Abstract
As judged by a single publication metric, the activity in the protein folding field has been declining over the past 5 years, after enjoying a decade-long growth. Does this development indicate that the field is sunsetting or is this decline only temporary? Upon surveying a small territory of its landscape, we find that the protein folding field is still quite active and many important findings have emerged from recent experimental studies. However, it is also clear that only continued development of new techniques and methods, especially those enabling dissection of the fine details and features of the protein folding energy landscape, will fuel this old field to move forward.
Main Text
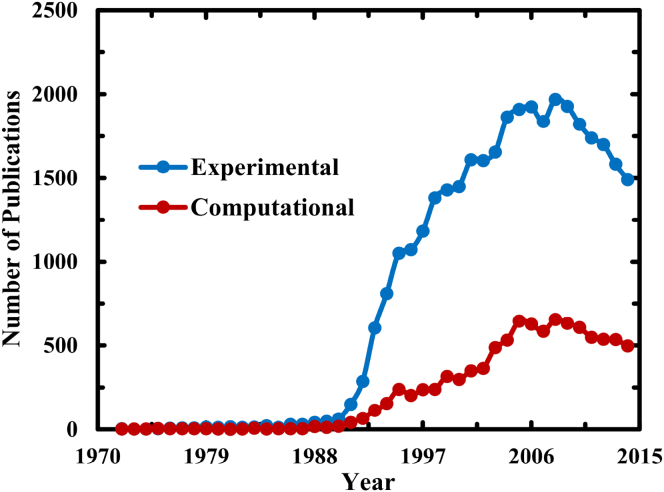
Ever since Anfinsen’s discovery in the early 1960s that a denatured protein can spontaneously and rapidly refold to its native conformation upon removal of denaturant (1), the question of how proteins fold has fascinated many people and inspired many studies. Part of this captivation stems from the notion that even the folding of small proteins (i.e., ∼100 amino acids in length) involves a vast number of conformational degrees of freedom and, hence, cannot be achieved through a random search process but rather through a well-defined folding pathway(s) (2). In other words, the linear amino acid sequence of a protein encodes not only its native structure but also the mechanism by which this structure is attained. Thus, the pursuit of this code, or more precisely the underlying principles that govern the thermodynamics and dynamics of protein folding, which are at the core of the protein folding problem, has become a major undertaking in many laboratories around the world. As shown (Fig. 1), the development of the protein folding field has been quite dynamic in the past 50 years, as judged by the total number of relevant articles published per year. Based on this indicator, it is clear that the protein folding field as a whole had enjoyed an enormous and exponential-like growth in the early 1990s, followed by a decade-long period of high activity and productivity. However, the trend in the past decade seems to indicate that the field is going through a declining phase, with a decreasing rate of ∼110 articles per year. If this declining trend were to continue with this rate, the field faces a dim future.
Figure 1.
Plot of the number of computational and experimental protein folding articles found on the ISI Web of Science database per year. The number of computational articles per year was produced by using the search, “protein folding” and (comput∗ or simulat∗ or theor∗), whereas the number of experimental articles per year was obtained by subtracting this value from that of the search using protein folding. To see this figure in color, go online.
Advancing a research field requires new ideas, new findings, new theoretical models, and new techniques. The rapid take off of the protein folding field in the early 1990s was fueled by such driving forces, for example, the development of the folding energy landscape theory (3, 4), introduction of the Φ-value analysis method (5), discovery of two-state folders (6), and advancements in various biological, chemical, physical, and computational techniques that made it possible to study previously inaccessible questions and protein systems. For example, the advent of ultrafast triggering methods, such as the laser-induced temperature-jump (T-jump) technique (7), has greatly enhanced the time resolution of kinetic studies, enabling investigation of protein folding dynamics on the nanosecond and microsecond timescale. Similarly, application of single-molecule-based techniques allowed for elucidation of conformational transitions and dynamics that were not accessible by traditional ensemble measurements (8). In this Perspective, we attempt to show that although the prevalence of publications in the literature related to protein folding has been decreasing over the last several years, the field is still quite active, and continued development of new ideas and new methods will keep it moving forward. Below, we will first provide an account of the major achievements since 2010, focusing mainly on experimental studies of protein folding dynamics and mechanisms in vitro. We will then offer some suggestions for future directions, in the context of the studies discussed. Although equally important, we can only provide a short overview of the recent advancements in theoretical and computational studies of protein folding dynamics. For readers interested in these and other types of protein folding studies, including intrinsically disordered and in vivo protein folding, we direct their attention to several recent reviews (9, 10, 11, 12, 13, 14, 15, 16, 17).
Recent progress
Experimental studies in the past 5 years have made significant progress in characterizing and understanding the dynamics and mechanism of protein folding. In the first three sections below, we describe some of the key findings that have emerged from those studies, which are organized based on the techniques they used, namely single-molecule fluorescence spectroscopy, single-molecule force spectroscopy, and ensemble spectroscopic methods. In the last section, we provide a brief summary of the major developments in computer simulations of protein folding dynamics.
Single-molecule fluorescence studies
Single-molecule fluorescence-based techniques, such as single-molecule fluorescence resonance energy transfer (smFRET), have become increasingly important in elucidating the fine features of the protein folding free energy landscape and mechanism. One distinct advantage of single-molecule-based spectroscopic techniques is that they can extract information that otherwise would be difficult to attain from ensemble measurements. In particular, the past 5 years have witnessed the rapid growth of using these techniques to assess the role of native and nonnative interactions, internal friction, unfolded state structure and dynamics, and transition-path time on protein folding pathways and kinetics. A few representative examples are given below.
Several recent studies have focused on the role of internal friction and frustration on the dynamics of protein folding (18, 19, 20, 21, 22, 23). For example, Schuler and co-workers (20, 21) used smFRET, fluorescence correlation spectroscopy (FCS), and microfluidic mixing to study the effect of internal friction on the folding energy landscape of the spectrin domains. They found that the internal friction localized at the early transition state plays a major role in determining the overall folding times of these proteins, which led them to suggest that the curvature of the transition state barrier is larger than that of the unfolded state potential well (20). Using a small cold shock protein (Csp) as a model, they further showed that internal friction, as measured by the conformational reconfiguration or relaxation time, is also a key determinant of the conformational dynamics in the unfolded potential well (21). Under native-like conditions where the polypeptide chain is more compact, the reconfiguration time of the unfolded ensemble of Csp is 100 ns but the reconfiguration time extrapolated to zero viscosity is nearly zero when the polypeptide chain becomes more extended (i.e., under high denaturant concentrations). Based on these finding, they suggested that internal friction is particularly important in determining the barrier crossing dynamics of microsecond folders. On the other hand, Sherman and Haran (18) found, based on the mean first-passage times extracted from their FCS experiments using the theory of Szabo, Schulten, and Schulten (24), that the intrachain diffusion coefficient of protein L remained approximately constant from a denaturant concentration of 3 to 7 M guanidinium chloride (GdmCl). Similarly, by using FCS to measure the intrachain motions of a set of unstructured peptides with and without side chains, Teufel et al. (19) concluded that side-chain interactions slow down loop formation while backbone-backbone hydrogen bonds accelerate intrachain interactions. Voelz et al. (22) also studied the protein unfolded state and concluded, based on smFRET measurements and molecular dynamics (MD) simulations, that there is a large network of metastable states in the unfolded ensemble of the ACBP protein, which interconvert on a timescale of ∼100 μs. Although previous studies had identified this kinetic phase as a folding intermediate, they attributed this time constant to the protein’s slow achievement of the unfolded state structure that leads to productive folding. Most recently, Chung et al. (23) showed that interresidue contacts, particularly nonnative salt bridges, were able to definitively decrease the folding rate for a designed α-helical protein, α3D (25). Although these protein interactions are able to create local minima along the folding coordinate thus increasing the roughness of the energy landscape and the time required to traverse the free energy barrier, they do not, however, significantly alter the folding free energy barrier height.
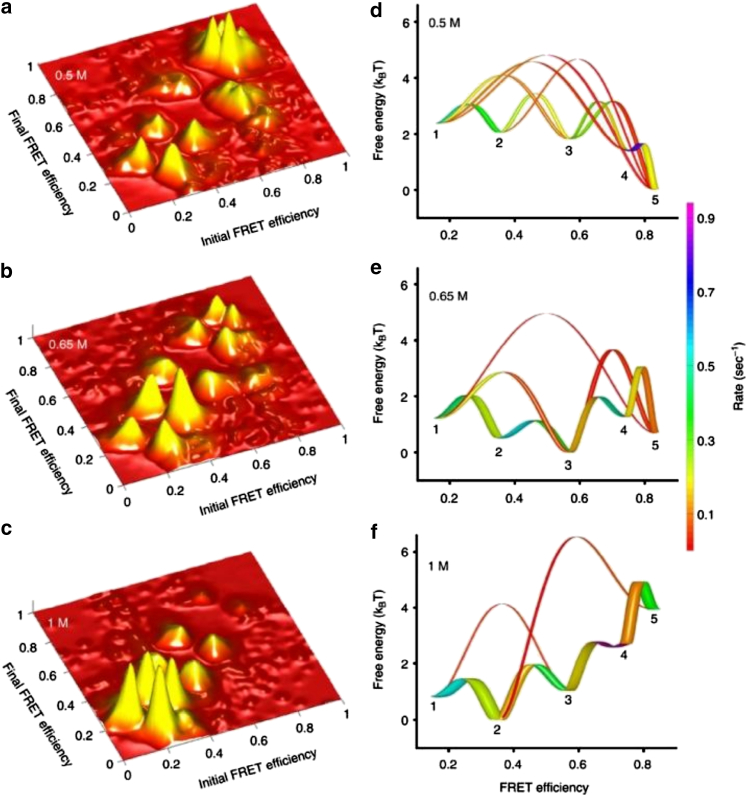
The power of smFRET in revealing the detailed features underlying the folding dynamics of large proteins was recently demonstrated by Haran and co-workers (26). By analyzing smFRET trajectories using a hidden Markov model, they showed that the folding landscape of adenylate kinase, a 214-residue, multidomain protein, encompasses six metastable states, with their connectivity depending on denaturant concentration (Fig. 2). Although many intersecting folding pathways were observed at low denaturant concentrations (∼0.5 M urea), one sequential mechanism became dominant when the denaturant concentration was increased to 1 M. Liu et al. (27) also showed that smFRET can be applied to study the dynamics of a downhill protein folding process. Additionally, Clarke and co-workers (28) used a single-molecule total internal reflection fluorescence microscopy technique as well as other computational and biophysical methods to study the SasG tandem repeat protein, which is known to form extended fibrils on the surface of Staphylococcus aureus bacteria. They found that the center domain, although intrinsically disordered, can mediate long-range folding cooperativity of the terminal domains as a result of the high stability of the interfaces.
Figure 2.
(a–c) Two-dimensional transition maps of the adenylate kinase protein at three different GdmCl concentrations, as indicated. (d–f) One-dimensional free energy maps of adenylate kinase showing five of the six metastable states as well as the transitions between them that contain at least 10% of the flux from beginning to end (or vice versa). The color bar indicates the rate of flux, whereas the line widths show the relative productive flux along each pathway. This figure is adapted from (26) with permission. To see this figure in color, go online.
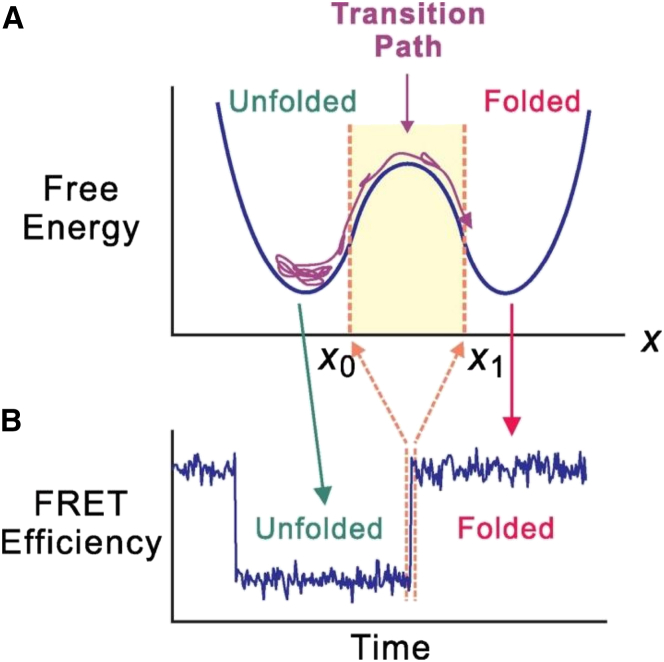
Perhaps one of the most significant advancements in the assessment of protein folding dynamics is the ability to determine the protein folding transition-path time, namely the time required for the successful crossing of the free energy barrier by a protein molecule (29), as shown in Fig. 3. Assuming an attempt frequency value of 105 to 106 s−1, the transition-path time is estimated to be between 0.6 and 6 μs. The very short nature of , as well as the fact that this quantity manifests itself in the barrier-crossing process of individual molecules, have made it very difficult to measure experimentally. By fully characterizing the smFRET time traces obtained with the GB1 protein, including the number of photons emitted, their polarizations, their relative and absolute arrival times, and their wavelengths, Eaton and co-workers (30) were able to isolate single-molecule barrier crossing events and used them to determine an upper bound of 200 μs for the transition-path time for this protein. Fundamentally, is relatively insensitive to the barrier height (30). Indeed, in a later study (29), they were not only able to more accurately determine , but also showed that the transition-path time (i.e., 10 μs) of a slow-folding protein (folding time = 1 s) is similar to that (i.e., 2 μs) of a fast-folding protein (folding time = 100 μs). This is an exciting finding because it experimentally illustrates that correct folding takes approximately the same time even for proteins that are drastically different in structure and size.
Figure 3.
(A) Representation of a one-dimensional free energy diagram for a two-state folding protein indicating the transition path region. (B) Sample FRET efficiency trajectory with the width of the highlighted jump region being the transition-path time. This figure is adapted from (29) with permission. To see this figure in color, go online.
Single-molecule force studies
Like single-molecule fluorescence studies, those based on single-molecule force measurements have also made outstanding contributions toward our understanding of how proteins fold. A distinct advantage of using force spectroscopy to study protein folding dynamics is that the reaction coordinate can be defined as the extension of the molecule, thus allowing for characterization of the folding energy landscape, identification of parallel folding pathways, and observation of anisotropic folding behaviors. Below, we highlight several recent studies, showcasing the advancement in this area.
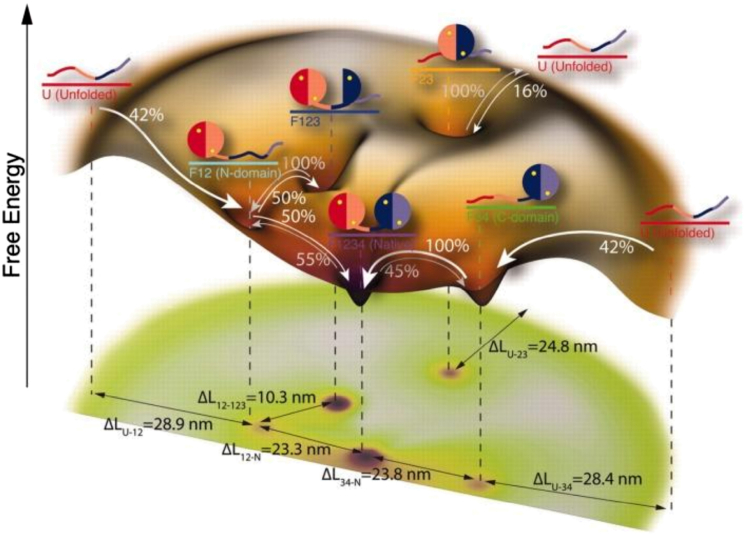
In one study, Zhang and co-workers (31) used optical tweezers to investigate the heterogeneity in the folding-unfolding process of the coiled coil GCN4 and found that the force-induced transition rate was highly anisotropic when the folded and unfolded states were equally populated, depending greatly on the pulling direction. This finding was corroborated by the study of Marquesee and co-workers (32) who found that a two-state folding protein, the src SH3 domain, was more resistant to a low force applied in the longitudinal direction than the perpendicular direction. Moreover, they observed biphasic behavior along the longitudinal pulling axis, indicating that the protein is capable of accessing parallel unfolding pathways. In a more recent study (33), they showed that for the same protein, it is possible to use mutations and denaturant to modulate the flux among the different pathways. Similarly, using atomic force microscopy, Li and co-workers (34) demonstrated that the mechanical unfolding and untying process of a slipknot protein, AFV3-109, also occurs via multiple pathways that are either two-state or three-state in nature. Furthermore, as shown in Fig. 4, the high-resolution force measurements of Rief and co-workers (35) revealed that the folding and unfolding transitions of single calmodulin molecules involve two on-pathway and two off-pathway intermediates and identified cooperative and anticooperative interactions between the domains.
Figure 4.
Schematic of the free energy diagram of the calmodulin protein with arrows indicating the observed transitions and the percentages giving the fraction of transitions along each pathway at zero force. Distances are differences in the contour length of calmodulin, and the cartoon representations of the protein highlight the regions of calmodulin that are folded at each state. This figure is adapted from (35) with permission. To see this figure in color, go online.
In the past 5 years, single-molecule force spectroscopy has also been actively applied to characterize other aspects of the folding free energy landscape of interest (36, 37, 38). For example, by using a high-resolution optical trap to apply tension to the prion protein, Yu et al. (38) were able to determine the free energy barrier height and position along the reaction coordinate and further used this information to determine the transition-path time and also the conformational diffusion coefficient. Similarly, Fernández and co-workers (37) employed force clamp spectroscopy to examine the energy landscape of an engineered I27 protein undergoing two separate reactions, namely unfolding and disulfide bond reduction. In the framework of static disorder theory, they showed that the disulfide-containing mutants had a high degree of heterogeneity in their unfolding pathways; however, the disulfide cleavage event itself followed a rather homogeneous reactive pathway. By performing a force-quench experiment (36), they also showed that the heterogeneous collapse trajectories observed for ubiquitin and I27 arise from a force-dependent free energy barrier.
Ensemble spectroscopic studies
Ensemble measurements based on various spectroscopic techniques continue to play a major role in the investigation of the protein folding problem. In particular, new insights into the dynamics and mechanism of protein folding have been generated from studies that have used new triggering and probing methods, as well as new strategies to enhance the time and structural resolution of the experiments, manipulate the folding energy landscape of interest, and extract detailed mechanistic information from conventional kinetic measurements. Although many outstanding studies have contributed to this area of research, we are unable to describe all of them due to space limitations. Below we highlight a few examples, with a focus on studies that employ light-based triggering and detection methods.
Unlike single-molecule fluorescence studies, which rely on spontaneous conformational fluctuations of individual protein molecules, measurements of protein folding kinetics at the ensemble level require a well-defined triggering event to define the zero time. Although flow- and mixing-based triggering methods continue to play an important role in this regard (39, 40, 41), recent years have seen an increased interest in applying various photo-induced processes to initiate and control protein folding reactions. In comparison to commonly used chemical and thermal triggering methods, those based on photo-induced isomerization or bond cleavage have the advantage of being site-specific. In addition, they allow interrogation of ultrafast protein folding events. Of course, the disadvantage of using a photoliable group or photoswitchable trigger is the possibility that it may introduce an undesirable structural perturbation to the protein/peptide system in question.
Currently, the most commonly used phototriggers are azobenzene and its derivatives (42), whose cis- to trans- (and vice versa) isomerization can be controlled with ultraviolet (UV) or visible light. Because this isomerization process occurs on a picosecond timescale, it is suitable for interrogation of ultrafast protein folding events. This has been nicely demonstrated by the works of Zinth and co-workers (43), Hamm and co-workers (44, 45), and Kliger and co-workers (46). More recently, Abaskharon et al. used an azobenzene cross-linker to manipulate the rigidity of the transition state structure and hence the attempt frequency of a protein folding reaction (47). Specifically, they strategically placed an azobenzene moiety in the α-helix of the miniprotein Trp-cage, which was previously shown to be involved in the major folding transition state. This cross-linker, upon photoisomerization, limits the degrees of freedom of the transition state structure thus causing the curvature of the free energy barrier to increase without changing its height. As a result, the folding rate is increased by an order of magnitude, manifesting as an increase in the aforementioned k0 value (i.e., the attempt frequency). A relatively less used phototriggering method due to the very fast geminate recombination rate of the underlying photocleavage reaction is based on disulfide bond cleavage via UV excitation (48). However, Volk and co-workers (49) took advantage of this geminate recombination and used it to monitor the conformational dynamics of the N-terminal domain of phosphoglycerate kinase, initiated by UV excitation of an aromatic disulfide bond.
Besides those well-established phototriggering strategies, the past 5 years have also seen the development of new photochemical methods. For example, Zinth and co-workers (50) demonstrated the feasibility of using hemithioindigo-hemistilbene derivatives as ultrafast protein folding triggers. These stilbene-based chromophores were shown to isomerize in tens of picoseconds and to induce strong structural changes in a model β-hairpin where the phototrigger was covalently linked to the termini of the peptide. Another very promising and useful phototriggering strategy is based on photodissociation of an S,S-tetrazine moiety. Brown and Smith (51) showed that this moiety can be easily incorporated into unprotected peptides and proteins via two cysteine residues, and Tucker et al. (52) demonstrated that the underlying photodissociation process, in response to an excitation with 330–400 nm light, occurs on the picoseconds timescale. They further illustrated the use of this phototrigger by using it to trigger a local conformational relaxation event in an α-helix and found, via two-dimensional infrared (2D IR) spectroscopy, that the dynamics of this reorganization process, which involves only a single helix turn, occurs in ∼100 ps (53). Finally, we note that Zewail and co-workers (54) have expanded the time resolution of the T-jump technique to the picosecond regime, allowing for the study of ultrafast protein folding processes, including helix nucleation.
Cross-linking is not only the basis for photo-induced structural transitions, but it can also be exploited to perform other novel applications in protein folding studies. For example, in one such application, Markiewicz et al. (55) used a strategically placed m-xylene cross-linker to assess how local friction or frustration affects protein folding dynamics, a topic that has also been explored in a recent study by Matthews, Brooks, and co-workers (56). By placing this cross-linker in a congested region of Trp-cage, Markiewicz et al. were able to decrease both the folding and unfolding rates of this miniprotein without significantly affecting its stability. They attributed this phenomenon to an increase in local internal friction, due to the presence of the m-xylene cross-linker, which acts as a local mass crowding agent (57). Using their kinetic results and a theoretical model developed by Thirumalai, Straub, and co-workers (58), they were able to further show that this local crowder increases the roughness of the folding free energy surface of Trp-cage by 0.4–1.0 kBT. In yet another study, Markiewicz et al. (59) hypothesized that it is possible to use cross-linking strategies to create structural analogs of protein-folding transition states. To verify this hypothesis, they employed a disulfide bond to enforce the β-turn of the Trpzip4 β-hairpin to be in a native-like configuration, which had been shown to be formed in the transition state. They found that this disulfide-bonded version of Trpzip4 folds 10 times faster than the uncross-linked peptide, supporting the notion that it is possible to engineer a mimic of the transition state structure of interest via cross-linking. In a different application, Sosnick and co-workers (60) employed cross-linking and ψ-analysis to probe the transition state heterogeneity of ubiquitin and found that despite the incorporation of a cross-linker, the structural content of the transition state was not substantially altered.
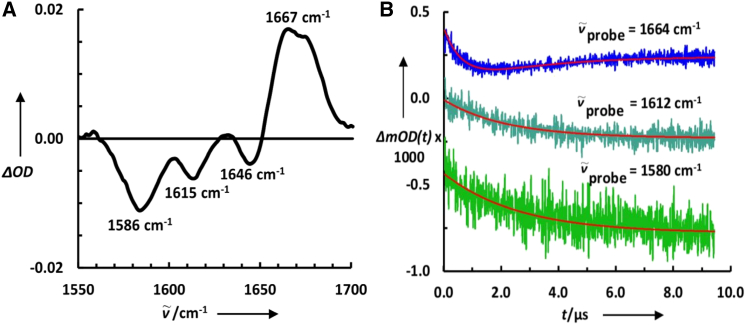
Another distinct effort of the field in the past 5 years has been to enhance the structural resolution and site-specificity in protein folding kinetic studies using various spectroscopic probes (61). In one example, Culik et al. (62) demonstrated that it is possible to monitor the formation of individual secondary structural elements in protein folding via isotopic labeling of selective amide carbonyls, as shown in Fig. 5. This isotope editing method was also used by Keiderling, Kubelka, and co-workers (63) to study the hydrogen-bonding pattern in the aggregation kinetics of polyglutamic acid peptides. In other examples, Tokmakoff and co-workers (64, 65), Zanni and co-workers (66), and Hochstrasser and co-workers (67) showed that more detailed structural content can be obtained on the folding reactions of interest by monitoring couplings or interactions between two or more isotopic amide labels using 2D IR spectroscopy. This is because distinct interactions between two specific groups in a protein reveal distance and thus structural information. Although similar ideas, for instance those based on FRET, have long been used in fluorescence-based protein folding studies, the past 5 years have seen a renewed interest in developing amino acid-based FRET or fluorophore-quencher pairs for this purpose. Examples include the p-cyanophenylalanine-tryptophan FRET pair (68), amino acid fluorophore-thioamide quencher pair (69), and p-cyanophenylalanine-selenomethionine fluorophore-quencher pair (70, 71). A recent example highlighting the use of such structure-sensitive probes is nicely illustrated by Gruebele and co-workers (72), who used three tryptophan-tyrosine quencher pairs to measure contact formation between three helices during the folding of a fast-folding protein, λ6–85.
Figure 5.
(A) FTIR temperature difference spectrum (65°C−25°C) of 13C-labeled Trp-Cage 10b. (B) Conformational relaxation kinetics of 13C-labeled Trp-Cage 10b, probed at the frequencies indicated, in response to a temperature jump from 5°C to 10°C. Smooth lines are fits of these data to either single exponential (1580 cm−1 and 1612 cm−1) or double exponential (1664 cm−1) functions. The traces are offset for clarity. This figure is adapted from (62) with permission. To see this figure in color, go online.
Acquiring site-specific folding kinetic information is also actively pursued in recent studies (73). For example, Dyer and co-workers (74) used the aspartic acid side chain as a local probe to show that the formation of the first hairpin in the WW domain is tightly linked to the protonation state of this charged residue and used an azide IR probe to specifically monitor side-chain reordering events in the folding of the N-terminal domain of the L9 protein (75). Similarly, Kiefhaber and co-workers (76) showed that by replacing a native oxoamide with a thioamide, it is possible to probe backbone-backbone H-bond formation in a site-specific manner, a notion also verified by the study of Culik et al. (77).
Although not discussed in this Perspective due to space limitations, we note that new insights into the protein folding problem have also resulted from many other outstanding studies, especially those employing NMR spectroscopy (78), hydrogen exchange methods (79), and hydroxyl radical footprinting techniques (80).
Computational studies
In parallel, computer simulation of protein folding has also made significant progress in the past 5 years. However, an in-depth discussion of all of the important studies and contributions in this area is beyond the scope of this Perspective. Instead, we only highlight a few key technological advancements and direct interested readers to several recent reviews for more information on this subject (9, 11, 13, 14, 17). First, the ability to perform long-time (e.g., millisecond timescale) MD simulations with atomic-level resolution has made it possible to directly fold a protein in silico, allowing visualization of its folding process (81, 82) and, perhaps more importantly, a direct comparison with experiment (83). Second, the development of Markov state models, which use a statistical approach to group structures observed in a simulation into microstates and connect them by a transition matrix, has allowed a more physical-based and mechanistic interpretation of MD trajectories (84, 85). Third, continued efforts in the improvement of the molecular force fields, such as those on protein secondary structures and hydration (86, 87), have led to more accurate characterization of various interactions underlying protein folding.
Outlooks
The examples discussed previously cover a wide variety of topics related to protein folding dynamics and mechanism, which include, but are not limited to 1) the role of internal friction and frustration, 2) the role of metastable states, 3) the transition-path time, 4) the curvature of the folding free energy barrier and attempt frequency, 5) the location of the folding free energy barrier, 6) sequential and parallel folding pathways, (7) specific contact formation, 8) strategies to enhance structural resolution in folding kinetics studies, and 9) manipulation of folding free energy surfaces. These examples clearly indicate that the protein folding field is still very active and new findings are constantly emerging from experimental studies. In addition, they demonstrate the enormous importance of refining initiation and detection methods to achieve a more detailed description of the underlying features of the protein folding energy landscape. Nonetheless, the decrease in overall activity of the field in recent years also suggests that, to keep the momentum going, new ideas and new technological developments are needed. In particular, we need new methods that will allow researchers to address previously unapproachable questions. In this regard, below we offer our opinions on how experimental studies of protein folding dynamics and mechanisms, especially those using spectroscopic techniques, can keep moving forward.
It is inherently difficult to acquire sufficient information from a single type of experiment to arrive at a molecular-level understanding of how a protein folds. This is because the folding process is intrinsically complex and fast, involving a large number of degrees of freedom that evolve quickly. Most, if not all, experimental techniques employed thus far are largely incapable of generating a “folding movie” detailing all relevant conformational motions, due to either a low structural resolution or a low temporal resolution. Thus, one of the future directions, we believe, is to develop new experimental capabilities that can help alleviate these limitations. In the context of the studies discussed previously, some potential areas of interest are to
-
1)
develop/use multiple site-specific spectroscopic probes, each with a unique environmental and/or structural sensitivity, to increase the content obtained in a single kinetic experiment
-
2)
develop/use multistep FRET systems or multiple fluorophore-quencher pairs to broaden the range of motions and interactions that can be observed, to probe correlated motions, and to potentially distinguish between parallel and sequential folding pathways using fluorescence-based spectroscopic techniques
-
3)
similarly, employ multiple vibrational transitions to increase the structural resolution of IR studies, via vibrational coupling or energy transfer
-
4)
combine smFRET and force spectroscopy to increase the structural resolution of force-based protein folding measurements
-
5)
develop fluorophores with higher brightness than those currently available to increase the sensitivity and time resolution of smFRET protein folding studies
-
6)
use well-chosen structural constraints, particularly those that can be triggered with light, to manipulate and control the starting point of the folding reaction in question
-
7)
devise new and backbone compatible cross-linking strategies to engineer protein folding transition state analogs
-
8)
combine an ultrafast phototrigger with a structure-sensitive technique, such as solution x-ray scattering methods (88, 89), to monitor protein folding events with high spatial and temporal resolution.
Author Contributions
R.M.A. and F.G. wrote this article together.
Acknowledgments
We gratefully acknowledge financial support from the National Institutes of Health (GM-065978 and P41-GM104605). R.M.A. is a National Science Foundation Graduate Research Fellow (DGE-1321851).
Editor: James Shorter.
References
- 1.Anfinsen C.B., Haber E., White F.H., Jr. The kinetics of formation of native ribonuclease during oxidation of the reduced polypeptide chain. Proc. Natl. Acad. Sci. USA. 1961;47:1309–1314. doi: 10.1073/pnas.47.9.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levinthal C. Are there pathways for protein folding. J. Chim. Phys. Physico-Chemie Biol. 1968;65:44–45. [Google Scholar]
- 3.Bryngelson J.D., Onuchic J.N., Wolynes P.G. Funnels, pathways, and the energy landscape of protein folding: a synthesis. Proteins. 1995;21:167–195. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- 4.Dill K.A., Chan H.S. From Levinthal to pathways to funnels. Nat. Struct. Biol. 1997;4:10–19. doi: 10.1038/nsb0197-10. [DOI] [PubMed] [Google Scholar]
- 5.Matouschek A., Kellis J.T., Jr., Fersht A.R. Mapping the transition state and pathway of protein folding by protein engineering. Nature. 1989;340:122–126. doi: 10.1038/340122a0. [DOI] [PubMed] [Google Scholar]
- 6.Jackson S.E., Fersht A.R. Folding of chymotrypsin inhibitor 2. 1. Evidence for a two-state transition. Biochemistry. 1991;30:10428–10435. doi: 10.1021/bi00107a010. [DOI] [PubMed] [Google Scholar]
- 7.Williams S., Causgrove T.P., Dyer R.B. Fast events in protein folding: helix melting and formation in a small peptide. Biochemistry. 1996;35:691–697. doi: 10.1021/bi952217p. [DOI] [PubMed] [Google Scholar]
- 8.Schuler B., Lipman E.A., Eaton W.A. Probing the free-energy surface for protein folding with single-molecule fluorescence spectroscopy. Nature. 2002;419:743–747. doi: 10.1038/nature01060. [DOI] [PubMed] [Google Scholar]
- 9.Thirumalai D., O’Brien E.P., Hyeon C. Theoretical perspectives on protein folding. Annu. Rev. Biophys. 2010;39:159–183. doi: 10.1146/annurev-biophys-051309-103835. [DOI] [PubMed] [Google Scholar]
- 10.Uversky V.N., Dunker A.K. Understanding protein non-folding. Biochim. Biophys. Acta. 2010;1804:1231–1264. doi: 10.1016/j.bbapap.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bowman G.R., Voelz V.A., Pande V.S. Taming the complexity of protein folding. Curr. Opin. Struct. Biol. 2011;21:4–11. doi: 10.1016/j.sbi.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gershenson A., Gierasch L.M. Protein folding in the cell: challenges and progress. Curr. Opin. Struct. Biol. 2011;21:32–41. doi: 10.1016/j.sbi.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Best R.B. Atomistic molecular simulations of protein folding. Curr. Opin. Struct. Biol. 2012;22:52–61. doi: 10.1016/j.sbi.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 14.Dror R.O., Dirks R.M., Shaw D.E. Biomolecular simulation: a computational microscope for molecular biology. Annu. Rev. Biophys. 2012;41:429–452. doi: 10.1146/annurev-biophys-042910-155245. [DOI] [PubMed] [Google Scholar]
- 15.Guzman I., Gruebele M. Protein folding dynamics in the cell. J. Phys. Chem. B. 2014;118:8459–8470. doi: 10.1021/jp501866v. [DOI] [PubMed] [Google Scholar]
- 16.Popot J.-L., Engelman D.M. Membranes do not tell proteins how to fold. Biochemistry. 2016;55:5–18. doi: 10.1021/acs.biochem.5b01134. [DOI] [PubMed] [Google Scholar]
- 17.Whitford P.C., Onuchic J.N. What protein folding teaches us about biological function and molecular machines. Curr. Opin. Struct. Biol. 2015;30:57–62. doi: 10.1016/j.sbi.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 18.Sherman E., Haran G. Fluorescence correlation spectroscopy of fast chain dynamics within denatured protein L. ChemPhysChem. 2011;12:696–703. doi: 10.1002/cphc.201000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teufel D.P., Johnson C.M., Neuweiler H. Backbone-driven collapse in unfolded protein chains. J. Mol. Biol. 2011;409:250–262. doi: 10.1016/j.jmb.2011.03.066. [DOI] [PubMed] [Google Scholar]
- 20.Borgia A., Wensley B.G., Schuler B. Localizing internal friction along the reaction coordinate of protein folding by combining ensemble and single-molecule fluorescence spectroscopy. Nat. Commun. 2012;3:1195–1203. doi: 10.1038/ncomms2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soranno A., Buchli B., Schuler B. Quantifying internal friction in unfolded and intrinsically disordered proteins with single-molecule spectroscopy. Proc. Natl. Acad. Sci. USA. 2012;109:17800–17806. doi: 10.1073/pnas.1117368109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Voelz V.A., Jäger M., Pande V.S. Slow unfolded-state structuring in Acyl-CoA binding protein folding revealed by simulation and experiment. J. Am. Chem. Soc. 2012;134:12565–12577. doi: 10.1021/ja302528z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chung H.S., Piana-Agostinetti S., Eaton W.A. Structural origin of slow diffusion in protein folding. Science. 2015;349:1504–1510. doi: 10.1126/science.aab1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szabo A., Schulten K., Schulten Z. First passage time approach to diffusion controlled reactions. J. Chem. Phys. 1980;72:4350–4357. [Google Scholar]
- 25.Zhu Y., Alonso D.O.V., Gai F. Ultrafast folding of alpha3D: a de novo designed three-helix bundle protein. Proc. Natl. Acad. Sci. USA. 2003;100:15486–15491. doi: 10.1073/pnas.2136623100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pirchi M., Ziv G., Haran G. Single-molecule fluorescence spectroscopy maps the folding landscape of a large protein. Nat. Commun. 2011;2:493–499. doi: 10.1038/ncomms1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J., Campos L.A., Muñoz V. Exploring one-state downhill protein folding in single molecules. Proc. Natl. Acad. Sci. USA. 2012;109:179–184. doi: 10.1073/pnas.1111164109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gruszka D.T., Whelan F., Clarke J. Cooperative folding of intrinsically disordered domains drives assembly of a strong elongated protein. Nat. Commun. 2015;6:7271–7279. doi: 10.1038/ncomms8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chung H.S., McHale K., Eaton W.A. Single-molecule fluorescence experiments determine protein folding transition path times. Science. 2012;335:981–984. doi: 10.1126/science.1215768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chung H.S., Louis J.M., Eaton W.A. Experimental determination of upper bound for transition path times in protein folding from single-molecule photon-by-photon trajectories. Proc. Natl. Acad. Sci. USA. 2009;106:11837–11844. doi: 10.1073/pnas.0901178106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao Y., Sirinakis G., Zhang Y. Highly anisotropic stability and folding kinetics of a single coiled coil protein under mechanical tension. J. Am. Chem. Soc. 2011;133:12749–12757. doi: 10.1021/ja204005r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jagannathan B., Elms P.J., Marqusee S. Direct observation of a force-induced switch in the anisotropic mechanical unfolding pathway of a protein. Proc. Natl. Acad. Sci. USA. 2012;109:17820–17825. doi: 10.1073/pnas.1201800109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guinn E.J., Jagannathan B., Marqusee S. Single-molecule chemo-mechanical unfolding reveals multiple transition state barriers in a small single-domain protein. Nat. Commun. 2015;6:6861–6868. doi: 10.1038/ncomms7861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He C., Genchev G.Z., Li H. Mechanically untying a protein slipknot: multiple pathways revealed by force spectroscopy and steered molecular dynamics simulations. J. Am. Chem. Soc. 2012;134:10428–10435. doi: 10.1021/ja3003205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stigler J., Ziegler F., Rief M. The complex folding network of single calmodulin molecules. Science. 2011;334:512–516. doi: 10.1126/science.1207598. [DOI] [PubMed] [Google Scholar]
- 36.Berkovich R., Garcia-Manyes S., Fernandez J.M. Collapse dynamics of single proteins extended by force. Biophys. J. 2010;98:2692–2701. doi: 10.1016/j.bpj.2010.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia-Manyes S., Kuo T.-L., Fernández J.M. Contrasting the individual reactive pathways in protein unfolding and disulfide bond reduction observed within a single protein. J. Am. Chem. Soc. 2011;133:3104–3113. doi: 10.1021/ja109865z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu H., Gupta A.N., Woodside M.T. Energy landscape analysis of native folding of the prion protein yields the diffusion constant, transition path time, and rates. Proc. Natl. Acad. Sci. USA. 2012;109:14452–14457. doi: 10.1073/pnas.1206190109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waldauer S.A., Bakajin O., Lapidus L.J. Extremely slow intramolecular diffusion in unfolded protein L. Proc. Natl. Acad. Sci. USA. 2010;107:13713–13717. doi: 10.1073/pnas.1005415107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu M., Beresneva O., Roder H. Microsecond folding dynamics of apomyoglobin at acidic pH. J. Phys. Chem. B. 2012;116:7014–7025. doi: 10.1021/jp3012365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aksel T., Barrick D. Direct observation of parallel folding pathways revealed using a symmetric repeat protein system. Biophys. J. 2014;107:220–232. doi: 10.1016/j.bpj.2014.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Markiewicz B.N., Culik R.M., Gai F. Tightening up the structure, lighting up the pathway: application of molecular constraints and light to manipulate protein folding, self-assembly and function. Sci. China Chem. 2014;57:1615–1624. doi: 10.1007/s11426-014-5225-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deeg A.A., Rampp M.S., Zinth W. Isomerization- and temperature-jump-induced dynamics of a photoswitchable β-hairpin. Chemistry. 2014;20:694–703. doi: 10.1002/chem.201303189. [DOI] [PubMed] [Google Scholar]
- 44.Bredenbeck J., Helbing J., Hamm P. Alpha-helix formation in a photoswitchable peptide tracked from picoseconds to microseconds by time-resolved IR spectroscopy. Proc. Natl. Acad. Sci. USA. 2005;102:2379–2384. doi: 10.1073/pnas.0406948102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ihalainen J.A., Paoli B., Hamm P. Alpha-Helix folding in the presence of structural constraints. Proc. Natl. Acad. Sci. USA. 2008;105:9588–9593. doi: 10.1073/pnas.0712099105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen E., Kumita J.R., Kliger D.S. The kinetics of helix unfolding of an azobenzene cross-linked peptide probed by nanosecond time-resolved optical rotatory dispersion. J. Am. Chem. Soc. 2003;125:12443–12449. doi: 10.1021/ja030277+. [DOI] [PubMed] [Google Scholar]
- 47.Abaskharon R.M., Culik R.M., Gai F. Tuning the attempt frequency of protein folding dynamics via transition-state rigidification: application to Trp-cage. J. Phys. Chem. Lett. 2015;6:521–526. doi: 10.1021/jz502654q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu H.S.M., Volk M., DeGrado W.F. Aminothiotyrosine disulfide, an optical trigger for initiation of protein folding. J. Am. Chem. Soc. 1997;119:7173–7180. [Google Scholar]
- 49.Milanesi L., Waltho J.P., Volk M. Measurement of energy landscape roughness of folded and unfolded proteins. Proc. Natl. Acad. Sci. USA. 2012;109:19563–19568. doi: 10.1073/pnas.1211764109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Regner N., Herzog T.T., Zinth W. Light-switchable hemithioindigo-hemistilbene-containing peptides: ultrafast spectroscopy of the Z → E isomerization of the chromophore and the structural dynamics of the peptide moiety. J. Phys. Chem. B. 2012;116:4181–4191. doi: 10.1021/jp300982a. [DOI] [PubMed] [Google Scholar]
- 51.Brown S.P., Smith A.B., 3rd Peptide/protein stapling and unstapling: introduction of s-tetrazine, photochemical release, and regeneration of the peptide/protein. J. Am. Chem. Soc. 2015;137:4034–4037. doi: 10.1021/ja512880g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tucker M.J., Courter J.R., Hochstrasser R.M. Tetrazine phototriggers: probes for peptide dynamics. Angew. Chem. Int. Ed. Engl. 2010;49:3612–3616. doi: 10.1002/anie.201000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tucker M.J., Abdo M., Hochstrasser R.M. Nonequilibrium dynamics of helix reorganization observed by transient 2D IR spectroscopy. Proc. Natl. Acad. Sci. USA. 2013;110:17314–17319. doi: 10.1073/pnas.1311876110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin M.M., Mohammed O.F., Zewail A.H. Speed limit of protein folding evidenced in secondary structure dynamics. Proc. Natl. Acad. Sci. USA. 2011;108:16622–16627. doi: 10.1073/pnas.1113649108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Markiewicz B.N., Jo H., Gai F. Assessment of local friction in protein folding dynamics using a helix cross-linker. J. Phys. Chem. B. 2013;117:14688–14696. doi: 10.1021/jp409334h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nobrega R.P., Arora K., Matthews C.R. Modulation of frustration in folding by sequence permutation. Proc. Natl. Acad. Sci. USA. 2014;111:10562–10567. doi: 10.1073/pnas.1324230111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hilaire M.R., Abaskharon R.M., Gai F. Biomolecular crowding arising from small molecules, molecular constraints, surface packing, and nano-confinement. J. Phys. Chem. Lett. 2015;6:2546–2553. doi: 10.1021/acs.jpclett.5b00957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sagnella D.E., Straub J.E., Thirumalai D. Time scales and pathways for kinetic energy relaxation in solvated proteins: application to carbonmonoxy myoglobin. J. Chem. Phys. 2000;113:7702–7711. [Google Scholar]
- 59.Markiewicz B.N., Yang L., Gai F. How quickly can a β-hairpin fold from its transition state? J. Phys. Chem. B. 2014;118:3317–3325. doi: 10.1021/jp500774q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shandiz A.T., Baxa M.C., Sosnick T.R. A “Link-Psi” strategy using cross-linking indicates that the folding transition state of ubiquitin is not very malleable. Protein Sci. 2012;21:819–827. doi: 10.1002/pro.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Serrano A.L., Waegele M.M., Gai F. Spectroscopic studies of protein folding: linear and nonlinear methods. Protein Sci. 2012;21:157–170. doi: 10.1002/pro.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Culik R.M., Serrano A.L., Gai F. Achieving secondary structural resolution in kinetic measurements of protein folding: a case study of the folding mechanism of Trp-cage. Angew. Chem. Int. Ed. Engl. 2011;50:10884–10887. doi: 10.1002/anie.201104085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chi H., Welch W.R.W., Keiderling T.A. Insight into the packing pattern of β2 fibrils: a model study of glutamic acid rich oligomers with 13C isotopic edited vibrational spectroscopy. Biomacromolecules. 2013;14:3880–3891. doi: 10.1021/bm401015f. [DOI] [PubMed] [Google Scholar]
- 64.Chung H.S., Khalil M., Tokmakoff A. Transient two-dimensional IR spectrometer for probing nanosecond temperature-jump kinetics. Rev. Sci. Instrum. 2007;78:063101. doi: 10.1063/1.2743168. [DOI] [PubMed] [Google Scholar]
- 65.Jones K.C., Peng C.S., Tokmakoff A. Folding of a heterogeneous β-hairpin peptide from temperature-jump 2D IR spectroscopy. Proc. Natl. Acad. Sci. USA. 2013;110:2828–2833. doi: 10.1073/pnas.1211968110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moran S.D., Woys A.M., Zanni M.T. Two-dimensional IR spectroscopy and segmental 13C labeling reveals the domain structure of human γD-crystallin amyloid fibrils. Proc. Natl. Acad. Sci. USA. 2012;109:3329–3334. doi: 10.1073/pnas.1117704109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Remorino A., Korendovych I.V., Hochstrasser R.M. Residue-specific vibrational echoes yield 3D structures of a transmembrane helix dimer. Science. 2011;332:1206–1209. doi: 10.1126/science.1202997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rogers J.M.G., Lippert L.G., Gai F. Non-natural amino acid fluorophores for one- and two-step fluorescence resonance energy transfer applications. Anal. Biochem. 2010;399:182–189. doi: 10.1016/j.ab.2009.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Goldberg J.M., Wissner R.F., Petersson E.J. Thioamide quenching of intrinsic protein fluorescence. Chem. Commun. (Camb.) 2012;48:1550–1552. doi: 10.1039/c1cc14708k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mintzer M.R., Troxler T., Gai F. p-Cyanophenylalanine and selenomethionine constitute a useful fluorophore-quencher pair for short distance measurements: application to polyproline peptides. Phys. Chem. Chem. Phys. 2015;17:7881–7887. doi: 10.1039/c5cp00050e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peran I., Watson M.D., Raleigh D.P. Selenomethionine, p-cyanophenylalanine pairs provide a convenient, sensitive, non-perturbing fluorescent probe of local helical structure. Chem. Commun. (Camb.) 2016;52:2055–2058. doi: 10.1039/c5cc08232c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Prigozhin M.B., Chao S.-H., Gruebele M. Mapping fast protein folding with multiple-site fluorescent probes. Proc. Natl. Acad. Sci. USA. 2015;112:7966–7971. doi: 10.1073/pnas.1422683112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ma J., Pazos I.M., Gai F. Site-specific infrared probes of proteins. Annu. Rev. Phys. Chem. 2015;66:357–377. doi: 10.1146/annurev-physchem-040214-121802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Davis C.M., Dyer R.B. The role of electrostatic interactions in folding of β-proteins. J. Am. Chem. Soc. 2016;138:1456–1464. doi: 10.1021/jacs.5b13201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nagarajan S., Taskent-Sezgin H., Dyer R.B. Differential ordering of the protein backbone and side chains during protein folding revealed by site-specific recombinant infrared probes. J. Am. Chem. Soc. 2011;133:20335–20340. doi: 10.1021/ja2071362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bachmann A., Wildemann D., Kiefhaber T. Mapping backbone and side-chain interactions in the transition state of a coupled protein folding and binding reaction. Proc. Natl. Acad. Sci. USA. 2011;108:3952–3957. doi: 10.1073/pnas.1012668108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Culik R.M., Jo H., Gai F. Using thioamides to site-specifically interrogate the dynamics of hydrogen bond formation in β-sheet folding. J. Am. Chem. Soc. 2012;134:8026–8029. doi: 10.1021/ja301681v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sanchez-Medina C., Sekhar A., Kay L.E. Probing the free energy landscape of the fast-folding gpW protein by relaxation dispersion NMR. J. Am. Chem. Soc. 2014;136:7444–7451. doi: 10.1021/ja502705y. [DOI] [PubMed] [Google Scholar]
- 79.Kan Z.-Y., Walters B.T., Englander S.W. Protein hydrogen exchange at residue resolution by proteolytic fragmentation mass spectrometry analysis. Proc. Natl. Acad. Sci. USA. 2013;110:16438–16443. doi: 10.1073/pnas.1315532110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Calabrese A.N., Ault J.R., Ashcroft A.E. Using hydroxyl radical footprinting to explore the free energy landscape of protein folding. Methods. 2015;89:38–44. doi: 10.1016/j.ymeth.2015.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shaw D.E., Chao J.C., Bowers K.J. Anton, a special-purpose machine for molecular dynamics simulation. Commun. ACM. 2008;51:91–97. [Google Scholar]
- 82.Shaw D.E., Maragakis P., Wriggers W. Atomic-level characterization of the structural dynamics of proteins. Science. 2010;330:341–346. doi: 10.1126/science.1187409. [DOI] [PubMed] [Google Scholar]
- 83.Lindorff-Larsen K., Piana S., Shaw D.E. How fast-folding proteins fold. Science. 2011;334:517–520. doi: 10.1126/science.1208351. [DOI] [PubMed] [Google Scholar]
- 84.Pande V.S., Beauchamp K., Bowman G.R. Everything you wanted to know about Markov State Models but were afraid to ask. Methods. 2010;52:99–105. doi: 10.1016/j.ymeth.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Voelz V.A., Bowman G.R., Pande V.S. Molecular simulation of ab initio protein folding for a millisecond folder NTL9(1-39) J. Am. Chem. Soc. 2010;132:1526–1528. doi: 10.1021/ja9090353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Best R.B., Zhu X., Mackerell A.D., Jr. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Best R.B., Zheng W., Mittal J. Balanced protein-water interactions improve properties of disordered proteins and non-specific protein association. J. Chem. Theory Comput. 2014;10:5113–5124. doi: 10.1021/ct500569b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cho H.S., Dashdorj N., Anfinrud P. Protein structural dynamics in solution unveiled via 100-ps time-resolved x-ray scattering. Proc. Natl. Acad. Sci. USA. 2010;107:7281–7286. doi: 10.1073/pnas.1002951107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kim J.G., Kim T.W., Ihee H. Protein structural dynamics revealed by time-resolved x-ray solution scattering. Acc. Chem. Res. 2015;48:2200–2208. doi: 10.1021/acs.accounts.5b00198. [DOI] [PMC free article] [PubMed] [Google Scholar]