Abstract
Rice empty glumes, also named sterile lemmas or rudimentary lemmas according to different interpretations, are distinct from lemmas in morphology and cellular pattern. Consistently, the molecular mechanism to control the development of lemmas is different from that of empty glumes. Rice LEAFY HULL STERILE1(OsLHS1) and DROOPING LEAF(DL) regulate the cellular pattern and the number of vascular bundles of lemmas respectively, while LONG STERILE LEMMA1 (G1)/ELONGATED EMPTY GLUME (ELE) and PANICLE PHYTOMER2 (PAP2)/OsMADS34 determine identities of empty glumes. Though some progress has been made, identities of empty glumes remain unclear, and genetic interactions between lemma genes and glume genes have been rarely elucidated. In this research, a new G1/ELE mutant g1–6 was identified and the phenotype was analyzed. Similar to previously reported mutant lines of G1/ELE, empty glumes of g1–6 plants transform into lemma-like organs. Furthermore, Phenotypes of single and double mutant plants suggest that, in addition to their previously described gene-specific functions, G1/ELE and OsLHS1 play redundant roles in controlling vascular bundle number, cell volume, and cell layer number of empty glumes and lemmas. Meanwhile, expression patterns of G1/ELE in osmads1-z flowers and OsLHS1 in g1–6 flowers indicate they do not regulate each other at the level of transcription. Finally, down-regulation of the empty glume gene OsMADS34/PAP2 and ectopic expression of the lemma gene DL, in the g1–6 plants provide further evidence that empty glumes are sterile lemmas. Generally, our findings provided valuable information for better understanding functions of G1 and OsLHS1 in flower development and identities of empty glumes.
Keywords: rice, empty glume, lemma, G1/ELE, OsLHS1
Introduction
Flower development is the basis for seed development in angiosperms. Based on analyses of flower mutants in Arabidopsis thaliana and Antirrhinum majus, the ABCDE model was proposed to interpret the molecular mechanism in control of flower development (Coen and Meyerowitz, 1991; Pelaz et al., 2000; Theissen, 2001; Theissen and Saedler, 2001; Ditta et al., 2004).
Rice (Oryza sativa L.) is a model plant of monocots and one important cereal crop. The structural units of the rice flower are spikelets and florets. The spikelet is the primary unit of the rice inflorescence and contains a fertile floret and a pair of empty glumes (also called “sterile lemmas” or “rudimentary lemmas”), subtended by a pair of highly reduced glumes called rudimentary glumes. The floret consists of a pair of bract-like organs (lemma and palea), two lodicules (equivalent to eudicot petals), six stamens, and a carpel (Yoshida and Nagato, 2011; Lombardo and Yoshida, 2015).
Recently, functions of many homeotic genes have been elucidated in rice. There are four APETALA1 (AP1)/FRUITFULL (FUL) orthologs in the rice genome: OsMADS14, OsMADS15, OsMADS18, and OsMADS20 (Kater et al., 2006). Recent studies suggested that OsMADS14, OsMADS15, and OsMADS18 together with OsMADS34/PANICLE PHYTOMER2 (PAP2) regulate the transition from shoot apical meristem (SAM) to the inflorescence meristem (IM; Kobayashi et al., 2012). The AP3 homolog SUPERWOMAN1 (SPW1)/OsMADS16 is expressed in lodicules and stamens. Mutations in SPW1 results in homeotic transformation from stamens to carpels and lodicules into lemma/palea-like structures respectively (Nagasawa et al., 2003). OsMADS3, OsMADS58, OsMADS13, and OsMADS21 belong to the AGAMOUS (AG) subfamily. Among them, OsMADS3 and OsMADS58 are expressed in stamens and carpels. OsMADS3 mainly functions in regulating stamen development while OsMADS58 mainly determines the carpel development (Yamaguchi et al., 2006; Dreni et al., 2011; Li et al., 2011a). OsMADS13 is expressed in ovules and specifies ovule identity. When mutations occurred in OsMADS13, ovules were transformed into carpelloid organs (Dreni et al., 2007; Li et al., 2011b; Yamaki et al., 2011). Additionally, floral determinacy is redundantly determined by OsMADS3, OsMADS58, and OsMADS13 (Yamaguchi et al., 2006; Dreni et al., 2011; Li et al., 2011b). No obvious function of OsMADS21 has been found (Dreni et al., 2007).
The SEP subfamily of rice consists of five genes. OsMADS7 and OsMADS8, two SEP3 homologs, are expressed in the inner three whorls and function in floral development redundantly (Cui et al., 2010). In addition to OsMADS7 and OsMADS8, LEAFY HULL STERILE1 (OsLHS1), OsMADS5 and OsMADS34/PAP2 were divided into the LOFSEP subgroup of MADS-box genes (Kobayashi et al., 2010). OsLHS1 plays versatile roles in floral organ development. In OsLHS1 mutant plants, lemmas, and paleas are under-developed and they fail to interlock each other. Meanwhile, lodicules and stamens develop abnormally. Additionally, the identity of floral meristem is affected. As a result, one new floret is formed in the spikelet occasionally (Jeon et al., 2000; Agrawal et al., 2005; Chen et al., 2006; Gao et al., 2010). OsMADS5 does not have any obvious function in flower development (Agrawal et al., 2005). Like OsLHS1, another member of the rice LOFSEP subgroup OsMADS34/PAP2 has versatile functions in flower development. In addition to regulating spikelet meristem identity and ovule development, OsMADS34/PAP2 regulates the development of empty glumes. In OsMADS34/PAP2 mutant plants, empty glumes elongate to form leaf-like or lemma-like organs (Gao et al., 2010; Kobayashi et al., 2010; Lin et al., 2014). With evolutionary and sequence analyses of OsMADS34/PAP2, Lin et al. (2014) provided evidence to support the hypothesis that rice empty glumes originated from the lemmas of degenerate florets and named them as rudimentary lemmas.
In addition to ABCDE genes, there are other homeotic genes involved in the development of rice floral organs and/or meristems. Among them are DROOPING LEAF (DL) and OsMADS6/MOSAIC FLORAL ORGANS1 (MFO1). DL plays a key role in specifying carpel identity and regulates the number of vascular bundles in lemmas (Yamaguchi et al., 2004; Li et al., 2011a,b). OsMADS6/MFO1, a member of the AGL6 subfamily, determines the palea identity and has versatile functions in regulating flower development (Ohmori et al., 2009; Li et al., 2010; Zhang et al., 2010; Yadav et al., 2011; Duan et al., 2012).
LONG STERILE LEMMA1 (G1) /ELONGATED EMPTY GLUME (ELE) encodes a DUF640 domain protein and determines identities of empty glumes. When mutations occurred in G1/ELE, empty glumes transformed into lemma-like organs (Yoshida et al., 2009; Hong et al., 2010). Meanwhile, natural mutations in the genome of allotetraploid Oryza grandiglumis cause similar homeotic conversions in empty glumes, suggesting that empty glumes are serial lemma homologs that have been modified by the action of G1/ELE (Yoshida et al., 2009).
Despite the fact that the molecular mechanism controlling reproductive organ development in rice is well-understood, the control of empty glume identity remains unclear. In this study, g1–6, a new strong mutant allele of G1/ELE, was identified and double mutant g1–6 osmads1-z plants were analyzed. Additionally, the expression profile of DL, OsLHS1, G1/ELE, and OsMADS34/PAP2 was analyzed. Our findings provided valuable information for understanding functions of these genes and interpreting identities of empty glumes.
Materials and Methods
Plant Materials
A single recessive rice mutant, g1–6, displaying abnormal empty glumes was identified from a M2 population of 9311 (cultivar indica), obtained via ethylmethane sulfonate (EMS) mutagenesis (Ye et al., 2006); osmads1-z was identified previously (Gao et al., 2010). The 9311 cultivar was used as a wild type strain for phenotype observation and RNA extraction. All plants were planted in the greenhouse in Northwest A&F University or paddy fields in Yangling and Hang Zhou in China under natural conditions. In the greenhouse, the conditions were 14 h of light at 28°C, 10 h of dark at 22°C during the vegetative stage. The conditions were adjusted to 10 h of light at 28°C, and 14 h of dark at 22°C to induce the transition from the vegetative stage to the reproductive stage. The humidity was maintained at 70%.
Histological Analysis and Scanning Electron Microscopy (SEM)
Spikelets of rice plants grown in a paddy field in Yangling were fixed in 75% ethanol and 25% acetate and dehydrated in a series of graded ethanol. For histological analysis, materials were substituted by xylene and embedded in paraplast plus and cut into 8 μm-thick sections. Then, sections were stained with 0.2% toluidine blue and photographed using a Nikon E600 microscope and Nikon DXM1200 digital camera. For scanning electron microscopy (SEM), samples were dehydrated in a series of ethanol solutions, then dried at a critical point, sputter-coated with platinum, and observed under a JSM-6360LV (JEOL) scanning electron microscope, as described previously (Li et al., 2010).
RNA Extraction
Rice plants grown in a paddy field in Yangling under field conditions during the 2014 cultivation season were used. Empty glumes, lemmas, and paleas of wild type 9311, g1–6, osmads1-z, and g1–6 osmads1-z plants at the Sp6 stage defined by Ikeda were collected (Ikeda et al., 2004) and preserved at -80°C. Total RNA was isolated using a TRIZOL kit (Sangon Biotech) according to the manufacturer’s protocol and treated with DNaseI (Sangon Biotech). First-strand cDNA was synthesized using a Prime Script RT reagent kit (Fermentas) according to the protocol.
Statistical Analyses
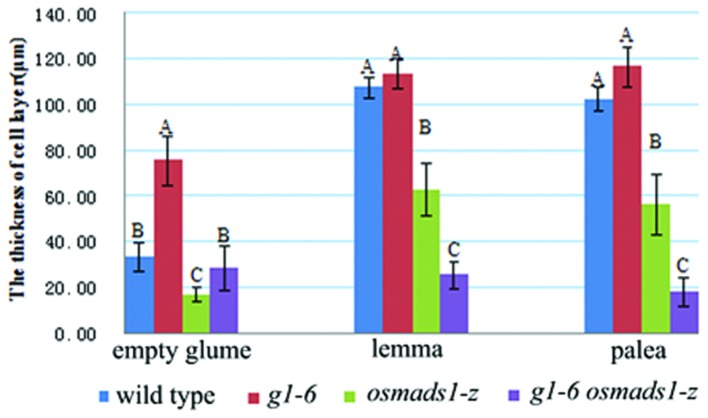
Stained 8 μm-thick sections of randomly selected samples at the Sp6 stage were observed using a Nikon E600 microscope. Data of the number of vascular bundles and thickness of different kinds of organs from different genetic backgrounds were analyzed using Duncan’s test (Duncan, 1955). Letters A, B, or C of every sample in Figures 5 and 7 indicates a significant difference between two analyzed samples at the level of P < 0.01. If the letters of two samples are the same, there is no significant difference between these two samples.
FIGURE 5.
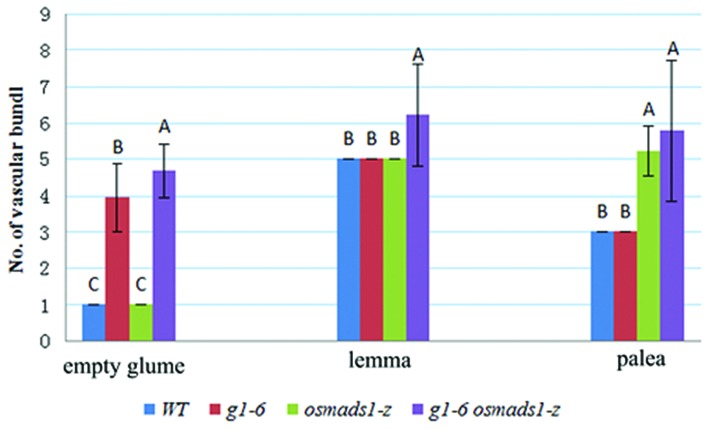
Statistical analyses of number of vascular bundles in empty glumes, lemmas, and paleas. For every kind of organs, 15 samples of wild-type (WT), g1–6, osmads1-z, g1–6 osmads1-z were examined with transverse sections, respectively. The means were statistically analyzed with the Duncan test and grouped (A, B, and C) according to significant differences at P < 0.01. Error bars indicate SD.
FIGURE 7.
Statistical analyses of the thickness of empty glumes, lemmas, and paleas. For every kind of organ, 13 samples of wild type, 14 samples of g1–6, 17 samples of osmads1-z, 22 samples of g1–6 osmads1-z plants were measured with transverse sections, respectively. The means were statistically analyzed with the Duncan test and grouped (A, B, and C) according to significant differences at P < 0.01. Error bars indicate SD.
Quantitative RT-PCR
Reverse transcription polymerase chain reaction (RT-PCR) was performed using CFX96 real-time polymerase chain reaction (PCR) detection systems (Roche Applied Science). A final volume of 15 μL reactions contained 7.5 μL SYBR® Premix Ex Taq (Takara, 2×), 0.75 μL forward and reverse primers (10 pM) respectively, 0.5 μL cDNA (5.0 ng/μL) and 5.5 μL ddH2O. PCR cycling conditions were 95°C for 30 s followed by 40 cycles of 95°C for 5 s, 55°C for 30 s and 61 cycles of 65°C for 10 s. Three biological replicates were sampled in each experiment and all samples were run with three technical replicates. Data acquisition and analyses were performed using the Roche Light Cycler software. Samples were normalized using ACTIN expression (Kim et al., 2003) and relative expression levels were determined using the 2(-Δ Ct) analysis method (Pfaffl, 2001). Primers used in this article are listed in Supplementary Table S1.
Results
Identification of the g1–6 Allele
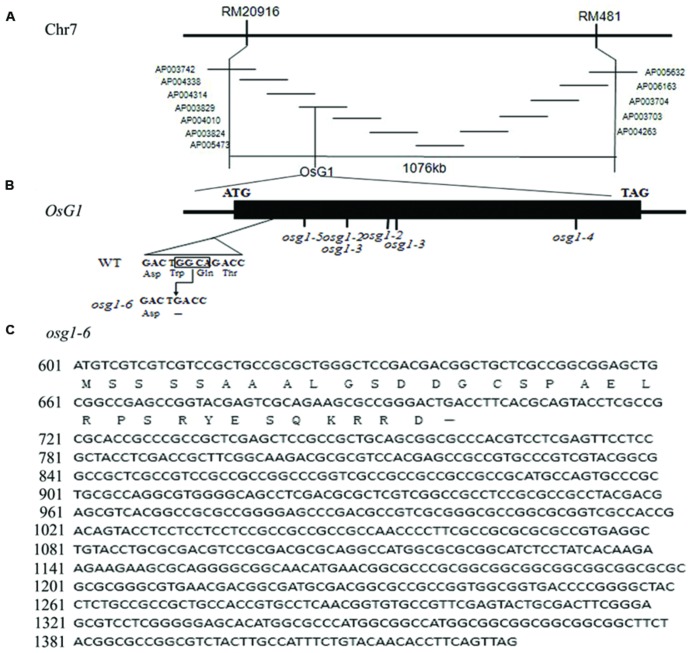
To identify rice mutants with floral defects, a population of rice mutants in the indica subspecies 9311 background treated by EMS was screened. A mutant line displaying an abnormal phenotype of empty glumes was isolated. When mutant plants were crossed with 9311 plants, the F1 progeny generated normal flowers, whereas the F2 plants yielded a ~3:1 ratio of phenotype segregation, indicating that this phenotype was linked with one nuclear recessive locus mutation. To identify the mutant gene, a map-based cloning strategy was used. The homozygous mutant plants were crossed with one japonica cultivar 02428. Using a population of 484 F2 mutant plants, the mutation site was mapped between two simple sequence repeat (SSR) molecular markers, RM20916 and RM481, on the short arm of chromosome 7 (Figure 1A).
FIGURE 1.
Positional cloning of g1–6 and sequence analysis. (A) The candidate gene was mapped in BAC AP003829 on the short arm of Chromosome 7. (B) Schematic representation of G1 and mutation site of g1–6. (C) Full cDNA of g1–6. The translation is terminated prematurely at the mutation site.
Since plants of long sterile lemma1 and elongated empty glume displayed similar phenotypes and the G1/ELE gene was mapped in the same region (Yoshida et al., 2009; Hong et al., 2010), we considered if a mutation of G1/ELE occurred in our mutant plants. Therefore, we designed a pair of primers to amplify a fragment including the encoding sequence, partial 5′ UTR and 3′ UTR of G1/ELE. Then PCR products using genome DNA extracted from wild-type and mutant plants as templates were sequenced respectively. Compared with the wild-type plants, a 4-bp (+98 to +101) deletion was found in G1/ELE of our mutant plants (Figure 1B). This result proved that our mutant was an allele of G1/ELE. The G1/ELE gene only contains one exon and encodes one DUF640 domain protein which is localized to the nucleus (Yoshida et al., 2009; Hong et al., 2010). In combination with its transcriptional activity, we speculated that G1/ELE played a role as a transcription factor. The mutation in our plants resulted in a frame shift and caused a premature termination of translation at the 33rd amino acid immediately after the deletion, and a complete loss of the DUF640 domain (Figure 1C). Since five mutant lines of G1/ELE have been reported previously (Yoshida et al., 2009; Hong et al., 2010), we named our mutant g1–6.
Empty Glumes of g1–6 Plants Transform into Lemma-Like Organs
Compared with wild-type 9311 plants, mutant plants develop normally at the vegetative stage (data not shown). The flowering time of mutant plants is about 90 days after sowing (DAS), which is indistinct from the wild-type in Hangzhou. However, mutant plants displayed abnormal empty glumes during the reproductive stage (Figures 2A,B). Therefore, we analyzed phenotypes of empty glumes in detail. Firstly, the average length of abnormal empty glumes in g1–6 plants was >8 mm whereas that of wild type plants is about 3 mm (Figures 2C,D, Supplementary Figure S1). Secondly, whereas wild-type empty glumes form only one vascular bundle (Figure 2E), g1–6 empty glumes develop 3–5 vascular bundles (Figures 2G,H). Thirdly, similar to those of lemmas in wild type plants, two marginal regions of abnormal empty glumes in g1–6 plants curled inwardly (Figures 2F–H). Fourthly, like wild-type lemmas, abnormal empty glumes contain four types of cells: silicified cells (sc), fibrous sclerenchyma (fs), spongy parenchymatous cells (spc), and no silicified cells (nsc; Figures 2F–H). On the contrary, wild-type empty glumes only contain one type of cell, sclerenchymatous cells, between two epidermal layers (Figure 2E). Furthermore, the epidermal cells of abnormal empty glumes in g1–6 were similar to those of lemmas and paleas but differed from those of glumes of wild-type plants (Figure 3). In conclusion, empty glumes in g1–6 plants transformed into lemma-like organs. These phenotypes are similar to those of the long sterile lemmas (LSLin g1–1 plants, but stronger than those of ele plants (Yoshida et al., 2009; Hong et al., 2010). This is probably, because the mutation in ele was a one-nucleotide substitution of G to A, resulting in only one amino acid substitution of glycine to serine in the protein (Hong et al., 2010).
FIGURE 2.
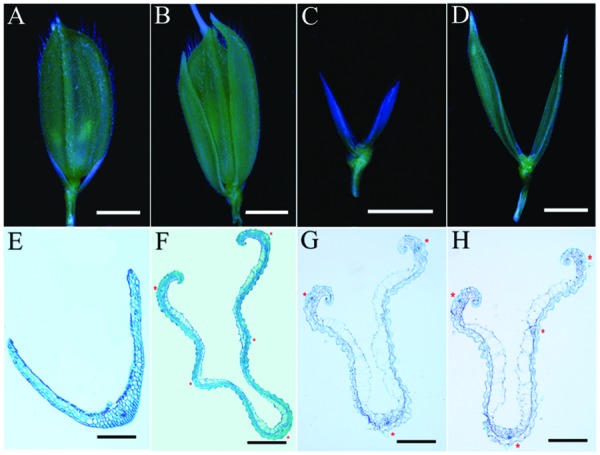
Phenotypes of spikelets and empty glumes. (A) One spikelet of wild type. (B) One g1–6 spikelet. (C) One empty glume of wild type. (D) One g1–6 empty glume. Transverse sections of one empty glume (E) and one lemma of wild type (F). Transverse sections of one g1–6 empty glume (G) and one g1–6 lemma (H). Red asterisks in (F–H) indicate vascular bundles. Bars = 2 mm in (A–D), 100 μm in (E), 200 μm in (F–H).
FIGURE 3.
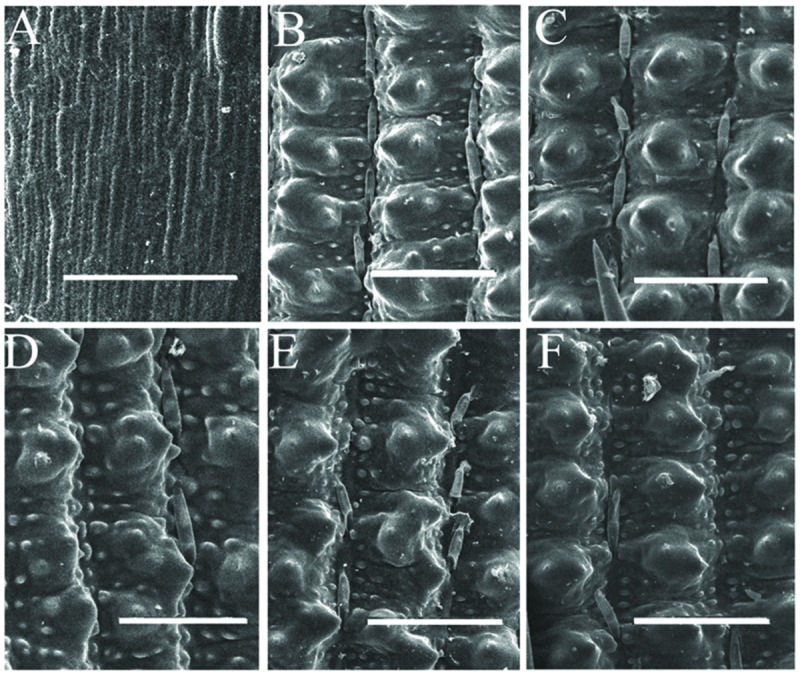
Scanning electron microscopy observation of empty glumes, lemmas, and paleas. Epidermis of one empty glume (A), one lemma (B), and one palea (C) of wild- type plants. Epidermis of one empty glume (D), one lemma (E), and one palea (F) of g1–6 plants. Bars = 50 μm.
g1–6 osmads1-z Plants Displayed Similar Defects in Inner Floral Organs and Floral Meristems to osmads1-z
LHS1-like genes are a grass specific clade of SEP-like genes (Malcomber and Kellogg, 2004). In grasses, homologs of OsLHS1 display distinct expression patterns in different species, implying that differences in LHS1 expression patterns may contribute to the morphological diversification of grass inflorescence architecture (Malcomber and Kellogg, 2004). The biological function of OsLHS1 has been elucidated. It is mainly strongly expressed in lemmas, paleas and pistils, as well as floral meristems at the early stage (Li et al., 2010). In general, it regulates the development of lemmas/paleas and affects the meristem determinacy of inner floral organs (Jeon et al., 2000; Agrawal et al., 2005; Chen et al., 2006; Gao et al., 2010). Previously, one null OsLHS1 mutant line osmads1-z, containing a 1312 bp deletion which spans the first exon and intron, was identified (Gao et al., 2010). Similar to other alleles of OsLHS1, lemmas and paleas in osmads1-z could not interlock closely (Figure 4A). Meanwhile, the development of lodicules and stamens was disrupted (Gao et al., 2010; Figure 4B), while the empty glumes developed normally (Figures 4A,B).
FIGURE 4.
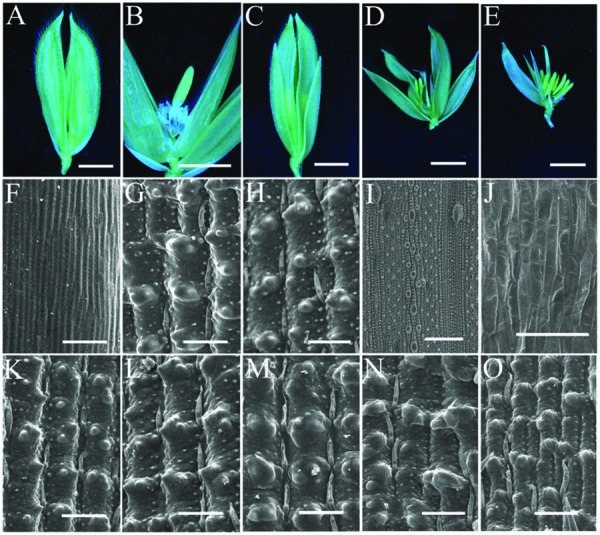
Flower Phenotypes of osmads1-z and osmads1-z g1–6 plants. (A,B) Spikelets of osmads1-z. (C–E) Spikelets of osmads1-z g1–6; the lemma and palea were removed in (E) to show the inner organs. Epidermal cells of one empty glume (F), one lemma (G) and one palea (H) of osmads1-z; Epidermal cells of one leaf (I) and one lodicule (J) of wild type plants. Epidermal cells of one empty glume (K), one lemma (L) and one palea (M) of g1–6 osmads1-z. (N), Epidermal cells of one lodicule of osmads1-z. (O) Epidermal cells of one lodicule of osmads1-z g1–6. Bars = 2 mm in (A–E), 50 μm in (F–O) except for (I), 100 μm in (I).
To analyze the genetic interaction between G1/ELE and OsLHS1, a double mutant was constructed and flower phenotypes of double mutant plants were analyzed. Primary observations indicated that double mutant plants exhibited additive phenotypes, i.e., the phenotype of g1–6 osmads1-z empty glumes was similar to that of g1–6 plants (Figure 4C), whereas phenotypes of inner floral organs mimicked osmads1-z (Figures 4D,E). Like g1–6 plants, empty glumes of g1–6 osmads1-z plants developed into lemma-like organs (Figure 4C). Correspondingly, similar to osmads1-z, lemmas and paleas of double mutant plants could not interlock each other (Figures 4A,C); meanwhile, phenotypes of the inner floral organs of g1–6 osmads1-z plants were the same as those of the osmads1-z plants mentioned above: the number of stamens decreased; development of lodicules was disrupted and the epidermal cells were different from those of wild type lodicules, but indistinct from those of osmads1-z plants (Figures 4B,D,J,N,O, Supplementary Table S2). As reported in osmads1-z, double mutant plants exhibited defects in floral meristems by generating ectopic florets occasionally (Gao et al., 2010; Figure 4E).
The Mutation of g1–6 Enhanced the Phenotype of Lemmas in osmads1-z Plants
To further elucidate the function of OsLHS1 and G1/ELE in lemma development, we analyzed phenotypes of abnormal empty glumes and lemmas in double mutant plants in detail. Epidermal (Epidermal) cells of abnormal empty glumes were indistinct from those of g1-6 or osmads1-z lemmas/paleas, but distinct from those of wild-type empty glumes (Figures 4F–H,K,L); similarly, epidermal cells of double mutant lemmas/paleas were indistinct from those of g1-6 or osmads1-z plants, but distinct from those of wild-type leaves (Figures 4G–I,L,M), indicating that they still maintained partial identities of lemmas or paleas. However, two differences were found, including the number of vascular bundles, and lemma thickness. Previously, we characterized one strong dl mutant line and found the function of DL in regulating the number of vascular bundles in lemmas (Li et al., 2011b). Since the ectopic expression of DL was detected in this research (see below, Figure 8), we investigated the number of vascular bundles in empty glumes and lemmas/paleas of double mutant plants. The number of vascular bundles of abnormal empty glumes was 3.94 ± 0.93 in g1–6 plants, whereas the number increased to 4.67 ± 0.72 in g1–6 osmads1-z plants. Meanwhile, the number of vascular bundles in lemmas increased from 5.00 ± 0.00 in osmads1-z plants to 6.2 ± 1.42 in g1–6 osmads1-z plants (Figure 5). Statistical analysis showed these differences were significant (Figure 5).
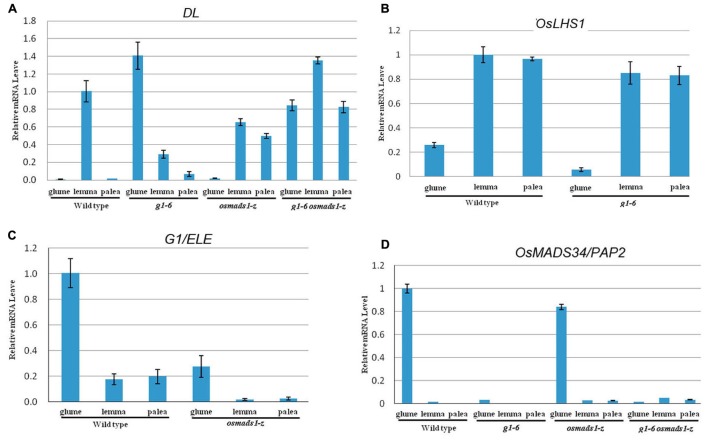
FIGURE 8.
Expression patterns of DL, OsLHS1, OsMADS34/PAP2, and G1/ELE. (A) Expression analyses of DL gene in empty glumes, lemmas, and paleas. (B) Expression analyses of OsLHS1 in empty glumes, lemmas, and paleas. (C) Expression analyses of G1/ELE in empty glumes, lemmas, and paleas. (D) Expression analyses of OsMADS34/PAP2 in empty glumes, lemmas, and paleas. Error bars indicate SD.
Previously, it was reported that OsLHS1 controls the differentiation of specific cell types in lemmas/paleas (Prasad et al., 2005). Therefore, we carefully analyzed the cellular pattern of lemma-like organs and lemmas/paleas of the wild-type, g1–6, osmads1-z, and g1–6 osmads1-z plants at same developmental stage. Lemmas of osmads1-z plants were thinner than those of wild type plants. The average thickness of lemmas of osmads1-z plants was 62.74 ± 11.27 μm, whereas that of wild-type plants was 107.20 ± 4.81 μm (Figures 6B,C,H,I and 7). We found the total size of the four cell types of lemma-like organs and lemmas/paleas in double mutant plants were further reduced in g1–6 osmads1-z plants than those in single mutant osmads1-z or g1–6 plants (Figures 6D–L). Spongy parenchymatous cells were not observed in some regions (Figures 6J–L). As a result, the thickness of lemma-like organs and lemmas/paleas in double mutant plants was much thinner than corresponding organs in g1–6 or osmads1-z plants (Figures 6 and 7). Statistically, these differences are meaningful (Figure 7).
FIGURE 6.
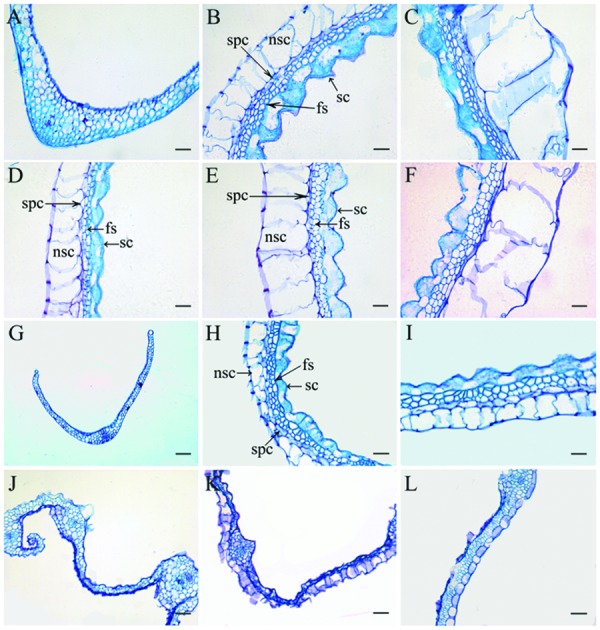
Histological observation of empty glumes, lemmas, and paleas. Transverse sections of one empty glume (A), one lemma (B), and one palea (C) in wild type plants. Transverse sections of one abnormal empty glume (D), one lemma (E), and one palea (F) in g1–6 plants. Transverse sections of one empty glume (G), one lemma (H), and one palea (I) in osmads1-z plants; Transverse sections of one empty glume (J), one lemma (K), and one palea (L) in osmads1-z g1–6 plants. Sc, silicified cells; fs, fibrous sclerenchyma; spc, spongy parenchymatous cells; nsc, no silicified cells. Bars = 50 μm.
Expression Profile of DL, OsLHS1, G1/ELE, and OsMADS34/PAP2
To further analyze the genetic interactions between genes associated with development of empty glumes and lemmas, expression patterns of DL, OsLHS1, G1/ELE, and OsMADS34 were analyzed under different genetic backgrounds by using quantitative RT-PCR.
In rice, DL is expressed in lemmas in addition to carpels, but not in paleas and empty glumes of wild-type plants (Nagasawa et al., 2003; Li et al., 2011b; Figure 8A). In addition to specifying carpel identity, DL regulates the development of vascular bundles in lemmas (Nagasawa et al., 2003; Li et al., 2011b). Based on these facts, we initially analyzed the expression pattern of DL. As shown in Figure 8A, strong ectopic expression of DL was detected in abnormal empty glumes of g1–6 and double mutant plants, further suggesting empty glumes acquire identities of lemmas. Meanwhile, consistent with the increased number of vascular bundles, ectopic expression of DL was detected in paleas of osmads1-z and double mutant plants (Figure 8A).
In wild-type plants, OsLHS1 was strongly expressed in lemmas and paleas, whereas the expression was weak in empty glumes (Figure 8B). Since empty glumes transform into lemma-like organs in g1–6 plants, we predicted OsLHS1 might be strongly expressed in abnormal empty glumes of g1–6 plants. Surprisingly, the expression level of OsLHS1 in abnormal glumes of g1–6 plants was lower than that of lemmas or paleas (Figure 8B). Similarly, mutation in osmads1-z did not change the expression domains of G1/ELE (Figure 8C). These results indicate OsLHS1 and G1/ELE do not regulate each other at the level of transcription. OsMADS34/PAP2 is another regulator of empty glumes. Mutations in OsMADS34/PAP2 also disrupt the development of empty glumes (Gao et al., 2010; Kobayashi et al., 2010; Lin et al., 2014). We considered if the expression of OsMADS34/PAP2 was altered in g1–6 plants and analyzed its expression. In wild-type plants, strong expression of OsMADS34/PAP2 was detected in empty glumes but excluded from lemmas and paleas (Figure 8D). However, no obvious expression of OsMADS34/PAP2 in abnormal glumes of either g1–6 or double mutant plants was detected (Figure 8D). These results suggested that G1/ELE regulate the expression of OsMADS34/PAP2 positively in empty glumes. It was reported that OsLHS1 represses the expression of OsMADS34/PAP2 (Khanday et al., 2013). However, obvious expression of OsMADS34/PAP2 was not found in lemmas or paleas of osmads1-z (Figure 8D). One probable reason is that other genes also repress the expression of OsMADS34/PAP2.
Discussion
Distinct Mechanism for Controlling the Development of Empty Glumes and Lemmas
According to one hypothesis, the spikelet is a reduced leaf branch comprising a series of bracts and florets. Similar to rudimentary glumes, empty glumes have been considered as metamorphic bract leaves (Chase, 1968; Bell, 1991; Takeoka et al., 1993). The alternative hypothesis is that empty glumes are remnants of two lower reduced florets that lost all their inner floral organs. Under this interpretation, a rice spikelet would possibly have been derived from an ancestral one that contained three florets, and empty glumes would then represent the sterile lemmas rather than the empty glumes (Yoshida and Nagato, 2011). Recently, Lin et al. (2014) proposed that empty glumes were rudimentary lemmas. Relatively, the hypothesis that empty glumes are sterile lemmas is more acceptable.
At the molecular level, the mechanism to determine lemmas is also different from that of empty glumes. It was reported that OsLHS1 controls the differentiation of specific cell types in lemmas and paleas (Prasad et al., 2005). The development of lemmas and paleas was disrupted and they could not interlock each other in OsLHS1 mutant plants (Jeon et al., 2000; Chen et al., 2006; Gao et al., 2010). These results implied that OsLHS1 regulates the development of lemmas. DL is expressed in lemmas and carpels, and regulates the number of vascular bundles of lemmas in addition to specifying identities of carpels (Yamaguchi et al., 2004; Li et al., 2011a,b). In dl-sup6, the number of vascular bundles in lemmas decreases (Li et al., 2011b). In osmads6-1 or osmads1-z plants, DL is ectopically expressed in abnormal paleas which form more than three vascular bundles (Li et al., 2010, 2011a; Figure 7). Together, we suggest that DL regulates lemma identity by determining the number of vascular bundles. Its expression in lemma-like organs in g1–6 is consistent with the transformation from empty glumes to lemmas (Figure 8A; Lin et al., 2014). However, mutations of neither OsLHS1 nor DL caused the severe loss of lemma identities. Mutations of OsMADS6/MFO1 and DEP/OsMADS15 retarded the development of paleas, but lemmas developed normally (Ohmori et al., 2009; Li et al., 2010; Wang et al., 2010; Zhang et al., 2010; Duan et al., 2012). The simultaneous suppression of three AP1/FUL-like genes OsMADS14, OsMADS15, and OsMADS18 did not affect the development of lemmas (Kobayashi et al., 2012), suggesting that they are not regulators of lemmas. Key regulators of lemmas might be identified in future research.
Two genes have been reported to determine the identities of empty glumes. G1/ELE, and OsMADS34/PAP2. Research (Yoshida et al., 2009; Hong et al., 2010) clearly showed G1/ELE was a key gene for maintaining the identities of empty glumes. Including natural mutations, empty glumes in all g1/ele plants lose identities. All features including the length, cell pattern, type of epidermal cells, inward curl of marginal tissues and the number of vascular bundles showed the transformation from empty glumes to lemmas (Yoshida et al., 2009; Hong et al., 2010; this research). Similarly, empty glumes of all osmads34/pap2 plants lose identities and develop into lemma-like or indeterminate organs, suggesting OsMADS34/PAP2 is also a key regulator for empty glume development (Gao et al., 2010; Kobayashi et al., 2010; Lin et al., 2014).
At present, there are several controversial interpretations regarding the identities of empty glumes; whether they are true glumes or lemmas. Yoshida et al. (2009) regarded empty glumes as remnants of two lower reduced florets and named them sterile lemmas. Indeed, formation of lemma-like organs at the position of empty glumes in the Oryza grandiglumis is probably resulted from the natural mutations in G1/ELE (Yoshida et al., 2009). Similarly, Lin et al. (2014) proposed empty glumes originated from lemmas and named them as rudimentary lemmas based on the analysis of OsMADS34/PAP2 mutant plants and other research. The repression of OsMADS34/PAP2 and ectopic expression of DL in lemma-like organs in g1–6 plants (Figure 8) provided further molecular evidence to support these hypotheses. Probably, research progress in the development of lemmas and further molecular evidence might provide clues to determine the identities of empty glumes. The cloning and identification of key genes for lemma identities and analyses of their expression in empty glumes are necessary.
The Relationship between G1/ELE, OsMADS34/PAP2, and OsLHS1
Previously, it was reported that OsLHS1 represses the expression of OsMADS34/PAP2. Further analysis showed OsMADS34/PAP2 is a direct target gene negatively regulated by OsLHS1 (Khanday et al., 2013). OsMADS34/PAP2 maintains the identity of the spikelet meristem, whereas OsLHS1 specifies the identity of the floral meristem. The probable mechanism is that OsLHS1 promotes the transition from spikelet meristem to floral meristem by repressing the expression of OsMADS34/PAP2. In the present research, we found that expression of OsMADS34/PAP2 was dramatically decreased in abnormal empty glumes in g1–6 plants (Figure 8D). Taking the transcriptional activity and nuclear location of the G1/ELE protein into account, we propose OsMADS34/PAP2 is downstream of G1/ELE. Additionally, although the expression of G1/ELE is not affected on osmads34-2, empty glumes still lose identities (Gao et al., 2010). These results suggest that G1/ELE might function in the development of empty glumes by regulating the expression of OsMADS34/PAP2. At present, it is unclear whether G1/ELE regulates OsMADS34/PAP2 directly or not; therefore, further analysis is needed for clarification.
In combination with their functions in empty glumes and lemmas respectively, phenotypes of empty glumes and inner floral organs in g1–6 osmads1-z plants indicate G1/ELE and OsLHS1 function independently.
Compared with osmads1-z plants, the increased number of vascular bundles and decreased thickness of lemmas in double mutant plants (Figures 6 and 7) showed that the abnormality becomes more severe. The enhancement of phenotypes suggests G1/ELE plays a role in the development of lemmas redundant with OsLHS1. Evidently, the redundancy is unequal, and although the expression of OsLHS1 is weak in abnormal glumes of g1–6 plants, the thickness is normal. This result implies low level expression of OsLHS1 is enough to maintain the normal cellular pattern (thickness) of lemma-like organs. On the contrary, in osmads1-z plants, although G1/ELE was still expressed in lemmas at low level, the thickness was decreased.
The participation of G1/ELE in lemma development further provided evidence to support the statement that empty glumes are sterile lemmas.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AB106553 (DL), AK100227 (OsMADS34/PAP2), AK070981 (OsLHS1), AB512480 (G1/ELE).
Author Contributions
Experimental design: HL, CS; Experiments: ML, HL, YS, WL, CS; Data analysis: HL, ML; Manuscript preparation: HL, ML; Supervision, funding and reagents: HL, CS.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Professor Qixin Sun from Northwest A&F University, for advice on this research project and critical reading of this manuscript. We thank Professor Dabing Zhang from Shanghai Jiaotong University for providing osmads1-z seeds kindly.
Footnotes
Funding. This work was supported by the Natural Science Foundation (31571657, 31200148), Program of Breeding for New Rice Variety with Higher Yield and Quality in Zhejiang Province (2012C12901-2).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01006
References
- Agrawal G., Abe K., Yamazaki M., Miyao A., Hirochika H. (2005). Conservation of the E-function for floral organ identity in rice revealed by the analysis of tissue culture-induced loss-of-function mutants of the OsMADS1 gene. Plant Mol. Biol. 59 125–135. 10.1007/s11103-005-2161-y [DOI] [PubMed] [Google Scholar]
- Bell A. (1991). Plant Form: An Illustrated Guide to Flowering Plant Morphology. New York, NY: Oxford University Press. [Google Scholar]
- Chase A. (1968). The Structure of Grasses Explained for Beginners. Washington, DC: Smithsonian Institution. [Google Scholar]
- Chen Z. X., Wu J. G., Ding W. N., Chen H. M., Wu P., Shi C. H. (2006). Morphogenesis and molecular basis on naked seed rice, a novel homeotic mutation of OsMADS1 regulating transcript level of AP3 homologue in rice. Planta 223 882–890. 10.1007/s00425-005-0141-8 [DOI] [PubMed] [Google Scholar]
- Coen E. S., Meyerowitz E. M. (1991). The war of the whorls: genetic interactions controlling flower development. Nature 353 31–37. 10.1038/353031a0 [DOI] [PubMed] [Google Scholar]
- Cui R., Han J., Zhao S., Su K., Wu F., Du X., et al. (2010). Functional conservation and diversification of class E floral homeotic genes in rice (Oryza sativa). Plant J. 61 767–781. 10.1111/j.1365-313X.2009.04101.x [DOI] [PubMed] [Google Scholar]
- Ditta G., Pinyopich A., Robles P., Pelaz S., Yanofsky M. F. (2004). The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 14 1935–1940. 10.1016/j.cub.2004.10.028 [DOI] [PubMed] [Google Scholar]
- Dreni L., Jacchia S., Fornara F., Fornari M., Ouwerkerk P. B., An G., et al. (2007). The D-lineage MADS-box gene OsMADS13 controls ovule identity in rice. Plant J. 52 690–699. 10.1111/j.1365-313X.2007.03272.x [DOI] [PubMed] [Google Scholar]
- Dreni L., Pilatone A., Yun D., Erreni S., Pajoro A., Caporali E., et al. (2011). Functional analysis of all AGAMOUS subfamily members in rice reveals their roles in reproductive organ identity determination and meristem determinacy. Plant Cell 23 2850–2863. 10.1105/tpc.111.087007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y., Xing Z., Diao Z., Xu W., Li S., Du X., et al. (2012). Characterization of Osmads6-5, a null allele, reveals that OsMADS6 is a critical regulator for early flower development in rice (Oryza sativa L.). Plant Mol. Biol. 80 429–442. 10.1007/s11103-012-9958-2 [DOI] [PubMed] [Google Scholar]
- Duncan D. B. (1955). Multiple range and multiple F tests. Biometrics 11 1–42. 10.2307/3001478 [DOI] [Google Scholar]
- Gao X., Liang W., Yin C., Ji S., Wang H., Su X., et al. (2010). The SEPALLATA-like gene OsMADS34 is required for rice inflorescence and spikelet development. Plant Physiol. 153 728–740. 10.1104/pp.110.156711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong L., Qian Q., Zhu K., Tang D., Huang Z., Gao L., et al. (2010). ELE restrains empty glumes from developing into lemmas. J. Genet. Genomics 37 101–115. 10.1016/S1673-8527(09)60029-1 [DOI] [PubMed] [Google Scholar]
- Ikeda K., Sunohara H., Nagato Y. (2004). Developmental course of inflorescence and spikelet in rice. Breed. Sci. 54 147–156. 10.1270/jsbbs.54.147 [DOI] [Google Scholar]
- Jeon J. S., Jang S., Lee S., Nam J., Kim C., Lee S. H., et al. (2000). leafy hull sterile1 is a homeotic mutation in a rice MADS box gene affecting rice flower development. Plant Cell 12 871–884. 10.1105/Tpc.12.6.871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kater M. M., Dreni L., Colombo L. (2006). Functional conservation of MADS-box factors controlling floral organ identity in rice and Arabidopsis. J. Exp. Bot. 57 3433–3444. 10.1093/jxb/erl097 [DOI] [PubMed] [Google Scholar]
- Khanday I., Yadav S. R., Vijayraghavan U. (2013). Rice LHS1/OsMADS1 controls floret meristem specification by coordinated regulation of transcription factors and hormone signaling pathways. Plant Physiol. 161 1970–1983. 10.1104/pp.112.212423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B. R., Nam H. Y., Kim S. U., Kim S. I., Chang Y. J. (2003). Normalization of reverse transcription quantitative-PCR with housekeeping genes in rice. Biotechnol. Lett. 25 1869–1872. 10.1023/A:1026298032009 [DOI] [PubMed] [Google Scholar]
- Kobayashi K., Maekawa M., Miyao A., Hirochika H., Kyozuka J. (2010). PANICLE PHYTOMER2 (PAP2), encoding a SEPALLATA subfamily MADS-box protein, positively controls spikelet meristem identity in rice. Plant Cell Physiol. 51 47–57. 10.1093/pcp/pcp166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K., Yasuno N., Sato Y., Yoda M., Yamazaki R., Kimizu M., et al. (2012). Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene. Plant Cell 24 1848–1859. 10.1105/tpc.112.097105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Liang W., Hu Y., Zhu L., Yin C., Xu J., et al. (2011a). Rice MADS6 interacts with the floral homeotic genes SUPERWOMAN1, MADS3, MADS58, MADS13, and DROOPING LEAF in specifying floral organ identities and meristem fate. Plant Cell 23 2536–2552. 10.1105/tpc.111.087262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Liang W., Jia R., Yin C., Zong J., Kong H., et al. (2010). The AGL6-like gene OsMADS6 regulates floral organ and meristem identities in rice. Cell Res. 20 299–313. 10.1038/cr.2009.143 [DOI] [PubMed] [Google Scholar]
- Li H., Liang W., Yin C., Zhu L., Zhang D. (2011b). Genetic interaction of OsMADS3, DROOPING LEAF, and OsMADS13 in specifying rice floral organ identities and meristem determinacy. Plant Physiol. 156 263–274. 10.1104/pp.111.172080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., Wu F., Du X., Shi X., Liu Y., Liu S., et al. (2014). The pleiotropic SEPALLATA-like gene OsMADS34 reveals that the ‘empty glumes’ of rice (Oryza sativa) spikelets are in fact rudimentary lemmas. New Phytol. 202 689–702. 10.1111/nph.12657 [DOI] [PubMed] [Google Scholar]
- Lombardo F., Yoshida H. (2015). Interpreting lemma and palea homologies: a point of view from rice floral mutants. Front. Plant Sci. 6:61 10.3389/Fpls.2015.00061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcomber S. T., Kellogg E. A. (2004). Heterogeneous expression patterns and separate roles of the SEPALLATA gene LEAFY HULL STERILE1 in grasses. Plant Cell 16 1692–1706. 10.1105/tpc.021576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasawa N., Miyoshi M., Sano Y., Satoh H., Hirano H., Sakai H., et al. (2003). SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development 130 705–718. 10.1242/dev.00294 [DOI] [PubMed] [Google Scholar]
- Ohmori S., Kimizu M., Sugita M., Miyao A., Hirochika H., Uchida E., et al. (2009). MOSAIC FLORAL ORGANS1, an AGL6-like MADS box gene, regulates floral organ identity and meristem fate in rice. Plant Cell 21 3008–3025. 10.1105/tpc.109.068742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaz S., Ditta G. S., Baumann E., Wisman E., Yanofsky M. F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405 200–203. 10.1038/35012103 [DOI] [PubMed] [Google Scholar]
- Pfaffl M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29 e45. 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad K., Parameswaran S., Vijayraghavan U. (2005). OsMADS1, a rice MADS-box factor, controls differentiation of specific cell types in the lemma and palea and is an early-acting regulator of inner floral organs. Plant J. 43 915–928. 10.1111/j.1365-313X.2005.02504.x [DOI] [PubMed] [Google Scholar]
- Takeoka Y., Shimizu M., Wada T. (1993). “Panicles,” in Science of the Rice Plant, Morphology Vol. 1 eds Matsuo T., Hoshikawa K. (Tokyo: Food and Agriculture Policy Research Center; ) 295–338. [Google Scholar]
- Theissen G. (2001). Development of floral organ identity: stories from the MADS house. Curr. Opin. Plant Biol. 4 75–85. 10.1016/S1369-5266(00)00139-4 [DOI] [PubMed] [Google Scholar]
- Theissen G., Saedler H. (2001). Plant biology, Floral quartets. Nature 409 469–471. 10.1038/35054172 [DOI] [PubMed] [Google Scholar]
- Wang K., Tang D., Hong L., Xu W., Huang J., Li M., et al. (2010). DEP and AFO regulate reproductive habit in rice. PLoS Genet. 6:e1000818 10.1371/journal.pgen.1000818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav S. R., Khanday I., Majhi B. B., Veluthambi K., Vijayraghavan U. (2011). Auxin-responsive OsMGH3, a common downstream target of OsMADS1 and OsMADS6, controls rice floret fertility. Plant Cell Physiol. 52 2123–2135. 10.1093/pcp/pcr142 [DOI] [PubMed] [Google Scholar]
- Yamaguchi T., Lee D. Y., Miyao A., Hirochika H., An G., Hirano H. Y. (2006). Functional diversification of the two C-class MADS box genes OSMADS3 and OSMADS58 in Oryza sativa. Plant Cell 18 15–28. 10.1105/tpc.105.037200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T., Nagasawa N., Kawasaki S., Matsuoka M., Nagato Y., Hirano H. Y. (2004). The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell 16 500–509. 10.1105/tpc.018044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaki S., Nagato Y., Kurata N., Nonomura K. (2011). Ovule is a lateral organ finally differentiated from the terminating floral meristem in rice. Dev. Biol. 351 208–216. 10.1016/j.ydbio.2010.12.006 [DOI] [PubMed] [Google Scholar]
- Ye J., Wu J. G., Du Q., Zheng X., Zhang Z., Shi C. H. (2006). The screening of mutants and construction of mutant population for cultivar “9311” in rice (Oryza sativa L.). Crop J. 32 1525–1529. [Google Scholar]
- Yoshida A., Suzaki T., Tanaka W., Hirano H. Y. (2009). The homeotic gene long sterile lemma (G1) specifies sterile lemma identity in the rice spikelet. Proc. Natl. Acad. Sci. U.S.A. 106 20103–20108. 10.1073/pnas.0907896106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H., Nagato Y. (2011). Flower development in rice. J. Exp. Bot. 62 4719–4730. 10.1093/jxb/err272 [DOI] [PubMed] [Google Scholar]
- Zhang J., Nallamilli B. R., Mujahid H., Peng Z. (2010). OsMADS6 plays an essential role in endosperm nutrient accumulation and is subject to epigenetic regulation in rice (Oryza sativa). Plant J. 64 604–617. 10.1111/j.1365-313X.2010.04354.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.