Abstract
Macroalgae belonging to the genus Padina are known to produce antibacterial compounds that may inhibit growth of human- and animal pathogens. Hitherto, it was unclear whether this antibacterial activity is produced by the macroalga itself or by secondary metabolite producing epiphytic bacteria. Here we report antibacterial activities of epiphytic bacteria isolated from Padina pavonica (Peacocks tail) located on northern coast of Tunisia. Eighteen isolates were obtained in pure culture and tested for antimicrobial activities. Based on the 16S rRNA gene sequences the isolates were closely related to Proteobacteria (12 isolates; 2 Alpha- and 10 Gammaproteobacteria), Firmicutes (4 isolates) and Actinobacteria (2 isolates). The antimicrobial activity was assessed as inhibition of growth of 12 species of pathogenic bacteria (Aeromonas salmonicida, A. hydrophila, Enterobacter xiangfangensis, Enterococcus faecium, Escherichia coli, Micrococcus sp., Salmonella typhimurium, Staphylococcus aureus, Streptococcus sp., Vibrio alginoliticus, V. proteolyticus, V. vulnificus) and one pathogenic yeast (Candida albicans). Among the Firmicutes, isolate P8, which is closely related to Bacillus pumilus, displayed the largest spectrum of growth inhibition of the pathogenic bacteria tested. The results emphasize the potential use of P. pavonica associated antagonistic bacteria as producers of novel antibacterial compounds.
Keywords: Padina pavonica, Bacillus pumilus, antibiotics, epibionts, seaweed
Introduction
In aquatic environments, planktonic microbial communities live as free-living forms or attached to biotic or abiotic surfaces (Martin et al., 2014). In the marine environment, competition for space and nutrients is intense; the surfaces of marine eukaryotes such as seaweeds or invertebrates represent a suitable nutrient-rich habitat for microbial colonization and biofilm formation (Egan et al., 2008; Goecke et al., 2010). As the surfaces of theses eukaryotes are highly complex and differentiated, marine microbial biofilms should constitute a huge source of diversity, and the microbial communities forming them are expected to differ considerably in composition from pelagic microbial communities (Martin et al., 2014). Microbial community living on seaweed surfaces are highly complex, dynamic and consist of bacteria, fungi, diatoms, protozoa, spores and larvae of marine invertebrates (Lachnit et al., 2009, 2011; Goecke et al., 2010; Burke et al., 2011). Because especially bacteria interact with their host in multiple and complex ways, they constitute an interesting source of novel bioactive compounds with biotechnological potential (Martin et al., 2014). These bacteria may protect their host from harmful entities present in pelagic areas (Singh and Reddy, 2014) but can also produce compounds important for human and animal health (Kanagasabhapathy et al., 2006; Penesyan et al., 2010).
Bacteria may degrade algal polysaccharides such as for instance fucoidan (Nedashkovskaia et al., 2002) or alginates (Preston et al., 1986; Brown and Preston, 1991). Secondary metabolites are increasingly recognized for their importance for industrial and medical applications (Faulkner, 2000; Kelecom, 2002). Various novel compounds with antibiotic activity have already been identified from seaweed-associated bacteria (Chellaram et al., 2013; Horta et al., 2014; Martin et al., 2014). Several species of marine bacteria have been isolated from brown algae such as the spot-wounded fronds of Laminaria japonica (Sawabe et al., 2000), the surface of Fucus serratus (Johansen et al., 1999) and from rotten thallus of Fucus evanescens (Ivanova et al., 2004) and Undaria pinnatifida (Lee et al., 2006).
During summer, the brown alga Padina pavonica is abundantly present along the Tunisian coast. Despite its accessibility, nothing is known about the diversity and biological potential of its epibionts. Seaweeds of the genus Padina were previously reported to harbor epiphytic bacteria with variety of biological activities such as antibacterial activities of epiphytic bacteria isolated from P. arborescens, which was collected from Awaji Island, Japan (Kanagasabhapathy et al., 2006). Bacterial isolates from a close relative of P. pavonica, P. tetrastromatica collected from southeastern India were previously studied for their antibacterial activities against a panel of human pathogens (Chellaram et al., 2013; Ravisankar et al., 2013). However, since the geographic location is amongst the factors that impact on a bacterial community's diversity, composition and secondary metabolites production, the study of the epiphytic bacteria of P. pavonica, which is abundant at the Tunisian coast, was considered to be promising for finding novel antibacterial compounds.
The present study aimed at assessing the culturable bacterial diversity of the community attached to P. pavonica, by isolating and identifying the associated strains, and by testing their potential antimicrobial activity using a variety of pathogens as target organisms.
Materials and methods
Sampling
The seaweed P. pavonica (Class: Phaeophyceae, Order: Dictyotales, Family: Dictyotaceae) was collected during two seasons: winter and summer of 2007 from Cap Zebib (37° 16.2′N, 10° 3.6′E) at the northern coast of Tunisia. The algae were transferred in dark sterile plastic bags filled with water from the same location.
Isolation of epiphytic bacteria
Seaweed samples were washed three times with autoclaved seawater in order to remove free-living and loosely attached bacteria (Burgess et al., 2003). Subsequently, the firmly attached epiphytic bacteria were extracted by vortexing 10 g of algal biomass in 90 ml autoclaved seawater for 6 min. Bacteria were isolated by serial dilution to 10−3 using autoclaved seawater. From each dilution 100 μl was spread-plated in triplicate on marine agar plates (MA: Pronadisa Laboratories, CONDA). The plates were incubated at 20°C until colonies appeared or at least for 7 days (Lemos et al., 1985). Visually distinct bacterial colonies were selected and further plated on MA until clonal cultures were obtained. The pure cultures were stored at −80°C in marine broth (Pronadisa Laboratories, CONDA) supplemented with 20% glycerol (v/v).
Extraction of DNA and PCR conditions
Single colonies from the agar plates were suspended in sterile Milli-Q water and used as template in the PCR reactions using the bacterial 16S rRNA gene primer B8F and the universal primer U1492R (Lane, 1991; Table 1). PCR reactions were performed using a Gene Biometra T1 DNA Thermal Cycler (Perkin-Elmer Co. Norwalk-CT, USA) in 25 μl (final volume) reaction mixtures containing 0.1 μl of Hot Star Taq DNA polymerase (Qiagen), each primer at a final concentration of 10 pmol μl−1, each deoxynucleoside triphosphate at a concentration of 200 μM, 1.25 μl of 100% DMSO (Dimethylsulfoxide), 2.5 μl of BSA (Bovine Serum Albumin) at 0.2 mg ml−1 final concentration, 2.5 μl of PCR buffer (incl. MgCl2) and 1 μl of DNA template. The PCR protocol consisted of 40 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s and extension at 72°C for 1 min 50 s. The cycles were preceded by 15 min of denaturation at 94°C and ended with a final extension for 7 min at 72°C. Negative controls contained all components of the PCR mixture except the DNA template.
Table 1.
Primers used for PCR and DNA sequencing.
| Primer | Sequence |
|---|---|
| B8F | AGAGTTTGATCMTGGCTCAG |
| U1492R | GGTTACCTTGTTACGACTT |
| C5 | AGAGTTTGATCCTGGCTCAGG |
| C26 | GGGCGGTGTGTACAAGG |
| C72 | CCGGAATIATTGGGCGTAA |
| C112 | CTCGTTGCGGGACTTAACCC |
Analysis of PCR products
PCR products (2 μl) were analyzed by agarose gel electrophoresis (1% agarose, 1 × TAE buffer containing 40 mM Tris acetate and 1 mM EDTA, pH 8). Electrophoresis was performed at 100 V for 45 min. The gels were stained for 45 min with SYBR Gold nucleic acid gel stain (Invitrogen Corp.) and photographed under UV illumination.
DNA sequencing
Approximately 100 ng DNA template was sequenced using the BigDye Terminator chemistry (Big Dye Terminator v3.1 Cycle Sequencing Kit, Applied Biosystems), according to the manufacturer's instructions. The sequence products were analyzed on an ABI 377 DNA sequencing system (Applied Biosystem) using the primers C5 (Gao et al., 2006), C26 (Ciancio et al., 2000), C72 (Gao et al., 2006), and C112 (Hauben et al., 1997; Table 1).
Phylogenetic analysis
The closest phylogenetic relatives of each isolate were identified by comparison of the 16S rRNA gene sequence to the National Center for Biotechnology Information (NCBI) GenBank database using the Basic Local Alignment Search Tool (BLAST) analysis tools (http://www.ncbi.nlm.nih.gov/BLAST). Phylogenetic analysis of the sequences of the isolates and those of their selected top five best hit blast close relatives was performed using the Neighbor Joining method as available in MEGA version 4.1 software (Tamura et al., 2007).
Nucleotide sequence accession numbers
Partial sequences of the 16S rRNA genes were submitted to GenBank and received the following accession numbers: P1 (FN652906), P2 (FN652907), P3 (FN652908), P4 (FN652909), P5 (FN652910), P6 (FN652911), P7 (FN652912), P8 (FN652913), P9 (FR695065), P11 (FR695066), P12 (FR695067), P14 (FR695068), P17 (FR695069), P18 (FR695072), P19 (FR695074), P20 (FR695070), P26 (FR695071), and P27 (FR695073).
Antimicrobial activity
Microorganisms used for antimicrobial tests
Bacterial type strains (18 strains) used for the evaluation of antimicrobial activity of all isolated epiphytes (and also for alga extracts) were: Aeromonas salmonicida LMG3780, A. hydrophila B3 (RVAU-Denmark), Enterococcus faecalis ATCC 29212, Escherichia coli O126-B16 (ATTC 14948), E. coli ATCC 25922, E. coli ATCC 8739, Micrococcus sp. (Pasteur Institute, Tunis), Pseudomonas cepacia (INSTM, Tunis), P. fluorescens AH2 (Danish Institute for Fisheries Research, Denmark), P. aeruginosa ATCC 27853, Salmonella typhimurium C52 (Laboratoire Hydrobiologie Marine et Continentale, Université de Montpellier II, France), Staphylococcus aureus (Pasteur Institute, Tunis), S. aureus ATCC 25923, S. aureus ATCC 6538, Streptococcus sp. (Pasteur Institute, Tunis), Vibrio tapetis CECT4600 (Department of Microbiology and Parasitology, University of Santiago de Compostela, Spain), V. anguillarum ATCC12964T, V. alginolyticus ATCC 17749T. In addition the yeast Candida albicans ATCC 10231 was used for the tests.
The selected most active isolate against these 19 pathogens (the Firmicutes isolate P8) was also tested for antagonistic activity against nine Vibrio spp. type strains (pathogens well represented in marine ecosystems): V. fluvialis ATCC 33809T, V. (Aliivibrio) logei LMG14011, V. parahaemolyticus ATCC 17802 T, V. pectenicida CIP105190T, V. pectenicida LMG19642T, V. proteolyticus ATCC15338T, V. salmonicida LMG3780T, V. splendidus ATCC33125T, V. vulnificus ATCC27562T, and few other enteric bacteria species: Enterobacter xiangfangensis (CCY15226), Enterococcus durans (CCY15220), Enterococcus faecium (CCY15234), Proteus mirabilis (CCY15230), and Morganella morganii (CCY15221), originally obtained from the IFREMER Laboratory of Microbiology of Invertebrates (Brest, France).
[ATCC: American Type Culture Collection; CCY: Culture Collection Yerseke; LMG: Laboratory for Microbiology Gent; RVAU: Royal Veterinary and Agricultural University (now University of Copenhagen, Faculty of Life Sciences, Denmark); INSTM: National Institute of Sciences and Marine Technologies (Salammbô, Tunisia)].
Antimicrobial assay
Bacterial isolates were screened for their antimicrobial activity against human and fish pathogenic bacteria. The following two methods were used. The drop test assay on Trypto-Casein-Soy agar (TSA, BIO RAD) plates containing 20 g l−1 NaCl was carried out as described by James et al. (1996) and Rao et al. (2005) with slight modifications. Briefly, drops of cell suspension of an overnight culture were spotted onto the agar plates containing a confluent lawn of the target strain (dried for 30 min at 30°C) and then incubated at 30°C. The overlay assay was carried out as described by Wiese et al. (2009) with some modifications. Briefly, drops (10 μl) of an overnight culture of the isolates were spotted onto TSA plates supplemented with 20 g l−1 NaCl and incubated for 24 h at 30°C before they were covered with an overlay containing the target strains in soft agar [3 g l−1 Tryptone Soy Broth- (TSB-BIO RAD, 9 g l−1 agar (PRS, Panreac), 20 g l−1 NaCl, pH 7.2]. The suspension with the target strains contained ~107 cells ml−1. Growth inhibition was evaluated after incubation (24 h at 30°C) by measuring the zone of inhibition around the spots with the test organism.
The antibacterial assay of algal crude extracts against its isolates was evaluated by disk diffusion method. Briefly, 500 μg of the total alga crude extract dissolved in dichloromethane and dichloromethane/methanol solvents (10 μl) was applied to sterile filter paper discs (6 mm). After solvent evaporation, the discs were placed on TSA plates, inoculated with an 18 h cultured associated strain (106 bacteria. ml−1) in TSB containing 20 g. l−1 NaCl. As control, a disc loaded with solvent was simultaneously prepared. Plates were incubated overnight at 30°C. The diameter (in millimeter) of growth inhibition halo was measured after 24 h incubation considering the disc diameter. Assays were carried out in triplicate. These extracts were also tested in our previous work (Ismail-Ben Ali et al., 2010a) against the 18 indicator bacterial pathogens used in this study and cited above in material and methods.
Preparing algal extracts
Fresh algae were dried during 15 days at room temperature in the dark and powdered. About 20 g of dried algae were extracted consecutively with two organic solvents with increasing polarity: dichloromethane (D) and dichloromethane/methanol (D/M) (1:1 v/v). Each extraction was carried out three times by maceration for 24 h at room temperature. The extracts were pooled, filtered, and concentrated under reduced pressure in a rotary evaporator. Algal extracts were stored at −20°C until use.
Test of antibiotic sensitivity of isolated strains
The sensitivity to various antibiotics was investigated on Muller Hinton agar (MH, BIO RAD) plates by the disc diffusion method (Barry and Thornsberry, 1980), using the following antimicrobial compounds (BIO RAD, France), amounts are given per disc: streptomycin (500 μg), amoxicillin (25 μg), tobramycin (10 μg), nalidixic acid (30 μg) oleandomycin (15 μg), cefoxitin (30 μg), furans (300 μg), trimethoprim sulfamid (25 μg), penicillin G (6 μg), chloramphenicol (30 μg), tetracyclin (30 μg), oxacillin (1 μg), ceftriaxon (30 μg). A strain was considered susceptible to an antimicrobial compound if any growth inhibition zone was observed around the disc. Interpretation was given according the guidelines of the French Society for Microbiology (Soussy, 2008).
Extracellular activity test
Preparation of the cell-free supernatant
Ten ml of culture of the isolates P9, P11, P12, and P20 grown for 48 h at 30°C in TSB was centrifuged at 5000 rpm for 15 min and the supernatants were transferred to new tubes. Activities of the cell-free supernatants were evaluated by the drop method on TSA plates previously inoculated with Micrococcus sp., A. salmonicida LMG3780T or S. aureus. The same protocol was used for isolate P8 using cultures taken after 24, 48, and 72 h of growth.
Effects of enzymatic and heat treatments on antibacterial activity
Protease effect
The enzymatic effect on the antibacterial activity was evaluated on cell-free supernatants of the active isolates. Samples of 1 ml were treated at 37°C for 2 h with 1 mg ml−1 final concentration of trypsin and pepsin. The residual activities of the samples were assayed by the drop method.
Heat effect
To analyze thermal stability, supernatants were heated to 60°C for 30 min, 80°C for 10 min, and 100°C for 5 min and then cooled to room temperature before being tested for antibacterial activity.
Results
Alga epiphytic bacteria identification
Epiphytic bacteria were extracted from P. pavonica thallus and plated on MA. A total number of 26 isolates was obtained after the first round of cultivation of algal samples. Colony morphology dereplication resulted in 18 bacterial isolates that were obtained in pure culture. Analysis of the 16S rRNA gene sequences showed that all isolates were affiliated to three major groups within the bacterial domain, Alpha- and Gammaproteobacteria, Firmicutes and Actinobacteria (Table 2). Most of the isolates belongs to the proteobacterial group (twelve isolates: P1–P5, P9, P11, P12, P14, P17, P20, and P26). Two isolates belong to Alphaproteobacteria: Paracoccus (P1) and Devosia (P2) and ten isolates affiliated with the Gammaproteobacteria which were closely related to Pseudoalteromonas sp. (P3, P12, and P14), Pseudomonas sp. (P9, P11 and P20), Pseudomonas putida (P5), Acinetobacter (P17), Erwinia billingiae (P26) and Vibrio sp. (P4). The phylum of the Gram-positive Firmicutes was represented by four isolates closely related to Bacillus pumilus (P8), Planomicrobium glaciei (P18), and Staphylococcus sp. (P7 and P27). Isolate P27 is related to Staphylococcus but with less than 90% identity. Two isolates (P6 and P19) were closely related to the genus Brevibacterium of the phylum Actinobacteria.
Table 2.
Identification of 18 bacterial strains associated with Padina pavonica based on 16S rRNA gene sequence identity obtained by BLAST analysis.
| Isolate | Accession number | Best hit | Identity (%) | Phylum |
|---|---|---|---|---|
| P1 | FN652906 | Paracoccus sp. | 98 | Alphaproteobacteria |
| P2 | FN652907 | Devosia sp. | 99 | Alphaproteobacteria |
| P3 | FN652908 | Pseudoalteromonas sp. | 99 | Gammaproteobacteria |
| P4 | FN652909 | Vibrio sp. | 96 | Gammaproteobacteria |
| P5 | FN652910 | Pseudomonas putida | 98 | Gammaproteobacteria |
| P6 | FN652911 | Brevibacterium iodinum | 96 | Actinobacteria |
| P7 | FN652912 | Staphylococcus sp. | 97 | Firmicutes |
| P8 | FN652913 | Bacillus pumilus | 99 | Firmicutes |
| P9 | FR695065 | Pseudomonas sp. | 93 | Gammaproteobacteria |
| P11 | FR695066 | Pseudomonas sp. | 95 | Gammaproteobacteria |
| P12 | FR695067 | Pseudoalteromonas sp. | 100 | Gammaproteobacteria |
| P14 | FR695068 | Pseudoalteromonas sp. | 94 | Gammaproteobacteria |
| P17 | FR695069 | Acinetobacter sp. | 95 | Gammaproteobacteria |
| P18 | FR695072 | Planomicrobium glaciei | 100 | Firmicutes |
| P19 | FR695074 | Brevibacterium sp. | 98 | Actinobacteria |
| P20 | FR695070 | Pseudomonas sp. | 100 | Gammaproteobacteria |
| P26 | FR695071 | Erwinia billingiae | 99 | Gammaproteobacteria |
| P27 | FR695073 | Staphylococcus sp. | 88 | Firmicutes |
Host and epibionts affinity
To verify the close relationship between host and isolated bacteria, antibacterial activity of P. pavonica organic crude extracts (polar and non-polar) were tested against the 18 isolates. The results revealed that extracts of the alga harvested in summer exhibited a weak activity (1 mm diameter) against three of the 18 isolates, P6, P7, and P8. While the same extracts had a pronounced inhibitory activity against human and animal pathogens indicator bacteria (9 amongst 18 were inhibited) (Ismail-Ben Ali et al., 2010a; Figure 4).
Epiphytic bacteria potential activity
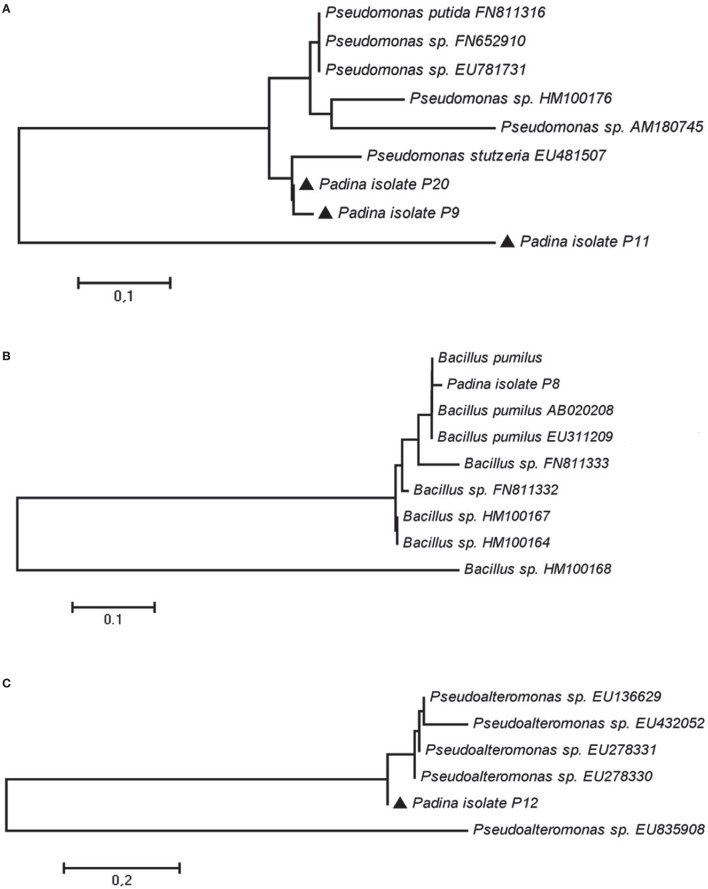
The 18 isolates were tested for their antimicrobial activities using two different methods and a number of selected pathogens. Five isolates: P8, P9, P11, P12, and P20 showed antibacterial activities against a broad range of pathogens (Table 3). Phylogenetic analysis of the 16S rRNA gene sequences of these active isolates was performed using the Neighbor Joining method (Figure 1).
Table 3.
Antibacterial and antifungal activities of isolate P8, P9, P11, P12, and P20 against pathogen bacteria.
| Test isolates | P8 | P9 | P11 | P12 | P20 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| D | O | D | O | D | O | D | O | D | O | |
| E. coli O126B16 ATTC 14948 | + | ++ | − | − | − | − | − | − | − | − |
| E. coli ATCC 25922 | ++ | +++ | − | − | − | − | − | − | − | − |
| E. coli ATCC 8739 | ++ | +++ | − | − | − | − | − | − | − | − |
| V. tapetis CECT4600 | + | + | + | ++ | + | ++ | + | ++ | + | ++ |
| V. anguillarum | − | − | − | − | − | − | − | − | − | − |
| V. alginoliticus | + | +++ | − | − | − | − | − | − | − | − |
| P. cepacia (INSTM) | − | − | − | − | − | − | − | − | − | − |
| P. fluorescens AH2 | + | ++ | − | − | − | − | − | − | − | − |
| P. aeruginosa ATCC 27853 | − | − | − | − | − | − | − | − | − | − |
| A. salmonicida LMG3780 | ++ | ++++ | − | − | − | − | − | − | − | − |
| A. hydrophila B3 | + | +++ | + | ++ | + | + | + | + | + | + |
| S. typhymurium C52 | + | +++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | +++ |
| Streptococcus sp. (Pasteur Inst) | + | +++ | + | + | + | + | + | + | + | ++ |
| S. aureus (Pasteur Inst) | +++ | ++++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| S. aureus ATCC 25923 | + | ++ | + | ++ | + | ++ | + | ++ | + | ++ |
| S. aureus ATCC 6538 | ++ | ++++ | + | + | + | + | + | + | + | + |
| E. faecalis ATCC 29212 | + | ++ | + | + | + | + | + | + | + | + |
| Micrococcus sp.(Pasteur Inst) | ++ | +++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | +++ |
| C. albicans ATCC 10231 | ++ | +++ | − | − | − | − | − | − | − | − |
D, Drop method; O, overlay method; Inhibition zones > to 25 mm were declared as very strong (++++), Inhibition zones from 15 to 25 mm as strong (+++), from 7 to 15 as moderate (++) and < 7 as weak activities (+), no activity (−).
Figure 1.
Neighbor-joining tree based on 16S rRNA gene sequences derived from P. pavonica isolates and top five blast best hit sequences identified via GenBank search. (A) P9, P11, and P20 isolates and Pseudomonas genus (B) P8 isolate and Bacillus genus (C) P12 isolate and Pseudoalteromonas genus.
As shown in Table 3, isolates P9, P11, P12, and P20 had the same antibacterial spectrum and were effective against nine indicator strains tested. The antibiotic properties were most effective against the pathogens S. aureus, S. typhymurium, and Micrococcus sp. Growth of the yeast C. albicans was not inhibited by these isolates. Isolate P8 (related to Bacillus pumilus) displayed the strongest antimicrobial activity and was effective against 15 tested bacteria and the yeast C. albicans. A weak inhibition of P8 was observed against V. tapetis and no inhibition of V. anguillarum, P. cepacia, and P. aeruginosa was observed.
Inhibition zones depended on the method applied. Generally larger inhibition zones were observed when using the overlay assays compared to the drop assay (Table 3). The strongest effect was recorded for isolate P8 against S. aureus (inhibition zones varied from 20 to 45 mm; Figure 2). On the basis of its strong activity, isolate P8 was selected to test its antagonistic activity against nine Vibrio species, which are well known marine (fish, shellfish) pathogens. The overlay method was chosen for this test, showing a higher effectiveness when compared to the drop method. As shown in Table 4, two strains of Vibrio species tested were inhibited (V. proteolyticus and V. vulnificus) and furthermore E. xiangfangensis and E. faecium were inhibited as well.
Figure 2.
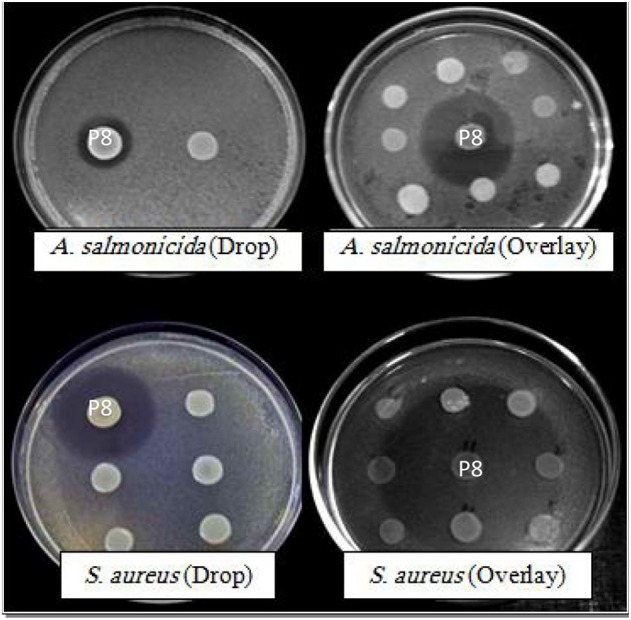
Plates testing antimicrobial activity of strain P8 isolated from P. pavonica by using the drop method and the overlay assay against A. salmonicida and Staphylococcus aureus.
Table 4.
Antagonistic activity of isolate P8 against sensitive Vibrio spp. and other bacteria using the overlay method (Wiese et al., 2009).
| Strain | Inhibition |
|---|---|
| V. proteolyticus | +++ |
| V. vulnificus ATCC 27562 T | + |
| E. xiangfangensis | ++ |
| E. faecium | +++ |
Inhibition zones from 15 to 25 mm were considered strong (+++), from 7 to 15 moderate (++) and < 7 weak activities (+).
Test of antibiotic sensitivity of isolated strains
The five isolates with clear antimicrobial activity were further characterized. Their own sensitivity against various antibiotics was investigated. Isolate P8 showed resistance to cefoxitin, oxacillin, and ceftriaxon. P9, P11, and P20 were resistant to amoxicillin, cefoxitin, streptomycin, tetracycline, penicillin G, oxacillin, furans, and ceftriaxone. P11 was also resistant to chloramphenicol. P12 was resistant to amoxicillin, cefoxitin, streptomycin, tobramycin, penicillin G and oxacillin.
Effects of enzymatic and heat treatments on antibacterial activity
The cell-free supernatants of cultures of the five active isolates were tested for presence of antimicrobial activity against Micrococcus sp. and S. aureus (the most sensitive species retained). Supernatants of cultures of the 5 isolates inhibited growth of both test strains (Table 5). The effect of incubation time on the production of active compounds was tested in isolate P8 after 24, 48, and 72 h of incubation. Antibacterial activity was detected for all three incubation periods, with the strongest activity after 48 h incubation, compared by the activity detected after 24 and 72 h.
Table 5.
Effects of enzymatic and heat treatments on antibacterial activity of Padina pavonica active isolates supernatant.
| Micrococcus sp. | S. aureus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S | T | P | T1 | T2 | T3 | S | T | P | T1 | T2 | T3 | |
| P8 | + | + | + | + | + | − | + | + | + | + | + | − |
| P9 | + | + | + | − | − | − | + | + | + | − | − | − |
| P11 | + | + | + | − | − | − | + | + | + | − | − | − |
| P12 | + | + | + | − | − | − | + | + | + | − | − | − |
| P20 | + | + | + | − | − | − | + | + | + | − | − | − |
S, supernatant; T, supernatant treated by trypsin; P, supernatant treated by pepsin; T1, supernatant heated at 60°C; T2, supernatant heated at 80°C; T3, supernatant heated at 100°C. +, active; −, not active.
Results of the sensitivity tests to proteases and heat treatment (Table 5) showed that bioactive substances produced by isolates P8, P9, P11, P12, and P20 were insensitive to trypsin and pepsin treatments. However, the bioactive substances were thermolabile. Antibacterial activity for isolates P9, P11, P12, and P20, was totally lost after being heated at 60°C for 30 min.
The active substance produced by isolate P8 was more resistant to increased temperatures since activity was not completely lost at 60°C but decreased with increasing temperature. About 50% of the diffusion zone remained after incubation at 60°C and 20% at 80°C, while the activity was totally lost after a treatment for 5 min at 100°C (Figure 3).
Figure 3.
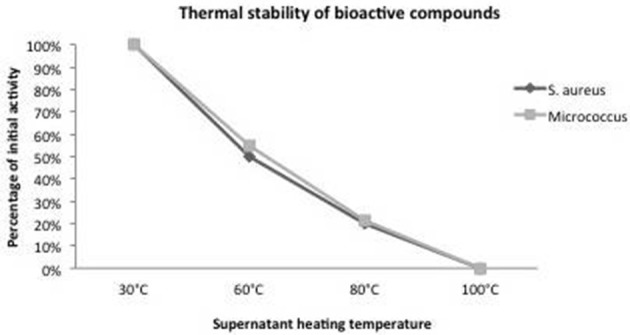
Influence of supernatant heating treatment on the P8 antibacterial activity against Staphylococcus aureus and Micrococcus sp.
Discussion
The brown alga P. pavonica is widespread along the Tunisian coastline, especially in the northern part (Cap Zebib). This alga can be found up to 10 m depth with a total estimated biomass of 112.5 tons of fresh matter (25 tons dry matter; Ksouri et al., 2008). A previous study by Ktari and Guyot (1999) highlighted the cytotoxic activity of a dichloromethane extract of P. pavonica against KERATIN-forming tumor cell line HeLa (KB cells). Besides, Omezzine et al. (2009), reported antifungal potential of this alga collected from Tunisian coast.
Several studies reported interactions between brown algae and their epibionts (Lee et al., 2006). Some brown algae-associated bacteria may degrade algal polysaccharides, such as fucoidan (Nedashkovskaia et al., 2002), or alginates (Preston et al., 1986; Brown and Preston, 1991). Brown algae may produce biologically active compounds that are capable of killing bacteria (bactericidal) or inhibiting bacterial growth (bacteriostatic; Nagayama et al., 2002).
In this study, a number of 26 isolates were firstly extracted from P. pavonica thallus at a first round cultivation on MA, then restricted to 18 after subculturing to obtain pure morphological different colonies since algal samples were subjected to a harsh pre-treatment in order to be sure that only attached bacteria will be isolated. Also in other previous study by Kanagasabhapathy et al. (2006), numbers of culturable isolates from some brown algae collected from Awaji Island Japan; were 10 for Padina arborescens and 21, 19, 17, 15, 12, 9, 8, and 5 for Ecklonia cava, Colpemenia sinusa, Sargassum serratifolium, S. fusiforme, Petalonia fascia, Scytosiphon lomentaria, S. filicinum, and Undaria pinnatifida respectively.
In other previous study by Albakosh et al. (2016) on the brown alga Splachnidium rugosum, a number of 41 isolates were obtained using 4 different medium; MA, nutrient sea water agar, nutrient agar and thiosulfate-citrate-bile-salts-sucrose agar. In this same study by Albakosh et al. (2016), initially 29 isolates were obtained on MA medium and after identification, the number was narrowed down to 22 unique isolates. From our results and these previous work and observations it seems that MA is a suitable medium to isolates algal epiphytic bacteria as compared to other medium. However, the number of culturable epiphytic bacteria will increase when using different culturing media.
After characterization, Gram-negative Alpha- and Gammaproteobacteria and Gram-positive Firmicutes and Actinobacteria were identified from P. pavonica. Most of the isolates belonged to Gram-negative bacteria, as was the case for epibionts of seaweeds (Lakshmanaperumalsamy and Purushothaman, 1982; Albakosh et al., 2016). Among the different P. pavonica isolates, isolates related to the genera Paracoccus, Acinetobacter, and Staphylococcus were reported in previous studies isolated from marine environments and seaweeds (Gallardo et al., 2004; Schulze et al., 2006; Li, 2009; Zeng et al., 2010). Bacteria of the genera Planomicrobium were previously isolated from seawater and sediment and have algicidal effect, affecting harmful algal blooms (Giudice et al., 2007; Zeng et al., 2010). The actinobacterial genus Brevibacterium (P6, P19) is found in marine and terrestrial environments (Jones and Keddie, 1953; Collins, 1992) and were previously also isolated from degraded thallus of the brown alga Fucus evanescens (Ivanova et al., 2004). Two of the P. pavonica associated isolates reported in this study, P2 and P26, closely related to the Proteobacteria: Devosia and Erwinia, respectively, were previously not known from the surfaces of marine seaweeds. Species of the genus Devosia have been isolated from beach sediment (Lee, 2007), from freshwater legume Neptunia natans (Rivas et al., 2003), as endosymbionts of the marine ciliate Euplotes magnicirratus (Vannini et al., 2004), from a nitrifying inoculum (Vanparys et al., 2005) and from soil and greenhouse soil (Yoo et al., 2006; Yoon et al., 2007). Bacteria of the genus Erwinia are known as plant pathogens (Paulert et al., 2007) and common inhabitants of the rhizosphere of cultivated crops and weeds (Burr and Schroth, 1977). Besides, in this study we notice that P9, P11, P14, P17, and P27 isolates may be considered as potentially novel species since they were related with less than 95% identity to Pseudomonas sp. (P9, P11), Pseudoalteromonas sp. (P14), and Acinetobacter sp. (P17). Isolate P27 may be a potentially novel genus, since it is less than 90% identical with its closest relative Staphylococcus. These isolates may be of biotechnological interest and need further characterization.
P. pavonica harbors several culturable epiphytic bacteria on its surface and our harsh isolation procedure indicates that these epibionts are intimately associated with their host. We tested the resistance of the isolates to crude extracts of the P. pavonica. Only extracts from P. pavonica harvested in summer revealed a low inhibitory effect toward three of the isolates. Organic crude extracts of P. pavonica were toxic against human and fish pathogens such as V. tapetis, V. anguillarum, V. alginoliticus, Ps. cepacia, A. salmonicida, A. hydrophila, E. faecalis ATCC 29212, and S. aureus (Ismail-Ben Ali et al., 2010a). In Figure 4 a diagram is depicted showing the number of pathogenic bacteria inhibited by P. pavonica extracts, which represent 50% of the total number of pathogens tested (Ismail-Ben Ali et al., 2010a), compared to P. pavonica isolates that are inhibited by their host extracts, which represent only 16.6% of the total isolates tested. Chbani et al. (2011) and Zerrin et al. (2011) also reported toxicity of P. pavonica against pathogenic indicator bacteria.
Figure 4.
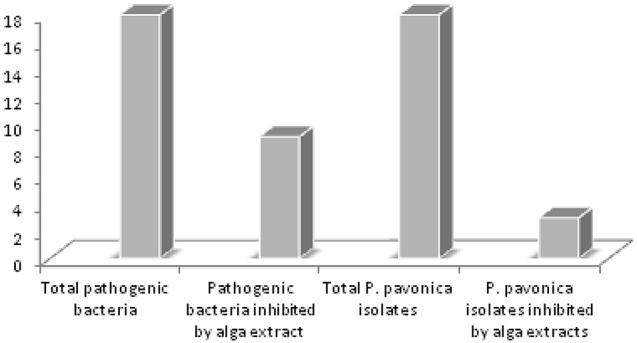
Comparison between P. pavonica crude extracts inhibition spectrum against sensitive alga isolates and pathogenic human and fish sensitive bacteria (18 pathogens).
In this study 5 of the 18 isolates possessed antimicrobial activities. In our previous work we isolated 10 bacteria from the green alga U. intestinalis with two strains showing antimicrobial potential and 19 from J. rubens with 7 strains displaying antibacterial activity against pathogenic indicator bacteria (Ismail-Ben Ali et al., 2010b, 2012).
Many Pseudoalteromonas species are associated with eukaryotic hosts in the marine environment (Holmström and Kjelleberg, 1999). Such species have been isolated from a variety of animals, such as mussels, tunicates and sponges, as well as from a range of marine plants (Holmström and Kjelleberg, 1999) and from the red alga Jania rubens collected from Tunisian coast (Ismail-Ben Ali et al., 2012). These Pseudoalteromonas species displayed different extracellular biological activities ranging from antibacterial, agarolytic, antiviral, antifouling, to algicidal activities (Sawabe et al., 1998; Holmström and Kjelleberg, 1999; Yuexin et al., 2010). Pseudoalteromonads may play an important role in the control of harmful algal blooms within the marine environment by the production of an extracellular inhibitor that causes rapid lyses of algal species within the genera Chattonella, Gymnodinium, and Heterosigma (Lovejoy et al., 1998). Pseudomonas, Pseudoalteromonas and Vibrio species have also been isolated from the brown alga Laminaria saccharina and possess antibacterial substances (Wiese et al., 2009). Previously, we isolated Pseudoalteromonas, Pseudomonas and B. pumilus species from the surface of the red alga J. rubens collected from the Tunisian coast. These isolates showed a varying spectrum of antimicrobial activity (Ismail-Ben Ali et al., 2012). Vibrio sp., Pseudomonas sp. and B. pumilus are known as probiotic bacteria used in aquaculture (Hill et al., 2009). Epiphytic Vibrio strains produced various antibacterial compounds (Oclarit et al., 1994). However, in our study 13 strains of bacteria that were isolated from the surface of P. pavonica did not show any antibacterial activity. These isolates may have another ecological function. For instance, they may have a protective role by releasing compounds into the surrounding seawater that would help preventing extensive fouling of the surface (Burgess et al., 1999; Kanagasabhapathy et al., 2008).
Bacteria of the genus Bacillus, to which our isolate strain P8 belongs, have been reported to be associated with various marine organisms including brown algae (Thakur and Anil, 2000). Lee et al. (2006) described B. psychrodurans as a representative strain associated with the brown alga Undaria pinnatifida. B. pumilus was isolated by Wiese et al. (2009) from the surfaces of the brown alga L. saccharina as well as from the surface of other brown algae such as Sargassum fusiforme, Colpomenia sinuosa, and Ecklonia cava (Kanagasabhapathy et al., 2006). Similarly, Parvathi et al. (2009) isolated B. pumilus strains from sediment, crab, fish, starfish and oyster. In the Table 6 the main studies on B. pumilus isolated from algae and other sources are summarized and the biological activities and compounds responsible for these inhibitory activities are reported. Bacillus is well known for producing antimicrobial compounds (Wiese et al., 2009). Since 1955, B. pumilus was described as antibiotic producer; Pumilin is an antibiotic isolated from this bacterial strain by Bhate (1955) and it is mainly active against gram positive microorganisms. Various metabolites with antibiotic activity, including the important group of antibiotic peptides like polymixin B, gramicidin and bacitracin are produced by Bacillus species (Crisley, 1964; Vandamme and Demain, 1976; Ishihara et al., 2002). Results obtained in this study are in agreement with these reports. P8 bacteria showed a broad inhibition spectrum against human and fish pathogens and the strongest inhibitory activity was observed against S. aureus and A. salmonicida. Bacillus sp. isolated from seaweeds and sediment by Suvega and Arunkumar (2014) had strong activities against bacteria plant pathogens (Xanthomonas axonopodis pv. citri, X. oryzae pv. oryzae) and fungi (Ustilaginoidea virens) causing diseases in rice paddy fields.
Table 6.
Biological activities of Bacillus pumilus isolated from marine algae and other sources.
| Origin | Inhibition of microorganisms/applications | References |
|---|---|---|
| Penaeus monodon (black tiger shrimp) | Marine bacterial pathogens Vibrio alginolyticus, V. mimicus and V. harveyi | Hill et al., 2009 |
| B. pumilus used as probiotic in aquaculture. | ||
| Ulva spp. (green alga) | S. aureus, E. coli and S. Typhimurium | Prieto et al., 2012 |
| Fucus vesiculosus (brown alga) | ||
| Polysiphonia lanosa (red alga) | ||
| Palmaria palmata (red alga) | ||
| Fucus serratus (brown alga) | ||
| Colpomenia sinuosa (brown alga) | Gram positive and Gram negative fouling bacteria; Vibrio and Photobacterium species: (V. fischeri V. harveyi V. alginolyticus P. phosphoreum P. damselae) and pathogens of the genus: Staphylococcus, Serratia, Klebsiella and Salmonella | Kanagasabhapathy et al., 2006 |
| Jania rubens (red alga) | Pseudomonas aeruginosa, Aeromonas salmonicida, A. hydrophila, Salmonella typhimurium, Streptococcus sp., S. aureus, Enterococcus feacalis, Micrococcus sp., Candida albicans. | Ismail-Ben Ali et al., 2012 |
| Soil | Micrococcus luteus ATCC 10240 and S. aureus ATCC 6538 | Hasan et al., 2009 |
| Soil | Bacitracin (peptide) active against Micrococcus luteus and S. aureus | Awais et al., 2007 |
| Endophytic bacteria from ethnovarieties of cassava | Active against the fungi Rhizoctonia solani, Pythium aphanidermatum and Sclerotium rolfsii. The antifungal compound was identified as pumilacidin | Pereira de Melo et al., 2009 |
| Lipopeptide biosurfactants (LPBSs) from Bacillus sp | Agriculture, chemical, food, and pharmaceutical industries | Roongsawang et al., 2010 |
| Soil and water samples | Pumilicin 4, inhibits several Gram-positive bacteria, including MRSA and VRE | Aunpad and Na-Bangchang, 2007; Abriouel et al., 2011 |
| Soil | E. coli ATCC 2939, Staphylococcus aureus ATCC 96, Bacillus subtilis ATCC 441, Aspergillus niger A1781 and Aspergillus flavous A873 | Sawale et al., 2014 |
| Clinical specimens | Fungicidal activity against Mucoraceae and Aspergillus spp | Bottone and Peluso, 2003 |
| Soil | S. aureus, S. pyogens, Klebsiella spp., S. typhi | Kuta et al., 2009 |
MRSA, Methicillin-Resistant Staphylococcus aureus; VRE, Vancomycin-Resistant-Enterococci; ATCC, American Type Culture Collection.
In our previous study (Ismail-Ben Ali et al., 2012) a B. pumilus related strain was isolated from the red alga J. rubens and had a different antibacterial spectrum than the isolate from P. pavonica, confirming that the algal species not only determines epiphytic diversity but also affects the type of secondary metabolite that is produced, even among the same species of epiphyte.
Kumar et al. (2014) reported antibacterial activities cell free supernatant of a culture of a Bacillus strain isolated from entomopathogenic nematode against E. coli. Only a few epiphytic bacteria isolated from algae are known to have inhibitory potential against the human pathogen E. coli. Inhibition of E. coli by isolates, P9, P11, P12, and P20 and especially by P8 suggests that these isolates may produce novel antimicrobial substances. P8 inhibits both Gram+ and Gram- pathogens. This finding is supported by other studies such as those of Chatterjee et al. (2008) and Prieto et al. (2012) who reported antibacterial activities of Bacillus strains isolated from reef sediments and marine alga against P. aeruginosa, S. aureus, Salmonella enterica, Bacillus sp., Proteus vulgaris, E. coli, and S. typhimurium. Gram negative isolates such as Pseudomonas and Pseudoalteromonas strains (P9, P10, P11, and P20) have variable antibacterial activities toward both Gram+ and Gram− tested pathogens. Qi et al. (2009) and Horta et al. (2014) reported Gram− marine isolates (from Bifurcaria bifurcata and deep see sediment) such as P. rhizosphaerae, Vibrio sp., Alteromonas sp., Shewanella sp., and Serratia sp. active against Gram+ pathogens.
Antimicrobial tests were carried out by two different methods, the drop technique and the overlay assay. The overlay assay was better suitable for the detection of antibacterial and antifungal activity of bacterial isolates. Spots of isolates that are grown overnight may already have secreted bioactive compounds in the agar before they were overlaid by the target strain. Therefore, it was concluded that the five active isolates closely related to Bacillus, Pseudomonas and Pseudoalteromonas, produced and excreted bioactive(s) inhibitory substance(s) during 24 h growth. When these organisms were dropped simultaneously with the target organism the zone of inhibition was much less apparent. Perhaps the target organism prevented growth of the test organism or its production or excretion of the bioactive compound.
Inhibitory compounds produced by the five active isolates were present in the cell-free spent medium. The inhibitory compounds produced by the Pseudomonas and Pseudoalteromonas isolates were resistant to proteolytic enzymes (trypsin and pepsin) and sensitive to heat treatment. Inhibitory compounds produced by isolate P8 were also resistant to proteolytic enzymes and the activity was only lost after heat treatment for 5 min at 100°C.
In summary, the five isolates P8, P9, P11, P12, and P20, isolated from the surface of Padina pavonica and identified as closely related to B. pumilus, Pseudomonas sp., and Pseudoalteromonas produce bioactive compounds that act against human pathogens such as S. aureus; the most pathogenic specie of the genus Staphylococcus and of which multi-resistant strains represent a major problem in hospital (Le Loir et al., 2003). They also act against V. alginolyticus, which is one of the major causes of bacterial infections on shrimp farms (Hill et al., 2009), and against the virulent Gram- bacterium V. vulnificus causing gastroenteritis, necrotic skin infections, meningitis and fatality (Mayer et al., 2014), as well as against a number of fish pathogens. Given the broad range of pathogens that are inhibited, these isolates may produce compounds that might prove promising candidates for potentially novel antibiotic and antifungal drugs.
Author contributions
AI performed all the experiments, analyzed and interpreted data, wrote and revised the manuscript. LK contributed in designation of the research plan, organization of the study, the development of work and corrected and evaluated the manuscript. MH assisted the molecular manipulations and sequencing, contributed to the blast, phylogentic analysis. HB was involved in design of the experiments, evaluation of outcome and involved in manuscript writing. MC did the Vibrio strains identification, contributed to data analyzes, interpretation and manuscript final finalization. AB and LS revised critically the manuscript and approved final version to be published. ME designed the research plan, organized and supervised the development of work, corrected and evaluated the manuscript, submitted the full sequences for assignment in GENBANK and supervised the whole work.
Funding
This work was co-funded and supported by the two projects “MOTOX” and “BIOMER” from the National Institute of Marine Sciences and Technologies (INSTM).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Mrs. Veronique Confurius-Guns, Department of Marine Microbiology, Royal Netherlands Institute for Sea Research (NIOZ), for her assistance and help with PCR and DNA sequencing. MC, HB, and LS, have received funding from the European Union Seven Frame Work Program (FP7/2007-2013) under grant agreement N°311975. This publication reflects the views of the authors and the European Union cannot be held responsible for any use which may be made of the information contained therein.
References
- Abriouel H., Franz C. M., Omar N. B., Gálvez A. (2011). Diversity and applications of Bacillus bacteriocins. FEMS Microbiol. Rev. 35, 201–232. 10.1111/j.1574-6976.2010.00244.x [DOI] [PubMed] [Google Scholar]
- Albakosh M. A., Naidoo R. K., Kirby B., Bauer R. (2016). Identification of epiphytic bacterial communities associated with the brown alga Splachnidium rugosum. J. Appl. Phycol. 28, 1891–1901. 10.1007/s10811-015-0725-z [DOI] [Google Scholar]
- Aunpad R., Na-Bangchang K. (2007). Pumilicin 4, a novel bacteriocin with anti-MRSA and anti-VRE activity produced by newly isolated bacteria Bacillus pumilus strain WAPB4. Curr. Microbiol. 55, 308–313. 10.1007/s00284-006-0632-2 [DOI] [PubMed] [Google Scholar]
- Awais M., Shah A. A., Hameed A., Hasan F. (2007). Isolation, identification and optimization of bacitracin produced by Bacillus sp. Pak. J. Bot. 39, 1303–1312. [Google Scholar]
- Barry A. L., Thornsberry C. (1980). Susceptibility testing, diffusion test procedures, in Manual of Clinical Microbiology, eds Lennette L. H., Balows A., Hausier W. J., Truant J. P. (Washington, DC: American Society for Microbiology; ), 463–474. [Google Scholar]
- Bhate D. S. (1955). Pumilin, a new antibiotic from Bacillus pumilus. Nature 175, 816–817. 10.1038/175816a0 [DOI] [PubMed] [Google Scholar]
- Bottone E. J., Peluso R. W. (2003). Production by Bacillus pumilus (MSH) of an antifungal compound that is active against Mucoraceae and Aspergillus species, preliminary report. J. Med. Microbiol. 52, 69–74. 10.1099/jmm.0.04935-0 [DOI] [PubMed] [Google Scholar]
- Brown B. J., Preston J. F., III. (1991). L-guluronan-specific alginate lyase from a marine bacterium associated with Sargassum. Carbohydr. Res. 211, 91–102. 10.1016/0008-6215(91)84148-8 [DOI] [PubMed] [Google Scholar]
- Burgess J. G., Boyd K. G., Armstrong E., Jiang Z., Yan L., Berggren M., et al. (2003). The development of a marine natural product-based antifouling paint. Biofouling 19, 197–205. 10.1080/0892701031000061778 [DOI] [PubMed] [Google Scholar]
- Burgess J. G., Mearns-Spragg A., Jordan E. M., Bregu M., Boyd K. G. (1999). Microbial antagonism, a neglected avenue of natural products research. J. Biotechnol. 70, 27–32. 10.1016/S0168-1656(99)00054-1 [DOI] [PubMed] [Google Scholar]
- Burke C., Thomas T., Lewis M., Steinberg P., Kjelleberg S. (2011). Composition, uniqueness and variability of the epiphytic bacterial community of the green alga Ulva australis. ISME J. 5, 590–600. 10.1038/ismej.2010.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr T. J., Schroth M. N. (1977). Occurrence of soft-rot Erwinia spp. in soil and plant material. Ecol. Epidemiol. 67, 1382–1387. 10.1094/phyto-67-1382 [DOI] [Google Scholar]
- Chatterjee S., Haldar S., Asakura M., Yamasaki S., Balasubramanian T. (2008). Molecular identification and phylogenetic status of marine bacillus associated with coral sediment, showing antibacterial effects against human pathogens. Ann. Microbiol. 58, 309–312. 10.1007/BF03175335 [DOI] [Google Scholar]
- Chbani A., Mawlawi H., Etahiri S. (2011). Antibacterial activity of brown seaweed extracts of Padina pavonica from the Mediterranean coast of Lebanon. Phytotherapie 9, 283–286. 10.1007/s10298-011-0634-5 [DOI] [Google Scholar]
- Chellaram C., Raja P., Alex John A., Krithika S. (2013). Antagonistic effect of epiphytic bacteria from marine algae, southeastern india. Pak. J. Biol. Sci. 16, 431–434. 10.3923/pjbs.2013.431.434 [DOI] [PubMed] [Google Scholar]
- Ciancio A., Leonetti P., Sialer M. M. F. (2000). Detection of nematode antagonistic bacteria by fluorogenic molecular probes. EPPO Bull. 30, 563–569. 10.1111/j.1365-2338.2000.tb00948.x [DOI] [Google Scholar]
- Collins M. D. (1992). The genus Brevibacterium, in The Prokaryotes, 2nd Edn., eds Balows A., Truper H. G., Dworkin M., Harder W., Schleifer K. H. (New York, NY: Springer; ), 1351–1354. [Google Scholar]
- Crisley F. D. (1964). Antibacterial interaction between bromothymol blue and polymyxin B. Nature 203, 211–213. 10.1038/203211b0 [DOI] [PubMed] [Google Scholar]
- Egan S., Thomas T., Kjelleberg S. (2008). Unlocking the diversity and biotechnological potential of marine surface associated microbial communities. Curr. Opinion. Microbiol. 11, 219–225. 10.1016/j.mib.2008.04.001 [DOI] [PubMed] [Google Scholar]
- Faulkner D. J. (2000). Marine natural products. Nat. Prod. Rep. 17, 7–55. 10.1039/a809395d [DOI] [PubMed] [Google Scholar]
- Gallardo A. A., Risso S., Fajardo M. A., Estevao Belchior S. (2004). Characterization of microbial population present in the edible seaweed, Monostroma undulatum, Wittrock. Arch. Latinoam. Nutr. 54, 337–345. [PubMed] [Google Scholar]
- Gao J., Bao H. Y., Xin M. X., Liu Y. X., Li Q., Zhang Y. F. (2006). Characterization of a bioflocculant from a newly isolated Vagococcus sp. W31. J. Zhej. Univ. Sci. B. 7, 186–192. 10.1631/jzus.2006.B0186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudice A. L., Brilli M., Bruni V., De Domenico M., Fani R., Michaud L. (2007). Bacterium-bacterium inhibitory interactions among psychrotrophic bacteria isolated from Antarctic seawater (Terra Nova Bay, Ross Sea). FEMS. Microbiol. Ecol. 60, 383–396. 10.1111/j.1574-6941.2007.00300.x [DOI] [PubMed] [Google Scholar]
- Goecke F., Labes A., Wiese J., Imhoff J. F. (2010). Chemical interactions between marine macroalgae and bacteria. Mar. Ecol. Prog. Ser. 409, 267–300. 10.3354/meps08607 [DOI] [Google Scholar]
- Hasan F., Khan S., Shah A. A., Hameed A. (2009). Production of antibacterial compounds by free and immobilized Bacillus pumilus SAF1. Pak. J. Bot. 41, 1499–1510. [Google Scholar]
- Hauben L., Vauterin L., Swings J., Moore E. R. B. (1997). Comparison of 16S ribosomal DNA sequences of all Xanthomonas species. Int. J. Syst. Evol. Microbiol. 47:328. 10.1099/00207713-47-2-328 [DOI] [PubMed] [Google Scholar]
- Hill J. E., Baiano J. C. F., Barnes A. C. (2009). Isolation of a novel strain of bacillus pumilus from penaeid shrimp that is inhibitory against marine pathogens. J. Fish. Dis. 32, 1007–1016. 10.1111/j.1365-2761.2009.01084.x [DOI] [PubMed] [Google Scholar]
- Holmström C., Kjelleberg S. (1999). Marine Pseudoalteromonas species are associated with higher organisms and produce biologically active extracellular agents. FEMS. Microbiol. Ecol. 30, 285–293. 10.1016/S0168-6496(99)00063-X [DOI] [PubMed] [Google Scholar]
- Horta A., Pinteus S., Alves C., Fino N., Silva J., Fernandez S., et al. (2014). Antioxidant and antimicrobial potential of the bifurcaria bifurcata epiphytic bacteria. Mar. Drugs 12, 1676–1689. 10.3390/md12031676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara H., Takoh M., Nishibayashi R., Sato A. (2002). Distribution and variation of bacitracin synthetase gene sequences in laboratory stock strains of Bacillus licheniformis. Curr. Microbiol. 45, 18–23. 10.1007/s00284-001-0041-5 [DOI] [PubMed] [Google Scholar]
- Ismail-Ben Ali A., El Bour M., Ktari L., Bolhuis H., Ahmed M., Boudabbous A., et al. (2012). Jania rubens associated bacteria, molecular identification and antimicrobial activity. J. Appl. Phycol. 24, 525–534. 10.1007/s10811-011-9758-0 [DOI] [Google Scholar]
- Ismail-Ben Ali A., Ktari L., Bolhuis H., Boudabbous A., Stal L. J., El Bour M. (2010b). Ulva intestinalis associated bacteria, molecular identification and antimicrobial potential. Rapp. Comm. Int. Mer. Médit. 39, 316. [Google Scholar]
- Ismail-Ben Ali A., Ktari L., Boudabbous A., El Bour M. (2010a). Seasonal variation of antibacterial activity of the brown alga Padina pavonica (l) thivy collected from northern coast of Tunisia. Bull. Institut. Natl. Des Sci. Technol. Mer. Salammbô 37, 127–132. [Google Scholar]
- Ivanova E. P., Christen R., Alexeeva Y. V., Zhukova N. V., Gorshkova N. M., Lysenko A. M., et al. (2004). Brevibacterium celere sp. nov., isolated from degraded thallus of a brown alga. Int. J. Syst. Evol. Microbiol. 54, 2107–2111. 10.1099/ijs.0.02867-0 [DOI] [PubMed] [Google Scholar]
- James S. G., Holmström C., Kjelleberg S. (1996). Purification and characterization of a novel antibacterial protein from the marine bacterium D2. Appl. Environ. Microbiol. 62, 2783–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen J. E., Nielsen P., Sjoholm C. (1999). Description of Cellulophaga baltica gen. nov., sp. nov. and Cellulophaga fucicola gen. nov., sp. nov. and reclassification of Cytophaga lytica to Cellulophaga lytica gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 49, 1231–1240. [DOI] [PubMed] [Google Scholar]
- Jones D., Keddie R. M. (1953). Genus Brevibacterium Breed, 13AL emend. Collins et al., 1980, in Bergey's Manual of Systematic Bacteriology, Vol. 6, eds Sneath P. H. A., Mair N. S., Sharpe M. E., Holt J. G. (Baltimore, MD: Williams & Wilkins 1986; ), 1301–1313. [Google Scholar]
- Kanagasabhapathy M., Sasaki H., Haldar S., Yamasaki S., Nagata S. (2006). Antibacterial activities of marine epibiotic bacteria isolated from brown algae of Japan. Ann. Microbiol. 56, 167–173. 10.1007/BF03175000 [DOI] [Google Scholar]
- Kanagasabhapathy M., Sasaki H., Nagata S. (2008). Phylogenetic identification of epibiotic bacteria possessing antimicrobial activities isolated from red algal species of Japan. World J. Microb. Biot. 24, 2315–2321. 10.1007/s11274-008-9746-y [DOI] [Google Scholar]
- Kelecom A. (2002). Secondary metabolites from marine microorganisms. Ann. Acad. Bras. Cienc. 74, 151–170. 10.1590/S0001-37652002000100012 [DOI] [PubMed] [Google Scholar]
- Ksouri J., Elferjani H., Mensi F. (2008). Estimation du stock naturel de l'algue brune Padina pavonica (L.) Thivy en Tunisie septentrionale (Cap Zébib). Bulletin de la Société Des Sciences Naturelles de Tunisie 35, 57–60. [Google Scholar]
- Ktari L., Guyot M. (1999). A cytotoxic oxysterol from the marine alga Padina pavonica (L.) Thivy. J. Appl. Phycol. 11, 511–513. 10.1023/A:1008162624027 [DOI] [Google Scholar]
- Kumar S. N., Nambisan B., Sundaresan A., Mohandas C., Anto R. J. (2014). Isolation and identification of antimicrobial secondary metabolites from bacillus cereus associated with a rhabditid entomopathogenic nematode. Ann. Microbiol. 64, 209–218. 10.1007/s13213-013-0653-6 [DOI] [Google Scholar]
- Kuta F. A., Nimzing L., Orka'A P. Y. (2009). Screening of Bacillus Species with Potentials of Antibiotics Production. Appl. Med. Informat. 24, 42–46. [Google Scholar]
- Lachnit T., Blumel M., Imhoff J. F., Wahl M. (2009). Specific epibacterial communities on macroalgae, phylogeny matters more than habitat. Aquat. Biol. 5, 181–186. 10.3354/ab00149 [DOI] [Google Scholar]
- Lachnit T., Meske D., Wahl M., Harder T., Schmitz R. (2011). Epibacterial community patterns on marine macroalgae are host-specific but temporally variable. Environ. Microbiol. 13, 655–665. 10.1111/j.1462-2920.2010.02371.x [DOI] [PubMed] [Google Scholar]
- Lakshmanaperumalsamy P., Purushothaman A. (1982). Heteroptrophic bacteria associated with seaweed. Proc. Plant Sci. 91, 487–493. [Google Scholar]
- Lane J. D. (1991). 16S/23S rRNA sequencing, in Nucleic Acid Techniques in Bacterial Systematics, eds Stackebrandt E., Goodfellow M. (Chichester: Wiley; ), 115–175 [Google Scholar]
- Lee S. D. (2007). Devosia subaequoris sp. nov., isolated from beach sediment. Int. J. Syst. Evol. Microbiol. 57, 2212–2215. 10.1099/ijs.0.65185-0 [DOI] [PubMed] [Google Scholar]
- Lee Y. K., Jung H. J., Lee H. K. (2006). Marine bacteria associated with the Korean brown alga, Undaria pinnatifida. J. Microbiol. 44, 694–698. [PubMed] [Google Scholar]
- Le Loir Y., Baron F, Gautier M. (2003). Staphylococcus aureus and food poisoning. Genet. Mol. Res. 2, 63–76. 10.3390/toxins2071751 [DOI] [PubMed] [Google Scholar]
- Lemos M. L., Toranzo A. E., Barja J. L. (1985). Antibiotic activity of epiphytic bacteria isolated from intertidal seaweeds. Microbial. Ecol. 11, 149–163. 10.1007/BF02010487 [DOI] [PubMed] [Google Scholar]
- Li Z. (2009). Advances in marine microbial symbionts in the China Sea and Related Pharmaceutical Metabolites. Mar. Drugs 7, 113–129. 10.3390/md7020113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovejoy C., Bowman J., Hallegraeff G. (1998). Algicidal effects of a novel marine Pseudoalteromonas isolate (class Proteobacteria, gamma subdivision) on harmful algal bloom species of the genera Chattonella, Gymnodinium, and Heterosigma. Appl. Environ. Microbiol. 64, 2806–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M., Portetelle D., Michel G., Vandenbol M. (2014). Microorganisms living on macroalgae, Diversity, interactions, and biotechnological applications. Appl. Microbiol. Biot. 98, 2917–2935. 10.1007/s00253-014-5557-2 [DOI] [PubMed] [Google Scholar]
- Mayer A. M. S., Hall M. L., Holland M., De Castro C., Molinaro A., Aldulescu M., et al. (2014). Vibrio vulnificus MO6-24/O lipopolysaccharide stimulates superoxide anion, thromboxane B2, matrix met alloproteinase-9, cytokine and chemokine release by rat brain microglia in vitro. Mar. Drugs 12, 1732–1756. 10.3390/md12041732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagayama K., Iwamura Y., Shibata T., Hirayama I., Nakamura T. (2002). Bactericidal activity of phlorotannins from the brown alga Ecklonia kurome. J. Antimicrob. Chemother. 50, 889–893. 10.1093/jac/dkf222 [DOI] [PubMed] [Google Scholar]
- Nedashkovskaia O. I., Ivanova E. P., Bakunina I. I., Svetashev V. I., Zviagintseva T. N., Mikhailov V. V. (2002). Characterization of marine bacteria Pseudoalteromonas citrea degradating fucoidan. Microbiol. 64, 3–10. [PubMed] [Google Scholar]
- Oclarit J. M., Okada H., Ohta S., Kaminura K., Yamaoka Y., Iizuka T., et al. (1994). Anti-bacillus substance in the marine sponge, Hyatella species, produced by an associated Vibrio species bacterium. Microbios 78, 7–16. [PubMed] [Google Scholar]
- Omezzine F., Haouala R., El Ayeb A., Boughanmi N. (2009). Allelopathic and antifungal potentialities of Padina pavonica (L.) extract. J. Plant. Breed. Crop. Sci. 1, 194–203. [Google Scholar]
- Parvathi A., Krishna K., Jose J., Joseph N., Nair S. (2009). Biochemical and molecular characterization of Bacillus pumilus isolated from coastal environment in Cochin, India. Braz. J. Microbiol. 40, 269–275. 10.1590/s1517-83822009000200012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulert R., Júnior A. S., Stadnik M. J., Pizzolatti M. G. (2007). Antimicrobial properties of extracts from the green seaweed Ulva fasciata Delile against pathogenic bacteria and fungi. Algol. Stud. 123, 123–130. 10.1127/1864-1318/2007/0123-0123 [DOI] [Google Scholar]
- Penesyan A., Kjelleberg S., Egan S. (2010). Development of novel drugs from marine surface associated microorganisms. Mar. Drugs 8, 438–459. 10.3390/md8030438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira de Melo F. M., Fiore M. F., Beraldo de Moraes L. A., Silva-Stenico M. E., Scramin S., Teixeira M. A., et al. (2009). Antifungal compound produced by the cassava endophyte Bacillus pumilus MAIIIM4A. Sci. Agric. 66, 583–592. 10.1590/S0103-90162009000500002 [DOI] [Google Scholar]
- Preston J. F., III., Romeo T., Bromley J. C. (1986). Selective alginate degradation by marine bacteria associated with the algal genus Sargassum. J. Ind. Microbiol. Biotechnol. 1, 235–244. [Google Scholar]
- Prieto M. L., O'sullivan L., Tan S. P., McLoughlin P., Hughes H., O'Connor P. M., et al. (2012). Assessment of the bacteriocinogenic potential of marine bacteria reveals lichenicidin production by seaweed-derived bacillus spp. Mar. Drugs 10, 2280–2299. 10.3390/md10102280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi S., Xu Y., Gao J., Qian P., Zhang S. (2009). Antibacterial and antilarval compounds from marine bacterium pseudomonas rhizosphaerae. Ann. Microbiol. 59, 229–233. 10.1007/BF03178321 [DOI] [Google Scholar]
- Rao D., Webb J. S., Kjelleberg S. (2005). Competitive interactions in mixed-species biofilms containing the marine bacterium Pseudoalteromonas tunicata. Appl. Environ. Microbiol. 71, 1729–1736. 10.1128/AEM.71.4.1729-1736.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravisankar A., Gnanambal M. E. K., Sundaram L. R. (2013). A newly isolated Pseudomonas sp., epibiotic on the seaweed, Padina tetrastromatica, off Southeastern Coast of India, Reveals Antibacterial Action. Appl. Biochem. Biotechnol. 171, 1968–1985. 10.1007/s12010-013-0473-y [DOI] [PubMed] [Google Scholar]
- Rivas R., Willems A., Subba-Rao N. S., Mateos P. F., Dazzo F. B., Kroppenstedt R. M., et al. (2003). Description of Devosia neptuniae sp. nov. that nodulates and fixes nitrogen in symbiosis with Neptunia natans, an aquatic legume from India. Syst. Appl. Microbiol. 26, 47–53. 10.1078/072320203322337308 [DOI] [PubMed] [Google Scholar]
- Roongsawang N., Washio K., Morikawa M. (2010). Diversity of nonribosomal peptide synthetases involved in the biosynthesis of lipopeptide biosurfactants. Int. J. Mol. Sci. 12, 141–172. 10.3390/ijms12010141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawabe T., Makino H., Tatsumi M., Nakano K., Tajima K., Iqbal M. M., et al. (1998). Pseudoalteromonas bacteriolytica sp. nov., a marine bacterium that is the causative agent of red spot disease of Laminaria japonica. Int. J. Syst. Bacteriol. 48, 769–774. 10.1099/00207713-48-3-769 [DOI] [PubMed] [Google Scholar]
- Sawabe T., Tanaka R., Iqbal M. M., Tajima K., Ezura Y., Ivanova E. P., et al. (2000). Assignment of Alteromonas elyakovii KMM 162T and five strains isolated from spot-wounded fronds of Laminaria japonica to Pseudoalteromonas elyakovii comb. nov. and the extended description of the species. Int. J. Syst. Evol. Microbiol. 50, 265–271. 10.1099/00207713-50-1-265 [DOI] [PubMed] [Google Scholar]
- Sawale A., Kadam T. A., Karale M. A., Kadam O. A. (2014). Antimicrobial Activity of Secondary Metabolites from Halophilic Bacillus pumilus sp. Int. J. Curr. Microbiol. App. Sci 3, 506–512. [Google Scholar]
- Schulze A. D., Alabi A. O., Tattersall-Sheldrake A. R., Miller K. M. (2006). Bacterial diversity in a marine hatchery, Balance between pathogenic and potentially probiotic bacterial strains. Aquaculture 256, 50–73. 10.1016/j.aquaculture.2006.02.008 [DOI] [Google Scholar]
- Singh R. P., Reddy C. R. K. (2014). Seaweed-microbial interactions, Key functions of seaweed-associated bacteria. FEMS Microbiol. Ecol. 88, 213–230. 10.1111/1574-6941.12297 [DOI] [PubMed] [Google Scholar]
- Soussy C. J. (2008). Antibiogram Committee of the French Society of Microbiology. Créteil: Centre Hospitalier Universitaire Henri Mondor. [Google Scholar]
- Suvega T., Arunkumar K. (2014). Antimicrobial activity of bacteria associated with seaweeds against plant pathogens on par with bacteria found in seawater and sediments. Br. Microbiol. Res. J. 4, 841–855. 10.9734/BMRJ/2014/8678 [DOI] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4, Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599. 10.1093/molbev/msm092 [DOI] [PubMed] [Google Scholar]
- Thakur N. L., Anil A. C. (2000). Antibacterial activity of the sponge Ircinia ramose, importance of its surface associated bacteria. J. Chem. Ecol. 26, 57–72. 10.1023/A:1005485310488 [DOI] [Google Scholar]
- Vandamme E. J., Demain A. L. (1976). Nutrition of Bacillus brevis ATCC 9999, the producer of gramicidin S. Antimicrob. Agents Chemother. 10, 265–273. 10.1128/AAC.10.2.265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini C., Rosati G., Verni F., Petroni G. (2004). Identification of the bacterial endosymbionts of the marine ciliate Euplotes magnicirratus (Ciliophora, Hypotrichia) and proposal of ‘Candidatus Devosia euplotis’. Int. J. Syst. Evol. Microbiol. 54, 1151–1156. 10.1099/ijs.0.02759-0 [DOI] [PubMed] [Google Scholar]
- Vanparys B., Heylen K., Lebbe L., De Vos P. (2005). Devosia limi sp. nov., isolated from a nitrifying inoculum. Int. J. Syst. Evol. Microbiol. 55, 1997–2000. 10.1099/ijs.0.63714-0 [DOI] [PubMed] [Google Scholar]
- Wiese J., Thiel V., Nagel K., Staufenberger T., Imhoff J. F. (2009). Diversity of antibiotic-active bacteria associated with the brown alga Laminaria saccharina from the Baltic Sea. Mar. Biotechnol. 11, 287–300. 10.1007/s10126-008-9143-4 [DOI] [PubMed] [Google Scholar]
- Yoo S. H., Weon H. Y., Kim B. Y., Hong S. B., Kwon S. W., Cho Y. H., et al. (2006). Devosia soli sp. nov., isolated from greenhouse soil in Korea. Int. J. Syst. Evol. Microbiol. 56, 2689–2692. 10.1099/ijs.0.64214-0 [DOI] [PubMed] [Google Scholar]
- Yoon J. H., Kang S. J., Park S., Oh T. K. (2007). Devosia insulae sp. nov., isolated from soil, and emended description of the genus Devosia. Int. J. Syst. Evol. Microbiol. 57, 1310–1314. 10.1099/ijs.0.65028-0 [DOI] [PubMed] [Google Scholar]
- Yuexin M. A., Liu P., Zhang Y., Cao S., Li D., Chen W. (2010). Inhibition of spore germination of Ulva pertusa by the marine bacterium Pseudoalteromonas haloplanktis CI4. Acta Oceanol. Sin. 29, 69–78. 10.1007/s13131-010-0009-z [DOI] [Google Scholar]
- Zeng X., Xiao X., Li D., Gu Q., Wang F. (2010). Isolation, identification and screening of microorganisms for cytotoxic activities from deep sea sediments at different pacific stations. World J. Microb. Biot. 26, 2141–2150. 10.1007/s11274-010-0396-5 [DOI] [Google Scholar]
- Zerrin C., Mehmet O., Evrim T., Ergun T. (2011). Antioxidative and antimicrobial activities of some marine algae. Fresenius Environ. Bull. 20, 1001–1006. [Google Scholar]