Abstract
Counting chromosomes is not just simple math. Although normal males and females differ in sex chromosome content (XY vs. XX), X chromosome imbalance is tolerated because dosage compensation mechanisms have evolved to ensure functional equivalence. In mammals this is accomplished by two processes—X chromosome inactivation that silences most genes on one X chromosome in females, leading to functional X monosomy for most genes in both sexes, and X chromosome upregulation that results in increased gene expression on the single active X in males and females, equalizing dosage relative to autosomes. This review focuses on genes on the X chromosome, and how gene content, organization and expression levels can be influenced by these two processes. Special attention is given to genes that are not X inactivated, and are not necessarily fully dosage compensated. These genes that “escape” X inactivation are of medical importance as they explain phenotypes in individuals with sex chromosome aneuploidies and may impact normal traits and disorders that differ between men and women. Moreover, escape genes give insight into how X chromosome inactivation is spread and maintained on the X.
Keywords: dosage compensation, epigenetics, X inactivation, escape gene, x-upregulation, transgenics
Introduction
Male and female therian mammals (marsupials and placental eutherians) differ in sex chromosome composition; most males are XY and females are XX. With very few exceptions, the SRY gene on the Y chromosome underlies sex determination by initiating testis formation (Wilson and Makova 2009b). Yet, sex chromosome differences are much more extensive. In humans, the 159-megabase (Mb) X chromosome comprises 5% of the genome and includes more than 1,400 transcripts (RefSeq RNA entries from hg18, build 36.1). In contrast, the largely heterochromatic 58-Mb Y chromosome is gene poor with only 195 RefSeq RNAs annotated to its euchromatic sequence. While transcript numbers for both the X and Y include multicopy genes that are expressed during spermatogenesis (Skaletsky et al. 2003; Mueller et al. 2008), most genes on the X do not have a Y chromosome counterpart and do not have sex-specific functions (Ross et al. 2005). As a result, X copy number differences in males and females have several consequences. Perhaps the most apparent is that males lack a “back-up copy” and can manifest many X-linked recessive disorders. However, an additional repercussion is that X imbalance requires mechanisms to equalize gene dosage between the sexes and relative to levels on autosomes. This review focuses on how genes on the X chromosome in placental mammals respond to these dosage compensation processes.
Central to this discussion is the rich evolutionary history of the X that has impacted X-chromosome gene content, function and organization. Therian sex chromosomes evolved from an identical pair of autosomes at least 165 million years ago (mya) (Ohno 1967; Veyrunes et al. 2008). Chromosome rearrangements suppressed recombination allowing the X and Y to evolve independently, and only small XY identical pseudoautosomal regions (PARs) remain at the ends of each chromosome to pair and recombine during male meiosis. In the absence of interchromosomal recombination, the Y has rapidly degraded; outside of the PARs, only 16 functional single-copy human XY pairs (gametologs) remain (Ross et al. 2005). X evolution has been heavily influenced by dosage regulation, and these processes ensure that gene content remains highly conserved (Ohno 1967). As a consequence, even the 76 Mb marsupial X retains considerable homology with a large proportion of the human X (Spencer et al. 1991; Mikkelsen et al. 2007). Of note, monotreme (platypus) X chromosomes have an independent origin and are not discussed here (Veyrunes et al. 2008). Comparative sequence analysis and measurements of divergence between remaining XY gametologs suggest that much of the human X short arm is a recent acquisition to the human X, added in sequential segments, or strata, after the split of the marsupial and eutherian lineages ~150 mya (Spencer et al. 1991; Lahn and Page 1999; Delbridge et al. 2009). Most remaining XY pairs map to these recent short arm segments. Therefore, how formerly autosomal sequences acquire dosage compensation gives mechanistic insight into these processes.
Dosage compensation I: X chromosome inactivation
X dosage compensation between male and female mammals involves chromosome-wide transcriptional silencing of most genes on one X chromosome in females. As a result of X chromosome inactivation (XCI), both males and females are functionally monosomic for most X-linked genes. Since XCI was initially hypothesized almost 50 years ago (Lyon 1961), much progress has been made in understanding this complex long-range chromosome silencing process.
X-linked gene expression patterns in the adult female reflect, for the most part, events that initiate in early development, at about the time of implantation. At this time point, XCI counting and choice steps initiate inactivation of the maternally or paternally inherited X chromosome in cells with more than one X (Monkhorst et al. 2008; Payer and Lee 2008). Inactivation then spreads in cis along the length of the chromosome, gene silencing is established, and the X is extensively epigenetically modified. These epigenetic changes, described in more detail below, include physical, temporal and spatial differences that distinguish the inactive X chromosome from its active counterpart and serve as a chromosomal memory to ensure that a particular X remains silenced throughout all successive somatic cell divisions. As a result of XCI, females are mosaic with cells that differ in the parental origin of the X that is inactivated. Nonetheless, because of the stochastic nature of XCI, the percentage of cells that inactivate a particular X can be quite skewed, in rare cases revealing female carriers that manifest recessive X-linked disorders (Orstavik 2009). Alternatively skewed XCI patterns can result from post-XCI loss of cells expressing mutations that result in a proliferative disadvantage.
Mechanistically, the initial stages of XCI are governed by a locus on the X, known as the X inactivation center, that includes the XIST gene (Xist in mouse) and sequences and transcripts that regulate Xist expression (reviewed in more detail elsewhere in this issue (Okamoto and Heard 2009)). XIST/Xist is a large, 17-kb non-coding functional RNA that is expressed only from the inactive X in adult female cells (Brockdorff et al. 1991; Brown et al. 1991) and closely associates with or “coats” the inactive X (Brown et al. 1992; Clemson et al. 1996). From studies in mouse, Xist is necessary for random XCI to occur (Penny et al. 1996; Marahrens et al. 1997; Wutz and Jaenisch 2000). An antisense transcript, Tsix, negatively regulates Xist, and additional cis sequences and transcripts are involved in the complex counting and choice steps of the process (Lee et al. 1999; Ogawa and Lee 2003; Augui et al. 2007; Zhao et al. 2008; Okamoto and Heard 2009). Via these cis sequences, both X chromosomes closely associate just prior to initiating XCI, and such cross-chromosomal communication may be necessary to select a single X to undergo XCI (Bacher et al. 2006; Xu et al. 2006b; Augui et al. 2007). Humans do not have a similar TSIX counterpart and regulation of XIST at the onset of X inactivation is less well understood (Migeon et al. 2002; Chow and Brown 2003).
Epigenetic modification of the inactive X chromosome
The inactive X is easily distinguishable from the active X chromosome by widespread epigenetic modifications. Such alterations originally led to the identification of the inactive X chromosome as the darkly-staining, condensed Barr body at the periphery of interphase nuclei in female cells (Barr and Bertram 1949). It is now known that Xist RNA forms a nuclear compartment that excludes RNA polymerase II and other transcription factors (Chaumeil et al. 2006; Clemson et al. 2006). DNA within this compact inactive X structure is non-randomly distributed; noncoding and repetitive DNA sequences are internalized, whereas coding sequences remain at the periphery of the Xist RNA compartment (Chaumeil et al. 2006; Clemson et al. 2006), but are reported to move inward upon gene silencing (Chaumeil et al. 2006).
Concomitant with this spatial restriction, inactive X chromatin is heavily remodeled. One of the earliest events following Xist upregulation includes loss of histone acetylation marks that typically associate with active chromatin (Keohane et al. 1996). Additionally, components of polycomb complexes PRC2 and PRC1 are targeted to the inactive X and are responsible for the enrichment of H3K27 trimethylation and H2AK119 ubiquitination (Silva et al. 2003; Plath et al. 2004; de Napoles et al. 2004). Other chromatin changes at this early time point include the incorporation of H3K9 methylation and H4K20 monomethylation (Heard et al. 2001; Mermoud et al. 2002; Kohlmaier et al. 2004). A second wave of epigenetic remodeling follows these early chromatin changes and gene silencing. Replication timing changes; the inactive X replicates later than the active X during S phase of the cell cycle (Taylor 1960; Morishima et al. 1962). There are DNA methylation differences between the active and inactive X chromosomes. CpG islands are methylated on the inactive X, although the rest of the inactive X is globally hypomethylated compared to the active X (Mohandas et al. 1981; Pfeifer et al. 1989; Hellman and Chess 2007). The histone variant, macroH2A also becomes associated with the inactive X (Costanzi and Pehrson 1998). Although these modifications are present at other silent loci throughout the genome, how they are specifically utilized on the X to promote silencing remains unclear, as XCI is unprecedented in scale and involves the unique aspect of Xist RNA incorporation.
Work from many laboratories continues to reveal the complex nature of inactive X heterochromatin. Despite extensive analysis, no one individual epigenetic feature has proven critical for the establishment or maintenance of random X inactivation (Csankovszki et al. 2001; Hernandez-Munoz et al. 2005; Kalantry and Magnuson 2006; Blewitt et al. 2008) with the exception of Xist, and even then, only during a slim window of early development (Csankovszki et al. 1999; Wutz and Jaenisch 2000). These many epigenetic layers create redundancy to ensure that global reactivation of the X does not occur (Csankovszki et al. 2001). With this in mind, it is particularly intriguing that at least some modifications do not homogenously cover the inactive X, but are spatially separated (Duthie et al. 1999; Chadwick and Willard 2004; Valley et al. 2006), indicating that the inactive X is clearly not just a single, uniform block of heterochromatin.
Many conclusions about enriched epigenetic modifications along the X are based on cytological observations. How specific marks are distributed across the whole chromosome at the sequence level is just beginning to emerge, although efforts have focused on sequences at the X inactivation center (Rougeulle et al. 2004; Marks et al. 2009). Large-scale genome-wide approaches using array-based techniques or massively parallel sequencing strategies are underway to capture and decipher the complex chromatin code that directs regulatory sequences throughout the genome (Heintzman et al. 2009). Similar analyses of the X have been limited, and it is not yet completely clear how the inactive X compares to other silenced domains throughout the genome. A complication for such X studies is that the two transcriptionally-distinct Xs in females each carry their own compilation of epigenetically-encoded regulatory instructions. Approaches that distinguish the active X from the inactive X are optimal (Brinkman et al. 2006; Valley et al. 2006; Chadwick 2007; Marks et al. 2009). Such studies continue to reveal the heterogeneous nature of inactive X heterochromatin as the distribution of marks specific to gene bodies, promoters, and non-coding sequences is probed. No single chromatin modification has been identified that differentiates either silenced or expressed genes on the inactive X (Brinkman et al. 2006; Valley et al. 2006; Chadwick 2007; Marks et al. 2009; Mietton et al. 2009). Like autosomal loci that were simultaneously queried, acetyl H3/H4 or trimethylated H3K4, generally thought to be active histone marks, do not exclusively associate with transcription on the inactive X, nor do the presumptive inactive marks, trimethylated H3K27 or methylated H3K9, specifically associate with silenced regions (Brinkman et al. 2006; Valley et al. 2006; Marks et al. 2009). While these studies have begun to outline a picture of heterochromatin along the inactive X chromosome, they highlight the importance of further high-resolution analysis to fully reveal the location and combination of key functional marks that comprise the inactive X histone code.
Despite our current appreciation for the epigenomic environment of the X, there is still very little known about how Xist/XIST and specific chromatin modifications are targeted in cis along the chromosome to promote gene silencing. Additional insight into spreading and maintenance of XCI may come from genes that are not X inactivated.
Genes that escape X chromosome inactivation
Although most X-linked genes are silenced, the idea that some are expressed from the inactive X chromosome was originally hypothesized for genes with Y gametologs that should be dosage compensated with two functional copies in males and females (Lyon 1962). Further, aberrant gene dosage of such “escape” genes explains phenotypes in individuals with X chromosome aneuploidies, such as Turner Syndrome (most commonly due to a 45,X karyotype) and Klinefelter Syndrome (XXY). The existence of escape genes has been established for some time (Shapiro et al. 1979), although it is now known that not all have copies on the Y.
Given the small number of X genes with Y gametologs, it is somewhat surprising that human escape genes are not rare. This conclusion is based on large-scale analysis that directly measured inactive X expression using two methods: qualitative assessment of escape genes in rodent-human somatic cell hybrids that retain human inactive X chromosomes and quantitative analysis of inactive X expression relative to active X expression by measuring transcribed polymorphisms in primary fibroblast cell lines (Carrel and Willard 2005). Both approaches established that approximately 15% of genes escape XCI, although in most cases inactive expression levels are much lower than active X levels (Carrel and Willard 2005). Microarrays can also indirectly identify escape genes that are expressed at higher levels in females compared to males (Sudbrak et al. 2001; Craig et al. 2004; Talebizadeh et al. 2006; Johnston et al. 2008). Altogether, these data are consistent; microarray studies report lower estimates of human escape genes (Talebizadeh et al. 2006; Johnston et al. 2008), but are not able to detect genes with low levels of inactive X expression.
Intriguingly, XCI escape is variable for some genes. 10% of human loci escape XCI in only a subset of female lines tested yet are X inactivated in other lines (Carrel and Willard 2005). Inactive X expression levels can vary between different individuals and between different tissues (Anderson and Brown 1999; Carrel and Willard 1999; Talebizadeh et al. 2006). The mechanistic basis and the phenotypic consequences of variable escape are not yet well understood. XCI skewing differences are frequently proposed to explain phenotype spectrums in female carriers of X-linked mutations (Orstavik 2009), but heterogeneous escape could impact traits as well. The prevalence of genome-wide transcript-level variation and its potential to impact complex traits has only been recently appreciated (Cookson et al. 2009). That inactive X expression levels for some genes could also influence traits that show differences amongst females or between males and females adds additional complexities. At first glance, it seems unclear how low escape levels could affect such traits, yet one possibility is amelioration of carrier phenotypes. Low levels of autosomal genes can function in such a manner (e.g. (Bhuiyan et al. 2008)). On the X, partial escape of the OFD1 gene in some females is proposed to contribute to variation in Oral-facial-digital type I syndrome (Morleo and Franco 2008).
The organization and distribution of human escape genes gives insight into their regulation. The chromosome location of human escape genes is non-random. Most escape genes map to the youngest evolutionary strata on the X short arm, as do almost all remaining XY gene pairs (Carrel and Willard 2005; Ross et al. 2005). Acquisition of XCI is proposed to be linked to Y gene degradation, and these formerly autosomal genes may escape XCI because they are at an intermediate stage in this process, not yet acquiring dosage compensation (Jegalian and Page 1998). Local escape gene organization is also revealing; most escape genes are found in clusters that include at least one gene with Y homology, suggesting that they are organized in domains and are controlled by regional mechanisms (Miller and Willard 1998; Carrel and Willard 2005).
In contrast to the human X, very few mouse genes have been identified that escape XCI, and most have functional Y gametologs (Heard and Disteche 2006). This difference could explain why Turner Syndrome females have more severe phenotypes than XO mice (Ashworth et al. 1991), although confirmation must await the availability of large-scale analysis of mouse escape genes. There are clear differences in the organization of mouse and human escape genes. Genes adjacent to four mouse escape genes are X inactivated, and therefore clustering does not appear to be a common feature in mouse (Tsuchiya et al. 2004; Heard and Disteche 2006). Differences between mouse and human escape domains argue that aspects of escape gene regulation are species specific, although common features have been identified and are discussed below.
From the standpoint of gene dosage, escape genes use many strategies to solve the complex equation towards male/female parity, although many of these solutions prove unsuccessful. As originally predicted (Lyon 1962), most genes with functional XY pairs do escape XCI (Heard and Disteche 2006; Carrel and Willard 2005). Nonetheless, escape does not necessarily lead to dosage compensation, as there is increasing evidence that X and Y gametologs are not equivalent. Surprisingly, in both mouse and human, many XY pairs vary widely in expression levels and tissue distribution (Wilson and Makova 2009a). Using an elegant mouse system that can distinguish between phenotypic sex and sex chromosome contribution, expression of at least five XY gametologs is sexually dimorphic; females have higher expression, but Y genes in males do not compensate, particularly in brain (Xu et al. 2002). Intriguingly, while expression of the Eif2s3x gene is sexually dimorphic, protein levels are not (Xu et al. 2006a) and therefore, post-transcriptional regulation is a novel X strategy to equalize gene dosage. Furthermore, translational regulation can also disrupt dosage equivalence, as both Ddx3x and Ddx3y are widely expressed in many tissues, but in this case, translation of Ddx3y is quite restricted (Ditton et al. 2004). There are also X genes with Y gametologs that do not escape XCI. SYBL1 and SPRY3 map to the human Xq PAR and have come up with a unique mechanism of dosage compensation that involves silencing on both the inactive X and Y chromosome (De Bonis et al. 2006). Finally, many genes, particularly on the human X, escape XCI but lack a functional Y partner (Carrel and Willard 2005). This suggests that a substantial number of X-linked genes are expressed at higher levels in females than males, although those with low levels of inactive X expression are expected to be largely dosage compensated (Carrel and Willard 2005; Johnston et al. 2008). Altogether, escape genes demonstrate that absolute dosage compensation is not a strict requirement for all genes on the X.
Genomic influences on XCI propagation and escape gene expression
How XCI spreads in cis and maintains silencing of this 159-Mb X chromosome is still poorly understood, but models must incorporate escape gene regulation as well. Initial clues come from analyses of X:autosome translocations and X-linked transgenes (Rastan 1983; Lee and Jaenisch 1997) indicating that autosomal sequences can be at least partially inactivated. Therefore, sequences necessary for spreading XCI are not restricted to the X chromosome, but could be enriched or uniquely organized on the X to promote stable, chromosome-wide inactivation. Such observations led to the hypothesis that specific X sequences, “way stations”, propagate an XCI signal (Gartler and Riggs 1983; Riggs 1990) and these elements could serve as docking sites for XIST RNA or heterochromatin proteins. The repetitive element LINE-1 (L1) has been proposed for such a function (Lyon 1998). Chromosome-wide analysis of L1s and motifs within L1s support this hypothesis, particularly on the human X; such sequences are enriched relative to levels on autosomes and depleted within escape gene regions (Bailey et al. 2000; Ross et al. 2005; Carrel et al. 2006; Wang et al. 2006). Nonetheless, unique features of sex chromosome biology could partially explain L1 enrichment and distribution on the X (Boissinot et al. 2001). However, insertional bias does not preclude L1s from playing a functional role in XCI. In fact, a sequence that rapidly accumulates on the X fits with the hypothesis that formerly autosomal genes must acquire certain sequence characteristics to be X inactivated (Jegalian and Page 1998).
Additional repeats and sequence features have been identified that correlate with escape or inactivated genes (Ke and Collins 2003; Tsuchiya et al. 2004; Carrel et al. 2006; McNeil et al. 2006; Wang et al. 2006). Among the sequences identified, escape genes are enriched for Alu repetitive elements (Wang et al. 2006) and (GATA)n simple repeats (McNeil et al. 2006), and depleted for LTRs (Tsuchiya et al. 2004) and MIRs (Wang et al. 2006). Similar to the way station model that requires such elements for silencing, sequences enriched in escape domains could be necessary for escape gene expression. Remarkably, two groups successfully used a subset of these genomic features, including motifs within L1s, to predict XCI status for a large proportion of genes on the X (Carrel et al. 2006; Wang et al. 2006). The high XCI status prediction rate for each study, despite using different classifiers, strongly supports the role of genomic sequence environment in regulating inactive X expression. In specific support of the L1 hypothesis, we found motifs with high XCI status prediction capabilities mapped to L1s, even though they were initially identified within a region of relatively low L1 density (Carrel et al. 2006). Combined, these data strongly suggest that L1s, or sequences within L1, influence aspects of inactive X regulation, yet indicate that a chromatin environment consisting of multiple genomic sequence elements influences XCI status.
If L1s function as way stations to seed heterochromatin, one logical prediction is that markers of XCI may localize to L1s just prior to accumulation elsewhere on the X. This hypothesis was recently tested by performing chromatin immunoprecipitation sequencing (ChIP-Seq) to measure trimethylated H3K27 during the onset of XCI in mouse embryonic stem cells (Marks et al. 2009). Trimethylated H3K27 was not enriched at L1s relative to the rest of the X at any time point of differentiation. Do these data rule out the role of L1s as way stations? Potentially, although a way station may not be epigenetically distinguishable from the rest of the X for enough time to be adequately captured by such approaches. Alternatively, computational strategies point towards specific motifs within L1s in XCI regulation (Carrel et al. 2006), and this subset may not be differentiated by ChIP-Seq. It is also possible that way stations are not marked by trimethylated H3K27, and therefore it will also be important to survey additional chromatin marks linked to the onset of XCI.
Interestingly, repeat sequences are internalized within the Xist compartment (Chaumeil et al. 2006; Clemson et al. 2006) suggesting an alternative role for L1s in XCI regulation that could be independent of way station function. As described above, coding sequences are peripheral and move inward upon XCI. Three-dimensional positioning is proposed to influence XCI status as an escape gene remained outside of the Xist boundary (Chaumeil et al. 2006) (Fig. 1b). A direct role for nuclear localization in inactive X expression will need further investigation, particularly in light of recent autosomal gene studies demonstrating that localization exterior to chromosome territories is not sufficient to upregulate gene expression (Morey et al. 2009).
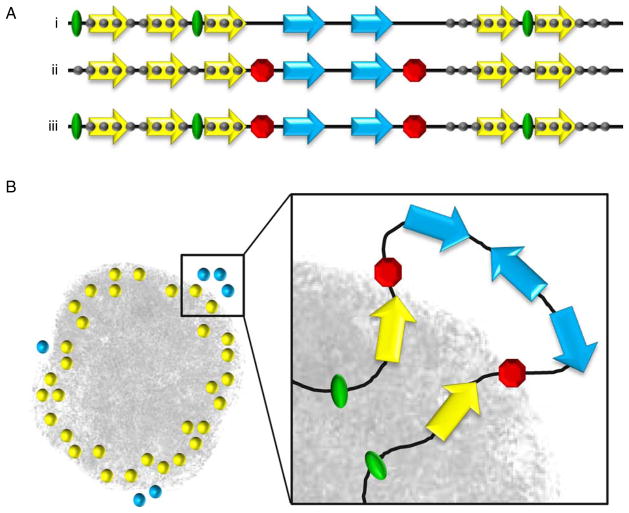
Fig. 1.
XCI models to incorporate escape gene regulation. a Genomic sequences or boundary elements may regulate inactive X expression and differ in their proximity to escape gene domains. i) Inactive X heterochromatin (gray circles) is propagated from “way stations” (green ovals) and encompasses X inactivated genes (yellow). In this model, escape genes (blue) reside too far from way stations to be inactivated. Although not shown, a variation of this model is that specific escape sequences could lie too far from inactivated genes to impart an effect (Carrel et al. 2006; McNeil et al. 2006). Nevertheless, regulatory elements that rely on distance to distinguish genes cannot account for the close juxtaposition of some escape and inactivated genes (Tsuchiya et al. 2004; Carrel and Willard 2005), but could explain large Mb sized domains with large transition regions elsewhere on the X. ii) Sequences such as insulators, boundary elements or barriers (red octagon) flank coordinately regulated genes and protect them from silencing. This model, or at least this model with respect to the CTCF protein (Filippova et al. 2005), also cannot fully explain escape gene expression (Ciavatta et al. 2006). iii) Incorporation of both models; way stations propagate XCI which is prevented from reaching escape genes by boundary elements. b Three-dimensional organization of X-linked genes can further affect escape gene expression (Chaumeil et al. 2006). A cross-section of the inactive X is enlarged. Both active (blue) and inactive (yellow) genes lie at the periphery of the XIST-delineated inactive X territory (gray). Non-genic and repeat sequences (black line) reside largely within the XIST compartment. Exterior positioning of escape genes is facilitated by boundary elements (red) and/or excessive distance from way stations (green)
A limitation of the way station model is that it cannot account for the close juxtaposition of some escape and inactivated genes (Tsuchiya et al. 2004; Carrel and Willard 2005), as escape genes are predicted to simply lie too far from such sequences to be properly inactivated (Fig. 1a, model i). Instead, appealing candidates are sequences such as barriers or insulators that block the spread of heterochromatin into escape domains (Gaszner and Felsenfeld 2006) (Fig. 1a, model ii). Several boundaries between escape and inactivated genes, in both mice and humans, have insulators bound on the inactive X by the CCCTC binding factor protein (CTCF) (Filippova et al. 2005). CTCF is a multifunctional protein that has widespread roles as a repressor, activator, and insulator (Phillips and Corces 2009). Of the boundaries identified, a role in inactive X regulation, as opposed to gene-specific regulation, is supported at the Jarid1c gene, as the orthologous human JARID1C gene lacks CTCF binding but is also embedded within a multigene escape domain (Filippova et al. 2005). CTCF sites at Jarid1c are unmethylated throughout development (Filippova et al. 2005). Since methylation precludes CTCF binding (Phillips and Corces 2009), the unmethylated sites can serve as a platform for CTCF to bind at the onset of XCI.
While the evidence above supports a role for CTCF in escape gene regulation, it is not known whether CTCF is necessary to establish escape domains. At a minimum, the story is clearly not so simple. CTCF binding sites alone are not sufficient for escape expression, as a reporter gene, flanked by insulators that included CTCF binding sites, was still silenced by XCI (Ciavatta et al. 2006). Nonetheless, this result may not be completely surprising, since CTCF has many functions, binds many locations, and the distribution of sites on the X is inconsistent with a role solely in escape gene regulation (Kim et al. 2007). What additional factors could direct CTCF to escape gene boundaries to specify a role for inactive X regulation remain to be seen.
As discussed above, some escape genes are found in large clusters, particularly on the human X, whereas others abut X-inactivated genes. Regulation may differ for different escape genes, potentially in a species-specific manner. Therefore, different escape genes and domains likely incorporate aspects of both models (way stations and boundary elements) (Fig. 1a). Further, the role of gene positioning, within and outside of the Xist territory, adds additional dimension, literally and figuratively, to escape gene regulation (Fig. 1b). Approaches to differentiate these models and functionally test candidate sequences are necessary. One such system is discussed below.
A transgene system to study escape gene expression
An approach to functionally evaluate how specific chromosome features or sequences impact inactive X expression is to analyze X-linked transgenes. Most transgenes are X inactivated, although a number escape XCI (e.g. (Goldman et al. 1987; Wu et al. 1992; Chong et al. 2002; Farivar et al. 2004)). It is difficult to conclude mechanistically why some escape, as the transgenes differ in composition, size, copy number, and insertion site. Recently we established a transgenic system to ask whether escape gene expression is autonomous or is instead influenced by genomic location (Li and Carrel 2008). Transgenes were tested in mouse embryonic stem cells, a well-characterized ex-vivo model system for studying XCI. X-linked transgenes were generated using BACs that contain the escape gene Jarid1c and adjacent X-inactivated genes (Tsuchiya et al. 2004). Upon ES cell differentiation and XCI, transgene expression patterns recapitulate endogenous expression patterns; at four locations transgenic Jarid1c escapes XCI whereas adjacent genes are X inactivated. These studies establish that escape is an intrinsic feature of the Jarid1c locus and delimit a 112-kb domain that directs inactive X expression in a position-independent manner. Whether this is because the region contains the previously identified CTCF binding sites (Filippova et al. 2005) and/or other critical regulatory sequences can now be tested. This approach will be useful for addressing the models proposed above. For example, if nuclear localization is an absolute requirement for escape gene expression, a prediction is that these transgenes will be positioned outside of the Xist compartment.
Dosage compensation II: Active X upregulation
As discussed above, XCI ensures equal dosage for most X-linked genes between males and females, yet an additional X gene dosage problem remains. Because of XCI, both males and females have a single functional X chromosome, but two copies of each autosome. The solution to this problem was derived by Susumu Ohno who recognized that upregulation of genes on the active X chromosome would equalize X and autosome gene dosage (Ohno 1967). Whole-genome microarray studies in multiple tissues for several mammals confirmed this hypothesis as a general phenomenon (Nguyen & Disteche 2006; Lin et al. 2007; Johnston et al. 2008). One intriguing exception was observed; X-linked genes in brain tissues have higher overall expression compared to their autosomal counterparts (Nguyen and Disteche 2006). A large number of genes on the X are involved in cognition and are associated with mental retardation (Zechner et al. 2001). Such enrichment is likely due to selection (Zechner et al. 2001), and perhaps also as a result of their increased expression due to active X upregulation.
One of the most pressing questions about Xa upregulation is whether this phenomenon is an active mechanism. To begin to address this possibility, expression was analyzed in cells in which the X to autosome ratio (X:A) differs. Importantly, dosage is essentially balanced in cells with different X:A ratios: 2X:2A (primary oocytes), and 1X:1A (secondary oocytes), and 1X:2A (somatic cells in males or females with a single active X) (Nguyen and Disteche 2006). However, since results are based on global expression levels, it seems important to question whether they could be due simply to the specific subset of genes that are expressed in any given cell type. This possibility seems unlikely given that the onset of dosage compensation occurs progressively in cultured male stem cells upon differentiation (Lin et al. 2007). Similar results were seen in XX female stem cells, although such measurements are somewhat complicated by the onset of XCI (Lin et al. 2007). Together, these data appear to support an active mechanism that ensures X:autosome dosage is achieved in multiple cell types. How could such upregulation on the active X occur? Chromatin alterations are obvious candidates, but none have been identified that specifically decorate the active X chromosome. DNA methylation signatures on the active X are different than on the inactive X; the active X is globally hypermethylated except at CpG islands (Hellman and Chess 2007). Nonetheless, how this could translate to upregulation of expression is still unclear. Could the transgene system described above aid in dissecting aspects of this dosage compensation? If this is a chromosome-wide phenomenon, one prediction is that transgenes integrated on the active X would be expressed at higher levels than an autosomal transgene.
Concluding thoughts
XCI and active X upregulation are extraordinary examples of the efforts that are taken to overcome dosage imbalance. On the inactive X chromosome, genomic and epigenomic influences on the complex pattern of gene expression and heterochromatin composition are emerging. Active X upregulation has only recently been demonstrated, and we await insight into the mechanism and the role that genomics and epigenetics plays. As the necessary cis and trans regulatory factors involved in both XCI and active X upregulation are determined, it is likely that new variables and unknowns will be added to the growing equations.
Acknowledgments
We apologize for work that could not be cited due to space limitations. We are grateful to Kateryna Makova and Melissa Wilson for sharing results prior to publication. Work in the authors’ laboratory is supported by the NIH and the Pennsylvania Department of Health.
List of abbreviations
- BAC
Bacterial artificial chromosome
- ChIP-Seq
Chromatin immunoprecipitation sequencing
- CTCF
CCCTC binding factor protein
- Ddx3x/y
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3 gene on the X and Y
- Eif2s3x/y
Eukaryotic translation initiation factor 2, subunit 3, structural gene on the X and Y
- JARID1C/Jarid1c
Jumonji, AT-rich interactive domain 1c gene on the human and mouse X
- L1
Long interspersed repetitive element 1
- LTR
long terminal repeat element
- Mb
megabase
- mya
million years ago
- OFD1
Oral-facial-digital syndrome 1 gene
- PAR
pseudoautosomal region
- PRC1
polycomb repressive complex 1
- PRC2
polycomb repressive complex 2
- SPRY3
Sprouty homolog 3 gene
- SRY
Sex-determining region gene on the Y
- SYBL1
Synaptobrevin-like 1 gene
- TSIX/Tsix
X inactive specific transcript, antisense gene in human and mouse
- Utx/Uty
Ubiquitously transcribed tetratricopeptide repeat gene on the X and Y
- XCI
X chromosome inactivation
- XIST/Xist
X inactive specific transcript gene in human and mouse
References
- Anderson CL, Brown CJ. Polymorphic X-chromosome inactivation of the human TIMP1 gene. Am J Hum Genet. 1999;65:699–708. doi: 10.1086/302556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashworth A, Rastan S, Lovell-Badge R, Kay G. X-chromosome inactivation may explain the difference in viability of XO humans and mice. Nature. 1991;351:406–408. doi: 10.1038/351406a0. [DOI] [PubMed] [Google Scholar]
- Augui S, Filion GJ, Huart S, et al. Sensing X chromosome pairs before X inactivation via a novel X-pairing region of the Xic. Science. 2007;318:1632–1636. doi: 10.1126/science.1149420. [DOI] [PubMed] [Google Scholar]
- Bacher CP, Guggiari M, Brors B, et al. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat Cell Biol. 2006;8:293–299. doi: 10.1038/ncb1365. [DOI] [PubMed] [Google Scholar]
- Bailey JA, Carrel L, Chakravarti A, Eichler EE. Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: the Lyon repeat hypothesis. Proc Natl Acad Sci USA. 2000;97:6634–6639. doi: 10.1073/pnas.97.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr ML, Bertram EG. A morphological distinction between neurones of the male and female, and the behaviour of the nucleolar satellite during accelerated nucleoprotein synthesis. Nature. 1949;163:676. doi: 10.1038/163676a0. [DOI] [PubMed] [Google Scholar]
- Bhuiyan ZA, Momenah TS, Amin AS, et al. An intronic mutation leading to incomplete skipping of exon-2 in KCNQ1 rescues hearing in Jervell and Lange-Nielsen syndrome. Prog Biophys Mol Biol. 2008;98:319–327. doi: 10.1016/j.pbiomolbio.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Blewitt ME, Gendrel AV, Pang Z, et al. SmcHD1, containing a structural-maintenance-of-chromosomes hinge domain, has a critical role in X inactivation. Nat Genet. 2008;40:663–669. doi: 10.1038/ng.142. [DOI] [PubMed] [Google Scholar]
- Boissinot S, Entezam A, Furano AV. Selection against deleterious LINE-1-containing loci in the human lineage. Mol Biol Evol. 2001;18:926–935. doi: 10.1093/oxfordjournals.molbev.a003893. [DOI] [PubMed] [Google Scholar]
- Brinkman AB, Roelofsen T, Pennings SW, et al. Histone modification patterns associated with the human X chromosome. EMBO Rep. 2006;7:628–634. doi: 10.1038/sj.embor.7400686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockdorff N, Ashworth A, Kay GF, et al. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature. 1991;351:329–331. doi: 10.1038/351329a0. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Lafreniere RG, Powers VE, et al. Localization of the X inactivation centre on the human X chromosome in Xq13. Nature. 1991;349:82–84. doi: 10.1038/349082a0. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Hendrich BD, Rupert JL, et al. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- Carrel L, Willard HF. Heterogeneous gene expression from the inactive X chromosome: an X-linked gene that escapes X inactivation in some human cell lines but is inactivated in others. Proc Natl Acad Sci USA. 1999;96:7364–7369. doi: 10.1073/pnas.96.13.7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- Carrel L, Park C, Tyekucheva S, et al. Genomic environment predicts expression patterns on the human inactive X chromosome. PLoS Genet. 2006;2:e151. doi: 10.1371/journal.pgen.0020151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick BP. Variation in Xi chromatin organization and correlation of the H3K27me3 chromatin territories to transcribed sequences by microarray analysis. Chromosoma. 2007;116:147–157. doi: 10.1007/s00412-006-0085-1. [DOI] [PubMed] [Google Scholar]
- Chadwick BP, Willard HF. Multiple spatially distinct types of facultative heterochromatin on the human inactive X chromosome. Proc Natl Acad Sci USA. 2004;101:17450–17455. doi: 10.1073/pnas.0408021101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumeil J, Le Baccon P, Wutz A, Heard E. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 2006;20:2223–2237. doi: 10.1101/gad.380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong S, Kontaraki J, Bonifer C, Riggs AD. A Functional chromatin domain does not resist X chromosome inactivation: silencing of cLys correlates with methylation of a dual promoter-replication origin. Mol Cell Biol. 2002;22:4667–4676. doi: 10.1128/MCB.22.13.4667-4676.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow JC, Brown CJ. Forming facultative heterochromatin: silencing of an X chromosome in mammalian females. Cell Mol Life Sci. 2003;60:2586–2603. doi: 10.1007/s00018-003-3121-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciavatta D, Kalantry S, Magnuson T, Smithies O. A DNA insulator prevents repression of a targeted X-linked transgene but not its random or imprinted X inactivation. Proc Natl Acad Sci USA. 2006;103:9958–9963. doi: 10.1073/pnas.0603754103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, McNeil JA, Willard HF, Lawrence JB. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hall LL, Byron M, McNeil J, Lawrence JB. The X chromosome is organized into a gene-rich outer rim and an internal core containing silenced non-genic sequences. Proc Natl Acad Sci USA. 2006;103:7688–7693. doi: 10.1073/pnas.0601069103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson W, Liang L, Abecasis G, Moffatt M, Lathrop M. Mapping complex disease traits with global gene expression. Nat Rev Genet. 2009;10:184–194. doi: 10.1038/nrg2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzi C, Pehrson JR. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature. 1998;393:599–601. doi: 10.1038/31275. [DOI] [PubMed] [Google Scholar]
- Craig IW, Mill J, Craig GM, Loat C, Schalkwyk LC. Application of microarrays to the analysis of the inactivation status of human X-linked genes expressed in lymphocytes. Eur J Hum Genet. 2004;12:639–646. doi: 10.1038/sj.ejhg.5201212. [DOI] [PubMed] [Google Scholar]
- Csankovszki G, Panning B, Bates B, Pehrson JR, Jaenisch R. Conditional deletion of Xist disrupts histone macroH2A localization but not maintenance of X inactivation. Nat Genet. 1999;22:323–324. doi: 10.1038/11887. [DOI] [PubMed] [Google Scholar]
- Csankovszki G, Nagy A, Jaenisch R. Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J Cell Biol. 2001;153:773–784. doi: 10.1083/jcb.153.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bonis ML, Cerase A, Matarazzo MR, et al. Maintenance of X- and Y-inactivation of the pseudoautosomal (PAR2) gene SPRY3 is independent from DNA methylation and associated to multiple layers of epigenetic modifications. Hum Mol Genet. 2006;15:1123–1132. doi: 10.1093/hmg/ddl027. [DOI] [PubMed] [Google Scholar]
- de Napoles M, Mermoud JE, Wakao R, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Delbridge ML, Patel HR, Waters PD, McMillan DA, Marshall Graves JA. Does the human X contain a third evolutionary block? Origin of genes on human Xp11 and Xq28. Genome Res. 2009 May 15; doi: 10.1101/gr.088625.108. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditton HJ, Zimmer J, Kamp C, Rajpert-De Meyts E, Vogt PH. The AZFa gene DBY (DDX3Y) is widely transcribed but the protein is limited to the male germ cells by translation control. Hum Mol Genet. 2004;13:2333–2341. doi: 10.1093/hmg/ddh240. [DOI] [PubMed] [Google Scholar]
- Duthie SM, Nesterova TB, Formstone EJ, et al. Xist RNA exhibits a banded localization on the inactive X chromosome and is excluded from autosomal material in cis. Hum Mol Genet. 1999;8:195–204. doi: 10.1093/hmg/8.2.195. [DOI] [PubMed] [Google Scholar]
- Farivar S, Yamaguchi S, Sugimoto M, Takagi N. X-chromosome inactivation in differentiating mouse embryonic stem cells carrying X-linked GFP and lacZ transgenes. Int J Dev Biol. 2004;48:629–635. doi: 10.1387/ijdb.041898sf. [DOI] [PubMed] [Google Scholar]
- Filippova GN, Cheng MK, Moore JM, et al. Boundaries between chromosomal domains of X inactivation and escape bind CTCF and lack CpG methylation during early development. Dev Cell. 2005;8:31–42. doi: 10.1016/j.devcel.2004.10.018. [DOI] [PubMed] [Google Scholar]
- Gartler SM, Riggs AD. Mammalian X-chromosome inactivation. Annu Rev Genet. 1983;17:155–190. doi: 10.1146/annurev.ge.17.120183.001103. [DOI] [PubMed] [Google Scholar]
- Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- Goldman MA, Stokes KR, Idzerda RL, et al. A chicken transferrin gene in transgenic mice escapes X-chromosome inactivation. Science. 1987;236:593–595. doi: 10.1126/science.2437652. [DOI] [PubMed] [Google Scholar]
- Heard E, Disteche CM. Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev. 2006;20:1848–1867. doi: 10.1101/gad.1422906. [DOI] [PubMed] [Google Scholar]
- Heard E, Rougeulle C, Arnaud D, et al. Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell. 2001;107:727–738. doi: 10.1016/s0092-8674(01)00598-0. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellman A, Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315:1141–1143. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- Hernandez-Munoz I, Lund AH, van der Stoop P, et al. Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc Natl Acad Sci USA. 2005;102:7635–7640. doi: 10.1073/pnas.0408918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jegalian K, Page DC. A proposed path by which genes common to mammalian X and Y chromosomes evolve to become X inactivated. Nature. 1998;394:776–780. doi: 10.1038/29522. [DOI] [PubMed] [Google Scholar]
- Johnston CM, Lovell FL, Leongamornlert DA, et al. Large-scale population study of human cell lines indicates that dosage compensation is virtually complete. PLoS Genet. 2008;4:e9. doi: 10.1371/journal.pgen.0040009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantry S, Magnuson T. The Polycomb group protein EED is dispensable for the initiation of random X-chromosome inactivation. PLoS Genet. 2006;2:e66. doi: 10.1371/journal.pgen.0020066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke X, Collins A. CpG islands in human X-inactivation. Ann Hum Genet. 2003;67:242–249. doi: 10.1046/j.1469-1809.2003.00038.x. [DOI] [PubMed] [Google Scholar]
- Keohane AM, O'Neill LP, Belyaev ND, Lavender JS, Turner BM. X-Inactivation and histone H4 acetylation in embryonic stem cells. Dev Biol. 1996;180:618–630. doi: 10.1006/dbio.1996.0333. [DOI] [PubMed] [Google Scholar]
- Kim TH, Abdullaev ZK, Smith AD, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlmaier A, Savarese F, Lachner M, et al. A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol. 2004;2:e171. doi: 10.1371/journal.pbio.0020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahn BT, Page DC. Four evolutionary strata on the human X chromosome. Science. 1999;286:964–967. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- Lee JT, Jaenisch R. Long-range cis effects of ectopic X-inactivation centres on a mouse autosome. Nature. 1997;386:275–279. doi: 10.1038/386275a0. [DOI] [PubMed] [Google Scholar]
- Lee JT, Davidow LS, Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet. 1999;21:400–404. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- Li N, Carrel L. Escape from X chromosome inactivation is an intrinsic property of the Jarid1c locus. Proc Natl Acad Sci USA. 2008;105:17055–17060. doi: 10.1073/pnas.0807765105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Gupta V, Vermilyea MD, et al. Dosage compensation in the mouse balances up-regulation and silencing of X-linked genes. PLoS Biol. 2007;5:e326. doi: 10.1371/journal.pbio.0050326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- Lyon MF. Sex chromatin and gene action in the mammalian X-chromosome. Am J Hum Genet. 1962;14:135–148. [PMC free article] [PubMed] [Google Scholar]
- Lyon MF. X-chromosome inactivation: a repeat hypothesis. Cytogenet Cell Genet. 1998;80:133–137. doi: 10.1159/000014969. [DOI] [PubMed] [Google Scholar]
- Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 1997;11:156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- Marks H, Chow JC, Denissov S, et al. High-resolution analysis of epigenetic changes associated with X inactivation. Genome Res. 2009;19:1361–1373. doi: 10.1101/gr.092643.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil JA, Smith KP, Hall LL, Lawrence JB. Word frequency analysis reveals enrichment of dinucleotide repeats on the human X chromosome and [GATA]n in the X escape region. Genome Res. 2006;16:477–484. doi: 10.1101/gr.4627606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermoud JE, Popova B, Peters AH, Jenuwein T, Brockdorff N. Histone H3 lysine 9 methylation occurs rapidly at the onset of random X chromosome inactivation. Curr Biol. 2002;12:247–251. doi: 10.1016/s0960-9822(02)00660-7. [DOI] [PubMed] [Google Scholar]
- Migeon BR, Lee CH, Chowdhury AK, Carpenter H. Species differences in TSIX/Tsix reveal the roles of these genes in X-chromosome inactivation. Am J Hum Genet. 2002;71:286–293. doi: 10.1086/341605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Wakefield MJ, Aken B, et al. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature. 2007;447:167–177. doi: 10.1038/nature05805. [DOI] [PubMed] [Google Scholar]
- Mietton F, Sengupta AK, Molla A, et al. Weak but uniform enrichment of the histone variant macroH2A1 along the inactive X chromosome. Mol Cell Biol. 2009;29:150–156. doi: 10.1128/MCB.00997-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AP, Willard HF. Chromosomal basis of X chromosome inactivation: identification of a multigene domain in Xp11.21–p11.22 that escapes X inactivation. Proc Natl Acad Sci USA. 1998;95:8709–8714. doi: 10.1073/pnas.95.15.8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohandas T, Sparkes RS, Shapiro LJ. Reactivation of an inactive human X chromosome: evidence for X inactivation by DNA methylation. Science. 1981;211:393–396. doi: 10.1126/science.6164095. [DOI] [PubMed] [Google Scholar]
- Monkhorst K, Jonkers I, Rentmeester E, Grosveld F, Gribnau J. X inactivation counting and choice is a stochastic process: evidence for involvement of an X-linked activator. Cell. 2008;132:410–421. doi: 10.1016/j.cell.2007.12.036. [DOI] [PubMed] [Google Scholar]
- Morey C, Kress C, Bickmore WA. Lack of bystander activation shows that localization exterior to chromosome territories is not sufficient to up-regulate gene expression. Genome Res. 2009;19:1184–1194. doi: 10.1101/gr.089045.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishima A, Grumbach MM, Taylor JH. Asynchronous duplication of human chromosomes and the origin of sex chromatin. Proc Natl Acad Sci USA. 1962;48:756–763. doi: 10.1073/pnas.48.5.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morleo M, Franco B. Dosage compensation of the mammalian X chromosome influences the phenotypic variability of X-linked dominant male-lethal disorders. J Med Genet. 2008;45:401–408. doi: 10.1136/jmg.2008.058305. [DOI] [PubMed] [Google Scholar]
- Mueller JL, Mahadevaiah SK, Park PJ, et al. The mouse X chromosome is enriched for multicopy testis genes showing postmeiotic expression. Nat Genet. 2008;40:794–799. doi: 10.1038/ng.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DK, Disteche CM. Dosage compensation of the active X chromosome in mammals. Nat Genet. 2006;38:47–53. doi: 10.1038/ng1705. [DOI] [PubMed] [Google Scholar]
- Ogawa Y, Lee JT. Xite, X-inactivation intergenic transcription elements that regulate the probability of choice. Mol Cell. 2003;11:731–743. doi: 10.1016/s1097-2765(03)00063-7. [DOI] [PubMed] [Google Scholar]
- Ohno S. Sex chromosomes and sex-linked genes. Springer-Verlag; Berlin: 1967. [Google Scholar]
- Okamoto I, Heard E. Lessons from comparative analysis of the X-chromosome inactivation in mammals. Chromosome Res. 2009 doi: 10.1007/s10577-009-9057-7. [DOI] [PubMed] [Google Scholar]
- Orstavik KH. X chromosome inactivation in clinical practice. Hum Genet. 2009 Apr 28; doi: 10.1007/s00439-009-0670-5. 2009. [DOI] [PubMed] [Google Scholar]
- Payer B, Lee JT. X chromosome dosage compensation: how mammals keep the balance. Annu Rev Genet. 2008;42:733–772. doi: 10.1146/annurev.genet.42.110807.091711. [DOI] [PubMed] [Google Scholar]
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- Pfeifer GP, Steigerwald SD, Mueller PR, Wold B, Riggs AD. Genomic sequencing and methylation analysis by ligation mediated PCR. Science. 1989;246:810–813. doi: 10.1126/science.2814502. [DOI] [PubMed] [Google Scholar]
- Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath K, Talbot D, Hamer KM, et al. Developmentally regulated alterations in Polycomb repressive complex 1 proteins on the inactive X chromosome. J Cell Biol. 2004;167:1025–1035. doi: 10.1083/jcb.200409026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastan S. Non-random X-chromosome inactivation in mouse X-autosome translocation embryos–location of the inactivation centre. J Embryol Exp Morphol. 1983;78:1–22. [PubMed] [Google Scholar]
- Riggs AD. Marsupials and mechanisms of X chromosome inactivation. Aust J Zool. 1990;37:419–441. [Google Scholar]
- Ross MT, Grafham DV, Coffey AJ, et al. The DNA sequence of the human X chromosome. Nature. 2005;434:325–337. doi: 10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougeulle C, Chaumeil J, Sarma K, et al. Differential histone H3 Lys-9 and Lys-27 methylation profiles on the X chromosome. Mol Cell Biol. 2004;24:5475–5484. doi: 10.1128/MCB.24.12.5475-5484.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro LJ, Mohandas T, Weiss R, Romeo G. Non-inactivation of an x-chromosome locus in man. Science. 1979;204:1224–1226. doi: 10.1126/science.156396. [DOI] [PubMed] [Google Scholar]
- Silva J, Mak W, Zvetkova I, et al. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev Cell. 2003;4:481–495. doi: 10.1016/s1534-5807(03)00068-6. [DOI] [PubMed] [Google Scholar]
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- Spencer JA, Sinclair AH, Watson JM, Graves JA. Genes on the short arm of the human X chromosome are not shared with the marsupial X. Genomics. 1991;11:339–345. doi: 10.1016/0888-7543(91)90141-z. [DOI] [PubMed] [Google Scholar]
- Sudbrak R, Wieczorek G, Nuber UA, et al. X chromosome-specific cDNA arrays: identification of genes that escape from X-inactivation and other applications. Hum Mol Genet. 2001;10:77–83. doi: 10.1093/hmg/10.1.77. [DOI] [PubMed] [Google Scholar]
- Talebizadeh Z, Simon SD, Butler MG. X chromosome gene expression in human tissues: male and female comparisons. Genomics. 2006;88:675–681. doi: 10.1016/j.ygeno.2006.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JH. Asynchronous duplication of chromosomes in cultured cells of Chinese hamster. J Biophys Biochem Cytol. 1960;7:455–464. doi: 10.1083/jcb.7.3.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya KD, Greally JM, Yi Y, et al. Comparative sequence and x-inactivation analyses of a domain of escape in human xp11.2 and the conserved segment in mouse. Genome Res. 2004;14:1275–1284. doi: 10.1101/gr.2575904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valley CM, Pertz LM, Balakumaran BS, Willard HF. Chromosome-wide, allele-specific analysis of the histone code on the human X chromosome. Hum Mol Genet. 2006;15:2335–2347. doi: 10.1093/hmg/ddl159. [DOI] [PubMed] [Google Scholar]
- Veyrunes F, Waters PD, Miethke P, et al. Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes. Genome Res. 2008;18:965–973. doi: 10.1101/gr.7101908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Willard HF, Mukherjee S, Furey TS. Evidence of influence of genomic DNA sequence on human X chromosome inactivation. PLoS Comput Biol. 2006;2:e113. doi: 10.1371/journal.pcbi.0020113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MK, Makova KD. Evolution and survival on eutherian sex chromosomes. PLoS Genet. 2009a;5:e1000568. doi: 10.1371/journal.pgen.1000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MK, Makova KD. Genomic analyses of sex chromosome evolution. Annu Rev Genomics Hum Genet. 2009b;10:25.1–25.22. doi: 10.1146/annurev-genom-082908-150105. [DOI] [PubMed] [Google Scholar]
- Wu H, Fassler R, Schnieke A, et al. An X-linked human collagen transgene escapes X inactivation in a subset of cells. Development. 1992;116:687–695. doi: 10.1242/dev.116.3.687. [DOI] [PubMed] [Google Scholar]
- Wutz A, Jaenisch R. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell. 2000;5:695–705. doi: 10.1016/s1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]
- Xu J, Burgoyne PS, Arnold AP. Sex differences in sex chromosome gene expression in mouse brain. Hum Mol Genet. 2002;11:1409–1419. doi: 10.1093/hmg/11.12.1409. [DOI] [PubMed] [Google Scholar]
- Xu J, Watkins R, Arnold AP. Sexually dimorphic expression of the X-linked gene Eif2s3x mRNA but not protein in mouse brain. Gene Expr Patterns. 2006a;6:146–155. doi: 10.1016/j.modgep.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Xu N, Tsai CL, Lee JT. Transient homologous chromosome pairing marks the onset of X inactivation. Science. 2006b;311:1149–1152. doi: 10.1126/science.1122984. [DOI] [PubMed] [Google Scholar]
- Zechner U, Wilda M, Kehrer-Sawatzki H, et al. A high density of X-linked genes for general cognitive ability: a run-away process shaping human evolution? Trends Genet. 2001;17:697–701. doi: 10.1016/s0168-9525(01)02446-5. [DOI] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]