non-invasive means to characterize the topography of metabolic disorders in neurological diseases and its relationship with clinical deficit. This opens a new era for the characterization of brain metabolic alteration associated with neurological/neuropsychiatric diseases, and to study its impact on function, especially through the topography of NAA decrease.
Supplementary Material
Acknowledgments
The authors thank Ben Ridley for his help in editing the English version of the manuscript.
Grant support: The first author is the recipient of a PhD Grant (CIFRE) supported by Siemens France and the French ‘Association Nationale Recherche et Technologie’ (ANRT).
The fourth author is the recipient of a NIH grant R01EB016064.
REFERENCES
- 1.Bertholdo D, Watcharakorn A, Castillo M. Brain proton magnetic resonance spectroscopy: introduction and overview. Neuroimaging Clin N Am. 2013;23:359–380. doi: 10.1016/j.nic.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 2.Gonen O, Catalaa I, Babb JS, et al. Total brain N-acetylaspartate: a new measure of disease load in MS. Neurology. 2000;54:15–19. doi: 10.1212/wnl.54.1.15. [DOI] [PubMed] [Google Scholar]
- 3.Truong T-K, Clymer BD, Chakeres DW, Schmalbrock P. Three-dimensional numerical simulations of susceptibility-induced magnetic field inhomogeneities in the human head. Magn Reson Imaging. 2002;20:759–770. doi: 10.1016/s0730-725x(02)00601-x. [DOI] [PubMed] [Google Scholar]
- 4.Ebel A, Soher BJ, Maudsley AA. Assessment of 3D proton MR echo-planar spectroscopic imaging using automated spectral analysis. Magn Reson Med. 2001;46:1072–1078. doi: 10.1002/mrm.1301. [DOI] [PubMed] [Google Scholar]
- 5.Maudsley AA, Darkazanli A, Alger JR, et al. Comprehensive processing, display and analysis for in vivo MR spectroscopic imaging. NMR Biomed. 2006;19:492–503. doi: 10.1002/nbm.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sabati M, Sheriff S, Gu M, et al. Multivendor implementation and comparison of volumetric whole-brain echo-planar MR spectroscopic imaging: Standardization of MRSI. Magn Reson Med. 2014 doi: 10.1002/mrm.25510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niddam DM, Tsai S-Y, Lin Y-R. Statistical mapping of metabolites in the medial wall of the brain: A proton echo planar spectroscopic imaging study: Statistical Mapping of Brain Metabolites. Hum Brain Mapp. 2015;36:852–861. doi: 10.1002/hbm.22669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lecocq A, Le Fur Y, Maudsley AA, et al. Whole-brain quantitative mapping of metabolites using short echo three-dimensional proton MRSI. J Magn Reson Imaging. 2015;42:280–289. doi: 10.1002/jmri.24809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davie CA, Hawkins CP, Barker GJ, et al. Serial proton magnetic resonance spectroscopy in acute multiple sclerosis lesions. Brain. 1994;117:49–58. doi: 10.1093/brain/117.1.49. [DOI] [PubMed] [Google Scholar]
- 10.Fernando KT, McLean MA, Chard DT, et al. Elevated white matter myo-inositol in clinically isolated syndromes suggestive of multiple sclerosis. Brain. 2004;127:1361–1369. doi: 10.1093/brain/awh153. [DOI] [PubMed] [Google Scholar]
- 11.Geurts JJG, Reuling IEW, Vrenken H, et al. MR spectroscopic evidence for thalamic and hippocampal, but not cortical, damage in multiple sclerosis. Magn Reson Med. 2006;55:478–483. doi: 10.1002/mrm.20792. [DOI] [PubMed] [Google Scholar]
- 12.Stromillo ML, Giorgio A, Rossi F, et al. Brain metabolic changes suggestive of axonal damage in radiologically isolated syndrome. Neurology. 2013;80:2090–2094. doi: 10.1212/WNL.0b013e318295d707. [DOI] [PubMed] [Google Scholar]
- 13.Tur C, Wheeler-Kingshott CA, Altmann DR, Miller DH, Thompson AJ, Ciccarelli O. Spatial variability and changes of metabolite concentrations in the cortico-spinal tract in multiple sclerosis using coronal CSI: Characterization Metabolic Changes Along the CST. Hum Brain Mapp. 2014;35:993–1003. doi: 10.1002/hbm.22229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benedetti B, Rovaris M, Rocca M, et al. In-vivo evidence for stable neuroaxonal damage in the brain of patients with benign multiple sclerosis. Mult Scler. 2009;15:789–794. doi: 10.1177/1352458509103714. [DOI] [PubMed] [Google Scholar]
- 15.Kirov, Tal A, Babb JS, Herbert J, Gonen O. Serial proton MR spectroscopy of gray and white matter in relapsing-remitting MS. Neurology. 2013;80:39–46. doi: 10.1212/WNL.0b013e31827b1a8c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muhlert N, Atzori M, De Vita E, et al. Memory in multiple sclerosis is linked to glutamate concentration in grey matter regions. J Neurol Neurosurg Psychiatry. 2014;85:833–839. doi: 10.1136/jnnp-2013-306662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hattingen E, Magerkurth J, Pilatus U, Hübers A, Wahl M, Ziemann U. Combined 1H and 31P spectroscopy provides new insights into the pathobiochemistry of brain damage in multiple sclerosis. NMR Biomed. 2011;24:536–546. doi: 10.1002/nbm.1621. [DOI] [PubMed] [Google Scholar]
- 18.Mathiesen HK, Tscherning T, Sorensen PS, et al. Multi-slice echo-planar spectroscopic MR imaging provides both global and local metabolite measures in multiple sclerosis. Magn Reson Med. 2005;53:750–759. doi: 10.1002/mrm.20407. [DOI] [PubMed] [Google Scholar]
- 19.Soher BJ, Young K, Govindaraju V, Maudsley AA. Automated spectral analysis III: application to in vivo proton MR spectroscopy and spectroscopic imaging. Magn Reson Med Off J Soc. 1998;40:822–831. doi: 10.1002/mrm.1910400607. [DOI] [PubMed] [Google Scholar]
- 20.Lassmann H. Mechanisms of white matter damage in multiple sclerosis: Mechanisms of White Matter Damage in Multiple Sclerosis. Glia. 2014;62:1816–1830. doi: 10.1002/glia.22597. [DOI] [PubMed] [Google Scholar]
- 21.Chard DT, Griffin CM, McLean MA, et al. Brain metabolite changes in cortical grey and normal-appearing white matter in clinically early relapsing–remitting multiple sclerosis. Brain. 2002;125:2342–2352. doi: 10.1093/brain/awf240. [DOI] [PubMed] [Google Scholar]
- 22.Kapeller P, Brex P, Chard D, et al. Quantitative H MRS imaging 14 years after presenting with a clinically isolated syndrome suggestive of multiple sclerosis. Mult Scler. 2002;8:207–210. doi: 10.1191/1352458502ms822oa. [DOI] [PubMed] [Google Scholar]
- 23.Vrenken H, Barkhof F, Uitdehaag BMJ, Castelijns JA, Polman CH, Pouwels PJW. MR spectroscopic evidence for glial increase but not for neuro-axonal damage in MS normal-appearing white matter. Magn Reson Med. 2005;53:256–266. doi: 10.1002/mrm.20366. [DOI] [PubMed] [Google Scholar]
- 24.Lassmann H. Axonal and neuronal pathology in multiple sclerosis: What have we learnt from animal models. Exp Neurol. 2010;225:2–8. doi: 10.1016/j.expneurol.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 25.Witte ME, Mahad DJ, Lassmann H, van Horssen J. Mitochondrial dysfunction contributes to neurodegeneration in multiple sclerosis. Trends Mol Med. 2014;20:179–187. doi: 10.1016/j.molmed.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 26.Lassmann H. The architecture of inflammatory demyelinating lesions: implications for studies on pathogenesis: Active lesions in multiple sclerosis. Neuropathol Appl Neurobiol. 2011;37:698–710. doi: 10.1111/j.1365-2990.2011.01189.x. [DOI] [PubMed] [Google Scholar]
- 27.Kutzelnigg A, Lucchinetti CF, Stadelmann C, et al. Cortical demyelination and diffuse white matter injury in multiple sclerosis. Brain. 2005;128:2705–2712. doi: 10.1093/brain/awh641. [DOI] [PubMed] [Google Scholar]
- 28.Minagar A, Barnett MH, Benedict RH, et al. The thalamus and multiple sclerosis Modern views on pathologic, imaging, and clinical aspects. Neurology. 2013;80:210–219. doi: 10.1212/WNL.0b013e31827b910b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haider L, Simeonidou C, Steinberger G, et al. Multiple sclerosis deep grey matter: the relation between demyelination, neurodegeneration, inflammation and iron. J Neurol Neurosurg Psychiatry. 2014;85:1386–1395. doi: 10.1136/jnnp-2014-307712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weier K, Banwell B, Cerasa A, et al. The Role of the Cerebellum in Multiple Sclerosis. Cerebellum. 2015;14:364–374. doi: 10.1007/s12311-014-0634-8. [DOI] [PubMed] [Google Scholar]
- 31.Vingara LK, Yu HJ, Wagshul ME, et al. Metabolomic approach to human brain spectroscopy identifies associations between clinical features and the frontal lobe metabolome in multiple sclerosis. NeuroImage. 2013;82:586–594. doi: 10.1016/j.neuroimage.2013.05.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Stefano N, Filippi M. MR Spectroscopy in Multiple Sclerosis. J Neuroimaging. 2007;17:31S–35S. doi: 10.1111/j.1552-6569.2007.00134.x. [DOI] [PubMed] [Google Scholar]
- 33.Rothstein JD, Martin L, Levey AI, et al. Localization of neuronal and glial glutamate transporters. Neuron. 1994;13:713–725. doi: 10.1016/0896-6273(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 34.Lassmann H. Multiple sclerosis: Lessons from molecular neuropathology. Exp Neurol. 2014;262:2–7. doi: 10.1016/j.expneurol.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 35.Audoin B, Zaaraoui W, Reuter F, et al. Atrophy mainly affects the limbic system and the deep grey matter at the first stage of multiple sclerosis. J Neurol Neurosurg Psychiatry. 2010;81:690–695. doi: 10.1136/jnnp.2009.188748. [DOI] [PubMed] [Google Scholar]
- 36.Calabrese M, Atzori M, Bernardi V, et al. Cortical atrophy is relevant in multiple sclerosis at clinical onset. J Neurol. 2007;254:1212–1220. doi: 10.1007/s00415-006-0503-6. [DOI] [PubMed] [Google Scholar]
- 37.Srinivasan R, Sailasuta N, Hurd R, Nelson S, Pelletier D. Evidence of elevated glutamate in multiple sclerosis using magnetic resonance spectroscopy at 3 T. Brain. 2005;128:1016–1025. doi: 10.1093/brain/awh467. [DOI] [PubMed] [Google Scholar]
- 38.Audoin B, Guye M, Reuter F, et al. Structure of WM bundles constituting the working memory system in early multiple sclerosis: a quantitative DTI tractography study. NeuroImage. 2007;36:1324–1330. doi: 10.1016/j.neuroimage.2007.04.038. [DOI] [PubMed] [Google Scholar]
- 39.Cavallari M, Ceccarelli A, Wang G-Y, et al. Microstructural Changes in the Striatum and Their Impact on Motor and Neuropsychological Performance in Patients with Multiple Sclerosis. PLoS ONE. 2014;9:e101199. doi: 10.1371/journal.pone.0101199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kern KC, Sarcona J, Montag M, Giesser BS, Sicotte NL. Corpus callosal diffusivity predicts motor impairment in relapsing–remitting multiple sclerosis: A TBSS and tractography study. NeuroImage. 2011;55:1169–1177. doi: 10.1016/j.neuroimage.2010.10.077. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.