Abstract
Serum- and glucocorticoid-regulated protein kinase 3 (SGK3) is critical for tumor survival, proliferation and invasion. In the present study we evaluated SGK3 expression in breast tissues and investigated alterations in SGK3 levels in tumor multiplication, progression and apoptosis. Tissue microarray analyses were performed to examine SGK3 expression in breast cancer samples, as well as in adjacent noncancerous and normal tissues. The pEGFP-N1-SGK3 plasmid was transfected into MDA-MB-231 cells to generate SGK3-overexpressing cells. Cell growth assays, colony formation assays, cell cycle analyses, horizontal and vertical migration tests, reverse transcription-polymerase chain reaction and western blot assays were employed to investigate the biological behavior of SGK3-overexpressing cells. SGK3 levels were significantly higher in breast cancer samples compared with adjacent noncancerous and normal tissues. Cell growth curves revealed increased proliferation and decreased apoptosis in SGK3-overexpressing cells. SGK3-overexpressing cells also demonstrated enhanced invasion and migration abilities. SGK3-overexpressing cells had high levels of an apoptosis-related gene (bcl-xl) and invasion-related genes (mmp2 and mmp9), and decreased levels of an anti-apoptosis gene (bad). Phosphorylation of GSK-3β, which is downstream in the phosphoinositide 3-kinase signaling pathway, was activated by SGK3 overexpression. β-catenin phosphorylation did not differ between the SGK3-overexpressing and non-SGK3-overexpressing cells. SGK3 overexpression induces GSK-3β phosphorylation, enhancing apoptosis- and invasion-related genes and proteins and thereby leading to tumor development and aggression in breast cancer cells.
Keywords: serum- and glucocorticoid-regulated protein kinase 3, apoptosis, tumor invasion, cell migration, breast cancer
Introduction
Breast cancer is the most common malignancy in females in the United States, and the disease is expected to cause 40,430 mortalities in 2014 according to the National Cancer Institute's Surveillance, Epidemiology and End Results (SEER) Program (1). Although breast cancer is one of the most life-threatening diseases, the molecular processes involved in its pathogenesis have not yet been fully elucidated. Various risk factors, including family history, genetic mutations, reproductive history and environmental alterations, correspond to cancer development (2,3). Advances in modern biochemistry provide numerous technical strategies for illustrating the etiology of breast malignancies. However, the complexity of cancer evolution, which involves a number of genes and molecular pathways, prevents further investigation of underlying mechanisms.
Over the past few decades, developments in early cancer detection and the combination of multiple treatments have significantly reduced breast cancer mortality (1). Among these, targeted therapy is one of the most promising strategies. However, with ever-increasing new cases and the development of drug resistance, there is an urgent requirement to identify more effective methods to deliver biochemical therapies.
Previous studies have demonstrated that phosphoinositide 3-kinase (PI3K) signaling plays a critical role in cell proliferation. Within this pathway, key components including PIK and AKT have emerged as potential targets for drug development (4,5). However, a closely related protein, serum- and glucocorticoid-regulated protein kinase (SGK), which shares 54% of its identity with the kinase domains of AKT, has received little attention (6). Vasudevan et al (7) reported that a number of cancer cell lines are SGK3-dependent but exhibit only minimal AKT activation, indicating that SGK3 is an essential independent factor in the PI3K pathway and providing an alternative explanation for tumor behavior. SGK is active in various cell signaling pathways and serves as a significant factor in cell phosphorylation, which subsequently triggers downstream substrates to stimulate cellular responses, including cell survival, differentiation and material transduction. SGK3, first described by Kobayashi et al (6), differs significantly in terms of enzyme structure, function, corresponding gene locus and regulation of protein expression compared with its isoform SGK1, which has been more frequently investigated. Current animal experimental data provide limited information with regard to the function of SGK3, and do not address the application of this enzyme in humans. However, previous studies have revealed that increased SGK3 expression in various types of tumor cells contributes to cancer development as well as tumor cell infiltration and migration, suggesting that this enzyme may be a potential target in disease prognosis and drug development (8).
In this study, we investigated SGK3 expression in human breast tissues and the potential effects of increased SGK3 levels on tumor cell multiplication, invasion, migration and apoptosis. We also examined the underlying mechanisms of these phenomena.
Materials and methods
Cells and plasmids
The human breast cancer cell line MDA-MB-231 (Cancer Institution, Chinese Academy of Medical Science, Beijing, China) was cultured in RPMI-1640 media containing 10% fetal bovine serum (FBS) at 37°C with 5% CO2. The pAcG-4T3-SKG3 plasmid, which contains 1.3 kb of human SGK3 cDNA with BamHI/XhoI sites, was provided by Dr. Xianming Xia (Johns Hopkins University School of Medicine, Baltimore, MD, USA). The pEGFP-N1 plasmid was a gift from Dr. Changzhi Huang (Cancer Institute and Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China).
Reagents and antibodies
The following reagents and antibodies were applied in this study: breast tissue chips BR725 (Alenabio Company, Xi'an, China), RPMI-1640 media (Gibco Life Technologies, Carlsbad, CA, USA), FBS (Invitrogen Life Technologies, Carlsbad, CA, USA), trypsin (CAS number 9002-07-7, Sigma-Aldrich, St. Louis, MO, USA), BamHI/XhoI and PrimeScript™ RT-PCR kit (Takara Biotechnology Co., Ltd., Dalian, China), PCR enzyme mix and Advantage PCR cloning kit (Clontech Laboratories, Inc., Mountain View, CA, USA), SV Total RNA isolation system (Promega Corporation, Madison, WI, USA), radioimmunoprecipitation assay (RIPA) buffer (Applygen Technologies, Inc., Beijing, China), anti-β-actin, anti-SGK3, anti-BAD, anti-Bcl-xL, anti-matrix metalloproteinase 2 (MMP2), anti-MMP9, anti-phospho-GSK-3β, anti-phospho-β-catenin (CTS, Boston, MA, USA), anti-breast cancer metastasis suppressor 1 (BRMS1) and horseradish peroxidase-labeled secondary antibody (Santa Cruz Biotechnology, Inc., Dallas, Texas, USA). Sangon Biotech (Shanghai, China) synthesized all primers.
Specimens and tissue microarray analyses
Breast tissue microarray analyses were performed to assess SGK3 expression in formalin-fixed, paraffin-embedded specimens from various breast tissues, including 24 normal breast samples, 24 breast carcinoma specimens and 24 adjacent noncancerous tissues. The clinicopathological data are presented in Table I. Visible brown ink in cells using fluorescence microscopy indicated positive SGK3 expression. The integral optical density (IOD) of each image was analyzed using Image-Pro Plus software (Media Cybernetics, Rockville, MD, USA). The study was approved by the Institutional Review Board of Qiqihar Medical University, China, and patient consent was waived.
Table I.
Clinicopathological characteristics of enrolled breast cancer patients and tissues.
| Clinicopathological factors | Mean or number |
|---|---|
| Age (years) | 47.22±8.90 |
| Tumor type | |
| Invasive ductal carcinoma | 20 |
| Atypical medullary carcinoma | 2 |
| Mucinous adenocarcinoma | 2 |
| Histological grade | |
| 1 | 1 |
| 2 | 19 |
| 3 | 1 |
| N/A | 3 |
Reconstruction of pEGFP-N1-SGK3
A full-length clone encoding human SGK3 was obtained as follows: XhoI and BamHI sites were introduced via the forward and reverse primers, which were designed according to the SGK3 cDNA using Primer Premier 5.0 software (Premier Biosoft, Palo Alto, CA, USA). SGK3 was amplified by polymerase chain reaction (PCR) using pAcG-4T3-SKG3 as a template and the specific primers 5′-CCGCTCGAGACCATGGCCCTGAAGATTC-3′ (forward) and 5′-CGCGGATCCAAAAATAAGTCTTCTG-3′ (reverse). Purified PCR products and the pEGFP-N1 vector were digested with XhoI and BamHI, and subsequently ligated to construct pEGFP-N1-SGK3. Positive clones were analyzed by sequencing (Invitrogen, Shanghai, China) and cross-comparisons were performed using the results and data from GenBank [ClustalW software, European Molecular Biology Laboratory - European Bioinformatics Institute (EMBL-EBI)].
Plasmid transfection
Logarithmic phase MDA-MB-231 cells were seeded into six-well plates (Nunc, Roskilde, Denmark) at 24 h before transfection to achieve a density of 2×105 cells/well. Each well was transfected with 3 µg pEGFP-N1-SGK3 or pEGFP-N1 using GenEscort™ I (Isegen, Nanjing, China) according to the manufacturer's instructions. Forty-eight hours after transfection, the treated cells were harvested and SGK fusion protein analyses were performed. Cells successfully transfected with pEGFP-N1-SGK3 or pEGFP-N1 were termed group SGK3 and group N1 (negative control), respectively. MDA-MB-231 cells served as a blank control (group 231). All in vitro experiments were carried out for each group.
Cell growth assays
Logarithmic phase tumor cells were placed separately in four wells in a 96-well plate (500 cells/well in five plates). As described by Shi et al (9) one plate was used for MTT assays for five consecutive days. Briefly, 10 µl MTT reagent was added to each well and cultured for 2 h at 37°C. Media were discarded and cells were incubated with 150 µl dimethyl sulfoxide for 15 min with mild agitation. Cells were counted every day by measuring the absorbance at 5700 nm. Cell growth curves were subsequently generated.
Colony formation assays
Cells were seeded in six-well plates (500 cells/well) and cultured for 2 weeks at 37°C with 5% CO2. Each sample was performed in triplicate, and culture media were changed every 3 days. Cells were fixed in methanol for 30 min, stained with crystal violet, and air-dried. The colony formation rate = number of colonies / number of seeded cells.
Cell cycle analyses
Tumor cells from the three groups were seeded in six-well plates (1×105 cells per well) and incubated for 48 h before digestion with 0.25% trypsin. All cells were washed twice with pre-cooled phosphate-buffered saline (PBS) and incubated for 30 min at room temperature with propidium iodide (PI; Sigma-Aldrich). Cells were immediately examined by flow cytometry using a FACSCalibur (Becton-Dickinson, New York, NY, USA). Data were analyzed using ModFit LT (Verity Software House, Topsham, ME, USA).
Horizontal migration tests (scratch tests)
Tumor cells were placed in six-well plates (1×105 cells per well). Once a single layer of cells had formed, two scratches were made by swiping a small sterilized tip across the inner surface of the bottom of the plate. The scratch area was washed with PBS until the cells were removed. RPMI-1640 was added and cells were cultured for 24 h. The distance between cells at 0, 12 and 24 h in the scratch area was measured using microscopy. Horizontal cell migration rates were automatically analyzed using Image-Pro Plus software.
Vertical migration tests (Transwell tests)
Logarithmic growth phase cells were cultured in serum-free media for 12–24 h, and then collected and suspended at a density of 2×105 cells/ml in media containing 0.2% FBS. Cell suspensions (200 µl) were added to the upper Transwell chamber, and 600 µl RPMI-1640 media containing 10% FBS was added to the lower chamber. After 24 h, cells that remained in the upper membrane were gently removed with cotton and washed three times with PBS. Cells that migrated to the lower membrane were fixed in methanol for 30 min and stained for 20 min with 0.1% crystal violet. Cells were counted in five random fields of the lower membrane to identify the number of vertically migrating cells.
mRNA expression of apoptosis- and invasion-related genes
We isolated total RNA from tumor cells using the SV Total RNA isolation kit. cDNA was prepared using the RT-PCR kit as per the manufacturer's instructions. The sequences of primers are listed in Table II. Reactions were subjected to 30 cycles of 94°C denaturation for 4 min, 94°C melting for 30 sec, annealing for 30 sec (specific annealing temperatures are listed in Table II), 72°C extension for 60 sec, with a final elongation step at 72°C for 10 min. PCR products were separated by electrophoresis in 1% agarose gels, visualized using ethidium bromide, and analyzed using the AlphaImager HP system (ProteinSimple, San Jose, CA, USA).
Table II.
Primer sequences used for apoptosis- and invasion-related mRNA analyses.
| mRNA | Sequence | Annealing temperature (°C) | Length (bp) |
|---|---|---|---|
| bad | F 5′-CCATCCCTTCGTCGTCCTC-3′ | 51.0 | 162 |
| R 5′-GCTCCGGCAAGCATCATC-3′ | |||
| bcl-xl | F 5′-CGGGCATTCAGTGACCTGAC-3′ | 54.3 | 151 |
| R 5′-TCAGGAACCAGCGGTTGAAG-3′ | |||
| mmp2 | F 5′-GTATTTGATGGCATCGCTCA-3′ | 52.0 | 196 |
| R 5′-CATTCCCTGCAAAGAACACA-3′ | |||
| mmp9 | F 5′-GGAGCCGCTCTCCAAGAAGCTT-3′ | 58.6 | 522 |
| R 5′-CTCCTCCCTTTCCTCCAGAACAGAA-3′ | |||
| brms1 | F 5′-ACTGAGTCAGCTGCGGTTGCGG-3′ | 61.7 | 335 |
| R 5′-AAGACCTGGAGCTGCCTCTGGCGTGC-3′ | |||
| β-actin | F 5′-CATCTCTTGCTCGAAGTCCA-3′ | 52.8 | 263 |
| R 5′-ATCATGTTTGAGACCTTCAACA-3′ |
Western blot analysis
Cells were collected by centrifugation for 5 min at 800 rpm, and proteins were extracted following lysis in RIPA buffer. Protein concentrations were determined using protein assays (Amersham Biosciences, London, England). Purified proteins (50 µl) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes. Membranes were blocked for 2 h at room temperature in 5% non-fat milk and incubated overnight at 4°C with primary antibodies against BAD, Bcl-xL, MMP2, MMP9 and BRMS1 (1:1,000 dilution). Following incubation with the horseradish peroxidase-labeled secondary antibodies (1:5,000 dilution), membranes were washed and proteins were visualized using chemiluminescence reagents. Western blotting results were quantified using ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Statistical analyses
SPSS version 17.0 (SPSS Inc., Chicago, IL, USA) was used for data analyses. All results are presented as the means ± standard deviation. One-way analysis of variance and Student's t-tests were used to compare the difference in means within groups. P<0.05 was considered to indicate a statistically significant difference.
Results
SGK3 expression in breast tissues
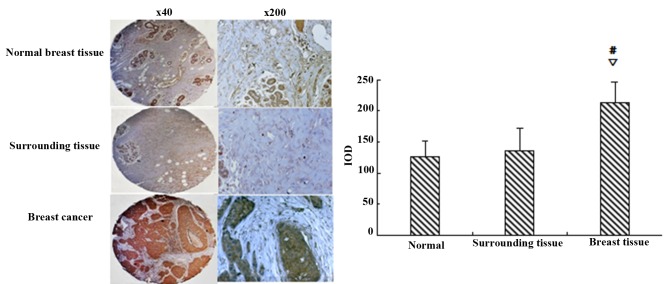
The IOD of each enrolled breast tissue revealed no significant differences in SGK3 expression between normal and adjacent noncancerous tissues (P>0.05). However, SGK3 levels were significantly higher in breast carcinoma samples compared with normal and adjacent noncancerous tissues (P<0.01, Fig. 1).
Figure 1.
Immunohistochemistry analyses reveal the expression of serum- and glucocorticoid-regulated protein kinase 3 (SGK3) in breast tissues. SGK3 levels were significantly higher in breast cancer specimens; #∇P<0.01. IOD, integral optical density.
Verification of SGK3 expression in transfected tumor cells
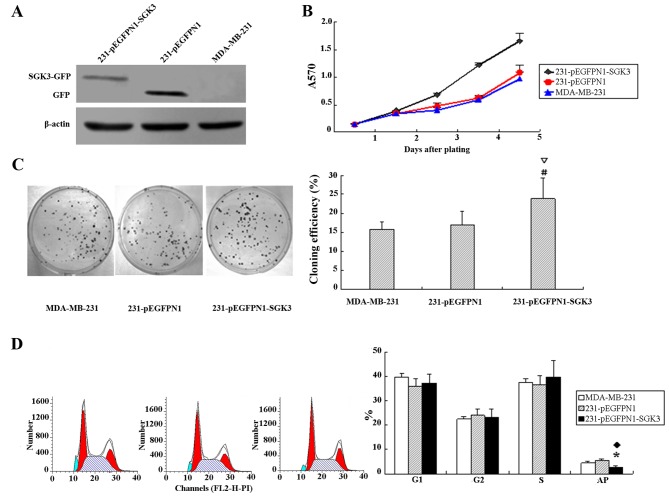
pEGFP-N1-SGK3 and pEGFP-N1 were successfully transfected into MDA-MB-231 cells. Western blotting results revealed SGK3 expression in corresponding host cells; however, the level of SGK3-green fluorescent protein (GFP) fusion protein in cells from the SGK3 group was slightly lower than that of GFP in N1 group negative control cells (Fig. 2A).
Figure 2.
(A) Expression of serum- and glucocorticoid-regulated protein kinase 3 (SGK3)-green fluorescent protein (GFP) and SGK3 in cells from group SGK3, N1 and 231 cells (western blot analysis). (B) Effect of SGK3 overexpression on cell multiplication. (C) Colony formation assays, #∇P<0.01. (D) Effect of SGK3 overexpression on cell apoptosis and the cell cycle; *♦P<0.05.
SGK3 contributes to tumor cell proliferation
The cell growth curves of each group are presented in Fig. 2B. We observed accelerating tumor multiplication in cells from the SGK3 group, whereas the proliferation rate of N1 and 231 group cells remained low. Additionally, the doubling times were 27.6, 34.5 and 35.1 h, respectively, for SCK3, N1 and 231 group cells. These data demonstrated that SGK3 induced cell proliferation.
Increased colony formation in SGK3-expressing tumor cells
Significantly higher colony formation rates were detected in cells from the SGK3 group compared with those from groups N1 and 231 (16.3±1.96%, 17.1±2.12% and 22.5±1.6%, respectively; P<0.01). The colony formation rates in N1 and 231 cells were not significantly different (P>0.05, Fig. 2C).
SGK3 inhibits apoptosis
Figure 2D reveals a lower proportion of apoptotic cells in SGK3-overexpressing cells compared with cells in group N1 and 231 (P<0.05). No significant differences were observed with regard to the percentage of G1, G2 and S phase cells (P>0.05).
Cell migration in SGK3-overexpressing cells
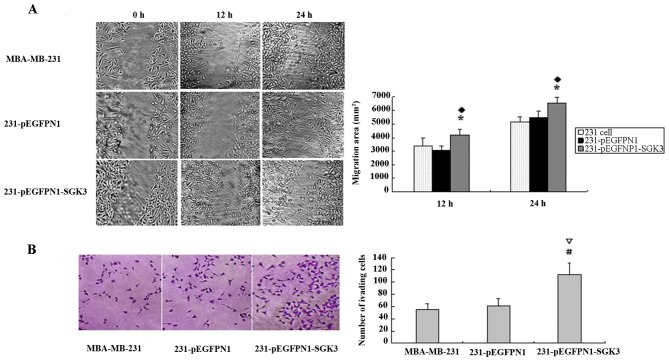
Horizontal migration tests revealed that scratches in the blank and negative control groups remained virtually unchanged after 12 h, whereas SGK3-overexpressing cells revealed higher migration rates (P<0.05). After 24 h, scratches were hardly visible in the SGK3 group cells; however, N1 and 231 cells had not yet recovered from the damage (P<0.05, Fig. 3A). We also observed similar results in vertical migration tests. Larger numbers of SGK3 group cells migrated to the lower membrane compared with N1 and 231 cells (P<0.01), but no differences were observed between the latter two groups (P>0.05, Fig. 3B).
Figure 3.
(A) Horizontal migration tests revealed significantly higher rates of migration in serum- and glucocorticoid-regulated protein kinase 3 (SGK3) cells; *♦P<0.05. (B) Vertical migration tests revealed that more cells migrated to the lower membrane in group SGK3; #∇P<0.01.
mRNA expression of apoptosis- and invasion-related genes
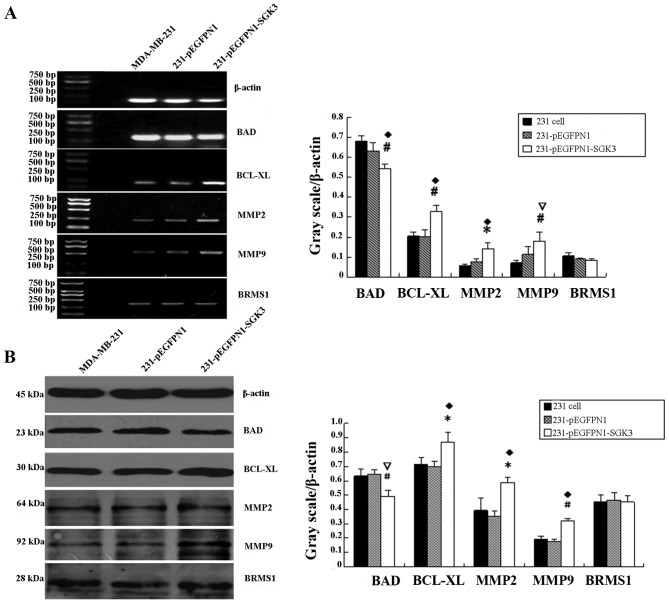
RT-PCR product analyses using the AlphaImager HP system revealed that the grayscale values of electrophoretic bands for bcl-xl, mmp2 and mmp9 in the SGK3 group were significantly higher compared with the other two groups (P<0.05, P<0.01). However, bad mRNA expression decreased in SGK3-overexpressing cells (P<0.05, Fig. 4A). brms1 mRNA levels were comparable among the three groups (P>0.05). No significant differences in these genes were noted between the N1 and 231 group cells (P>0.05).
Figure 4.
(A) Effect of serum- and glucocorticoid-regulated protein kinase 3 (SGK3) overexpression on bcl-xl, bad, mmp2, mmp9 and brms1 mRNA levels (reverse transcription-polymerase chain reaction); *♦P<0.05, #∇P<0.01. (B) Effect of SGK3 overexpression on Bcl-xL, BAD, MMP2, MMP9 and BRMS1 levels (western blot analysis); *♦P<0.05, #∇P<0.01.
Western blot analysis of apoptosis- and migration-related proteins
Bcl-xL, MMP2 and MMP9 expression was significantly higher in SGK3-overexpressing cells compared with the other two groups (P<0.05, P<0.01), whereas BAD expression was decreased (P<0.01, Fig. 4B). There was no difference in BRMS1 expression among the three groups (P>0.05). In the blank and negative control groups, no significant differences were detected with regard to these five proteins (P>0.05).
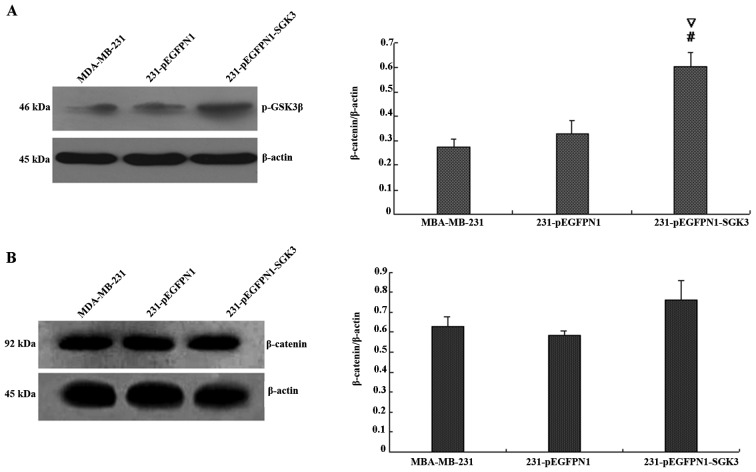
Effect of SGK3 overexpression on PI3K signaling
We next examined the expression of phospho-GSK3β and phospho-β-catenin (Fig. 5A and B). The grayscale value of phospho-GSK3β demonstrated that expression was higher in the SGK3 group cells (P<0.01), whereas no differences were detected between N1 and 231 group cells (P>0.05). In contrast, western blot analyses revealed that SGK3-overexpressing cells did not differ in phospho-β-catenin levels compared with the other two groups (P>0.05). Thus, SGK3 overexpression did not contribute to β-catenin phosphorylation.
Figure 5.
(A) Effect of serum- and glucocorticoid-regulated protein kinase 3 (SGK3) overexpression on GSK3β phosphorylation; #∇P<0.01. (B) Effect of SGK3 overexpression on β-catenin phosphorylation. # and * indicate comparison between groups SGK3 and 231; ∇ and ♦ indicate comparison between groups SGK3 and N1.
Discussion
In this study, we observed higher expression levels of SGK3 in breast cancer tissues compared with adjacent noncancerous and normal breast tissues using microarray assay. These data are similar to previous publications evaluating SGK3 expression in prostate, breast and liver cancers (8,10–12). Our findings confirm that SGK3 may be a specific target for drug development and delivery, since this protein is strictly expressed in tumor cells.
The results of gene analyses in our study indicated successful reconstruction of the eukaryotic expression vector pEGFP-N1-SGK3 using the GenEscort I system. This expression construct may be used for future investigations of breast cancer invasion and migration, along with their corresponding mechanisms. GFP labeling was also employed to achieve better visualization of the protein. The transfection system had an efficiency of greater than 70% when delivering the plasmid to MDA-MB-231 cells, and our transfection results were verified by western blot analysis in the different groups of cells.
Cell cycle analyses revealed that cells in the SGK3 group had lower rates of apoptosis compared with group N1 and 231 cells, indicating that this protein may alter the behavior of tumor cells. Additionally, as demonstrated by the cell growth curves, SGK3-overexpressing cells presented more robust proliferation rates in the same logarithmic phase as the blank and negative control groups. Moreover, migration tests verified that SGK3 contributed to both tumor cell recovery and invasion. Scratch tests illustrated that SGK3 cells were more resistant to external damage and regenerated within 24 h. Transwell assays revealed that higher numbers of SGK3-overexpressing cells migrated to the lower chamber compared with N1 and 231 group cells. These data demonstrate that expression of exogenous SGK3 cDNA significantly improved breast cancer cell multiplication and invasion.
Gene dysregulation is a critical biological process contributing to tumor development. This theory postulates that uncontrolled cell proliferation and decreased apoptosis are caused by gene dysregulation and an upset in the balance between cell growth and death, thereby leading to tumor formation. Apoptosis has become a major target for ameliorating drug sensitivity and enhancing drug delivery (13). Molecules in the BCL-2 family, including BAD and Bcl-xL, are notable factors in cell apoptosis and contribute to a so-called ‘apoptosis checkpoint’. Our study demonstrated that SGK3 upregulates bcl-xl mRNA and protein levels, which inhibits the apoptosis process. Simultaneously, the expression of bad mRNA and protein, which facilitates apoptosis, significantly decreased. Another pivotal malignant behavior is the ability of invasion and migration, which are considered together as the most critical cause of patient mortality in clinical practice. MMPs have great potential for improving tumor invasion and migration (14–18), and also serve as biomarkers for predicting cancer development (15,16). RT-PCR and western blot assays indicated that SGK3 enhanced the mRNA and protein levels of MMP2 and MMP9, which further advanced tumor progression. Previous studies have reported that BRMS1 plays a critical role in metastasis in various tumors, including breast, ovarian, melanoma and hepatocellular carcinoma (19–22). Notably, BRMS1 mRNA and protein levels in the SGK3 group cells were not significantly different compared with cells in groups N1 and 231. One possible explanation for this discrepancy might be that other critical molecules involved in the process between matrix invasion and distant metastasis were not fully examined, and they may not respond to altered SGK3 levels.
GSK-3β is a downstream component of SGK3 in the PI3K pathway, and as expected, SGK3 levels significantly affected GSK-3β phosphorylation. However, β-catenin phosphorylation did not change with changes in SGK3 expression. Phosphorylation of β-catenin at specific Ser/Thr residues targets it for degradation via the ubiquitin proteasome pathway. However, mutations in β-catenin that cause dephosphorylation increase accumulation of the non-degradable protein, ultimately leading to activation of cancerous genes. β-catenin is crucial for Wnt-Ca2+ signaling, and the absence of a response to SGK3 suggests that SGK3 is not a significant component in this pathway. We propose that the SGK3-mediated aggressive behavior of breast cancer cells is conducted via the PI3K pathway.
This study has certain limitations. First, we presented preliminary data of SGK3 function in a small group of breast tissues. Additionally, our samples were primarily limited to invasive ductal carcinoma. The behavior of different breast cancer subtypes and different histology grades when SGK levels are altered has not yet been examined. Finally, the aggressiveness of cancer cells following SGK gene silencing should be evaluated.
In conclusion, SGK3 overexpression induced phosphorylation of GSK-3β, which is downstream in PI3K signaling, and consequently enhanced the level of apoptosis- and invasion-related proteins, thereby leading to tumor development and aggression of breast cancer cells.
Acknowledgements
This study was supported by the Natural Science Foundation of Heilongjiang Province (QC2014C035).
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Li Y, Drabsch Y, Pujuguet P, Ren J, van Laar T, Zhang L, van Dam H, Clément-Lacroix P, Ten Dijke P. Genetic depletion and pharmacological targeting of αv integrin in breast cancer cells impairs metastasis in zebrafish and mouse xenograft models. Breast Cancer Res. 2015;17:28. doi: 10.1186/s13058-015-0537-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu D, Zhou X, Yao L, Liu C, Jin F, Wu Y. Clinical implications of the interleukin 27 serum level in breast cancer. J Investig Med. 2014;62:627–631. doi: 10.2310/JIM.0000000000000046. [DOI] [PubMed] [Google Scholar]
- 4.Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 5.Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov. 2009;8:627–644. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kobayashi T, Deak M, Morrice N, Cohen P. Characterization of the structure and regulation of two novel isoforms of serum- and glucocorticoid-induced protein kinase. Biochem J. 1999;344:189–197. doi: 10.1042/bj3440189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vasudevan KM, Barbie DA, Davies MA, Rabinovsky R, McNear CJ, Kim JJ, Hennessy BT, Tseng H, Pochanard P, Kim SY, et al. AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer Cell. 2009;16:21–32. doi: 10.1016/j.ccr.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Zhou D, Phung S, Masri S, Smith D, Chen S. SGK3 is an estrogen-inducible kinase promoting estrogen-mediated survival of breast cancer cells. Mol Endocrinol. 2011;25:72–82. doi: 10.1210/me.2010-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi L, Wang K, Zhao M, Yuan X, Huang C. Overexpression of PIP5KL1 suppresses the growth of human cervical cancer cells in vitro and in vivo. Cell Biol Int. 2010;34:309–315. doi: 10.1042/CBI20090040. [DOI] [PubMed] [Google Scholar]
- 10.Xu J, Wan M, He Q, Bassett RL, Jr, Fu X, Chen AC, Shi F, Creighton CJ, Schiff R, Huo L, Liu D. SGK3 is associated with estrogen receptor expression in breast cancer. Breast Cancer Res Treat. 2012;134:531–541. doi: 10.1007/s10549-012-2081-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu M, Chen L, Chan TH, Wang J, Li Y, Li Y, Zeng TT, Yuan YF, Guan XY. Serum and glucocorticoid kinase 3 at 8q13.1 promotes cell proliferation and survival in hepatocellular carcinoma. Hepatology. 2012;55:1754–1765. doi: 10.1002/hep.25584. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y, Zhou D, Chen S. SGK3 is an androgen-inducible kinase promoting prostate cancer cell proliferation through activation of p70 S6 kinase and up-regulation of cyclin D1. Mol Endocrinol. 2014;28:935–948. doi: 10.1210/me.2013-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gatti L, Cossa G, Tinelli S, Carenini N, Arrighetti N, Pennati M, Cominetti D, De Cesare M, Zunino F, Zaffaroni N, Perego P. Improved apoptotic cell death in drug-resistant non-small-cell lung cancer cells by tumor necrosis factor-related apoptosis-inducing ligand-based treatment. J Pharmacol Exp Ther. 2014;348:360–371. doi: 10.1124/jpet.113.210054. [DOI] [PubMed] [Google Scholar]
- 14.Zhang G, Miyake M, Lawton A, Goodison S, Rosser CJ. Matrix metalloproteinase-10 promotes tumor progression through regulation of angiogenic and apoptotic pathways in cervical tumors. BMC Cancer. 2014;14:310. doi: 10.1186/1471-2407-14-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Virós D, Camacho M, Zarraonandia I, García J, Quer M, Vila L, León X. Prognostic role of MMP-9 expression in head and neck carcinoma patients treated with radiotherapy or chemoradiotherapy. Oral Oncol. 2013;49:322–325. doi: 10.1016/j.oraloncology.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 16.Thorsen SB, Christensen SL, Würtz SO, Lundberg M, Nielsen BS, Vinther L, Knowles M, Gee N, Fredriksson S, Møller S, et al. Plasma levels of the MMP-9: TIMP-1 complex as prognostic biomarker in breast cancer: a retrospective study. BMC Cancer. 2013;13:598. doi: 10.1186/1471-2407-13-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang C, Ma HX, Jin MS, Zou YB, Teng YL, Tian Z, Wang HY, Wang YP, Duan XM. Association of matrix metalloproteinase (MMP)-2 and −9 expression with extra-gastrointestinal stromal tumor metastasis. Asian Pac J Cancer Prev. 2014;15:4187–4192. doi: 10.7314/APJCP.2014.15.10.4187. [DOI] [PubMed] [Google Scholar]
- 18.Yang B, Tang F, Zhang B, Zhao Y, Feng J, Rao Z. Matrix metalloproteinase-9 overexpression is closely related to poor prognosis in patients with colon cancer. World J Surg Oncol. 2014;12:24. doi: 10.1186/1477-7819-12-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang YL, Chen CZ, Jin LP, Ji QQ, Chen YZ, Li Q, Zhang XH, Qu JM. Effect and mechanism of the metastasis suppressor gene BRMS1 on the migration of breast cancer cells. Int J Clin Exp Med. 2013;6:908–916. [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Y, Jiang W, Wang Y, Wu J, Saiyin H, Qiao X, Mei X, Guo B, Fang X, Zhang L, et al. Breast cancer metastasis suppressor 1 regulates hepatocellular carcinoma cell apoptosis via suppressing osteopontin expression. PLoS One. 2012;7:e42976. doi: 10.1371/journal.pone.0042976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ventura BV, Quezada C, Maloney SC, Fernandes BF, Antecka E, Martins C, Bakalian S, di Cesare S, Burnier MN., Jr Expression of the metastasis suppressor BRMS1 in uveal melanoma. Ecancermedicalscience. 2014;8:410. doi: 10.3332/ecancer.2014.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheng XJ, Zhou YQ, Song QY, Zhou DM, Liu QC. Loss of breast cancer metastasis suppressor 1 promotes ovarian cancer cell metastasis by increasing chemokine receptor 4 expression. Oncol Rep. 2012;27:1011–1018. doi: 10.3892/or.2011.1596. [DOI] [PMC free article] [PubMed] [Google Scholar]