Abstract
Background
So far, more than 170 loci have been associated with circulating lipid levels through genome-wide association studies (GWAS). These associations are largely driven by common variants, their function is often not known, and many are likely to be markers for the causal variants. In this study we aimed to identify more new rare and low-frequency functional variants associated with circulating lipid levels.
Methods
We used the 1000 Genomes Project as a reference panel for the imputations of GWAS data from ∼60 000 individuals in the discovery stage and ∼90 000 samples in the replication stage.
Results
Our study resulted in the identification of five new associations with circulating lipid levels at four loci. All four loci are within genes that can be linked biologically to lipid metabolism. One of the variants, rs116843064, is a damaging missense variant within the ANGPTL4 gene.
Conclusions
This study illustrates that GWAS with high-scale imputation may still help us unravel the biological mechanism behind circulating lipid levels.
Keywords: Complex traits, Epidemiology, Genetics, Genome-wide, circulating lipid levels
Introduction
Genome-wide association studies (GWAS) for circulating lipid levels (high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), total cholesterol (TC) and triglycerides (TG)) have identified over 170 loci.1–3 These studies have been based on imputations to the HapMap reference panel2 or primary versions of the 1000 Genomes Project (1kG)1 or genotyping on the Illumina Exome Chip.3 None has used imputations with the Phase 1 integrated release v3 of the 1kG which allows the imputation of rare and low-frequency functional variants and structural variations with more precision. Evidence of rare and low-frequency functional variants associated with circulating lipid levels comes from recent studies in which exome sequencing of the NPC1L1 gene identified rare variants associated with reduced LDL-C levels and reduced risk of coronary heart disease.4 Moreover, exome sequencing of LDLR and APOA5 identified rare variants associated with an increased LDL-C and increased TG levels5 and exome sequencing of APOC3 identified rare variants associated with reduced TG levels and reduced risk of coronary heart disease.6
Our goal in this study was to identify rare and low-frequency functional variants associated with circulating lipid levels in a larger sample size compared with the exome sequencing of candidate gene approach. To this end, we imputed genotypes for study samples participating in the cohorts of the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium using the Phase 1 integrated release V. 3 of the 1kG and conducted a meta-analysis of about approximately 60 000 individuals, followed by a replication in an independent set of 90 000 individuals.
Methods
Please see online supplementary methods for complete descriptions of the methods. In summary, for the discovery stage of this project, we used the data from 20 cohorts of the CHARGE consortium (see online supplementary methods). All cohorts were imputed with reference to the 1kG reference panel (version Phase 1 integrated release V.3). The total number of individuals in the discovery stage was 59 409 for HDL-C, 48 780 for LDL-C, 60 024 for TC and 49 549 for TG. Online supplementary tables S1 and S2 contain the baseline characteristics per cohort and more details about SNP genotyping and genotype imputations. Within each cohort, each variant was tested for association with each of the lipid traits, assuming an additive genetic model. The association results of all cohorts for all variants were combined using inverse variance weighting. We used the following filters for the variants: 0.3<R2 (measurement for the imputation quality) ≤1.0 and expected minor allele count (expMAC=2×MAF (minor allele frequency)×R2×sample size) >10 prior to meta-analysis. After meta-analysis of all available variants, we excluded the variants that were not present in at least four cohorts, to prevent false positive findings. In order to select only variants that were independently associated with each of the lipid traits, we used the genome-wide complex trait analysis (GCTA)7 tool, V.1.13. To identify novel loci we selected from the list of variants identified by GCTA, those variants located more than 0.5 Mb away from previously identified loci of the corresponding trait2 3 and which were significant (p value<5×10−8) in the initial discovery stage. To prevent the identification of false positive loci, we added a second replication stage within 23 independent cohorts. The experiment-wide significance threshold required to keep type I error rate within the replication stage at 5% is 2.63×10−3 (Bonferroni correction based on 19 variants). We also meta-analysed the individuals of the discovery and replication stage together and per ethnicity using a fixed-effect approach. We also repeated this analysis with genome-wide association meta analysis (GWAMA) (V.2.0.5) using a random effect approach as the individuals in discovery and replication stages come from multiple ethnicities.
jmedgenet-2015-103439supp.pdf (1.8MB, pdf)
Results
The association of all variants with HDL-C, LDL-C, TC and TG was tested in all discovery cohorts (see online supplementary figures S1 and S2). The association results of all discovery cohorts for all variants were combined in a fixed-effect meta-analysis using METAL (see online supplementary figures S3 and S4). We significantly replicated 88.1% of the loci described by Teslovich et al2 despite a sample size of about 80% (see online supplementary figure S5 and supplementary table S3). We also significantly replicated 43.4% of the loci described by the Global Lipids Genetics Consortium (GLGC)3 despite a sample size of about 30% (see online supplementary figure S6 and supplementary table S4).
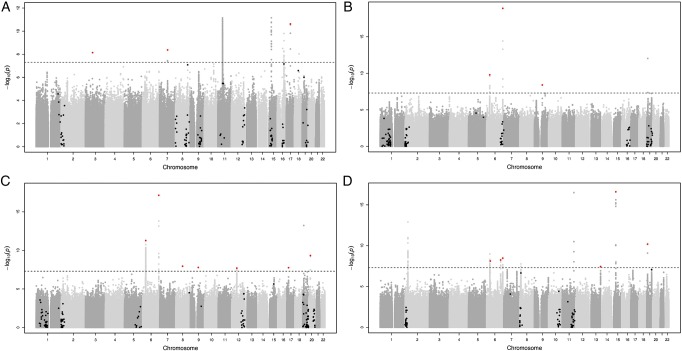
A conditional and joint analysis using GCTA identified 185 independent variants for HDL-C, 174 for LDL-C, 214 for TC and 119 for TG. Next, we excluded all variants that were not genome-wide significant (p value<5×10−8) in the initial discovery stage, which resulted in 56 variants for HDL-C, 50 for LDL-C, 66 for TC and 37 for TG. And we excluded all variants which are within 0.5 Mb of a loci previously published by Teslovich et al2 or GLGC,3 which resulted in three variants for HDL-C, three for LDL-C, seven for TC and six for TG. These variants are located at 17 different loci and include one deletion (figure 1 and table 1).
Figure 1.
Manhattan plots for HDL-C (A), LDL-C (B), TC (C) and TG (D) after the meta-analysis of all discovery cohorts. Variants that were present in at least four cohorts and that are not within 0.5 Mb of a previously published loci2 3 were included. The black line indicates the genome-wide significant line (5×10−8), the black and red dots the variants identified by GCTA which are not genome-wide significant and which are genome-wide significant, respectively. HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; TC, total cholesterol; TG, triglycerides.
Table 1.
The results for the 19 variants after the meta-analysis of all discovery cohorts, all replication cohorts and all cohorts combined
| Discovery cohorts | Replication cohorts | All cohorts combined | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trait | Chr:Position | rs identifier | nearest gene | A1/A2 | Freq | N | β | SEβ | p Value | Freq | N | β | SEβ | p Value | Freq | β | SEβ | p Value |
| HDL-C | 3:72 067 255 | rs75909755 | PROL2-EIF4E3 | T/C | 0.03 | 62 607 | 1.593 | 0.275 | 7.27E-09 | 0.03 | 86 252 | −0.019 | 0.031 | 5.45E-01 | 0.03 | 0.002 | 0.031 | 9.57E-01 |
| TC | 6:31 272 261 | rs6457374 | HLA-B | T/C | 0.75 | 46 839 | 2.339 | 0.339 | 5.32E-12 | 0.81 | 74 417 | 0.057 | 0.016 | 4.23E-04 | 0.81 | 0.062 | 0.016 | 1.18E-04 |
| LDL-C | 6:31 325 323 | rs9266229 | HLA-B | C/G | 0.53 | 37 981 | −2.201 | 0.344 | 1.62E-10 | 0.41 | 61 582 | −0.025 | 0.014 | 7.37E-02 | 0.41 | −0.029 | 0.014 | 4.04E-02 |
| TG | 6:36 648 275 | – | CDKN1A | CAG/C | 0.45 | 53 425 | −0.019 | 0.003 | 7.63E-09 | 0.49 | 59 018 | −0.003 | 0.004 | 5.20E-01 | 0.46 | −0.013 | 0.003 | 5.93E-07 |
| TG | 6:13 983 949 8 | rs608736 | – | C/G | 0.48 | 53 425 | −0.019 | 0.003 | 5.67E-09 | 0.49 | 73 512 | −0.008 | 0.003 | 2.67E-02 | 0.48 | −0.013 | 0.002 | 9.10E-09 |
| TG | 6:16 085 176 6 | rs376563 | SLC22A3 | T/C | 0.46 | 47 036 | −0.02 | 0.003 | 3.37E-09 | 0.46 | 73 512 | −0.001 | 0.003 | 8.22E-01 | 0.46 | −0.010 | 0.002 | 1.36E-05 |
| LDL-C | 6:16 111 170 0 | rs186696265 | LPA-PLG | T/C | 0.01 | 49 221 | 11.247 | 1.241 | 1.31E-19 | 0.01 | 59 497 | 0.263 | 0.076 | 5.42E-04 | 0.01 | 0.304 | 0.076 | 6.17E-05 |
| TC | 6:16 111 170 0 | rs186696265 | LPA-PLG | T/C | 0.01 | 59 859 | 10.004 | 1.162 | 7.20E-18 | 0.01 | 75 821 | 0.238 | 0.075 | 1.46E-03 | 0.01 | 0.278 | 0.075 | 1.93E-04 |
| HDL-C | 7:80 492 357 | rs60839105 | SEMA3C | T/C | 0.07 | 7882 | 3.355 | 0.571 | 4.26E-09 | 0.08 | 4971 | 1.067 | 1.228 | 3.85E-01 | 0.07 | 2.948 | 0.518 | 1.25E-08 |
| TC | 8:68 351 787 | rs151198427 | CPA6 | A/G | 0.11 | 17 361 | 6.552 | 1.147 | 1.12E-08 | 0.13 | 1419 | −2.858 | 2.396 | 2.33E-01 | 0.11 | 4.797 | 1.035 | 3.56E-06 |
| LDL-C | 9:78 728 065 | rs146369471 | PCSK5 | T/C | 0.99 | 43 398 | 8.529 | 1.449 | 3.99E-09 | 0.99 | 51 367 | 0.068 | 0.103 | 5.11E-01 | 0.99 | 0.110 | 0.103 | 2.84E-01 |
| TC | 9:78 728 065 | rs146369471 | PCSK5 | T/C | 0.99 | 53 787 | 7.978 | 1.413 | 1.64E-08 | 0.99 | 70 241 | 0.015 | 0.103 | 8.84E-01 | 0.99 | 0.057 | 0.103 | 5.79E-01 |
| TC | 12:51 207 704 | rs829112 | ATF1 | A/G | 0.68 | 56 924 | 1.448 | 0.258 | 2.02E-08 | 0.73 | 87 659 | 0.009 | 0.012 | 4.63E-01 | 0.73 | 0.012 | 0.012 | 3.18E-01 |
| TG | 13:11 454 402 4 | rs7140110 | GAS6 | T/C | 0.71 | 48 221 | −0.021 | 0.004 | 3.65E-08 | 0.72 | 60 437 | −0.006 | 0.005 | 2.68E-01 | 0.72 | −0.015 | 0.003 | 5.13E-07 |
| TG | 15:43 726 625 | rs150844304 | TP53BP1 | A/C | 0.97 | 52 720 | −0.083 | 0.01 | 2.52E-17 | 0.95 | 63 884 | −0.026 | 0.015 | 8.85E-02 | 0.96 | −0.066 | 0.008 | 9.52E-16 |
| TC | 17:18 046 290 | rs8065026 | MYO15A | T/C | 0.79 | 56 924 | −1.644 | 0.292 | 1.76E-08 | 0.81 | 76 913 | −0.026 | 0.013 | 4.93E-02 | 0.81 | −0.029 | 0.013 | 2.66E-02 |
| HDL-C | 17:41 840 849 | rs77697917 | SOST-DUSP3 | T/C | 0.02 | 45 052 | −2.717 | 0.407 | 2.38E-11 | 0.03 | 67 843 | −0.222 | 0.036 | 4.27E-10 | 0.03 | −0.241 | 0.035 | 1.04E-11 |
| TG | 19:8 429 323 | rs116843064 | ANGPTL4 | A/G | 0.03 | 35 643 | −0.101 | 0.016 | 6.46E-11 | 0.03 | 44 194 | −0.065 | 0.019 | 4.53E-04 | 0.03 | −0.087 | 0.012 | 3.83E-13 |
| TC | 20:17 844 684 | rs2618566 | BANF2-SNX5 | T/G | 0.65 | 63 300 | −1.566 | 0.251 | 4.68E-10 | 0.60 | 88 946 | −0.024 | 0.011 | 2.83E-02 | 0.60 | −0.027 | 0.011 | 1.38E-02 |
The variants in bold are the significantly replicated variants.
A1 is allele 1 and A2 is allele 2, Freq is the frequency of A1, β is the effect of A1.
HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; TC, total cholesterol; TG, triglycerides.
These 19 variants were selected for replication. The total number of individuals in the replication stage was 84 598, 72 486, 83 739 and 73 519 for HDL-C, LDL-C, TC and TG, respectively (see online supplementary tables S1 and S2 for baseline characteristics and information about SNP genotyping and imputation details). The sample size in the replication stage was larger than the initial discovery sample for 17 out of the 19 variants. The frequencies of the variants were similar between the discovery and replication cohorts. The directions of effect were the same in the discovery and replication cohorts for 16 out of the 19 variants (see online supplementary figure S7). We used a Bonferroni corrected threshold for significance (p value<2.63×10−3). Five out of the 19 variants were significantly replicated (table 1): rs6457374 (TC), rs186696265 (LDL-C and TC), rs77697917 (HDL-C) and rs116843064 (TG). The frequency of these variants ranged between 0.012 and 0.249 within the discovery sample. Online supplementary table S5 shows the heterogeneity for the 19 variants after the meta-analysis of all discovery cohorts and of all replication cohorts. We also meta-analysed all variants in the individuals of the discovery cohorts and replication cohorts combined (table 1 and see online supplementary tables S5 and S6) and per ethnicity (see online supplementary table S6) using a fixed-effect meta-analysis approach. We found that the five significantly replicated variants we identified in this study are only significant within the European samples, thereby noticing that there are much more European samples in this study, compared with the African and Asian samples. When using a random-effect meta-analysis to account for the multiple ethnicities in our sample (see online supplementary table S7), we found that of the five replicated variants, one attained genome-wide significance (p value<5×10−8) and the other four nominal significance (p value<0.05).
Discussion
We conducted a GWAS that included GWAS data imputed to the 1kG to identify rare and low-frequency, potentially functional, variants associated with circulating lipid levels. To this end, we imputed genotypes in approximately 60 000 individuals from 20 cohorts in the CHARGE consortium with the 1kG reference panel. The meta-analysis, followed by GCTA analysis revealed 19 associations with MAF ranging from 0.01 to 0.48. Of the 19 associations, we were able to replicate five in an independent sample of approximately 90 000 individuals.
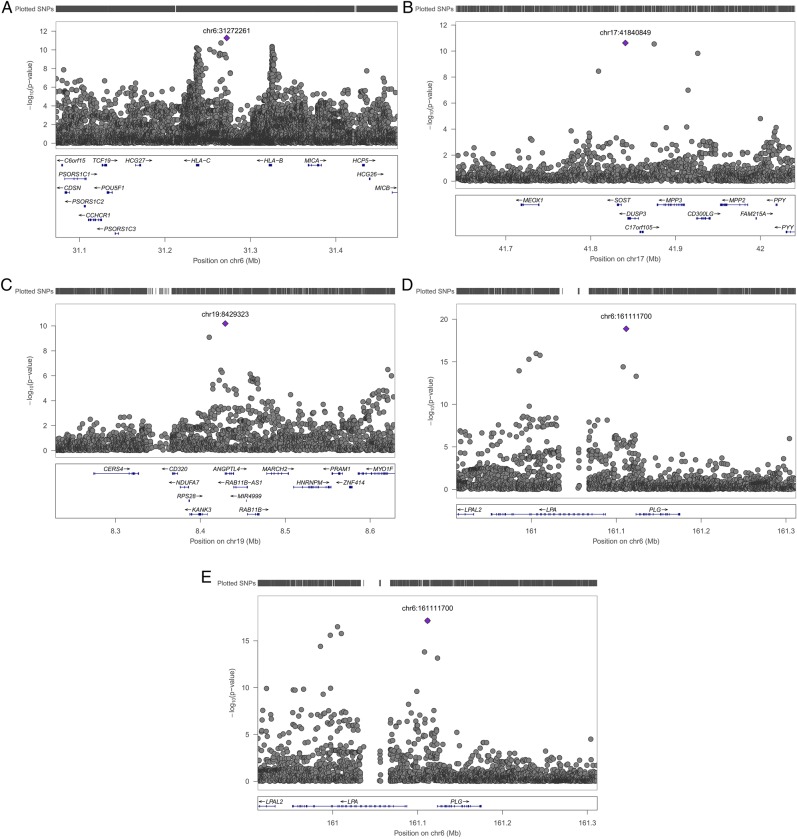
One of the five associations we identified is between TG and rs116843064, an exonic variant in the ANGPTL4 gene on chromosome 19 (figure 2C). This missense variant changes the amino acid glutamic acid into lysine (Glu40Lys) and is predicted to be damaging for the structure and function of the protein by Polyphen2,8 MutationTaster9 and likelihood ratio test (LRT).10 ANGPTL4 is significantly associated with the Kyoto Encyclopedia of Genes and Genomes (KEGG) term fatty acid metabolism, the GO process lipid storage and the gene ontology (GO) cellular component lipid particle (p value of 1.10×10−6, 1.31×10−10 and 2.87×10−18, respectively, genenetwork.nl). ANGPTL4 has been associated with HDL-C before using the GWAS approach2 and with TG before using an exome sequencing approach11 and more recently using the GWAS approach.1 We therefore do not claim this finding as novel, though this is the smallest study in which this variant was genome-wide significantly associated with TG and replicated in an independent sample.
Figure 2.
The regional association results of the initial meta-analysis of all discovery cohorts for (A) TC on chromosome 6, (B) HDL-C on chromosome 17, (C) TG on chromosome 19, (D) LDL-C on chromosome 6 and (E) TC on chromosome 6. HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; TC, total cholesterol; TG, triglycerides.
The second new finding we identified is the association between TC and rs6457374, an intergenic variant located on chromosome 6 between the genes HLA-C and HLA-B (figure 2A). Both genes are associated with the KEGG term ATP binding cassette (ABC) transporters (p value of 4.29×10−5 and 3.84×10−5 for HLA-C and HLA-B, respectively, genenetwork.nl) which is in line with, among others, a previously published association between TC and an exonic variant in the ABCA6 gene which is also an ABC transporter.12 ABC transporters transport a wide variety of substrates across extracellular and intracellular membranes, including lipids.13
The third finding of this study is the association between HDL-C and rs77697917, an intergenic variant on chromosome 17 between the genes SOST and DUSP3 (figure 2B). DUSP3 is associated with the regulation and function of carbohydrate-responsive element-binding protein (ChREBP) in the liver (p value=3.03×10−5, genenetwork.nl). ChREBP mediates the activation of several regulatory enzymes involved in lipogenesis.14–18 This variant is in high linkage disequilibrium (D′=0.936) in the 1 kG with rs72836561, an exonic variant in the gene CD300LG (MAF=0.027, β=−2.437, seβ=0.381, p value=1.51×10−10 in the discovery stage). This missense variant changes the amino acid arginine into cysteine (Arg82Cys) and is predicted to be damaging for the structure and function of the protein by Polyphen2,8 MutationTaster9 and LRT.10 This amino acid polymorphism has been associated with HDL-C in exome-wide association studies19 and TG in GWAS1 before.
The fourth variant we identified is rs186696265, which is located on chromosome 6 and associated with LDL-C and TC (figure 2D, E). This intergenic variant is between the LPA (Lipoprotein, Lp(A)) gene and the PLG (Plasminogen) gene. The LPA gene has been associated before with LDL-C and TC before.2 The reported lead SNP was rs1564348, which in the newer human genome versions is annotated to the SLC22A1 (Solute Carrier Family 22 (Organic Cation Transporter), Member 1) gene instead of the LPA gene. This explains why we again identified a locus near the LPA gene, which has been identified by others as well.1
Fourteen out of the 19 variants were not replicated despite similar sample sizes and similar frequencies within the replication stage as compared with the discovery stage. Of those 14 variants, 11 exhibited effect sizes in the same direction in both stages. A possible explanation might be that the replication sample size is much larger compared with that of the discovery sample size. Two variants might have lacked significant replication due to small sample size, rs60839105 and rs151198427. Both variants only pass quality control in the cohorts in the discovery stage that contain individuals of African ancestry (see online supplementary figure S7). Although there are several cohorts with individuals of African ancestry in the replication stage, both variants did not pass quality control in most cohorts which leads to the conclusion that these variants might be population-specific. This is also suggested by the 1 kG data (Phase 3) as the frequency of the C-allele is 92% in African samples and 100% in the European samples for rs60839105 and the frequency of the G-allele is 86% in the African samples and 100% in the European samples for rs151198427. Imputations of cohorts with individuals of African ancestry with the African Genome Variation Project20 might confirm the association of rs60839105 with HDL-C and rs151198427 with TC.
To our knowledge, this is the first GWAS of circulating lipid levels using the Phase 1 integrated release V.3 of the 1 kG, therefore we cannot compare the positive replication rate with other studies. However, we did replicate 88.1% of the findings of Teslovich et al2 and 43.4% of the findings of GLGC3 despite our smaller sample. A high replication rate is expected based on the high overlap of our samples with the samples of Teslovich et al2 and with the samples of GLGC3 though it indicates that when using the 1000 Genomes instead of the HapMap reference panel, we can achieve a high replication rate using a smaller sample size. We also tried to replicate findings from exome sequencing of candidate genes. The p.Arg406X mutation in the NPC1L1 gene (rs145297799), which was reported to be associated with reduced LDL-C levels and reduced risk of coronary heart disease,4 is not available in the 1kG reference panel and, therefore, we were not able to replicate this finding. Do et al5 described the exome sequencing of the genes LDLR and APOA5 and identified rare variants associated with an increased risk of myocardial infarction, increased LDL-C and TG levels. Of those rare variants, only two in the LDLR gene and seven in the APOA5 gene exist in our discovery meta-analysis. Both LDLR variants are associated with TG in our discovery meta-analysis (rs34282181, β=−0.093, SEβ=0.023, p value=4.827×10−5 and rs2075291, β=0.219, SEβ=0.046, p value=2.092×10−6), but not significantly associated with LDL-C (rs34282181, β=−3.939, SEβ=1.861, p value=0.034 and rs2075291, β=−2.316, SEβ=3.001, p value=0.440). None of the seven APOA5 variants were significantly associated with TG or LDL-C in our discovery meta-analysis (lowest p value is for LDL-C with rs72658860, β=−18.430, SEβ=7.140, p value=9.848×10−3). The third published finding we tried to replicate, was the association between APOC3 and TG levels.6 Of the seven variants reported, only one existed in our discovery meta-analysis (chromosome 11, position 116 701 354), which is associated with TG (β=−0.343, SEβ=0.113, p value=2.311×10−3). Those authors also reported an association between an APOA5 variant (rs3135506) and TG as the most significant finding. This variant was also significantly associated with TG in our discovery meta-analysis (β=0.129, SEβ=0.007, p value=1.099×10−87). These replication efforts demonstrate that many of the published results of exome sequencing can be replicated through the use of 1 kG imputations.
In conclusion, we identified and replicated five variants associated with circulating lipid levels. These variants are in genes that can be linked biologically to lipid metabolism. Although there were a large number of variants that did not replicate at the accepted genome-wide significance threshold, the low-cost, hypothesis-free approach that we applied uncovered five variants. This study, therefore, illustrates that GWAS may still help us unravel the biological mechanisms behind circulating lipid levels.
Acknowledgments
The authors especially thank all volunteers who participated in the study.
The authors thank the staff and participants of the ARIC study for their important contributions. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research.
We are grateful to all study participants and their relatives, general practitioners and neurologists for their contributions to the ERF study and to P Veraart for her help in genealogy, J Vergeer for the supervision of the laboratory work and P Snijders for his help in data collection.
The authors thank Behrooz Alizadeh, Annemieke Boesjes, Marcel Bruinenberg, Noortje Festen, Pim van der Harst, Ilja Nolte, Lude Franke, Mitra Valimohammadi for their help in creating the GWAS database, and Rob Bieringa, Joost Keers, René Oostergo, Rosalie Visser, Judith Vonk for their work related to data collection and validation. The authors are grateful to the study participants, the staff from the LifeLines Cohort Study and the contributing research centres delivering data to LifeLines and the participating general practitioners and pharmacists.
MESA and the MESA SHARe project are conducted and supported by contracts N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-001079 and UL1-TR-000040 from the National Heart, Lung, and Blood Institute (NHLBI). Funding for MESA SHARe genotyping was provided by NHLBI Contract N02-HL6-4278. The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, CTSI grant UL1TR000124, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center. This publication was developed under a STAR research assistance agreement, No. RD831697 (MESA Air), awarded by the U.S Environmental protection Agency. It has not been formally reviewed by the EPA. The views expressed in this document are solely those of the authors and the EPA does not endorse any products or commercial services mentioned in this publication.
The authors of the NEO study thank all individuals who participated in the Netherlands Epidemiology in Obesity study, all participating general practitioners for inviting eligible participants and all research nurses for collection of the data. The authors thank the NEO study group, Petra Noordijk, Pat van Beelen and Ingeborg de Jonge for the coordination, lab and data management of the NEO study. The genotyping in the NEO study was performed at the Centre National de Génotypage (Paris, France), headed by Jean-Francois Deleuze.
The authors of the ORCADES study would like to acknowledge the invaluable contributions of Lorraine Anderson and the research nurses in Orkney, the administrative team in Edinburgh and the people of Orkney.
On behalf of the Rotterdam Study, the authors thank Pascal Arp, Mila Jhamai, Marijn Verkerk, Lizbeth Herrera and Marjolein Peters for their help in creating the GWAS database, and Karol Estrada and Maksim V. Struchalin for their support in creation and analysis of imputed data. The authors are grateful to the study participants, the staff from the Rotterdam Study and the participating general practitioners and pharmacists.
Footnotes
Collaborators: The LifeLines Cohort Study: see online supplementary appendix 1. CHARGE Lipids Working Group: see online supplementary appendix 2.
Contributors: EMvL organised the study and designed the study with substantial input of AI, LAC and CMvD. EMvL drafted the manuscript with substantial input from CMvD. All authors had the opportunity to comment on the manuscript. Data collection, GWAS and statistical analysis was done by SWvdL, HMdR, GP (AEGS); AVS, VG, TBH (AGES); EE, MPS, PE (Airwave); AS, DEA, ACM, EB (ARIC); JCB, JAB, KMC, BMP (CHS); AI, EMvL, CMvD (ERF); MFF, IBB (FamHS); LPL, KN, MK (FINCAVAS); IS, VS, SR (FINRISK); SD, CCW, LAC (FHS); JEH, AC, LJH, SP (GS); QD, LAL, JGW (JHS); JEH, IK, PN, OP (CROATIA Korcula); IMN, MS (Lifelines); JD, AJMdC, PES (LLS); WZ, JSK, BL, WRS, STT, JCC (LOLIPOP); BOT, TF, CAM, RSC (Loyola); AM, JCM, SSR, JIR (MESA); RLG, RdM, DOMK (NEO); HM, EJdG, YM, BWJHP, GW, DIB (NTR-NESDA); PKJ, HC, JFW (ORCADES); NV, RTG, PvdH (PREVEND); ST, IF, JWJ (PROSPER); EMvL, AD, OHF, AH, FR, EJS, AGU, CMvD (RS); JEH, TZ, VV (CROATIA Split); PJvdM, AJO, HS (TRAILS); JEH, JM, CH, IR (CROATIA Vis); JSV, OTR, TL (YFS). EMvL performed the meta-analysis and all follow-up steps. Biological association of loci and bioinformatics were carried out by EMvL, JBvK, KWvD and CMvD.
Funding: The AGES Study has been funded by NIH contracts N01-AG-1-2100 and HHSN271201200022C, the NIA Intramural Research Program, Hjartavernd (the Icelandic Heart Association), and the Althingi (the Icelandic Parliament).
The Airwave Study is funded by the Home Office (grant number 780-TETRA) with additional support from the National Institute for Health Research (NIHR) Imperial College Healthcare NHS Trust (ICHNT) and Imperial College Biomedical Research Centre (BRC). PE is an NIHR Senior Investigator and is supported by the ICHNT and Imperial College BRC, the MRC-PHE Centre for Environment and Health and the NIHR Health Protection Research Unit on Health Impact of Environmental Hazards.
The ARIC Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute (NHLBI) contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C), R01HL087641, R01HL59367 and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C.
Cardiovascular Health Study: This CHS research was supported by NHLBI contracts HHSN268201200036C, HHSN268200800007C, N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086; and NHLBI grants U01HL080295, R01HL087652, R01HL105756, R01HL103612, and R01HL120393 with additional contribution from the National Institute of Neurological Disorders and Stroke (NINDS). Additional support was provided through R01AG023629 from the National Institute on Aging (NIA). A full list of principal CHS investigators and institutions can be found at CHS-NHLBI.org. The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, CTSI grant UL1TR000124, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
CROATIA-Korcula, CROATIA-Split and CROATIA-Vis (CR-Korcula, CR-Split, CR-Vis) were funded by the Medical Research Council UK, The Croatian Ministry of Science, Education and Sports (grant 216-1080315-0302), the European Union framework program 6 EUROSPAN project (contract no. LSHG-CT-2006-018947) and the Croatian Science Foundation (grant 8875).
The ERF study as a part of EUROSPAN (European Special Populations Research Network) was supported by European Commission FP6 STRP grant number 018947 (LSHG-CT-2006-01947) and also received funding from the European Community's Seventh Framework Programme (FP7/2007-2013)/grant agreement HEALTH-F4-2007-201413 by the European Commission under the programme “Quality of Life and Management of the Living Resources” of 5th Framework Programme (no. QLG2-CT-2002-01254). The ERF study was further supported by ENGAGE consortium and CMSB. High-throughput analysis of the ERF data was supported by joint grant from Netherlands Organisation for Scientific Research and the Russian Foundation for Basic Research (NWO-RFBR 047.017.043). Exome sequencing in ERF was supported by the ZonMw grant (project 91111025).
The Family Heart Study was supported by the by grants R01-HL-087700, R01-HL-088215 and R01-HL-117078 from the National Heart, Lung, and Blood Institute.
Generation Scotland received core funding from the Chief Scientist Office of the Scottish Government Health Directorate CZD/16/6 and the Scottish Funding Council HR03006. Genotyping of the GS:SFHS samples was carried out by the Genetics Core Laboratory at the Wellcome Trust Clinical Research Facility, Edinburgh, Scotland and was funded by the UK's Medical Research Council.
The LifeLines Cohort Study, and generation and management of GWAS genotype data for the LifeLines Cohort Study is supported by the Netherlands Organization of Scientific Research NWO (grant 175.010.2007.006), the Economic Structure Enhancing Fund (FES) of the Dutch government, the Ministry of Economic Affairs, the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the Northern Netherlands Collaboration of Provinces (SNN), the Province of Groningen, University Medical Center Groningen, the University of Groningen, Dutch Kidney Foundation and Dutch Diabetes Research Foundation.
The Leiden Longevity Study (LLS) has received funding from the European Union's Seventh Framework Programme (FP7/2007–2011) under grant agreement no 259679. This study was supported by a grant from the Innovation-Oriented Research Program on Genomics (SenterNovem IGE05007), the Centre for Medical Systems Biology, and the Netherlands Consortium for Healthy Ageing (grant 050-060-810), all in the framework of the Netherlands Genomics Initiative, Netherlands Organization for Scientific Research (NWO), UnileverColworth and by BBMRI-NL, a Research Infrastructure financed by the Dutch government (NWO 184.021.007).
The LOLIPOP study is supported by the National Institute for Health Research (NIHR) Comprehensive Biomedical Research Centre Imperial College Healthcare NHS Trust, the British Heart Foundation (SP/04/002), the Medical Research Council (G0601966,G0700931), the Wellcome Trust (084723/Z/08/Z) the NIHR (RP-PG-0407-10371),European Union FP7 (EpiMigrant, 279143) and Action on Hearing Loss (G51). The work was carried out in part at the NIHR/Wellcome Trust Imperial Clinical Research Facility.
The NEO study is supported by the participating Departments, the Division and the Board of Directors of the Leiden University Medical Center, and by the Leiden University, Research Profile Area ‘Vascular and Regenerative Medicine’. Dennis Mook-Kanamori is supported by Dutch Science Organization (ZonMW-VENI Grant 916.14.023).
ORCADES was supported by the Chief Scientist Office of the Scottish Government, the Royal Society, the MRC Human Genetics Unit, Arthritis Research UK and the European Union framework program 6 EUROSPAN project (contract no. LSHG-CT-2006-018947). DNA extractions were performed at the Wellcome Trust Clinical Research Facility in Edinburgh.
The PROSPER study was supported by an investigator initiated grant obtained from Bristol-Myers Squibb. JWJ is an Established Clinical Investigator of the Netherlands Heart Foundation (grant 2001 D 032). Support for genotyping was provided by the seventh framework program of the European commission (grant 223004) and by the Netherlands Genomics Initiative (Netherlands Consortium for Healthy Aging grant 050-060-810).
The generation and management of GWAS genotype data for the Rotterdam Study is supported by the Netherlands Organisation of Scientific Research NWO Investments (nr. 175.010.2005.011, 911-03-012). This study is funded by the Research Institute for Diseases in the Elderly (014-93-015; RIDE2), the Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO) project nr. 050-060-810. The Rotterdam Study is funded by Erasmus Medical Center and Erasmus University, Rotterdam, Netherlands Organization for the Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII), and the Municipality of Rotterdam.
Participating centres of TRAILS include the University Medical Center and University of Groningen, the Erasmus University Medical Center Rotterdam, the University of Utrecht, the Radboud Medical Center Nijmegen, and the Parnassia Bavo group, all in the Netherlands. TRAILS has been financially supported by various grants from the Netherlands Organization for Scientific Research NWO (Medical Research Council program grant GB-MW 940-38-011; ZonMW Brainpower grant 100-001-004; ZonMw Risk Behavior and Dependence grants 60-60600-97-118; ZonMw Culture and Health grant 261-98-710; Social Sciences Council medium-sized investment grants GB-MaGW 480-01-006 and GB-MaGW 480-07-001; Social Sciences Council project grants GB-MaGW 452-04-314 and GB-MaGW 452-06-004; NWO large-sized investment grant 175.010.2003.005; NWO Longitudinal Survey and Panel Funding 481-08-013 and 481-11-001), the Dutch Ministry of Justice (WODC), the European Science Foundation (EuroSTRESS project FP-006), Biobanking and Biomolecular Resources Research Infrastructure BBMRI-NL (CP 32), and the participating universities.
Competing interests: BMP serves on the DSMB of a clinical trial of a device funded by the manufacturer (Zoll LifeCor) and on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. OHF works in ErasmusAGE, a centre for aging research across the life course funded by Nestlé Nutrition (Nestec); Metagenics; and AXA. Nestlé Nutrition (Nestec); Metagenics; and AXA had no role in design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review or approval of the manuscript.
Patient consent: Obtained.
Ethics approval: All studies were performed with the approval of the Local Medical Ethics Committees.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: All meta-analysis results and the results of the follow-up analysis of this project are available.
References
- 1.Surakka I, Horikoshi M, Mägi R, Sarin AP, Mahajan A, Lagou V, Marullo L, Ferreira T, Miraglio B, Timonen S, Kettunen J, Pirinen M, Karjalainen J, Thorleifsson G, Hägg S, Hottenga JJ, Isaacs A, Ladenvall C, Beekman M, Esko T, Ried JS, Nelson CP, Willenborg C, Gustafsson S, Westra HJ, Blades M, de Craen AJ, de Geus EJ, Deelen J, Grallert H, Hamsten A, Havulinna AS, Hengstenberg C, Houwing-Duistermaat JJ, Hyppönen E, Karssen LC, Lehtimäki T, Lyssenko V, Magnusson PK, Mihailov E, Müller-Nurasyid M, Mpindi JP, Pedersen NL, Penninx BW, Perola M, Pers TH, Peters A, Rung J, Smit JH, Steinthorsdottir V, Tobin MD, Tsernikova N, van Leeuwen EM, Viikari JS, Willems SM, Willemsen G, Schunkert H, Erdmann J, Samani NJ, Kaprio J, Lind L, Gieger C, Metspalu A, Slagboom PE, Groop L, van Duijn CM, Eriksson JG, Jula A, Salomaa V, Boomsma DI, Power C, Raitakari OT, Ingelsson E, Järvelin MR, Thorsteinsdottir U, Franke L, Ikonen E, Kallioniemi O, Pietiäinen V, Lindgren CM, Stefansson K, Palotie A, McCarthy MI, Morris AP, Prokopenko I, Ripatti S, ENGAGE Consortium. The impact of low-frequency and rare variants on lipid levels. Nat Genet 2015;47:589–97. 10.1038/ng.3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, Johansen CT, Fouchier SW, Isaacs A, Peloso GM, Barbalic M, Ricketts SL, Bis JC, Aulchenko YS, Thorleifsson G, Feitosa MF, Chambers J, Orho-Melander M, Melander O, Johnson T, Li X, Guo X, Li M, Shin Cho Y, Jin Go M, Jin Kim Y, Lee JY, Park T, Kim K, Sim X, Twee-Hee Ong R, Croteau-Chonka DC, Lange LA, Smith JD, Song K, Hua Zhao J, Yuan X, Luan J, Lamina C, Ziegler A, Zhang W, Zee RY, Wright AF, Witteman JC, Wilson JF, Willemsen G, Wichmann HE, Whitfield JB, Waterworth DM, Wareham NJ, Waeber G, Vollenweider P, Voight BF, Vitart V, Uitterlinden AG, Uda M, Tuomilehto J, Thompson JR, Tanaka T, Surakka I, Stringham HM, Spector TD, Soranzo N, Smit JH, Sinisalo J, Silander K, Sijbrands EJ, Scuteri A, Scott J, Schlessinger D, Sanna S, Salomaa V, Saharinen J, Sabatti C, Ruokonen A, Rudan I, Rose LM, Roberts R, Rieder M, Psaty BM, Pramstaller PP, Pichler I, Perola M, Penninx BW, Pedersen NL, Pattaro C, Parker AN, Pare G, Oostra BA, O'Donnell CJ, Nieminen MS, Nickerson DA, Montgomery GW, Meitinger T, McPherson R, McCarthy MI, McArdle W, Masson D, Martin NG, Marroni F, Mangino M, Magnusson PK, Lucas G, Luben R, Loos RJ, Lokki ML, Lettre G, Langenberg C, Launer LJ, Lakatta EG, Laaksonen R, Kyvik KO, Kronenberg F, König IR, Khaw KT, Kaprio J, Kaplan LM, Johansson A, Jarvelin MR, Janssens AC, Ingelsson E, Igl W, Kees Hovingh G, Hottenga JJ, Hofman A, Hicks AA, Hengstenberg C, Heid IM, Hayward C, Havulinna AS, Hastie ND, Harris TB, Haritunians T, Hall AS, Gyllensten U, Guiducci C, Groop LC, Gonzalez E, Gieger C, Freimer NB, Ferrucci L, Erdmann J, Elliott P, Ejebe KG, Döring A, Dominiczak AF, Demissie S, Deloukas P, de Geus EJ, de Faire U, Crawford G, Collins FS, Chen YD, Caulfield MJ, Campbell H, Burtt NP, Bonnycastle LL, Boomsma DI, Boekholdt SM, Bergman RN, Barroso I, Bandinelli S, Ballantyne CM, Assimes TL, Quertermous T, Altshuler D, Seielstad M, Wong TY, Tai ES, Feranil AB, Kuzawa CW, Adair LS, Taylor HA Jr, Borecki IB, Gabriel SB, Wilson JG, Holm H, Thorsteinsdottir U, Gudnason V, Krauss RM, Mohlke KL, Ordovas JM, Munroe PB, Kooner JS, Tall AR, Hegele RA, Kastelein JJ, Schadt EE, Rotter JI, Boerwinkle E, Strachan DP, Mooser V, Stefansson K, Reilly MP, Samani NJ, Schunkert H, Cupples LA, Sandhu MS, Ridker PM, Rader DJ, van Duijn CM, Peltonen L, Abecasis GR, Boehnke M, Kathiresan S. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 2010;466:707–13. 10.1038/nature09270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Global Lipids Genetics Consortium Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J, Buchkovich ML, Mora S, Beckmann JS, Bragg-Gresham JL, Chang HY, Demirkan A, Den Hertog HM, Do R, Donnelly LA, Ehret GB, Esko T, Feitosa MF, Ferreira T, Fischer K, Fontanillas P, Fraser RM, Freitag DF, Gurdasani D, Heikkilä K, Hyppönen E, Isaacs A, Jackson AU, Johansson A, Johnson T, Kaakinen M, Kettunen J, Kleber ME, Li X, Luan J, Lyytikäinen LP, Magnusson PK, Mangino M, Mihailov E, Montasser ME, Müller-Nurasyid M, Nolte IM, O'Connell JR, Palmer CD, Perola M, Petersen AK, Sanna S, Saxena R, Service SK, Shah S, Shungin D, Sidore C, Song C, Strawbridge RJ, Surakka I, Tanaka T, Teslovich TM, Thorleifsson G, Van den Herik EG, Voight BF, Volcik KA, Waite LL, Wong A, Wu Y, Zhang W, Absher D, Asiki G, Barroso I, Been LF, Bolton JL, Bonnycastle LL, Brambilla P, Burnett MS, Cesana G, Dimitriou M, Doney AS, Döring A, Elliott P, Epstein SE, Eyjolfsson GI, Gigante B, Goodarzi MO, Grallert H, Gravito ML, Groves CJ, Hallmans G, Hartikainen AL, Hayward C, Hernandez D, Hicks AA, Holm H, Hung YJ, Illig T, Jones MR, Kaleebu P, Kastelein JJ, Khaw KT, Kim E, Klopp N, Komulainen P, Kumari M, Langenberg C, Lehtimäki T, Lin SY, Lindström J, Loos RJ, Mach F, McArdle WL, Meisinger C, Mitchell BD, Müller G, Nagaraja R, Narisu N, Nieminen TV, Nsubuga RN, Olafsson I, Ong KK, Palotie A, Papamarkou T, Pomilla C, Pouta A, Rader DJ, Reilly MP, Ridker PM, Rivadeneira F, Rudan I, Ruokonen A, Samani N, Scharnagl H, Seeley J, Silander K, Stancáková A, Stirrups K, Swift AJ, Tiret L, Uitterlinden AG, van Pelt LJ, Vedantam S, Wainwright N, Wijmenga C, Wild SH, Willemsen G, Wilsgaard T, Wilson JF, Young EH, Zhao JH, Adair LS, Arveiler D, Assimes TL, Bandinelli S, Bennett F, Bochud M, Boehm BO, Boomsma DI, Borecki IB, Bornstein SR, Bovet P, Burnier M, Campbell H, Chakravarti A, Chambers JC, Chen YD, Collins FS, Cooper RS, Danesh J, Dedoussis G, de Faire U, Feranil AB, Ferrières J, Ferrucci L, Freimer NB, Gieger C, Groop LC, Gudnason V, Gyllensten U, Hamsten A, Harris TB, Hingorani A, Hirschhorn JN, Hofman A, Hovingh GK, Hsiung CA, Humphries SE, Hunt SC, Hveem K, Iribarren C, Järvelin MR, Jula A, Kähönen M, Kaprio J, Kesäniemi A, Kivimaki M, Kooner JS, Koudstaal PJ, Krauss RM, Kuh D, Kuusisto J, Kyvik KO, Laakso M, Lakka TA, Lind L, Lindgren CM, Martin NG, März W, McCarthy MI, McKenzie CA, Meneton P, Metspalu A, Moilanen L, Morris AD, Munroe PB, Njølstad I, Pedersen NL, Power C, Pramstaller PP, Price JF, Psaty BM, Quertermous T, Rauramaa R, Saleheen D, Salomaa V, Sanghera DK, Saramies J, Schwarz PE, Sheu WH, Shuldiner AR, Siegbahn A, Spector TD, Stefansson K, Strachan DP, Tayo BO, Tremoli E, Tuomilehto J, Uusitupa M, van Duijn CM, Vollenweider P, Wallentin L, Wareham NJ, Whitfield JB, Wolffenbuttel BH, Ordovas JM, Boerwinkle E, Palmer CN, Thorsteinsdottir U, Chasman DI, Rotter JI, Franks PW, Ripatti S, Cupples LA, Sandhu MS, Rich SS, Boehnke M, Deloukas P, Kathiresan S, Mohlke KL, Ingelsson E, Abecasis GR. Discovery and refinement of loci associated with lipid levels. Nat Genet 2013;45:1274–83. 10.1038/ng.2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Myocardial Infarction Genetics Consortium Investigators. Stitziel NO, Won HH, Morrison AC, Peloso GM, Do R, Lange LA, Fontanillas P, Gupta N, Duga S, Goel A, Farrall M, Saleheen D, Ferrario P, König I, Asselta R, Merlini PA, Marziliano N, Notarangelo MF, Schick U, Auer P, Assimes TL, Reilly M, Wilensky R, Rader DJ, Hovingh GK, Meitinger T, Kessler T, Kastrati A, Laugwitz KL, Siscovick D, Rotter JI, Hazen SL, Tracy R, Cresci S, Spertus J, Jackson R, Schwartz SM, Natarajan P, Crosby J, Muzny D, Ballantyne C, Rich SS, O'Donnell CJ, Abecasis G, Sunyaev S, Nickerson DA, Buring JE, Ridker PM, Chasman DI, Austin E, Ye Z, Kullo IJ, Weeke PE, Shaffer CM, Bastarache LA, Denny JC, Roden DM, Palmer C, Deloukas P, Lin DY, Tang ZZ, Erdmann J, Schunkert H, Danesh J, Marrugat J, Elosua R, Ardissino D, McPherson R, Watkins H, Reiner AP, Wilson JG, Altshuler D, Gibbs RA, Lander ES, Boerwinkle E, Gabriel S, Kathiresan S. Inactivating mutations in NPC1L1 and protection from coronary heart disease. N Engl J Med 2014;371:2072–82. 10.1056/NEJMoa1405386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Do R, Stitziel NO, Won HH, Jørgensen AB, Duga S, Angelica Merlini P, Kiezun A, Farrall M, Goel A, Zuk O, Guella I, Asselta R, Lange LA, Peloso GM, Auer PL; NHLBI Exome Sequencing Project, Girelli D, Martinelli N, Farlow DN, DePristo MA, Roberts R, Stewart AF, Saleheen D, Danesh J, Epstein SE, Sivapalaratnam S, Hovingh GK, Kastelein JJ, Samani NJ, Schunkert H, Erdmann J, Shah SH, Kraus WE, Davies R, Nikpay M, Johansen CT, Wang J, Hegele RA, Hechter E, Marz W, Kleber ME, Huang J, Johnson AD, Li M, Burke GL, Gross M, Liu Y, Assimes TL, Heiss G, Lange EM, Folsom AR, Taylor HA, Olivieri O, Hamsten A, Clarke R, Reilly DF, Yin W, Rivas MA, Donnelly P, Rossouw JE, Psaty BM, Herrington DM, Wilson JG, Rich SS, Bamshad MJ, Tracy RP, Cupples LA, Rader DJ, Reilly MP, Spertus JA, Cresci S, Hartiala J, Tang WH, Hazen SL, Allayee H, Reiner AP, Carlson CS, Kooperberg C, Jackson RD, Boerwinkle E, Lander ES, Schwartz SM, Siscovick DS, McPherson R, Tybjaerg-Hansen A, Abecasis GR, Watkins H, Nickerson DA, Ardissino D, Sunyaev SR, O'Donnell CJ, Altshuler D, Gabriel S, Kathiresan S. Exome sequencing identifies rare LDLR and APOA5 alleles conferring risk for myocardial infarction. Nature 2015;518:102–6. 10.1038/nature13917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The TG and HDL Working Group of the Exome Sequencing Project, National Heart, Lung, and Blood Institute Crosby J, Peloso GM, Auer PL, Crosslin DR, Stitziel NO, Lange LA, Lu Y, Tang ZZ, Zhang H, Hindy G, Masca N, Stirrups K, Kanoni S, Do R, Jun G, Hu Y, Kang HM, Xue C, Goel A, Farrall M, Duga S, Merlini PA, Asselta R, Girelli D, Olivieri O, Martinelli N, Yin W, Reilly D, Speliotes E, Fox CS, Hveem K, Holmen OL, Nikpay M, Farlow DN, Assimes TL, Franceschini N, Robinson J, North KE, Martin LW, DePristo M, Gupta N, Escher SA, Jansson JH, Van Zuydam N, Palmer CN, Wareham N, Koch W, Meitinger T, Peters A, Lieb W, Erbel R, Konig IR, Kruppa J, Degenhardt F, Gottesman O, Bottinger EP, O'Donnell CJ, Psaty BM, Ballantyne CM, Abecasis G, Ordovas JM, Melander O, Watkins H, Orho-Melander M, Ardissino D, Loos RJ, McPherson R, Willer CJ, Erdmann J, Hall AS, Samani NJ, Deloukas P, Schunkert H, Wilson JG, Kooperberg C, Rich SS, Tracy RP, Lin DY, Altshuler D, Gabriel S, Nickerson DA, Jarvik GP, Cupples LA, Reiner AP, Boerwinkle E, Kathiresan S. Loss-of-function mutations in APOC3, triglycerides, and coronary disease. N Engl J Med 2014;371:22–31. 10.1056/NEJMoa1307095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 2011;88:76–82. 10.1016/j.ajhg.2010.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods 2010;7:248–9. 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods 2010;7:575–6. 10.1038/nmeth0810-575 [DOI] [PubMed] [Google Scholar]
- 10.Chun S, Fay JC. Identification of deleterious mutations within three human genomes. Genome Res 2009;19:1553–61. 10.1101/gr.092619.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Romeo S, Pennacchio LA, Fu Y, Boerwinkle E, Tybjaerg-Hansen A, Hobbs HH, Cohen JC. Population-based resequencing of ANGPTL4 uncovers variations that reduce triglycerides and increase HDL. Nat Genet 2007;39:513–16. 10.1038/ng1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Leeuwen EM, Karssen LC, Deelen J, Isaacs A, Medina-Gomez C, Mbarek H, Kanterakis A, Trompet S, Postmus I, Verweij N, van Enckevort DJ, Huffman JE, White CC, Feitosa MF, Bartz TM, Manichaikul A, Joshi PK, Peloso GM, Deelen P, van Dijk F, Willemsen G, de Geus EJ, Milaneschi Y, Penninx BW, Francioli LC, Menelaou A, Pulit SL, Rivadeneira F, Hofman A, Oostra BA, Franco OH, Mateo Leach I, Beekman M, de Craen AJ, Uh HW, Trochet H, Hocking LJ, Porteous DJ, Sattar N, Packard CJ, Buckley BM, Brody JA, Bis JC, Rotter JI, Mychaleckyj JC, Campbell H, Duan Q, Lange LA, Wilson JF, Hayward C, Polasek O, Vitart V, Rudan I, Wright AF, Rich SS, Psaty BM, Borecki IB, Kearney PM, Stott DJ, Adrienne Cupples L; Genome of The Netherlands Consortium, Jukema JW, van der Harst P, Sijbrands EJ, Hottenga JJ, Uitterlinden AG, Swertz MA, van Ommen GJ, de Bakker PI, Eline Slagboom P, Boomsma DI, Wijmenga C, van Duijn CM. Genome of the Netherlands population-specific imputations identify an ABCA6 variant associated with cholesterol levels. Nat Commun 2015;6:6065 10.1038/ncomms7065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones PM, George AM. The ABC transporter structure and mechanism: perspectives on recent research. Cell Mol Life Sci 2004;61:682–99. 10.1007/s00018-003-3336-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Towle HC. Glucose as a regulator of eukaryotic gene transcription. Trends Endocrinol Metab 2005;16:489–94. 10.1016/j.tem.2005.10.003 [DOI] [PubMed] [Google Scholar]
- 15.Dentin R, Pégorier JP, Benhamed F, Foufelle F, Ferré P, Fauveau V, Magnuson MA, Girard J, Postic C. Hepatic glucokinase is required for the synergistic action of ChREBP and SREBP-1c on glycolytic and lipogenic gene expression. J Biol Chem 2004;279:20314–26. 10.1074/jbc.M312475200 [DOI] [PubMed] [Google Scholar]
- 16.Dentin R, Girard J, Postic C. Carbohydrate responsive element binding protein (ChREBP) and sterol regulatory element binding protein-1c (SREBP-1c): two key regulators of glucose metabolism and lipid synthesis in liver. Biochimie 2005;87:81–6. 10.1016/j.biochi.2004.11.008 [DOI] [PubMed] [Google Scholar]
- 17.Ma L, Robinson LN, Towle HC. ChREBP*Mlx is the principal mediator of glucose-induced gene expression in the liver. J Biol Chem 2006;281:28721–30. 10.1074/jbc.M601576200 [DOI] [PubMed] [Google Scholar]
- 18.Uyeda K, Repa JJ. Carbohydrate response element binding protein, ChREBP, a transcription factor coupling hepatic glucose utilization and lipid synthesis. Cell Metab 2006;4:107–10. 10.1016/j.cmet.2006.06.008 [DOI] [PubMed] [Google Scholar]
- 19.Albrechtsen A, Grarup N, Li Y, Sparsø T, Tian G, Cao H, Jiang T, Kim SY, Korneliussen T, Li Q, Nie C, Wu R, Skotte L, Morris AP, Ladenvall C, Cauchi S, Stančáková A, Andersen G, Astrup A, Banasik K, Bennett AJ, Bolund L, Charpentier G, Chen Y, Dekker JM, Doney AS, Dorkhan M, Forsen T, Frayling TM, Groves CJ, Gui Y, Hallmans G, Hattersley AT, He K, Hitman GA, Holmkvist J, Huang S, Jiang H, Jin X, Justesen JM, Kristiansen K, Kuusisto J, Lajer M, Lantieri O, Li W, Liang H, Liao Q, Liu X, Ma T, Ma X, Manijak MP, Marre M, Mokrosiński J, Morris AD, Mu B, Nielsen AA, Nijpels G, Nilsson P, Palmer CN, Rayner NW, Renström F, Ribel-Madsen R, Robertson N, Rolandsson O, Rossing P, Schwartz TW; D.E.S.I.R. Study Group, Slagboom PE, Sterner M; DIAGRAM Consortium, Tang M, Tarnow L, Tuomi T, van't Riet E, van Leeuwen N, Varga TV, Vestmar MA, Walker M, Wang B, Wang Y, Wu H, Xi F, Yengo L, Yu C, Zhang X, Zhang J, Zhang Q, Zhang W, Zheng H, Zhou Y, Altshuler D, 't Hart LM, Franks PW, Balkau B, Froguel P, McCarthy MI, Laakso M, Groop L, Christensen C, Brandslund I, Lauritzen T, Witte DR, Linneberg A, Jørgensen T, Hansen T, Wang J, Nielsen R, Pedersen O. Exome sequencing-driven discovery of coding polymorphisms associated with common metabolic phenotypes. Diabetologia 2013;56:298–310. 10.1007/s00125-012-2756-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gurdasani D, Carstensen T, Tekola-Ayele F, Pagani L, Tachmazidou I, Hatzikotoulas K, Karthikeyan S, Iles L, Pollard MO, Choudhury A, Ritchie GR, Xue Y, Asimit J, Nsubuga RN, Young EH, Pomilla C, Kivinen K, Rockett K, Kamali A, Doumatey AP, Asiki G, Seeley J, Sisay-Joof F, Jallow M, Tollman S, Mekonnen E, Ekong R, Oljira T, Bradman N, Bojang K, Ramsay M, Adeyemo A, Bekele E, Motala A, Norris SA, Pirie F, Kaleebu P, Kwiatkowski D, Tyler-Smith C, Rotimi C, Zeggini E, Sandhu MS. The African Genome Variation Project shapes medical genetics in Africa. Nature 2015;517:327–32. 10.1038/nature13997 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jmedgenet-2015-103439supp.pdf (1.8MB, pdf)