Abstract
Neural stem progenitor cells (NSPCs) in the human subventricular zone (SVZ) potentially contribute to life-long neurogenesis, yet subtypes of glioblastoma multiforme (GBM) contain NSPC signatures that highlight the importance of cell fate regulation. Among numerous regulatory mechanisms, the post-translational methylations onto histone tails are crucial regulator of cell fate. The work presented here focuses on the role of two repressive chromatin marks tri-methylations on histone H3 lysine 27 (H3K27me3) and histone H4 lysine 20 (H4K20me3) in the adult NSPC within the SVZ. To best model healthy human NSPCs as they exist in vivo for epigenetic profiling of H3K27me3 and H4K20me3, we utilized NSPCs isolated from the adult SVZ of baboon brain (Papio anubis) with brain structure and genomic level similar to human. The putative role of H3K27me3 in normal NSPCs predominantly falls into the regulation of gene expression, cell cycle, and differentiation, whereas H4K20me3 is involved in DNA replication/repair, metabolism, and cell cycle. Using conditional knock-out mouse models to diminish Ezh2 and Suv4-20h responsible for H3K27me3 and H4K20me3, respectively, we found that both repressive marks have irrefutable function for cell cycle regulation in the NSPC population. While both EZH2/H3K27me3 and Suv4-20h/H4K20me3 have implication in cancers, our comparative genomics approach between healthy NSPCs and human GBM specimens revealed that substantial sets of genes enriched with H3K27me3 and H4K20me3 in the NSPCs are altered in the human GBM. In sum, our integrated analyses across species highlight important roles of H3K27me3 and H4K20me3 in normal and disease conditions in the context of NSPC.
Keywords: Glioblastoma Multiforme (GBM); Neural Stem Progenitor Cells (NSPCs); Chromatin Immunoprecipitation (ChIP); Cre recombinant protein; Epigenetic Repression; Stereotaxic injection; tri-methylation at histone 3 lysine 27 (H3K27me3) and histone 4 lysine 20 (H4K20me3).; Enhancer of zeste (Human- Gene: EZH2, Protein: EZH2) (Mouse- Gene: Ezh2, Protein: Histone-lysine N-methyltransferase EZH2); Suppressor of variegation homolog 1 (Human- Gene: KMT5B or SUV420H1, Protein: lysine methyltransferase 5B, synonym Suv4-20h1) (Mouse- Gene: Suv4-20h1, synonym Kmt5b, Protein: Histone-lysine N-methyltransferase KMT5B, synonym Suv4-20h1); Suppressor of variegation homolog 2 (Human- Gene: KMT5C or SUV420H2, Protein: lysine methyltransferase 5C, synonym Suv4-20h2) (Mouse- Gene: Suv4-20h2, synonym Kmt5c, Protein: Histone-lysine N-methyltransferase KMT5C, synonym Suv4-20h2)
1. Introduction
Epigenetic regulation such as histone modifications is one of the most influential players in broad aspects of biology including cell fate determination during neurogenesis and tumorigenesis. Histones are evolutionarily conserved proteins that interact with genomic DNA and directly impact chromatin structure and transcriptional activity. Various amino acid residues on the tails of histones provide a substrate for chemical modifications, such as addition of methyl groups. This catalytic mechanism involves transferring a methyl group from S-adenosyl-L-methionine (SAM) to the lysine residue of a histone tail (Trievel et al., 2002). Two such methylation sites include lysine 27 on histone H3 and lysine 20 on histone H4 capable of accepting up to three methyl groups that result in trimethylated forms of histone H3 lysine 27 (H3K27me3) and histone H4 lysine 20 (H4K20me3). Both H3K27me3 and H4K20me3 are associated with chromatin compaction and transcriptional repression, which have been implicated in tumorigenesis upon dysregulation (Varambally et al., 2002; Bracken et al., 2003; Kleer et al., 2003; Ting et al., 2006; Shen and Laird, 2013; Evertts et al., 2013). H3K27me3 and H4K20me3 are catalyzed independently by the SET-domain superfamily of histone methyltransferases (HMTs), the enhancer of zeste homolog 2 (EZH2) and the suppressor of variegation 4–20 homologs (KMT5B/Suv4-20h1 and KMT5C/Suv4-20h2), respectively (Sakaguchi et al., 2008; Yang et al., 2008; Schotta et al., 2008). EZH2 is present in undifferentiated cells of the murine subventricular zone (SVZ), the largest neurogenic niche in adult mammals. Ezh2 deletion by crossing to GFAP-Cre mouse line results in deficit of postnatal neurogenesis in the mouse SVZ (Hwang et al., 2014). Suv4-20h1 is ubiquitously present in adult tissues including brain, whereas Suv4-20h2 displays restricted abundance in some adult tissues. Double knock-out of Suv4-20h1 and Suv4-20h2 results in compromised genomic integrity via impaired double strand break repair and perinatal lethality. (Schotta et al., 2008). The molecular targets of H3K27me3 and H4K20me3 in the adult neural stem and progenitor cells (NSPCs) have yet to be identified. As epigenetic landscapes are vulnerable to culture artifact and obtaining endogenous cells freshly from healthy human brain is ethically unacceptable, we purified in vivo NSPCs from baboon brain for genome-wide approach to identify putative targets of H3K27me3 and H4K20me3. Given the extensive correlation of brain structure (Rogers et al., 2010; Kochunov et al., 2010) and genomic similarity between baboon and human (Cox et al., 2013), findings regarding epigenetic regulation in baboon models may hold significant relevance to healthy and neuropathological conditions in human. Therefore, we utilized a previously described technique to purify subpopulations of in vivo SVZ cells from baboon brain (Sandstrom et al., 2014) for genome-wide analyses including chromatin immunoprecipitation followed by sequencing (ChIP-Seq) and RNA-Seq. The current study is the first to align in vivo baboon brain-derived ChIP-Seq and RNA-Seq reads to newly annotated baboon genome. Through integrated analyses of ChIP-Seq and RNA-Seq, genes enriched with H3K27me3 were found to function primarily in cell cycle and differentiation. In addition to the known role of H4K20me3 in DNA replication licensing, we found that putative targets of H4K20me3 function in cell cycle, metabolism, cellular organization, and immune response.
To glean the in vivo function of H3K27me3 and H4K20me3 in this adult neurogenic niche, we utilized conditional mouse model of Ezh2 and Suv4-20h1/Suv4-20h2. Because H3K27me3 and H4K20me3 are ubiquitously present in NSPCs within the adult SVZ, we employed stereotaxic injection of Cre protein into the adult mouse SVZ to knock down the expression of Ezh2 and Suv4-20h in NSPC populations and subsequently assess the phenotypic outcome. Our innovative approach allows the assessment of loss of function in multiple populations simultaneously and provides broad insight into the role of EZH2/H3K27me3 and Suv4-20h/H4K20me3 in this adult neurogenic niche. We found that loss of H3K27me3 or H4K20me3 consistently affects S phase of cell cycle, suggesting that Ezh2/H3K27me3 and Suv4-20h/H4K20me3 play distinct mode in replication during adult neurogenesis.
While the SVZ harbors NSPCs with great proliferative potential and the niche microenvironment within the SVZ is permissive to growth and proliferation, this neurogenic niche is also suspected to be a vulnerable site for the origin of subtypes of GBM. Compelling evidence has shown that human glioblastoma multiforme (GBM) is composed of tumor cells and a portion of stemness signature expressing markers of GFAP, Vimentin, and DCX (Sanai et al., 2004; Haskins et al., 2013). In support of this notion, MRI has characterized that group I and group II GBM (GBMI and GBMII) contacts the SVZ intimately (Lim et al., 2007). The heterogeneous nature of GBM manifests in mixed SVZ cell types within the tumor, suggesting that disruption of cell fate transition within the SVZ is involved in tumorigenesis. Importantly, dysregulation of EZH2/H3K27me3 and SUV420H/H4K20me3 has been associated with human cancers (Varambally et al., 2002; Bracken et al., 2003; Kleer et al., 2003; Ting et al., 2006; Shen and Laird, 2013). Perhaps, the balance between self-renewal and differentiation during adult neurogenesis is in part maintained and regulated by these repressive marks, and disruption of this balance may be responsible for tumorigenesis. In line with this speculation, we carried out integrated analyses across genome-wide data and found that numerous genes enriched with H3K27me3 and H4K20me3 without detectable transcripts in normal adult NSPCs are altered in the SVZ-associated GBMI and GBMII. These genes are involved in cell death and survival, development, and cell cycle. Our findings provide an integrated view that heterogeneous tumors potentially arises under aberrant epigenetic regulation in part through H3K27me3 and H4K20me3. Additionally, our use of multiple animal models has allowed an integrated perspective of highly related biological processes in vivo which are not easily attained using a single model organism. Such insights include the roles of H3K27me3 and H4K20me3 in endogenous NSPC (i.e. baboons), genetic manipulation (i.e. mouse models), and correlation with pathobiology (i.e. human GBM). Further in vivo studies modulating the enrichment of H3K27me3 and H4K20me3 in the SVZ will decipher the mechanisms of this correlation. In summary, our approach will be of considerable interest to those applying cutting edge techniques to phenomena with tight temporal and spatial regulation for cell fate transition and disease associated with NSPCs across model organisms.
2. Materials and Methods
All experiments involving animals and animal use guidelines were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Texas at San Antonio and Texas Biomedical Research Institute/Southwest National Primate Research Center. All GBM specimens used in this study are non-identifiable (no patient identifiers connection).
2.1 Flow Cytometry
Rostral and caudal baboon SVZ was micro-dissected from forebrain tissue, digested with Accutase for 10 min, and strained with a 40 μm cell strainer (BD #352340) to yield single cell suspension. Cells were stained with viability dye (BD Biosciences #564406; 1:1000), subsequently fixed in 4% PFA, permeabilized, and blocked using FC Block (BD Pharmigen #553141; 1:200). Following blocking, cells were incubated for 60 min. at 4°C with primary antibodies anti- GFAP (mouse – Millipore #MAB3402; 1:500; rat – Invitrogen #130300; 1:500), Nestin (rabbit – Abcam #AB27952; 1:500, mouse – DHSB #rat-401, 1:500), DCX (Santa Cruz #sc-8066, 1:500), H3K27me3 (rabbit – Millipore #07-449; 1:500; mouse – Active Motif #61017;1:500), and/or H4K20me3 (Abcam #ab9053; 1:500). Flourochrome labeling utilized secondary antibodies conjugated with either AlexaFlour® 488 (Life Technologies anti-mouse #A21200/anti-goat #A11055; 1:300), AlexaFlour® 647 (Life Technologies anti-mouse #A21463/anti-rat #A21247; 1:300), and PE (Cell Signaling #8885S; 1:300) for 30 min at 4° C. Staining controls included single color positive controls labeled with each separate antibody, viability control, and unstained cells. Flow cytometry was run on a LSRII (BD Biosciences) configured with violet (405 nm with 450/50 BP), argon (488 nm with 530/30 BP), green (561 nm with 660/20 BP), and red (633 nm with 780/30 BP) lasers. Data analysis was performed using FlowJo software (FlowJo, LLC.). Final reported baboon SVZ populations were averaged across rostral and caudal baboon flow cytometry replicates.
2.2 Immunohistochemistry and Confocal Imaging
Fresh baboon forebrain was fixed in 4% paraformaldehyde overnight, and cryoprotected in 30% sucrose before embedding in OCT. 60 μm floating coronal sections were incubated with antibodies against H3K27me3 (Active Motif #39155; 1:1000) or H4K20me3 (Upstate #07-463; 1:1000) plus cell type markers Glial Fibrillary Acidic Protein (GFAP) – Clone GA5 (Millipore #MAB3402; 1:500), Nestin (Abcam #ab134017; 1:500), or Doublecortin (DCX) – clone C-18 (Santa Cruz #sc-8066; 1:200). Fluorescent labeling utilized secondary antibodies AlexaFlour® 488 (Life Technologies #A21200 1:1000) and AlexaFlour® 594 (Life Technologies #A21207; 1:2000). Sections were counterstained by DAPI in Vectashield (Vector Labratories; #H-1200). SVZ sections were imaged on a Ziess 710 confocal microscope (40X and 100X oil immersion objective). Images were processed using Zeiss Zen Black 2011, Zen v2 (Carl Zeiss Microscopy) and ImageJ (NIH) software.
2.3 Chromatin Immunoprecipitation
Neural Stem Progenitor Cells (NSPCs) were purified using Dynabeads-conjugated antibodies against cell type-specific markers (GFAP, Nestin, Vimentin, PSA-NCAM, or Doublecortin) as previously described (Sandstrom et al., 2014). Briefly, cells from fresh dissected baboon SVZ were immediately dissociated with Accutase, equilibrated in binding buffer containing phosphate-buffered saline (PBS), 0.05 % TritonX-100 (or saponin, detergent choice depends upon antibody), and subsequently subjected to Dynabeads-conjugated antibody purification. After elution with high salt and pH-gradient buffer (citrate buffer base), the purified populations were crosslinked in 1.1% formaldehyde before chromatin shearing by Diagenode Bioruptor. The sheared chromatin obtained from purified NSPCs were incubated with Protein A Dynabeads (Life Technologies #10001D) conjugated with antibodies against either H3K27me3 (Active Motif #39155; 1:1000) or H4K20me3 (Upstate #07-463; 1:1000). For normalization, aliquots of sheared chromatin were incubated with antibodies against either total H3 (Millipore #05-499; 1:1000) or total H4 (Cell Signaling #2592; 1:1000). Enriched chromatin was eluted and de-crosslinked, prior to library preparation with TruSeq ChIP sample prep kit (Illumina #IP-202-2012 and #IP-202-1024) and deep sequencing to > 200 million tags on an Illumina HiSeq2000 sequencer.
2.4 Sequence alignment and peak calling
The initial ≥200 million reads were quality filtered resulting in 123,893,167 H3K27me3, 175,382,266 Total H3, 160,188,013 H4K20me3, and 202,275,106 Total H4 pass filtered reads which were assessed for read quality using FastQC v0.11.2 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Pass filtered reads were aligned to Papio anubis (PapAnu2.0) reference genome (ftp://ftp.ensembl.org/pub/release-78/fasta/papio_anubis/dna/) lacking mitochondrial genome with Bowtie v1.1.1 (http://bowtie-bio.sourceforge.net/index.shtml) aligner before sorting with Samtools v1.1 (http://www.htslib.org/). Genomic regions enriched with target histone modifications were detected using MACS2 v2.1.0.20140616 (https://github.com/taoliu/MACS/) callpeak function (with a false discovery rate (FDR) < 0.05 for each peak). Total H3 and total H4 alignments were used as input controls when calling peaks for HK27me3 and H4K20me3, respectively. MACS2 was run with default parameters with the exception of setting fold change range to 2–50 (-m 2 50) for all data sets and broad peak detection (--broad) for H3K27me3 enrichment. ChIP-Seq peaks were associated to a nearest transcription start site using baboon transcript annotation (ftp://ftp.ensembl.org/pub/release-78/gtf/papio_anubis/) and the closest-features function included in BEDOPS v2.4.3 toolkit.
2.5 RNA-Seq Analysis
Total RNA was extracted from purified baboon SVZ cells as described previously (Sandstrom et al., 2014) and sequencing libraries were generated with Illumina Tru-Seq stranded total RNA library prep kit (Illumina; RS-122-2301; RS-122-2302). RNA libraries were deep sequenced using paired-end sequencing, (2×36 bp, >300 million reads) on an Illumina HiSeq2000 sequencer. Reads were quality filtered resulting in 148,304,589 total pass filtered reads which were assessed for read quality using FastQC v0.11.2. Pass filtered reads from individual flow cells were aligned to PapAnu2.0 containing mitochondrial genome (ftp://ftp.ensembl.org/pub/release-78/fasta/papio_anubis/dna/) using Tophat2 v2.0.13 (https://ccb.jhu.edu/software/tophat/index.shtml), plus PapAnu2.0 transcript annotation (ftp://ftp.ensembl.org/pub/release-78/gtf/papio_anubis/) to guide alignment. Transcripts were assembled by Cufflinks v2.2.1 (http://cole-trapnell-lab.github.io/cufflinks/) using PapAnu2.0 transcript annotation to guide assembly, and perform bias correction and multi-read rescue prior to merging assembled transcripts into a single sorted BAM file using Samtools v1.1. Genes with a read density of >1 fragment per kilobase of exon per million fragments mapped (FPKM) over the entire gene were considered detectable via RNA-Seq and used in our integrative analysis. The GBM samples were snap-frozen surgical resection tumor (~ 200 mg tissues for each RNA-Seq), which were not expanded in culture. Expression data for GBM and control was generated as previously described (Sandstrom et al., 2014). Genes with expression values >1 FPKM were considered for subsequent analyses. Cuffdiff was applied to analyze the differential gene expression between GBM and control. Integrated analysis of ChIP-Seq and RNA-Seq data across species was performed with R (https://cran.r-project.org/). Area-proportional Venn diagrams were generated using the R package Vennerable v3.0 (http://r-forge.r-project.org/projects/vennerable) and heatmaps were rendered using heatmap.3 function (https://github.com/obigriffith/biostar-tutorials/blob/master/Heatmaps/heatmap.3.R).
Sample sizes
For ChIP-Seq, we had 2 independent sets of ChIP performed with 2 antibodies from different vendors (i.e. Millipore and Active Motif for H3K27me3; Abcam and Active Motif for H4K20me3). The summary of H3K27me3 (or H4K20me3) putative targets in Supplementary Tables are common between 2 independent ChIP-Seq, and each set has obtained 120~200 million pass-filter reads). For RNA-Seq, we also had at least 2 sample sets for baboon SVZ cells (n=3), GBMI (n=2), GBMII (n=2), and control region specimens (n=3) to obtain at least 300 million pass-filtered reads for each sample set. The summary in Supplementary Tables is common across independent samples after standard pipeline of RNA-Seq analysis (Cufflink and Cuffdiff).
2.6 GO, Network, and Pathway analysis
Duplicate gene references were removed prior to Gene Ontology (GO) or functional pathway and network analyses. DAVID Functional Annotation Tool (DAVID Bioinformatics Resources 6.7, NIAID/NIH) was utilized to perform (GO) analysis. For GO, a significance cutoff was set at p-value < 0.05, including Bonferroni multiple test correction. Functional pathway and network analyses of enriched loci were performed using Ingenuity Pathway Analysis (IPA) (Ingenuity Systems, Redwood City, CA, USA). Data for up to 10 of the top networks and pathways predicted by IPA are presented in supplementary tables, with significance cut-off set at p-value < 0.05 (calculated by IPA using right-tailed Fisher’s exact test).
2.7 Conditional knock-down of EZH2 and Suv4-20h in NSPCs within the adult SVZ by stereotaxic injection of recombinant Cre protein
For stereotaxic administration of purified Cre Recombinase protein into the SVZ of Ezh2flox/flox:ROSA+/+ mice or Suv4-20h1flox/flox/Suv4-20h2−/−:ROSA+/+ mice, the gas/oxygen mixture (isoflurane 1–3%) was delivered to the animal via a nose cone during anesthesia maintenance. A one-centimeter incision was made on the head, and 2–8 small bore holes (0.53 diameter) was drilled in the skull with a KOPF® 1911 Drill, after mounting to a KOPF® Model 1900 stereotax equipped with a Anilam Wizard 211 DRO. Prior to drilling bore holes, the skull was balanced on the sagittal and horizontal planes according to the manufacturer’s instructions. The SVZ injection with custom-made Hamilton syringe follows the coordinates: aneteroposterior (+0.97, +0.49; +0.25; −0.01), mediolateral (±0.95, ±1.32; ±1.34; ±1.49), and dorsoventral (−2.6, −2.2; −2.15; −2.0). All coordinates are measured from Bregma and the surface of the brain. A total of 350 nl of purified recombinant Cre protein (4 μg/μl in 20%glycerol/PBS) was delivered at 60 nl/min to each site. A vehicle injection used 350 nl/per injection of 20% glycerol/PBS. Five days post-injection, 5-ethynyl-2′-deoxyuridine and 5-bromo-2′-deoxyuridine (EdU and BrdU) were administrated into mice by intraperitoneal (IP) injection for 2 hours EdU and 5 days BrdU tracing (5 mice/each time point). The animals were anesthetized with Avertin prior to performing transcardial perfusion and fixed with PBS and 4% paraformaldehyde; and then brains were cryo-protected with 30% sucrose in PBS prior to cryosectioning for immunohistochemistry analyses. The experimental group of Cre injection and control group of vehicle injection for Ezh2 n=6/each group; for Suv4-20h1 n=5/each group.
3. RESULTS
3.1 H3K27me3 and H4K20me3 are ubiquitously enriched in the NSPC of adult primate SVZ
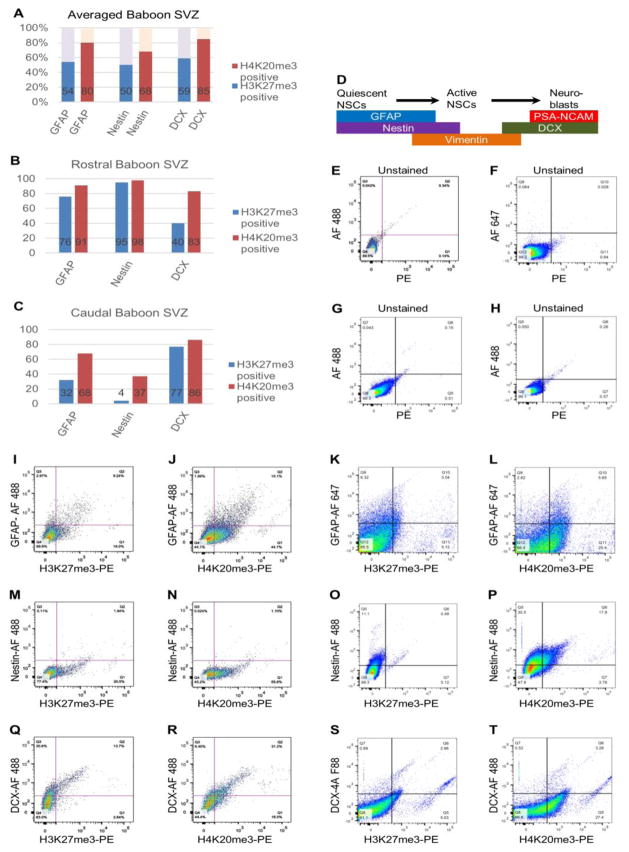
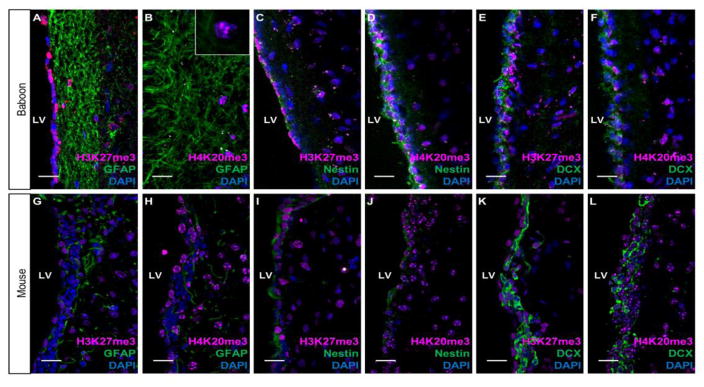
In mammals, the lateral ventricle is lined with neural stem and progenitor cells (NSPCs) including radial-glia like neural stem cells (NSCs), amplifying progenitor cells, and immature neuroblasts (NBs), as well as post-mitotic ependymal cells (Sandstrom et al., 2014; Jessberger et al., 2007; Ihrie and Alvarez-Buylla, 2008; Clelland et al., 2009; Ming and Song, 2011). To quantify mean enrichment ratios of H3K27me3 and H4K20me3 across NSPCs, rostral and caudal parts of baboon SVZ tissue was subjected to flow cytometry to measure the percent colocalization of H3K27me3, H4K20me3, and established cell type specific markers including glial fibrillary acidic protein (GFAP), Nestin, and doublecortin (DCX). H3K27me3-positve cells comprised approximately 54%, 50%, and 59% of GFAP-, Nestin-, and DCX-positive SVZ subpopulations, respectively (Fig. 1). H4K20me3-positive cells constituted approximately 80%, 68%, and 85% of GFAP, Nestin, and DCX-positive cells within the SVZ (Fig. 1). Co-immunostaining with antibodies for H3K27me3, H4K20me3, and established cell type specific markers including glial fibrillary acidic protein (GFAP), Nestin, and DCX revealed that H3K27me3 and H4K20me3 are present in quiescent/active NSCs and migrating neuroblasts (Fig. 2). The presence of H3K27me3 and H4K20me3 is not mutually exclusive to any subpopulation within the adult SVZ.
Fig. 1. Quantification of H3K27me3 or H4K20me3 co-localization with cell type markers in baboon SVZ by flow cytometry.
(A) Summary of co-localization between H3K27me3 or H4K20me3 and SVZ subpopulations by flow cytometry. Bar chart shows the mean percentages of H3K27me3 or H4K20me3 enriched neural stem and progenitor populations purified from baboon rostral and caudal SVZ regions. GFAP labels quiescent NSCs, Nestin is a pan-NSC marker, while DCX marks early and migrating neuroblasts. Percent of H3K27me3 or H4K20me3 indicate mean proportion across rostral and caudal baboon SVZ regions.
(B) Bar chart shows the relative percentages of H3K27me3 or H4K20me3 enriched neural stem and progenitor populations purified from baboon rostral SVZ.
(C) Bar chart shows the relative percentages of H3K27me3 or H4K20me3 enriched neural stem and progenitor populations purified from baboon caudal SVZ.
(D) Scheme dictates cell type specific markers expressed in NSPCs in the SVZ.
(E) Scatterplot of whole unstained cells isolated from rostral baboon SVZ using Alexa Fluor 488 and PE channels (Unstained control for Figure 1I, 1J, 1M, 1N, 1Q, 1R).
(F) Scatterplot of whole unstained cells isolated from caudal baboon SVZ using Alexa Fluor 647 and PE channels (Unstained control for Figure 1K, 1L).
(G) Scatterplot of whole unstained cells isolated from caudal baboon SVZ using Alexa Fluor 488 and PE channels (Unstained control for Figure 1O and Figure 1P).
(H) Scatterplot of whole unstained cells isolated from caudal baboon SVZ using Alexa Fluor 488 and PE channels (Unstained control for Figure 1S and Figure 1T).
(I) Dual labeling by GFAP and H3K27me3 for undifferentiated cells of the rostral baboon SVZ.
(J) Dual labeling by GFAP and H4K20me3 for undifferentiated cells of the rostral baboon SVZ.
(K) Cells labeled with GFAP and H3K27me3 isolated from caudal baboon SVZ.
(L) Cells labeled with GFAP and H4K20me3 isolated from caudal baboon SVZ.
(M) Cells labeled with Nestin and H3K27me3 isolated from rostral baboon SVZ.
(N) Cells labeled with Nestin and H4K20me3 isolated from rostral baboon SVZ.
(O) Cells labeled with Nestin and H3K27me3 isolated from caudal baboon SVZ.
(P) Cells labeled with Nestin and H4K20me3 isolated from caudal baboon SVZ.
(Q) Cells labeled with DCX and H3K27me3 isolated from rostral baboon SVZ.
(R) Cells labeled with DCX and H4K20me3 isolated from rostral baboon SVZ.
(S) Cells labeled with DCX and H3K27me3 isolated from caudal baboon SVZ.
(T) Cells labeled with DCX and H4K20me3 isolated from caudal baboon SVZ.
Fig. 2.
H3K27me3 and H4K20me3 distributions across SVZ subpopulations. Co-immunostaining of H3K27me3 or H4K20me3 with cell-type specific markers GFAP, Nestin, and DCX. Left panel presents coronal section of baboon brain and right panel presents coronal section of mouse brain. 40× magnification; Scale bar = 20 um. Inset shows 100× of H4K20me3 staining pattern.
3.2 Molecular targets of H3K27me3 and H4K20me3 in the endogenous NSPCs of adult SVZ
To assess the role of H3K27me3 and H4K20me3 within NSPCs in vivo without resorting to cultural NSPC expansion required a multispecies approach utilizing baboon for endogenous NSPCs, human GBM specimens for pathobiology, and mice for genetic manipulations (experimental strategy is outlined in Supplementary Fig. 1A). To interrogate the roles of H3K27me3 and H4K20me3 in endogenous NSPCs, we initially carried out genome-wide analyses including chromatin immunoprecipitation followed by sequencing (ChIP-Seq) and RNA-Seq on purified baboon SVZ cells. Because histone modifications are vulnerable to cultured condition (i.e. methyl donors from metabolism of cultured medium can alter histone methylation) (Black et al., 2012), we utilized in vivo NSPCs purified from non-human primate to avoid culture artifacts and the ethical obstacle of obtaining fresh brain tissue from healthy humans. To identify the molecular targets of H3K27me3 and H4K20me3 in cell populations of the adult SVZ, in vivo NSPCs were purified from baboon brain as previously described (Sandstrom et al., 2014) and subjected to ChIP-Seq using antibodies against H3K27me3 or H4K20me3 and parallel ChIP-Seq with antibodies against total H3 or H4 for normalization. ChIP-Seq reads were aligned to the newly annotated baboon genome maintained by the Ensembl project (PapAnu2.0) with Bowtie aligner prior to MACS2 peak calling (Supplementary Fig. 1B). Chromatin enriched with histone modifications identified by MACS2 peak calling (FDR < 0.05) were then associated to the nearest transcription start sites (TSS) using BEDOPS Closest Features tool and baboon TSS coordinates obtained from PapAnu2.0 annotation. H3K27me3 and H4K20me3 enriched peaks and genome-wide ChIP “read density” can be visualized on the robust UCSC genome browser (Supplementary Fig. 2). Our work presented here is the first to align brain-derived baboon ChIP-Seq reads to baboon genome. We identify a total of 709 loci are enriched with H3K27me3 (Supplementary Table 1i) corresponding to 271 unique genes (Supplementary Fig. 2A; Supplementary Table 1ii). Additionally, 10,000 loci corresponding to H4K20me3 peaks were detected (Supplementary Table 1iii); resulting in 763 unique baboon genes associated with H4K20me3 enrichment (Supplementary Fig. 2B; Supplementary Table 1iv).
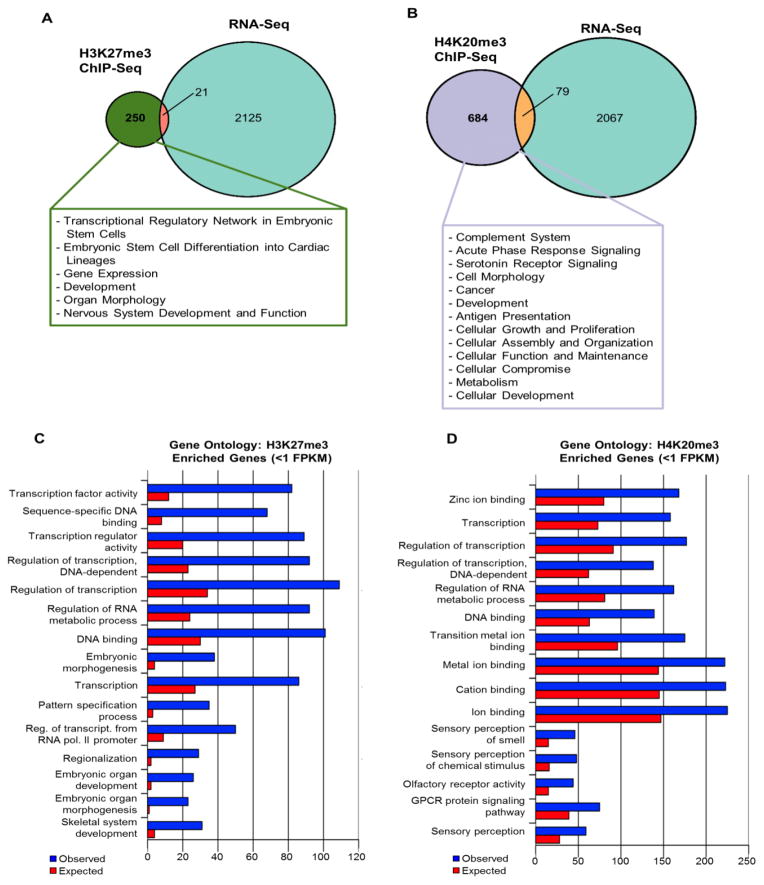
As H3K27me3 and H4K20me3 are associated with transcriptional repression, we compared our ChIP-Seq data with gene expression data of baboon NSPCs obtained from deep RNA sequencing (RNA-Seq) (Fig. 3). RNA-Seq reads were aligned to baboon genome with splice aware Tophat aligner then assembled into transcripts using Cufflinks (Supplementary Table 2i–2ii). Subsequently, integrated analysis was performed for ChIP-Seq loci (ChIP by H3K27me3 or H4K20me3) and RNA transcripts either detectable or undetectable determined by cutoff of 1 fragment per kilobase of exon per million fragments mapped (FPKM = 1) (Supplementary Table 2iii–2vi). Genes enriched with either H3K27me3 or H4K20me3 are largely associated with low or undetectable expression levels (FPKM ≤ 1), as 92.25% (250/271) of H3K27me3 (Fig. 3A; Supplementary Table 2iii) associated genes had no detectable RNA transcripts (FPKM ≤ 1), and 89.65% (684/763) of H4K20me3 (Fig. 3B; Supplementary Table 2v) associated genes did not produce a detectable transcript. To predict the role of genes enriched with H3K27me3, we used gene ontology (GO) to examine the 250 genes enriched by H3K27me3 with FPKM ≤ 1 and found these genes are correlated with transcriptional activity, particularly via transcription factors, as well as various processes related to differentiation (Top 15 GO categories had Bonferroni adjusted p-values ranging from 1.89×10−43 to 2.22×10−14) (Fig. 3C; Supplementary Table 2vii). Using GO to assess the cellular functions of the 684 genes enriched by H4K20me3 (FPKM ≤ 1) revealed that H4K20me3 enrichment is correlated with transcriptional regulation, mainly via zinc ion binding and DNA binding (Top 15 GO categories had Bonferroni adjusted p-values between 1.57×10−27 and 3.09×10−4) (Fig. 3D; Supplementary Table 2viii) in addition to olfaction and GPCR signaling pathway. Using Ingenuity pathway analysis (IPA), the 250 H3K27me3 enriched genes (FPKM ≤ 1) were predicted to function in transcriptional regulatory networks, lineage differentiation, and development (Fig. 3A (Box); Supplementary Table 2ix and 2x). The roles of these genes are largely relegated to coordination and commitment of differentiation by stem cells. Additionally, IPA predicted the 684 genes enriched with H4K20me3 and lacking detectable transcripts (FPKM ≤ 1) function in cellular assembly/organization/maintenance, innate immune responses, and metabolism (Fig. 3B (Box); Supplementary Table 2xi and 2xii).
Fig. 3. H3K27me3 and H4K20me3 genome-wide enrichment patterns in baboon SVZ.
(A) Proportional Venn diagram representations of genes enriched with H3K27me3 compared to RNA-Seq detectable genes as determined using expression threshold of FPKM > 1.0. Text box indicates IPA predicted functions correlated to genes enriched with H3K27me3 but lacking transcripts (≤1 FPKM). Green portion indicates H3K27me3 enriched genes with no detectable transcripts (≤1 FPKM) (n=250), orange portion indicates H3K27me3 genes with detectable RNA levels (>1 FPKM) (n=21) and light blue portion represents RNA-Seq genes with >1 FPKM but lacking H3K27me3 modifications (n=2125).
(B) Proportional Venn diagram representations of genes enriched with H4K20me3 compared to RNA-Seq detectable genes as determined using expression threshold of FPKM > 1.0. Text box indicates IPA predicted functions correlated to genes enriched with H4K20me3 but lacking transcripts (≤1 FPKM). Purple portion indicates H4K20me3 enriched genes with no detectable transcripts (≤1 FPKM) (n=684); orange portion indicates H4K20me3 genes with detectable RNA levels (n=79); light blue depicts RNA-Seq genes with >1 FPKM but no H4K20me3 enrichment (n=2067).
(C) Gene Ontology (GO) analysis of H3K27me3 enriched genes lacking detectable transcripts (≤1 FPKM) categorizes genes based on known biological processes or molecular functions and top 15 significant GO terms associated with H3K27me3 enriched genes with no expression (Top 15 GO categories had Bonferroni corrected p-values ranging from 1.89×10−43 to 2.22×10−14). Significance of GO terms was calculated based on binomial distribution model plus Bonferroni correction.
(D) Gene Ontology (GO) analysis of H4K20me3 enriched genes lacking detectable transcripts (≤1 FPKM) for top 15 significant GO terms in either biological processes of molecular function categories (Top 15 GO categories had Bonferroni adjusted p-values between 1.57×10−27 and 3.09×10−4). Significance of GO terms was calculated based on binomial distribution model plus Bonferroni correction.
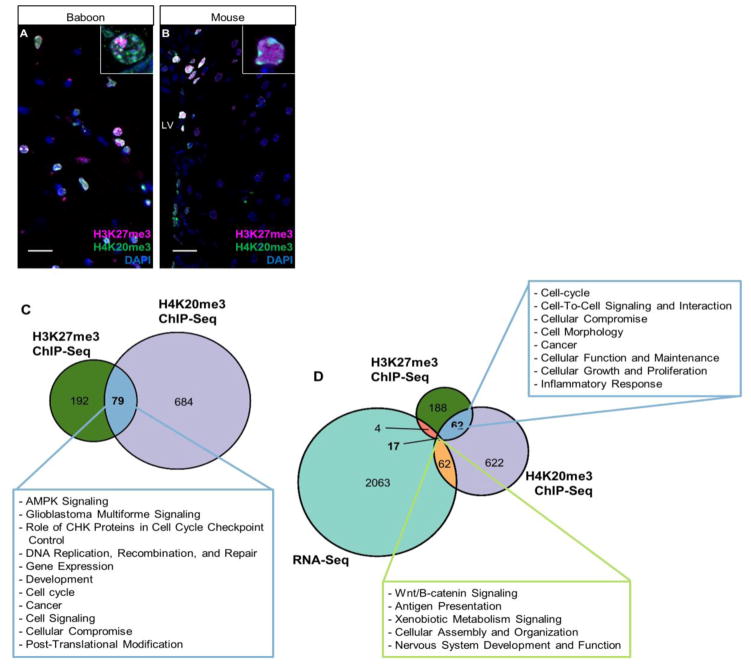
The SET-domain family proteins have been implicated in cancers and can interact with non-SET proteins to form chromatin modifying complexes (Ciferri et al., 2012; Dou et al., 2005). Additionally, we found co-localization of H3K27me3 and H4K20me3 in the baboon SVZ (Fig. 4), suggesting that spatial overlap of modifications have common genomic enrichment of a subclass of genes in the NSPCs. Therefore, we carried out comparison of H3K27me3 and H4K20me3 enriched loci and found 79 genes to be commonly enriched with both H3K27me3 and H4K20me3 (Fig. 4C; Supplementary Fig. 2C; Supplementary Table 3i), comprising 8.27% (79/955) of all genes enriched by either H3K27me3 or H4K20me3. However, many of the H3K27me3 and H4K20me3 peaks in this co-enriched subset are located at different genomic intervals of a specific gene. Through ChIP-Seq and RNA-Seq overlap comparison, of the 79 H3K27me3/H4K20me3 co-enriched genes identified by ChIP-Seq (from Fig. 4C), 62 have no detectable transcript (FPKM ≤ 1) (Fig. 4D; Supplementary Table 3iv), suggesting the co-repression of a set of genes by H3K27me3 and H4K20me3 in the NSPCs. Further, these 62 genes were classified by IPA with potential functions in cell-to-cell signaling and interaction, various metabolic processes, cancer, cell-cycle, cellular growth and proliferation, cellular compromise, and cellular maintenance (Fig. 4D (Blue Box); Supplementary Fig. 3; Supplementary Table 3v and 3vi). Our results suggest a protective role of H3K27me3/H4K20me3 against improper differentiation or aberrant cell-cycle progression. Lastly, the 17 H3K27me3/H4K20me3 co-enriched genes with detectable expression levels (FPKM>1) are associated with xenobiotic metabolism, antigen presentation, and cellular assembly and organization (Fig. 4D (Green Box); Supplementary Table 3viii and 3ix). Taken together, our data implicate that H3K27me3 and H4K20me3 plays common yet distinct roles in NSPCs of SVZ. We anticipate EZH2 and Suv4-20h1 interactions to function as a protective mechanism critical for the identity of adult NSPCs in the SVZ by preventing precocious lineage commitment and untimely cell cycle progression.
Fig. 4. Colocalization of H3K27me3 and H4K20me3 in NSPCs of baboon SVZ.
(A) Co-immunostaining of H3K27me3 and H4K20me3 in baboon brain. Region imaged corresponds to coronal section of the astrocytic ribbon within baboon SVZ. 40× magnification; Scale bar = 20 um. Inset shows 100× of H3K27me3 and H4K20me3 staining patterns.
(B) Co-immunostaining of H3K27me3 and H4K20me3 in mouse SVZ. 40× magnification; Scale bar = 20 um. Inset shows 100× of H3K27me3 and H4K20me3 staining patterns.
(C) Proportional Venn diagram generated by comparing numbers of genes enriched by H3K27me3, H4K20me3 or both histone modifications. Green area represents H3K27me3 enriched genes (n=192), purple represents H4K20me3 enriched genes (n=684), and blue area represents genes enriched by H3K27me3 and H4K20me3 (n=79). Text box describes functions of H3K27me3/H4K20me3 dual-enriched genes predicted by Ingenuity Pathway Analysis (IPA) software using known biochemical pathways and constructing de novo interaction networks.
(D) Proportional Venn diagram generated by comparing numbers of genes enriched by H3K27me3, H4K20me3 and genes detectable by RNA-Seq. Dark blue portion indicates genes enriched with H3K27me3 and H4K20me3 with no detectable transcription (≤1 FPKM) (n=62). Light green portion represents H3K27me3/H4K20me3 co-enriched genes with detectable RNA levels (>1 FPKM) (n=17).
3.3 EZH2/H3K27me3 and Suv4-20h/H4K20me3 influence cell cycle in the murine SVZ
Quiescent adult NSPCs divide very slowly during neurogenesis. The phenomenon of adult neurogenesis raises a fundamental question regarding epigenetic repression underlying the cell cycle of adult NSPCs. Importantly, our genome-wide analyses with baboon NSPCs implicated H3K27me3 and H4K20me3 co-regulating a subclass of genes associated with cell cycle including contact growth inhibition (i.e. SLC19A1, TTC5), cell cycle progression (i.e. ZIC1, MAP9) and checkpoint control (i.e. WNT9A). Yet, a number of genes are independently enriched by either H3K27me3 (i.e. CYP26B1) or H4K20me3 (i.e. MAP2K3, NRG1, DOT1L), suggesting their unique role in cell cycle regulation. To assess the in vivo effect of cell cycle upon loss of EZH2/H3K27me3 or Suv4-20h/H4K20me3, we utilized conditional knock-outs of the enzymatic domain (SET) essential for EZH2 and Suv4-20h activity because germ-line deletion of Ezh2 and Suv4-20h is embryonic lethal (Schotta et al., 2008; O’Carroll et al., 2001). These mouse models were generated with loxP sites flanking the SET domain of Ezh2 and Suv4-20h (Ezh2flox/flox and Suv4-20hflox/flox), respectively (Schotta et al., 2008; Su et al., 2003). Of note, although murine histone methyltransferases responsible for H4K20me3 are catalyzed by Suv4-20h1 and Suv4-20h2, Suv4-20h1 is ubiquitously present in adult tissues including brain. By contrast, Suv4-20h2 displays restricted abundance in some adult tissues though not in the brain, and the Suv4-20h2 null mice (Suv4-20h2−/−) have no apparent phenotype (Schotta et al., 2008). Nonetheless, to exclude compensation between Suv4-20h1 and Suv4-20h2, we chose to utilize floxed Suv4-20h1 allele and Suv4-20h2 null (Suv4-20h1flox/flox: Suv4-20h2−/−) for our studies. To simplify the genotype description, we denote Suv4-20hflox/flox representing Suv4-20h1flox/flox:Suv4-20h2−/− and Suv4-20h representing the subsequent phenotype at protein level. As H3K27me3 and H4K20me3 are ubiquitously enriched in NSPCs, we developed an alternative approach through stereotaxic injection of recombinant Cre protein into the SVZ that allows conditional loss of function of EZH2 and Suv4-20h1 activity in all cell types expressing Ezh2 and Suv4-20h within the SVZ (Supplementary Fig. 4). Stereotaxic injection coordinates were verified by injection of 50 nL of red fluorescent retrobeads (Supplementary Fig. 4A, 4B). Instead of knock-out of a single subpopulation of SVZ cells, this strategy allows us to gain broad insight as to whether the H3K27me3 and H4K20me3 repressive marks are critical for cell cycle regulation within this neurogenic niche. Following stereotaxic injection, Cre protein enters the plasma membrane and nucleus via an engineered transduction peptide and a nuclear localization sequence, respectively (Chien et al., 2014). To monitor the knock-out efficiency, we have bred the Ezh2flox/flox and Suv4-20hflox/flox to Rosa reporter mice (R26RLacZ and R26YFP with 2 loxp sites flanking the stop cassette) to obtain the Ezh2flox/flox:ROSA+/+ and Suv4-20hflox/flox:ROSA+/+ colonies. After Cre- induced site-specific recombination at Ezh2, Suv4-20h, and the ROSA reporter loci simultaneously, the SET domain and stop cassette are excised. Subsequently, the expression of β-galactosidase or YFP is largely confined to recombination events and is clearly demarcated from tissue outside injected regions (Supplementary Fig. 4C, 4D). We confirmed cells displaying β-galactosidase activity did not contain H3K27me3 modifications as determined by immunostaining of H3K27me3 and β-galactosidase (Supplementary Fig. 4E). Thus, we demonstrate that recombination is successfully confined to injection sites of the SVZ in adult mouse brain. We are also able to generate mice with Suv4-20h knock-out at specific locations within the SVZ (along with mock injection and reporter mice as controls). Post-Cre injection on day-5, EdU and phosphorylated-histone H3 labeling were used to trace cell cycle at S phase and M phase, respectively, within the population reporting loss of EZH2 and Suv4-20h activity. We found conditional knock-down EZH2 or Suv4-20h decreased short-term DNA replication (S phase), but no substantial effect on M phase (Fig. 5, 6), suggesting that both EZH2/H3K27me3 or Suv4-20h/H4K20me3 is not dispensable in chromosome duplication during cell cycle of adult neurogenesis.
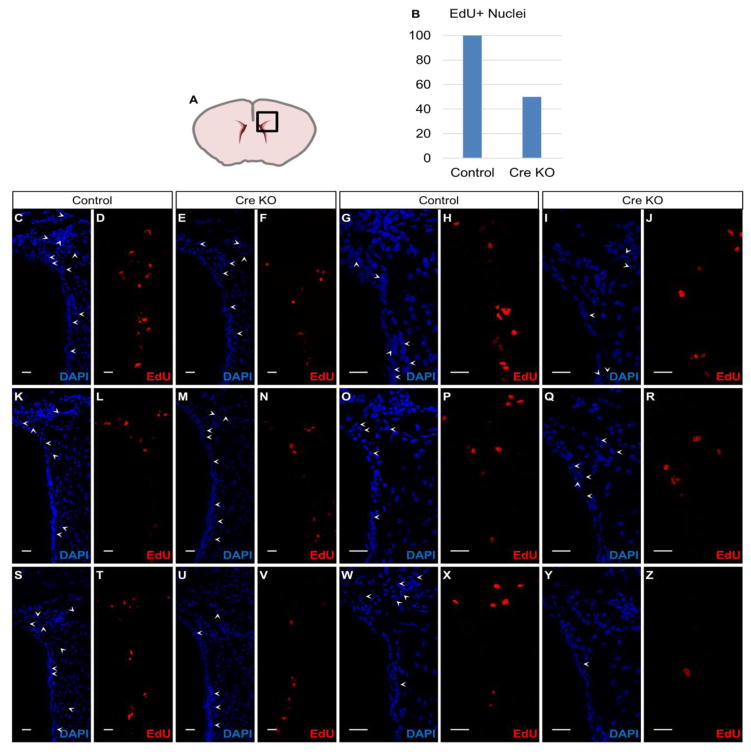
Fig. 5. EZH2/H3K27me3 influence cell cycle in the SVZ cells.
(A) Scheme of coronal sectioned mouse brain indicates region of immunostaining. Multiple images of dorsal SVZ and RMS were taken along rostro-caudal axis at 20X and 40X to obtain representative images of DAPI – positive and EdU-positive nuclei.
(B) Bargraph of EdU-positive nuclei within dorsal SVZ quantified using 40× magnification. Y-axis indicates percentage of cells which are EdU-positive compared to control.
(C) DAPI counterstain depicts nuclei at dorsal SVZ and rostral migratory stream in R26RLacZ reporter mouse with wildtype Ezh2 allele 10 days post-stereotaxic Cre administration. 20X, Scale bar = 20 um.
(D) 10 days post-stereotaxic Cre administration into a R26RLacZ reporter mouse with wildtype Ezh2 allele, EdU was detected 2 hours after intraperitoneal EdU injection. Cells undergoing DNA synthesis during the 2 hour window are detected as EdU -positive nuclei at dorsal SVZ and rostral migratory stream. 20X, Scale bar = 20 um.
(E) DAPI counterstain depicts nuclei at dorsal SVZ and rostral migratory stream 10 days post-stereotaxic Cre administration in an Ezh2flox/flox:ROSAL/L knockout mouse with 2 copies of a floxed Ezh2 gene. 20X, Scale bar = 20 um.
(F) 10 days post-stereotaxic Cre administration into an Ezh2flox/flox:ROSAL/L knockout mouse with 2 copies of a floxed Ezh2 gene, EdU was detected 2 hours after intraperitoneal EdU injection. The proportion of EdU-positive nuclei at the dorsal horn of the SVZ and RMS are diminished following stereotaxic injection into dorsal SVZ compared to Cre-injected R26RLacZ reporter mice. 20X, Scale bar = 20 um.
(G) DAPI counterstain depicts nuclei at dorsal SVZ and rostral migratory stream in R26RLacZ reporter mouse with wildtype Ezh2 allele 10 days post-stereotaxic Cre administration. 40X, Scale bar = 20 um.
(H) 10 days post-stereotaxic Cre administration into a R26RLacZ reporter mouse with wildtype Ezh2 allele, EdU was detected 2 hours after intraperitoneal EdU injection. Cells undergoing DNA synthesis during the 2 hour window are detected as EdU -positive nuclei at dorsal SVZ and rostral migratory stream. 40X, Scale bar = 20 um.
(I) DAPI counterstain depicts nuclei at dorsal SVZ and rostral migratory stream 10 days post-stereotaxic Cre administration in an Ezh2flox/flox:ROSAL/L knockout mouse with 2 copies of a floxed Ezh2 gene. 40X, Scale bar = 20 um.
(J) 10 days post-stereotaxic Cre administration into an Ezh2flox/flox:ROSAL/L knockout mouse with 2 copies of a floxed Ezh2 gene, EdU was detected 2 hours after intraperitoneal EdU injection. The proportion of EdU-positive nuclei at the dorsal horn of the SVZ and RMS are diminished following stereotaxic injection into dorsal SVZ compared to Cre-injected R26RLacZ reporter mice. 40X, Scale bar = 20 um.
(K) DAPI counterstain depicts nuclei at dorsal SVZ and rostral migratory stream in R26RLacZ reporter mouse 10 days post-stereotaxic Cre administration. 20X, Scale bar = 20 um.
(L) 10 days post-stereotaxic Cre administration into a R26RLacZ reporter mouse with wildtype Ezh2 allele, EdU was detected 2 hours after intraperitoneal EdU injection. 20X, Scale bar = 20 um.
(M) DAPI counterstain depicts nuclei at dorsal SVZ and rostral migratory stream 10 days post-stereotaxic Cre administration in an Ezh2flox/flox:ROSAL/L knockout mouse. 20X, Scale bar = 20 um.
(N) 10 days post-stereotaxic Cre administration into an Ezh2flox/flox:ROSAL/L knockout mouse with 2 copies of a floxed Ezh2 gene, EdU was detected 2 hours after intraperitoneal EdU injection. 20X, Scale bar = 20 um.
(O) DAPI counterstain depicts nuclei at dorsal SVZ and rostral migratory stream in R26RLacZ reporter mouse 10 days post-stereotaxic Cre administration. 40X, Scale bar = 20 um.
(P) 10 days post-stereotaxic Cre administration into a ROSA-LacZ reporter mouse with wildtype Ezh2 allele, EdU was detected 2 hours after intraperitoneal EdU injection. 40X, Scale bar = 20 um.
(Q) DAPI counterstain depicts nuclei at dorsal SVZ and rostral migratory stream 10 days post-stereotaxic Cre administration in an Ezh2flox/flox:ROSAL/L knockout mouse. 40X, Scale bar = 20 um.
(R) 10 days post-stereotaxic Cre administration into an Ezh2flox/flox:ROSAL/L knockout mouse with 2 copies of a floxed Ezh2 gene, EdU was detected 2 hours after intraperitoneal EdU injection. 40X, Scale bar = 20 um.
(S) DAPI counterstain depicts nuclei at dorsal SVZ and rostral migratory stream in R26RLacZ reporter mouse 10 days post-stereotaxic Cre administration. 20X, Scale bar = 20 um.
(T) 10 days post-stereotaxic Cre administration into a R26RLacZ reporter mouse with wildtype Ezh2 allele, EdU was detected 2 hours after intraperitoneal EdU injection. 20X, Scale bar = 20 um.
(U) DAPI counterstain depicts nuclei at dorsal SVZ and rostral migratory stream 10 days post-stereotaxic Cre administration in an Ezh2flox/flox:ROSAL/L knockout mouse. 20X, Scale bar = 20 um.
(V) 10 days post-stereotaxic Cre administration into an Ezh2flox/flox:ROSAL/L knockout mouse with 2 copies of a floxed Ezh2 gene, EdU was detected 2 hours after intraperitoneal EdU injection. 20X, Scale bar = 20 um.
(W) DAPI counterstain depicts nuclei at dorsal SVZ and rostral migratory stream in R26RLacZ reporter mouse 10 days post-stereotaxic Cre administration. 40X, Scale bar = 20 um.
(X) 10 days post-stereotaxic Cre administration into an R26RLacZ reporter mouse with wildtype Ezh2 allele, EdU was detected 2 hours after intraperitoneal EdU injection. 40X, Scale bar = 20 um.
(Y) DAPI counterstain depicts nuclei at dorsal SVZ and rostral migratory stream 10 days post-stereotaxic Cre administration in an Ezh2flox/flox:ROSAL/L knockout mouse. 40X, Scale bar = 20 um.
(Z) 10 days post-stereotaxic Cre administration into an Ezh2flox/flox:ROSAL/L knockout mouse with 2 copies of a floxed Ezh2 gene, EdU was detected 2 hours after intraperitoneal EdU injection. 40X, Scale bar = 20 um.
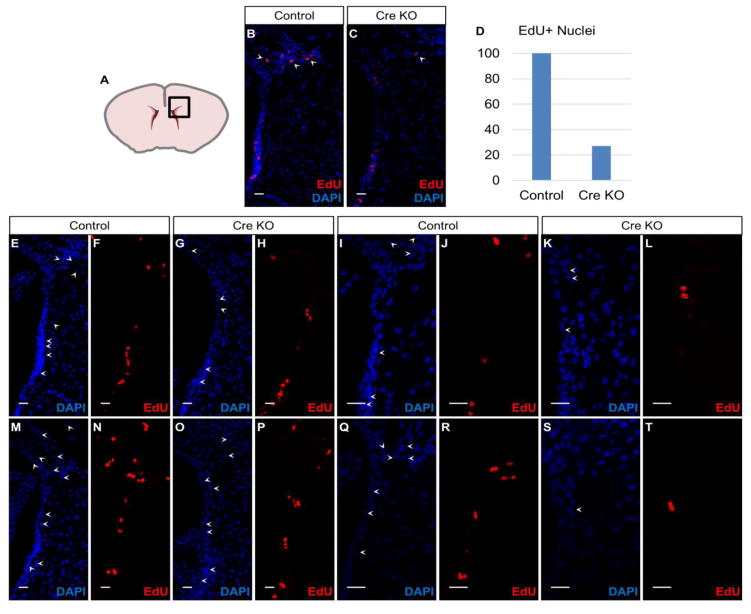
Fig. 6. Suv4-20h/H4K20me3 influence cell cycle in the SVZ cells.
(A) Scheme of coronal sectioned mouse brain indicates region of immunostaining.
(B) Co-localization of EdU and DAPI in non-injected SVZ in Suv4-20hflox/flox:RosaY/Y mouse. 20X, Scale bar = 20 um.
(C) Co-localization of EdU and DAPI in Cre injected SVZ in Suv4-20hflox/flox:RosaY/Y mouse, 5 days post-stereotaxic administration. 20X, Scale bar = 20 um.
(D) Bargraph of EdU-positive nuclei within dorsal SVZ quantified using 40x magnification. Y-axis indicates percentage of cells which are EdU –positive compared to control.
(E) Non-injected hemisphere in Suv4-20hflox/flox:RosaY/Y mouse depicting DAPI at dorsal SVZ and rostral migratory stream. 20X, Scale bar = 20 um.
(F) Non-injected hemisphere in a Suv4-20hflox/flox:RosaY/Y mouse. EdU was detected 2 hours after intraperitoneal EdU injection. 20X, Scale bar = 20 um.
(G) Cre injected hemisphere in Suv4-20hflox/flox:RosaY/Y mouse, 5 days post-stereotaxic Cre administration depicting DAPI at dorsal SVZ and rostral migratory stream. 20X, Scale bar = 20 um.
(H) Cre injected hemisphere in a Suv4-20hflox/flox:RosaY/Y mouse, 5 days post-stereotaxic Cre administration into. EdU was detected 2 hours after intraperitoneal EdU injection on last day of post-surgical rest. 20X, Scale bar = 20 um.
(I) Non-injected hemisphere in Suv4-20hflox/flox:RosaY/Y mouse depicting DAPI at dorsal SVZ and rostral migratory stream. 40X, Scale bar = 20 um.
(J) Non-injected hemisphere in a Suv4-20hflox/flox:RosaY/Y mouse. EdU was detected 2 hours after intraperitoneal EdU injection. 40X, Scale bar = 20 um.
(K) Cre injected hemisphere in Suv4-20hflox/flox:RosaY/Y mouse, 5 days post-stereotaxic Cre administration depicting DAPI at dorsal SVZ and rostral migratory stream. 40X, Scale bar = 20 um.
(L) Cre injected hemisphere in a Suv4-20hflox/flox:RosaY/Y mouse, 5 days post-stereotaxic Cre administration into. EdU was detected 2 hours after intraperitoneal EdU injection on last day of post-surgical rest. 40X, Scale bar = 20 um.
(M) Non-injected hemisphere in Suv4-20hflox/flox:RosaY/Y mouse depicting DAPI at dorsal SVZ and rostral migratory stream. 20X, Scale bar = 20 um.
(N) Non-injected hemisphere in a Suv4-20hflox/flox:RosaY/Y mouse. EdU was detected 2 hours after intraperitoneal EdU injection on last day of post-surgical rest. 20X, Scale bar = 20 um.
(O) Cre injected hemisphere in Suv4-20hflox/flox:RosaY/Y mouse, 5 days post-stereotaxic Cre administration depicting DAPI at dorsal SVZ and rostral migratory stream. 20X, Scale bar = 20 um.
(P) Cre injected hemisphere in a Suv4-20hflox/flox:RosaY/Y mouse, 5 days post-stereotaxic Cre administration into. EdU was detected 2 hours after intraperitoneal EdU injection on last day of post-surgical rest. 20X, Scale bar = 20 um.
(Q) Non-injected hemisphere in Suv4-20hflox/flox:RosaY/Y mouse depicting DAPI at dorsal SVZ and rostral migratory stream. 40X, Scale bar = 20 um.
(R) Non-injected hemisphere in a Suv4-20hflox/flox:RosaY/Y mouse. EdU was detected 2 hours after intraperitoneal EdU injection. 40X, Scale bar = 20 um.
(S) Cre injected hemisphere in Suv4-20hflox/flox:RosaY/Y mouse, 5 days post-stereotaxic Cre administration depicting DAPI at dorsal SVZ and rostral migratory stream. 40X, Scale bar = 20 um.
(T) Cre injected hemisphere in a Suv4-20hflox/flox:RosaY/Y mouse. EdU was detected 2 hours after intraperitoneal EdU injection on last day of post-surgical rest. 40X, Scale bar = 20 um.
3.4 A subset H3K27me3 and H4K20me3 enriched genes in NSPCs are altered in subtypes of glioblastoma multiforme
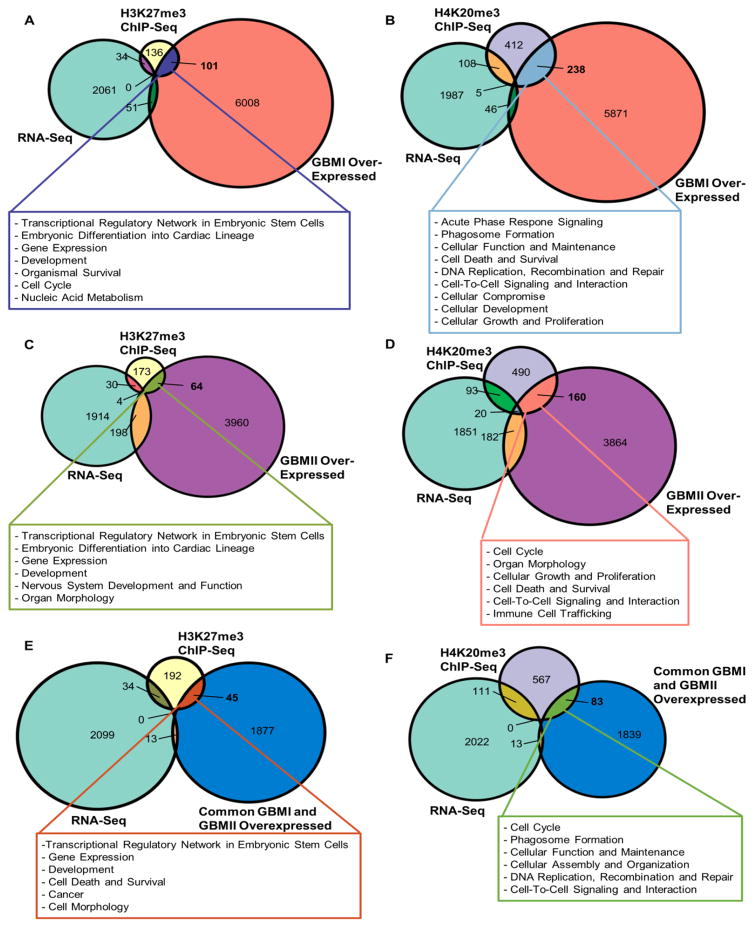
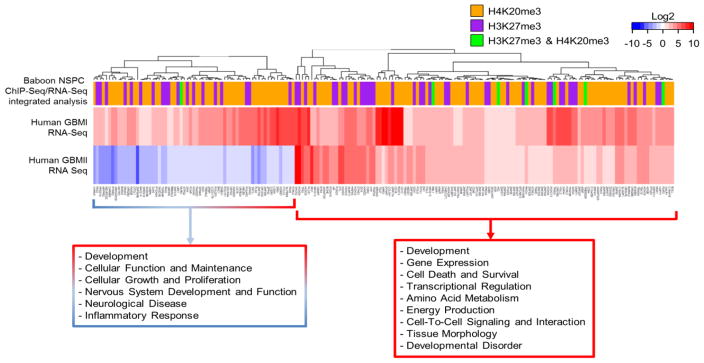
While considerable evidence reports EZH2 implication in human GBM (Kim et al., 2015; de Vries et al., 2015; Suva et al., 2009), our current study highlights several genes involved in cancer mechanism are co-enriched with H3K27me3 and H4K20me3. As MRI-classified group I and II GBM contain NSPCs signature (i.e. abundant GFAP, Vimentin, and DCX in tumor) (Haskins et al., 2013), poorly differentiated characteristics is apparent in subtypes of GBM. Given that i) epigenetic regulation by EZH2/H3K27me3 is known to suppress differentiation of stem cells and ii) both EZH2/H3K27me3 and Suv4-20h/H4K20me3 have implication in cancer mechanism prompted us to assess the relation of H3K27me3 and H4K20me3 in endogenous NSPCs and subtypes of GBM (GBMI and GBMII). Although the previous characterization of the genome and transcriptome of glioblastoma has provided the comprehensive data of molecular alterations that potentially drive glioblastoma pathogenesis and implicates glioblastoma as a heterogeneous brain tumor across each particular subtype (2008; Beroukhim et al., 2007; Parsons et al., 2008; Gunther et al., 2008; Phillips et al., 2006; Verhaak et al., 2010), there is no report thus far decipher the link between histone repressive marks and SVZ-associated GBM. Herein, we undertook comparative genomics approach between healthy NSPCs and GBM specimens that across i) molecular targets of H3K27me3 and/or H4K20me3 in the baboon NSPCs identified from ChIP-Seq; ii) expression data of in vivo NSPCs from baboon SVZ; iii) differential gene expression of GBMI and GBMII compared to controls on a genome-wide scale (Fig. 7). As obtaining fresh controls from live humans for GBM ChIP-Seq is ethically unacceptable, we used the current baboon ChIP-Seq data as a baseline epigenome. While not an exact match to adult human SVZ specimens, the current baboon data will undoubtedly shed light on epigenetic interactions in NSPCs of adult primates (including humans) and subsequent changes to gene expression in neuropathology. Because both H3K27me3 and H4K20me3 are known to suppress gene expression, we focus on targets of H3K27me3 and/or H4K20me3 identified from ChIP-Seq that are not detectable (FPKM <1) by RNA-Seq for purified NSPCs from baboon SVZ. Subsequently, differentially up-regulated genes in GBM cases were compared to corresponding H3K27me3 and H4K20me3 enriched genes in NSPCs without detectable transcripts. Using this integrated analysis of genome-wide data across species, we can uncover that genes aberrantly up-regulated in GBM are relevant to the dysregulation of H3K27me3 and/or H4K20me3 (Fig. 8). Of H3K27me3 enriched genes in baboon NSPCs with no detectable transcript (<1 FPKM), 101 and 64 are aberrantly up-regulated in GBMI and GBMII, respectively (Fig. 8A, 8C; Supplementary Table 4i and 4ii). These genes function in transcriptional regulation as well as nervous system and organismal development (Fig. 8A, 8C (Boxes); Supplementary Table 4iii – 4vi). Additionally, 238 and 160 of H4K20me3 enriched genes in baboon NSPCs with undetectable transcript (<1 FPKM) are aberrantly up-regulated in GBMI and GBMII, respectively (Fig. 8B, 8D; Supplementary Table 5i and 5ii). The 238 genes upregulated in GBMI are associated with cancer, cellular function and maintenance, cell death and survival, as well as DNA replication, recombination and repair (Fig. 8B (Box); Supplementary Table 5iii and 5iv). Similarly, the 160 genes aberrantly up-regulated in GBMII specimens have functions in cell cycle, cellular development, cellular growth and proliferation, cell death and survival, cell-to-cell signaling, and nervous system development (Fig. 8D (Box); Supplementary Table 5v and 5vi). Further, 45 of the H3K27me3 and 83 of the H4K20me3 enriched genes lacking expression in NSPCs have abnormally high expression levels in both GBMI and GBMII specimens (Fig. 8E, 8F; Supplementary Table 6i and 6ii). Functional analysis predicted by IPA reveals 45 genes function in transcriptional regulation, gene expression, development, cell death and survival, cancer and cell morphology (Fig. 8E (Box); Supplementary Table 6iii and 6iv); while 83 genes function in cell cycle, cellular maintenance/assembly/organization, DNA replication/repair/recombination, as well as cell-to-cell signaling and interaction (Fig. 8F (Box); Supplementary Table 6v and 6vi). We generated a heatmap to better illustrate the correlation among i) the enrichment of H3K27me3 or H4K20me3, ii) expression signature of normal NSPCs, and iii) aberrant gene expression of GBMI and GBMII (Fig. 9A and 9B present H3K27me3 and H4K20me3 separately; Supplementary Table 7i). A substantial proportion of upregulated genes in both GBMI and GBMII are enriched with H4K20me3 but lacking detectable transcripts in NSPCs (Fig. 9B; Supplementary Table 7iii). Assessing the presence of H3K27me3 and H4K20me3 simultaneously (Fig. 10), among the dendrogram cluster containing only upregulated GBMI and GBMII genes (Fig. 10 (Red Box); Supplementary Fig. 5A; Supplementary Table 7iv) have functions in development and cell death/survival predicted by IPA (Supplementary Fig. 7; Supplementary Table 7v and 7vi). Among genes upregulated in GBMI but downregulated in GBMII (Fig. 10 (Blue and Red Box); Supplementary Table 7vii) also are involved in similar category (i.e. development, cellular maintenance, growth and proliferation), yet different gene sets (Supplementary Table 7viii). As the heatmap dendrogram did not cluster genes into discernable groups of histone modifications (i.e. H3K27me3, H4K20me or both H3K27me3 and H4K20me3), we anticipate these two histone marks heterogeneously enrich for genes aberrantly expressed in SVZ-associated GBMI and GBMII.
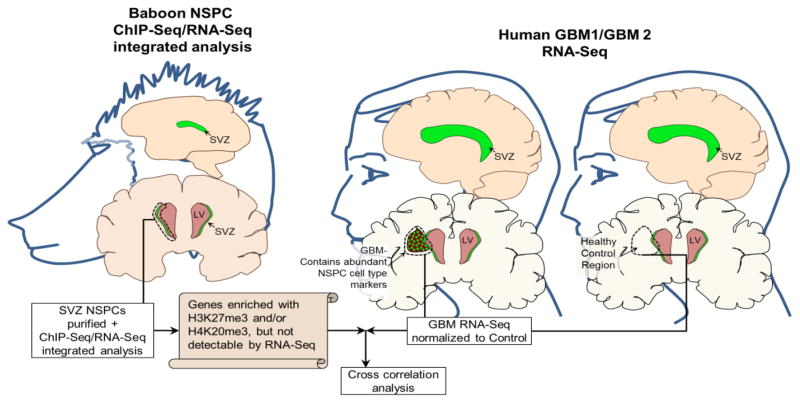
Fig. 7. Experimental Design for Correlation between genes in normal NSPCS enriched with H3K27me3 or H4K20me3 without detectable transcripts and genes altered in MRI-classified group I and group II GBM.
Graphical diagram illustrates the experimental design and analyses. Left panel: ChIP-Seq identified H3K27me3 and H4K20me3 enriched genes in normal NSPCs isolated from baboon SVZ and RNA-Seq analysis of normal NSPCs isolated from baboon SVZ. Right panel: RNA-Seq and Cuffdiff determined differential gene expression of human GBMI and GBMI compared to normal human specimens within correlated brain regions.
Fig. 8. Comparison among genes in normal NSPCS enriched with H3K27me3 or H4K20me3, genes without detectable transcripts in normal NSPCs, and genes elevated in MRI-classified group I and group II GBM.
(A) Proportional Venn diagram of three-way comparisons involving genes in NSPCS enriched with H3K27me3, undetectable genes in NSPCs by RNA-Seq (≤1 FPKM), and genes elevated in SVZ-associated human GBMI tumors. Text box indicates gene function as predicted by Ingenuity Pathway Analysis (IPA).
(B) Proportional Venn diagram of three-way comparisons involving genes in NSPCS enriched with H4K20me3, undetectable genes in NSPCs by RNA-Seq (≤1 FPKM), and genes elevated in SVZ-associated human GBMI tumors. Text box indicates gene function as predicted by IPA.
(C) Proportional Venn diagram of genes in NSPCs enriched with H3K27me3, undetectable genes in NSPCs by RNA-Seq (≤1 FPKM), and upregulated genes in GBMII.
(D) Proportional Venn diagram of genes in NSPCs enriched with H4K20me3, undetectable genes in NSPCs by RNA-Seq (≤1 FPKM), and upregulated genes in GBMII.
(E) Proportional Venn diagram of genes in NSPCs enriched with H3K27me3, undetectable genes in NSPCs by RNA-Seq (≤1 FPKM), and upregulated genes in GBMI and GBMII.
(F) Proportional Venn diagram of genes in NSPCs enriched with H4K20me3, undetectable genes in NSPCs by RNA-Seq (≤1 FPKM), and upregulated genes in GBMI and GBMII.
Fig. 9. Differential expression analysis of genes of human GBM specimens.
(A) A heatmap for differential expression of genes of human GBM specimens and corresponding H3K27me3 enrichment in endogenous NSPCs. Genes used for input are differentially expressed genes with greater than 2-fold change in human GBM corresponding to genes in normal NSPCs of baboon SVZ, which are lack of detectable transcript levels (≤1 FPKM) and enriched by H3K27me3. Inset shows symmetric color scale indicating differences in expression level as the (base 2) log of the fold change of GBM sample divided by control. Red indicates increased expression of genes in GBM relative to control, blue color indicates decreased expression of genes in GBM compared to control. Colored bars in column to left of heatmap indicate whether corresponding gene in baboon NSPC is not detectable by RNA-Seq (≤ 1 FPKM) or is expressed (> 1FPKM). Dendrogram was determined by hierarchical clustering using Euclidian distance and complete linkage.
(B) A heatmap for differential expression of genes of human GBM specimens and corresponding H4K20me3 enrichment in endogenous NSPCs. Genes used for input are differentially expressed genes with greater than 2-fold change in human GBM corresponding to genes in normal NSPCs of baboon SVZ, which are lack of detectable transcript levels (≤1 FPKM) and enriched by H4K20me3. Inset shows symmetric color scale indicating differences in expression level as the (base 2) log of the fold change of GBM sample divided by control. Red indicates increased expression of genes in GBM relative to control, blue color indicates decreased expression of genes in GBM compared to control. Colored bars in column to left of heatmap indicate whether corresponding gene in baboon NSPC is not detectable by RNA-Seq (≤ 1 FPKM) or is expressed (> 1FPKM). Dendrogram was determined by hierarchical clustering using Euclidian distance and complete linkage.
Fig. 10.
Correlation between genes in normal NSPCS enriched with H3K27me3 or H4K20me3 without detectable transcripts and genes altered in MRI-classified group I and group II GBM A heatmap for differential expression of genes of human GBM specimens. Genes used for input are 289 differentially expressed genes with greater than 2-fold change in human GBM corresponding to genes in normal NSPCs of baboon SVZ, which are lack of detectable transcript levels (≤1 FPKM) and enriched by either H3K27me3, H4K20me3, or co-enriched with H3K27me3/H4K20me3. Inset shows symmetric color scale indicating differences in expression level as the (base 2) log of the fold change of GBM sample divided by control. Red indicates increased expression of genes in GBM relative to control, blue color indicates decreased expression of genes in GBM compared to control. SVZ-associated GBM1 and GBM2 exhibit expression level changes in genes involved in multiple biological functions. Colored bars in column on top of heatmap indicate H3K27me3/H4K20me3 enrichment of corresponding genes in NSPCs of baboon SVZ. Dendrogram was determined by hierarchical clustering using Euclidian distance and complete linkage. Red text box indicates functions of clustered genes upregulated in both GBM1 and GBM2, as predicted by IPA. Red and blue box indicates functions of clustered genes upregulated in GBM1 and downregulated in GBM2, as predicted by IPA. While there is no evident pattern of clustering of histone modifications with respect to particular GBM genes, there is a substantial increase in the number of upregulated GBM genes which correspond to H3K27me3 and H4K20me3 enrichment, yet lacking detectable transcripts in the normal NSPCs of baboon SVZ.
4. Discussion
Multipotent NSPCs in adult SVZ must maintain a fine balance between self-renewal and differentiation (Doe, 2008). Improper control of this balance may shift otherwise normal neurogenesis towards oncogenesis or neurodegeneration. In line with this hypothesis, subtypes of SVZ-associated GBM represent similar signature of NSPCs (Sandstrom et al., 2014; Haskins et al., 2013) that further highlight the importance of cell fate regulation in health and disease conditions. While epigenetic modifications are increasingly appreciated for playing an intrinsic role in the cell fate determination of NSPCs (Hwang et al., 2014; Roman-Trufero et al., 2009; Molofsky et al., 2003; Gotz and Huttner, 2005; Ma et al., 2010; Zhang et al., 2014; Yao and Jin, 2014; Lunyak and Rosenfeld, 2008; Gao et al., 2011; Montalban-Loro et al., 2015), the importance of proper control of proliferation and differentiation in NSPCs is underscored by H3K27me3 and H4K20me3 in healthy NSPCs of adult SVZ. To better understand the potential function of these 2 repressive marks in endogenous NSPCs, we performed genome-wide approaches employing ChIP-Seq and RNA-Seq on purified NSPCs from the SVZ of baboon brain to determine their molecular targets. We found that 62 common genes co-enriched by H3K27me3 and H4K20me3 without detectable mRNA transcripts (<1 FPKM) are predominantly involved in cell-cycle, cellular maintenance, cellular compromise, cellular signaling, and cellular growth/proliferation. Our studies using conditional knock-out of Ezh2 or Suv4-20h further reveal their in vivo functions important to cell cycle regulation during adult neurogenesis, suggesting EZH2/H3K27me3 and Suv4-20h1/H4K20me3 may be a protective mechanism to minimize improper gene expression and modulate cellular proliferation, consequently preventing adult NSPCs from abnormal cell cycle re-entry or differentiation.
While DNA and RNA profiles of bulk tumors or single-cell from GBM have enabled genetic and transcriptional classification (2008; Patel et al., 2014), the relationships between epigenetic alteration and heterogeneity of GBM remain obscure. Such epigenetic alteration can change global gene expression and manifest intratumoral heterogeneity of GBM, wherein different stages or expression signatures are associated with distinct outcomes or therapeutic responses. Although conditional knock-down of Ezh2 and Suv4-20h in the NSPCs of rodent SVZ did not yield brain tumor over 46 weeks (unpublished data) and the levels of EZH2/H3K27me3 or Suv4-20h/H4K20me3 are not substantially changed in human specimens (Supplementary Fig. 5B, 5C), we reason their effects would be in a gene-specific manner. Through integrated analyses of genome-wide data across species, we found that a set of genes enriched with H3K27me3 and H4K20me3 in the in vivo NSPCs are altered in GBM specimens harboring a NSPC signature. We identified that 37% (101/271) and 24% (64/271) of H3K27me3 potentially repressed genes are upregulated in GBMI and GBMII, respectively. Similarly, 31% (238/763) and 21% (160/763) of H4K20me3 potentially repressed genes are aberrantly up-regulated in GBMI and GBMII, respectively. Our results suggest that changes in subsets of gene expression due to failure to respond to repression by H3K27me3 or H4K20me3 may contribute to tumorigenesis in GBM subtypes. One would argue this is simply a correlation; yet considering the heterogeneous nature of GBM tumors (Inda et al., 2014; Sottoriva et al., 2013) in which a set of genes are putative targets of these 2 repressive marks and are aberrantly up-regulated in both GBMI and GBMII, the role of EZH2/H3K27me3 and Suv4-20h/H4K20me3 is irrefutable. While extensive genomic characterization and transcriptome analysis have illustrated molecular landscape of glioblastoma (2008; Beroukhim et al., 2007; Parsons et al., 2008; Gunther et al., 2008; Phillips et al., 2006; Verhaak et al., 2010; Patel et al., 2014), the work presented here provides an emerging insight into critical networks and pathways central to GBM pathobiology, which harbor molecular heterogeneity in conjunction with epigenetic alteration.
Conclusion
Our result is the first report of across species analysis to unravel the epigenetic complexity in normal and disease conditions, which provides an integrated view of epigenetic regulation by H3K27me3 and H4K20me3 in cell cycle during adult neurogenesis and in heterogeneity in subtypes of GBM. In addition, our discovery as well as bioinformatics pipeline and scripts for analyses will have significant impact to the fields of stem cell and cancer biology. Further, as clinical impact, our data implicates that these epigenetic repressive marks and a set of their putative target genes may be potential candidates as biomarkers in monitoring cancer progression. Considering the heterogeneous nature of various GBM cases, aforementioned candidates from our findings would be valuable for further study pertinent to therapeutic application. Lastly, our cross-species approach will advance further study using a range of systems to delineate epigenetic regulation and beyond.
Supplementary Material
Highlight.
The first demonstration of analyses across species provides an integrated view of epigenetic repression in the adult neural stem progenitor cells under normal and disease conditions that advance our study using a range of systems to delineate epigenetic regulation and beyond.
A new direction for in vivo population-level analysis relevant to cell fate of stem cell and cancer biology using stereotaxically injected Cre recombinant protein.
A comparative genomics approach reveals epigenetic heterogeneity in subtype of glioblastoma multiforme.
The first alignment of baboon deep-sequencing reads to newly annotated baboon genome.
Acknowledgments
We acknowledge the UCSF, Department of Neurological Surgery, Brain Tumor Research Center tissue core as the resource of the human specimens. We thank Texas Biomedical research Institute/Southwest National Primate Research Center (SNPRC) at San Antonio for baboon tissues. The authors are grateful to Dr. Alexander Tarakhovsky at the Rockefeller University for providing EZH2 mouse model. We also thank Michael Foret, Gabriel Leal, and Bethany Zablotsky for their technical support. Sequencing was performed at the High Throughput Genomics Center in the Department of Genome Sciences, University of Washington, Seattle, WA. CTR is supported by scholarship from UTSA, Department of Biology PhD graduate program. This project is supported by the SPORE grant 5 P50 CA097257-13 to MSB, as well as the SCORE grant SC3GM112543 from the National Institutes of Health and TRAC award to CAL.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Trievel RC, Beach BM, Dirk LM, Houtz RL, Hurley JH. Structure and catalytic mechanism of a SET domain protein methyltransferase. Cell. 2002;111:91–103. doi: 10.1016/s0092-8674(02)01000-0. [DOI] [PubMed] [Google Scholar]
- 2.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 3.Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. The EMBO journal. 2003;22:5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ting AH, McGarvey KM, Baylin SB. The cancer epigenome--components and functional correlates. Genes & development. 2006;20:3215–3231. doi: 10.1101/gad.1464906. [DOI] [PubMed] [Google Scholar]
- 6.Shen H, Laird PW. Interplay between the Cancer Genome and Epigenome. Cell. 2013;153:38–55. doi: 10.1016/j.cell.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evertts AG, Manning AL, Wang X, Dyson NJ, Garcia BA, Coller HA. H4K20 methylation regulates quiescence and chromatin compaction. Molecular biology of the cell. 2013;24:3025–3037. doi: 10.1091/mbc.E12-07-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sakaguchi A, Karachentsev D, Seth-Pasricha M, Druzhinina M, Steward R. Functional characterization of the Drosophila Hmt4-20/Suv4-20 histone methyltransferase. Genetics. 2008;179:317–322. doi: 10.1534/genetics.108.087650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang H, Pesavento JJ, Starnes TW, Cryderman DE, Wallrath LL, Kelleher NL, Mizzen CA. Preferential dimethylation of histone H4 lysine 20 by Suv4–20. The Journal of biological chemistry. 2008;283:12085–12092. doi: 10.1074/jbc.M707974200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schotta G, Sengupta R, Kubicek S, Malin S, Kauer M, Callen E, Celeste A, Pagani M, Opravil S, De La Rosa-Velazquez IA, et al. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes & development. 2008;22:2048–2061. doi: 10.1101/gad.476008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hwang WW, Salinas RD, Siu JJ, Kelley KW, Delgado RN, Paredes MF, Alvarez-Buylla A, Oldham MC, Lim DA. Distinct and separable roles for EZH2 in neurogenic astroglia. eLife. 2014;3:e02439. doi: 10.7554/eLife.02439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rogers J, Kochunov P, Zilles K, Shelledy W, Lancaster J, Thompson P, Duggirala R, Blangero J, Fox PT, Glahn DC. On the genetic architecture of cortical folding and brain volume in primates. NeuroImage. 2010;53:1103–1108. doi: 10.1016/j.neuroimage.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kochunov P, Glahn DC, Fox PT, Lancaster JL, Saleem K, Shelledy W, Zilles K, Thompson PM, Coulon O, Mangin JF, et al. Genetics of primary cerebral gyrification: Heritability of length, depth and area of primary sulci in an extended pedigree of Papio baboons. NeuroImage. 2010;53:1126–1134. doi: 10.1016/j.neuroimage.2009.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cox LA, Comuzzie AG, Havill LM, Karere GM, Spradling KD, Mahaney MC, Nathanielsz PW, Nicolella DP, Shade RE, Voruganti S, et al. Baboons as a model to study genetics and epigenetics of human disease. ILAR journal/National Research Council, Institute of Laboratory Animal Resources. 2013;54:106–121. doi: 10.1093/ilar/ilt038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sandstrom RS, Foret MR, Grow DA, Haugen E, Rhodes CT, Cardona AE, Phelix CF, Wang Y, Berger MS, Lin CH. Epigenetic Regulation by Chromatin Activation Mark H3K4me3 in Primate Progenitor Cells within Adult Neurogenic Niche. Scientific reports. 2014;4:5371. doi: 10.1038/srep05371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanai N, Tramontin AD, Quinones-Hinojosa A, Barbaro NM, Gupta N, Kunwar S, Lawton MT, McDermott MW, Parsa AT, Manuel-Garcia Verdugo J, et al. Unique astrocyte ribbon in adult human brain contains neural stem cells but lacks chain migration. Nature. 2004;427:740–744. doi: 10.1038/nature02301. [DOI] [PubMed] [Google Scholar]
- 17.Haskins WE, Zablotsky BL, Foret MR, Ihrie RA, Alvarez-Buylla A, Eisenman RN, Berger MS, Lin CH. Molecular Characteristics in MRI-Classified Group 1 Glioblastoma Multiforme. Frontiers in oncology. 2013;3:182. doi: 10.3389/fonc.2013.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lim DA, Cha S, Mayo MC, Chen MH, Keles E, VandenBerg S, Berger MS. Relationship of glioblastoma multiforme to neural stem cell regions predicts invasive and multifocal tumor phenotype. Neuro-oncology. 2007;9:424–429. doi: 10.1215/15228517-2007-023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jessberger S, Nakashima K, Clemenson GD, Jr, Mejia E, Mathews E, Ure K, Ogawa S, Sinton CM, Gage FH, Hsieh J. Epigenetic modulation of seizure-induced neurogenesis and cognitive decline. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007;27:5967–5975. doi: 10.1523/JNEUROSCI.0110-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ihrie RA, Alvarez-Buylla A. Cells in the astroglial lineage are neural stem cells. Cell and tissue research. 2008;331:179–191. doi: 10.1007/s00441-007-0461-z. [DOI] [PubMed] [Google Scholar]
- 21.Clelland CD, Choi M, Romberg C, Clemenson GD, Jr, Fragniere A, Tyers P, Jessberger S, Saksida LM, Barker RA, Gage FH, et al. A functional role for adult hippocampal neurogenesis in spatial pattern separation. Science. 2009;325:210–213. doi: 10.1126/science.1173215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ming GL, Song H. Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron. 2011;70:687–702. doi: 10.1016/j.neuron.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Molecular cell. 2012;48:491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ciferri C, Lander GC, Maiolica A, Herzog F, Aebersold R, Nogales E. Molecular architecture of human polycomb repressive complex 2. eLife. 2012;1:e00005. doi: 10.7554/eLife.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 26.O’Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T. The polycomb-group gene Ezh2 is required for early mouse development. Molecular and cellular biology. 2001;21:4330–4336. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su IH, Basavaraj A, Krutchinsky AN, Hobert O, Ullrich A, Chait BT, Tarakhovsky A. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nature immunology. 2003;4:124–131. doi: 10.1038/ni876. [DOI] [PubMed] [Google Scholar]
- 28.Chien WM, Liu Y, Chin MT. Genomic DNA Recombination with Cell-Penetrating Peptide-Tagged Cre Protein in Mouse Skeletal and Cardiac Muscle. Genesis. 2014 doi: 10.1002/dvg.22782. [DOI] [PubMed] [Google Scholar]
- 29.Kim SH, Joshi K, Ezhilarasan R, Myers TR, Siu J, Gu C, Nakano-Okuno M, Taylor D, Minata M, Sulman EP, et al. EZH2 protects glioma stem cells from radiation-induced cell death in a MELK/FOXM1-dependent manner. Stem cell reports. 2015;4:226–238. doi: 10.1016/j.stemcr.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Vries NA, Hulsman D, Akhtar W, de Jong J, Miles DC, Blom M, van Tellingen O, Jonkers J, van Lohuizen M. Prolonged Ezh2 Depletion in Glioblastoma Causes a Robust Switch in Cell Fate Resulting in Tumor Progression. Cell reports. 2015 doi: 10.1016/j.celrep.2014.12.028. [DOI] [PubMed] [Google Scholar]
- 31.Suva ML, Riggi N, Janiszewska M, Radovanovic I, Provero P, Stehle JC, Baumer K, Le Bitoux MA, Marino D, Cironi L, et al. EZH2 is essential for glioblastoma cancer stem cell maintenance. Cancer research. 2009;69:9211–9218. doi: 10.1158/0008-5472.CAN-09-1622. [DOI] [PubMed] [Google Scholar]
- 32.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, Vivanco I, Lee JC, Huang JH, Alexander S, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gunther HS, Schmidt NO, Phillips HS, Kemming D, Kharbanda S, Soriano R, Modrusan Z, Meissner H, Westphal M, Lamszus K. Glioblastoma-derived stem cell-enriched cultures form distinct subgroups according to molecular and phenotypic criteria. Oncogene. 2008;27:2897–2909. doi: 10.1038/sj.onc.1210949. [DOI] [PubMed] [Google Scholar]
- 36.Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 37.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doe CQ. Neural stem cells: balancing self-renewal with differentiation. Development. 2008;135:1575–1587. doi: 10.1242/dev.014977. [DOI] [PubMed] [Google Scholar]
- 39.Roman-Trufero M, Mendez-Gomez HR, Perez C, Hijikata A, Fujimura Y, Endo T, Koseki H, Vicario-Abejon C, Vidal M. Maintenance of undifferentiated state and self-renewal of embryonic neural stem cells by Polycomb protein Ring1B. Stem Cells. 2009;27:1559–1570. doi: 10.1002/stem.82. [DOI] [PubMed] [Google Scholar]
- 40.Molofsky AV, Pardal R, Iwashita T, Park IK, Clarke MF, Morrison SJ. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gotz M, Huttner WB. The cell biology of neurogenesis. Nature reviews Molecular cell biology. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- 42.Ma DK, Marchetto MC, Guo JU, Ming GL, Gage FH, Song H. Epigenetic choreographers of neurogenesis in the adult mammalian brain. Nature neuroscience. 2010;13:1338–1344. doi: 10.1038/nn.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang J, Ji F, Liu Y, Lei X, Li H, Ji G, Yuan Z, Jiao J. Ezh2 regulates adult hippocampal neurogenesis and memory. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2014;34:5184–5199. doi: 10.1523/JNEUROSCI.4129-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yao B, Jin P. Unlocking epigenetic codes in neurogenesis. Genes & development. 2014;28:1253–1271. doi: 10.1101/gad.241547.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lunyak VV, Rosenfeld MG. Epigenetic regulation of stem cell fate. Human molecular genetics. 2008;17:R28–36. doi: 10.1093/hmg/ddn149. [DOI] [PubMed] [Google Scholar]
- 46.Gao Z, Ure K, Ding P, Nashaat M, Yuan L, Ma J, Hammer RE, Hsieh J. The master negative regulator REST/NRSF controls adult neurogenesis by restraining the neurogenic program in quiescent stem cells. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2011;31:9772–9786. doi: 10.1523/JNEUROSCI.1604-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Montalban-Loro R, Domingo-Muelas A, Bizy A, Ferron SR. Epigenetic regulation of stemness maintenance in the neurogenic niches. World journal of stem cells. 2015;7:700–710. doi: 10.4252/wjsc.v7.i4.700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Inda MM, Bonavia R, Seoane J. Glioblastoma multiforme: a look inside its heterogeneous nature. Cancers. 2014;6:226–239. doi: 10.3390/cancers6010226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sottoriva A, Spiteri I, Piccirillo SG, Touloumis A, Collins VP, Marioni JC, Curtis C, Watts C, Tavare S. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:4009–4014. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.