Abstract
From humble beginnings of a contaminated petri dish, β‐lactam antibiotics have distinguished themselves among some of the most powerful drugs in human history. The devastating effects of antibiotic resistance have nevertheless led to an “arms race” with disquieting prospects. The emergence of multidrug resistant bacteria threatens an ever‐dwindling antibiotic arsenal, calling for new discovery, rediscovery, and innovation in β‐lactam research. Here the current state of β‐lactam antibiotics from a structural perspective was reviewed.
Keywords: β‐lactam, β‐lactamase, penicillin‐binding protein, resistance, multidrug resistant bacteria
Introduction
In 1928, Alexander Fleming made the fortuitous observation that mold contaminating a petri dish was capable of destroying its surrounding bacteria. This was a monumental finding, setting the stage for the discovery of what has been termed “the miracle drug,” benzypenicillin (Fig. 1), as well as an ensuing plethora of β‐lactam based antibiotics. Nevertheless, in contrast to the golden age of antibiotic development culminating in the 1960s, recent decades have witnessed a continual decline in the discovery of novel antibacterial therapeutics.1 This void in discovery has been exacerbated by an accompanying surge of antibiotic resistance, leading to multidrug resistant (MDR) bacterial infections, such as those presented by ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.).2 This phenomenon is hardly surprising considering that bacteria have evolved along‐side antibiotics long before human discovery, and that the genetic exchange of resistance genes is inherent to bacterial survival.3 This is illustrated by the discovery of a bacterial penicillinase in 1940, several years before the introduction of penicillin into the clinic.4 Despite this resistance, β‐lactams nevertheless remain the most widely used class of antibiotics due to their ease of delivery, low toxicity, potent activity, and small cost that translates into wide availability. The unique targets of these drugs are the penicillin‐binding proteins (PBPs), enzymes that build and maintain the integrity of the bacterial cell wall. These enzymes have no human counterpart and prove lethal to the cell when compromised. It is, therefore, imperative that, despite the challenges, we continue to nurture the development of these powerful drugs. Here, we take a structural approach to address the modes of β‐lactam resistance and highlight the current progress and challenges that lay ahead in the design and development of these important antibacterials.
Figure 1.
The chemical structure of benzylpenicillin and various β‐lactamase inhibitors.
β‐Lactamases
A family of hydrolytic enzymes known as β‐lactamases collectively inactivates all current β‐lactams in clinical practice. Over half a century after their initial discovery, β‐lactamases are still recognized as the single most clinically prevalent mechanism of β‐lactam resistance amongst Gram‐negative pathogens. Our heavy reliance on the penicillins, cephalosporins, carbapenems, and β‐lactam‐inhibitor combinations has caused bacteria to rapidly evolve and exchange their repertoire of hydrolytic enzymes.5 β‐Lactamase genes can be chromosomally encoded under control of inducible promoters. However, the majority of β‐lactamase genes are located on readily transferable plasmids that often contain resistance genes for other antibacterial classes (e.g., aminoglycosides, fluoroquinolones, and tetracyclines). It is now a commonplace for a single bacterium to harbor up to eight distinct β‐lactamases, each tailored to hydrolyze a specific subset of β‐lactams.6 Of particular concern is the rapid global dissemination of Enterobacteriaceae harboring plasmid borne extended spectrum β‐lactamases (ESBLs) that confer resistance to the carbapenems, long heralded as the antibiotic of last resort. ESBLs include the emerging metallo‐β‐lactamases (MBLs) such as VIM, NDM, and IMP, and a subset of serine β‐lactamases (SBLs) including KPC, OXA, and GES.7, 8
Perhaps our greatest defense is to match the brilliant diversity of β‐lactamases with an ever‐growing arsenal of β‐lactam/β‐lactamase inhibitor combinations. However, due to the omnipresent threat of resistance, the pharmaceutical industry has become increasingly hesitant to develop new antibacterials.9 Therefore, the number of novel β‐lactams introduced into clinical practice is dwindling, a prospect that makes protecting our current pool of these compounds paramount.
β‐lactamases are commonly divided into four distinct classes (A–D), based on amino‐acid sequence identity. The class A, C, and D enzymes are SBLs, which evolved from the cellular target of the β‐lactams (the PBPs), and employ a catalytic serine during bond cleavage. SBL catalyzed β‐lactam hydrolysis proceeds via the formation and subsequent deacylation of a serine‐bound acyl enzyme intermediate.10 In contrast, the molecular class B enzymes are MBLs that utilize active site zinc ion(s) to coordinate a nucleophilic hydroxide to promote bond cleavage (mechanistic details reviewed in Ref. 11).
Despite having low overall sequence identity, the SBL classes share three active site amino acid sequence motifs in common: motif i (SXXK) contains the catalytic serine; motif ii (S/Y‐X‐N) is involved in protonation of the β‐lactam nitrogen leaving group; motif iii (K‐T/S‐G‐X) is involved in substrate stabilization and constitutes part of the oxyanion hole.12 The class A enzymes have a conserved E166 located in a region known as the Ω loop (residues 161–179) that is involved in activation of the hydrolytic water during deacylation.13, 14 In contrast, the class D (OXA) enzymes lack the aforementioned glutamic acid and instead have a unique N‐carboxylated lysine in motif i (i.e., the side chain N‐ε is reversibly modified with a CO2 group), which is thought to assume the role of general base during acylation/deacylation.15 The class C cephalosporinase enzymes have a tyrosine rather that serine in motif ii, which is thought to be involved in protonation of the β‐lactam nitrogen leaving group upon bond fission.16
To overcome SBL resistance, three β‐lactam based inhibitors have been introduced into clinical practice (sulbactam, clavulanic acid, and tazobactam). Sulbactam and tazobactam are penicillanic acid sulfones,17 while clavulanic acid is a clavam secondary metabolite from Streptomyces clavuligerus originally isolated in the early 1970s.18 These inhibitors form a long‐lived acyl‐enzyme intermediate with the catalytic serine, characterized by a very slow rate of hydrolytic deacylation (k3). Following acylation, a second ring‐opening event occurs leading to a stable imine that undergoes various chemical transformations. Eventually, the acyl‐enzyme hydrolytically deacylates either directly, or through a series of covalent intermediates to yield active enzyme and inactivated product.19 The β‐lactam based inhibitors were originally designed to target the molecular class A enzymes and are generally ineffective against strains harboring the emerging class C and D SBLs. Furthermore, there are now several class A enzymes that have evolved resistance to these β‐lactam based compounds, a prospect that makes the development of novel inhibitors paramount.19
The latest β‐lactam/β‐lactamase inhibitor combination to be approved for clinical use in humans by the U.S. Food and Drug Administration (FDA) was ceftazidime–avibactam in early 2015 for the treatment of complicated intra‐abdominal and urinary tract infections (IAI's, and UTI's). Additionally, there are four β‐lactam/inhibitor combinations in late stage clinical development; ceftaroline–avibactam, imipenem–cilastatin‐MK‐7655, ceftolozane–tazobactam, and meropenem‐RPX7009 (inhibitor chemical structures shown in Fig. 1).20, 21 Typically, new inhibitors are paired with either late generation cephalosporins or carbapenems in large part due to their broad‐spectrum activity and ease of protection when compared with their more hydrolytically susceptible penicillin counterparts.
Diazabicyclooctane SBL inhibitors
Avibactam and MK‐7655 are members of a novel non‐β‐lactam based class of inhibitors called diazabicyclooctanes (DBOs). These compounds contain a bridged bicyclic urea core and display potent inhibition of an unprecedented range of β‐lactamase targets that encompasses the class A, C, and a subset of class D SBLs.22, 23, 24 MK‐7655 is structurally very similar to avibactam with the addition of a piperidine ring attached to the C2 carboxamide (hereafter referred to as R1) (Fig. 1). The AztraZeneca and Activis combination therapies (avibactam–ceftaroline and avibactam–aztreonam) are currently in phase II and phase I clinical trials (http://www.clinicaltrials.gov). Merck has partnered MK‐7655 with imipenem and the human dehydropeptidase inhibitor “cilastatin” as part of a triple combination drug that is currently in phase II clinical trials for the treatment of cUTI's.20 The role of cilastatin is to prolong the half‐life of imipenem by inactivating human dehydropeptidase, which would otherwise readily degrade the carbapenem.
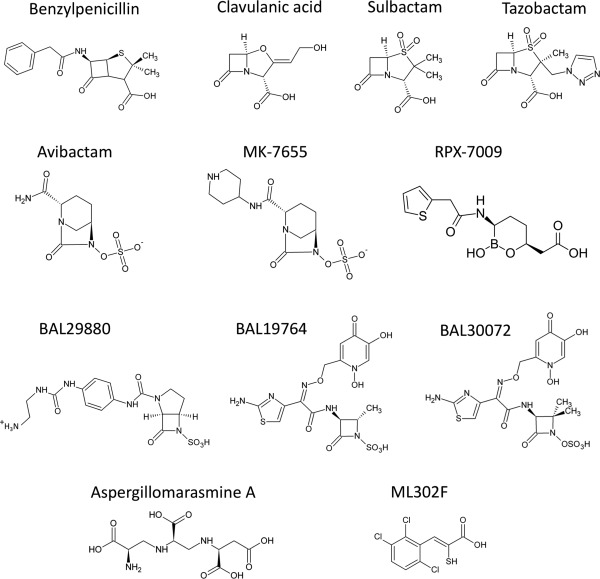
DBOs form a unique carbamyl linkage with the catalytic serine as opposed to the acyl‐enzyme observed for the β‐lactam based compounds [Fig. 2(A,B)].25 Importantly, avibactam does not decarbamylate via hydrolysis as is true for the β‐lactam based inhibitors, yet rather undergoes a reversible recyclization mechanism that re‐capitulates intact avibactam.26 Taken together, rapid carbamylation, slow recyclization and regeneration of intact avibactam during decarbamylation result in its unprecedented potency. Crystal structures of carbamyl‐avibactam bound to the class A, C, and D SBLs (CTX‐M15, AmpC, and OXA‐48) show that it acts as a substrate analog of the β‐lactam acyl enzyme [Fig. 2(A)].25, 27 The authors propose that upon carbamylation by the motif i serine, the C7‐N6 bond is cleaved liberating the N6 nitrogen which may be protonated by the motif i S/Y in an analogous fashion to the β‐lactam amide nitrogen. Similar to the analogous β‐lactam carboxylate, the avibactam N6 sulfate occupies an electropositive pocket formed in proximity to motif iii. Notably, the water involved in deacylation is present in the CTX‐M‐15‐avibactam carbamyl enzyme crystal structure suggesting that the inherent stability of the carbamyl bond and/or protonation of the E166 general base likely contribute to avibactam's avoidance of hydrolytic deacylation.25 Recyclization is proposed to be catalyzed by deprotonation of the N6 nitrogen by the motif ii S/Y resulting in an intramolecular decarbamylation and subsequent release from the catalytic serine.25 Future development of the DBOs should focus on extending their spectrum of activity to include a broader range of class D and B β‐lactamases.
Figure 2.
SBL inhibitors. (A) CTX‐M‐15 inhibition by avibactam. In the left panel is a general reaction scheme for the formation of an avibactam carbamyl‐enzyme intermediate. In the right panel is an active site close‐up of the carbamyl avibactam‐CTX‐M‐15 complex (PDB ID: 4HBU). The CTX‐M‐15 protein backbone is illustrated in green cartoon representation and the bound avibactam is shown as pink sticks. (B) Bacillus licheniformis BS3 inhibition by clavulanic acid. A general reaction scheme for clavulanic acid mediated SBL acylation is shown in the left panel. In the right panel is an active site close‐up of the acyl clavulanic acid‐BS3 complex (PDB ID: 2Y91). The BS3 protein backbone is displayed as a beige cartoon and clavulanic acid is represented as pink sticks. (C) Toho‐1 inhibition by the boronic acid based inhibitor BZB. In the left panel is a general reaction scheme for the formation of the SBL bound tetrahedral boron adduct. In the right panel is an active site close‐up of the tetrahedral BZB boronate adduct bound to the Toho‐1 catalytic serine (PDB ID: 4BD0). The Toho‐1 protein chain and bound BZB are displayed as an orange cartoon and pink sticks. In A–C, key active site residues are displayed in stick representation with all non‐carbon atoms colored by type (N; blue, O; red, S; yellow, B; beige). Hydrogen bonding interactions are represented as black dashes.
Boronic acid SBL inhibitors
Boron contains an empty p‐shell making it an excellent electrophile with a high propensity to form dative covalent bonds with active site serine nucleophiles (i.e., both electrons in the covalent bond originate from oxygen).28, 29 Since their initial discovery in the late 1970s, the boronates have proven to be effective SBL inhibitors in vitro. The development of SBL inhibitory boronates has largely focused on molecular mimicry with clinically approved β‐lactams to confer affinity and specificity.31 A novel heterocyclic boronate inhibitor RPX7009 is being developed by Rempex pharmaceuticals and displays strong potentiation of the carbapenem antibiotic biapenem against class A carbapenemase producing Enterobacteriaceae.21 RPX7009 contains a thiophene moiety in an analogous position to the R1 group of the nitrocefin and cefoxitin cephalosporins (Fig. 1). Currently, RPX7009 is in phase III clinical trials in combination with meropenem (carbavance) for the treatment of UTIs or acute pyelonephritis (http://www.clinicaltrials.gov). A major challenge for the future will be the design of boronic acid inhibitors that target the class B and D enzymes for which they are currently ineffective.
The boronates function as transition state analogues that form a reversible non‐hydrolyzable bond with the catalytic serine O‐γ, mimicking the sp3 hybridized anionic tetrahedral intermediate(s) formed during β‐lactam hydrolysis [Fig. 2(C)]. The anionic tetrahedral intermediate is stabilized by the oxyanion hole of the enzyme, which includes the backbone amides from the motif i serine and motif iii x. The boron atom is analogous to the β‐lactam electrophilic C7 carbon. One oxygen atom in the tetrahedral boron adduct occupies the oxyanion hole in a similar fashion to the β‐lactam acyl carbonyl oxygen in all boronate bound structures. However, the position of the second boronate oxygen varies between inhibitor bound structures. In some complexes, the second oxygen hydrogen bonds to the motif ii S/Y in an analogous fashion to the acylated β‐lactam N4 nitrogen and thereby mimics the acylation transition state [Fig. 2(C)].10, 30 In other structures, the oxygen displaces the catalytic water in proximity to the Ω loop, resulting in a mimic of the deacylation tetrahedral intermediate.31 Therefore, the boronic acid based inhibitors have provided insight into the transiently formed tetrahedral transition states during both β‐lactam acylation and deacylation. A major consideration in development of the boronic acid SBL inhibitors is their propensity for off‐target inhibition of serine proteases and the proteasome, such that structure‐guided functionalization conferring target specificity is of prime importance in their continued development.32
Overcoming MBL and class D SBL mediated resistance
There is a troubling paucity of inhibitors active against the rapidly emerging molecular class B and D β‐lactamases, creating an ominous unmet medical need. A current strategy to counteract MBL‐positive pathogens is to utilize monobactams, which are the only β‐lactam subclass that are immune to MBL catalyzed hydrolysis. Basilea Pharmaceutica is currently developing a triple combination antibacterial cocktail (BAL30376) containing the monobactam AmpC inhibitor BAL29880, monobactam‐siderophore BAL19764 and the clinically approved class A β‐lactamase inhibitor clavulanic acid (Fig. 1). The strategy is that BAL29880 and clavulanic acid will inactivate the class C and A enzymes, leaving the MBL stable BAL19764 to inhibit its PBP targets.33 Basilea is also developing a new siderophore monosulfactam “BAL30072” (Fig. 1), which has potent activity against carbapenemase carrying strains of Gram‐negative bacilli including Acinetobacter spp., Pseudomonas aeruginosa, Burkholderia cepacia, and a pan MDR Enterobacteriaceae34 (siderophore‐conjugated β‐lactams are discussed in further detail below). Furthermore, AztraZeneca has recently entered the aztreonam–avibactam combination therapy into phase I clinical trials, a cocktail that is impervious to the hydrolytic activity of the vast majority of β‐lactamases, with a notable exception being a subset of class D enzymes.35 A promising preclinical MBL inhibitor candidate is the fungal natural product aspergillomarasmine A (AMA) (Fig. 1), which is a potent inhibitor of the VIM and NDM MBLs, presumably via chelation of the catalytically essential active site zinc.36 AMA fully restored the antimicrobial activity of meropenem against VIM or NDM‐1 carrying clinical isolates of Enterobacteriaceae, Pseudomonas spp., and Acinetobacter spp. Most notably, however, is that in mice infected with NDM‐1 positive Klebsiella pneumoniae, AMA was able to effectively restore meropenem activity.36, 37 Recently, Brem and colleagues characterized rhodanine hydrolysis products (e.g., ML302F, Fig. 1) as potent inhibitors of purified MBLs and showed they complex with the active site zinc ions via thioenolate mediated zinc intercalation.38
The class D OXA SBLs, were originally named for their ability to hydrolyze oxacillin and are a diverse group of enzymes with substrate hydrolysis profiles spanning from narrow to broad. At present, the clinically available SBL inhibitors are ineffective against the class D enzymes. However, promising data is emerging with respect to certain penicillin sulfones and thio‐phenyl oxime phosphonates that display potent inhibition of OXA‐24/OXA‐40 and demonstrate efficacy at potentiating carbapenems against OXA carrying Acinetobacter baumannii.37, 39 Also of note is the encouraging ability of avibactam to inhibit certain class D enzymes. The observed variability is predominantly due to discrepancies in carbamylation rather than decarbamylation rates, an attribute that should be considered in future DBO drug design efforts.23
Modified Penicillin‐Binding Proteins
A common mechanism by which bacteria evade the onslaught of β‐lactams is through the acquisition of modified targets with reduced susceptibility (the peptidase domains of cellular PBPs involved in PG synthesis). Modified PBPs typically arise via gene mutations that alter the intended target in the presence of β‐lactam induced selective pressure, or by acquisition of resistant PBPs by horizontal gene transfer.40 The modified PBP must have reduced susceptibility to β‐lactams, yet still retain cellular function. Today, some of the most prominent nosocomial MDR bacterial infections owe their resistance to modified low‐affinity PBPs [e.g., methicillin‐resistant Staphylococcus aureus (MRSA) and Enterococcus faecium].41, 42
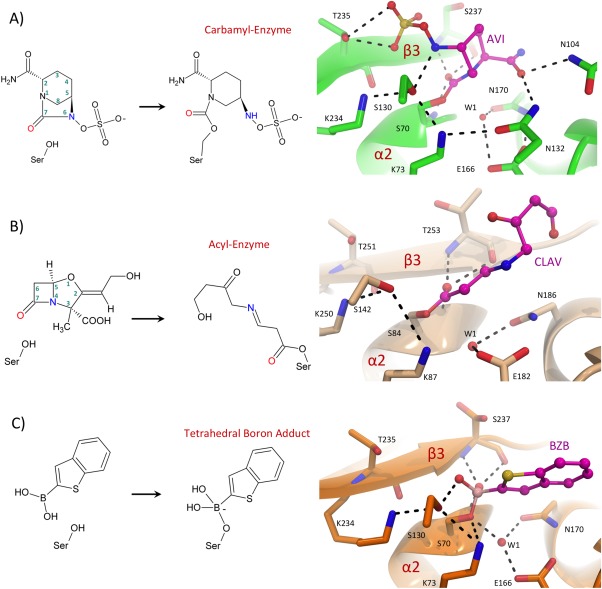
The typically C‐terminal, solvated penicillin‐binding domains of the array of cellular PBPs act as transpeptidases (TPases), carboxypeptidases (CPases), or endopeptidases (EPases) involved in peptidoglycan (PG) metabolism.43 Each bacterial species typically has several PBPs with unique roles during cell growth, division, rest, and infection.44 The TPases (typically membrane anchored with monofunctional or bifunctional variants coupled to glycosyl transfer and glycopolymerization of PG), catalyze the cross‐linking of adjacent peptide repeats to fortify the PG sacculus. The CPases remove the terminal D‐ala residue from the stem peptide via hydrolysis and thus, regulate the pool of “donor” peptides. The EPases hydrolyze the peptide bond connecting adjacent glycan strands and thereby regulate the degree of PG cross‐links.43 Somewhat analogous to the β‐lactamases, these domains harbor three highly conserved active site sequence motifs i–iii: SXXK, SXN, and KTGT/S. In brief, the catalytic serine nucleophile of motif i attacks the penultimate D‐ala–D‐ala peptide bond of the donor peptide, resulting in the formation of an acyl‐enzyme intermediate with concomitant release of the terminal D‐ala. In the CPases, this intermediate is hydrolyzed to liberate the terminal amino acid. In the TPases, the acyl‐enzyme intermediate undergoes attack from a primary amine nucleophile located on a separate “acceptor” stem peptide, thereby forming a continuous link between adjacent GlcNAc–MurNAc glycan backbones.43 The β‐lactams act as substrate analogues that mimic the terminal D‐ala–D‐ala on the donor stem peptide. The following kinetic model represents the inactivation of PBPs by β‐lactams. Firstly, a pre‐catalytic Michaelis complex (EI) is formed between the PBP (E) and β‐lactam (I), the affinity of which is represented by the dissociation constant (K D). Subsequently, acylation proceeds to form the long‐lived acyl enzyme complex (EI*) with the rate (k 2). The complex EI* is slowly hydrolyzed at the rate (k 3), which is generally negligible on the scale of bacterial generation time to regenerate E and hydrolytically inactivated product (P) [Fig. 3(A)].45
Figure 3.
Inhibition of the S. aureus PBP2a by Ceftobiprole. (A) Kinetic model for PBP inhibition by β‐lactams. (B) General model for SBL mediated ceftobiprole acylation. (C) Active site close‐up of acyl‐ceftobiprole bound S. aureus PBP2a (PDB ID: 4DKI). The acyl‐enzyme protein chain is displayed as a teal cartoon with key active site residues shown in stick representation with atoms colored by type. (D) Active site overlay of unbound (PDB ID: 1VQQ) and ceftobiprole bound S. aureus PBP2a. The bound and unbound protein backbones are shown as teal and yellow cartoons with key active site residues illustrated in stick representation with non‐carbon atoms colored according to atom type. In C and D, the bound ceftobiprole is shown in pink sticks with atoms colored by type and hydrogen‐bonding interactions are depicted as black dashes.
MRSA
Early reports of penicillin‐resistant Staphylococcus aureus in the mid‐1940s provoked the development of a new generation of penicillins (methicillin and oxacillin) that were effective against these early resistant strains. However, in the early 1960s, MRSA strains were identified with complete resistance to all β‐lactams in clinical practice.41 Today, MRSA remains a major global health problem, with 278,000 hospitalizations in the United States in 2005 alone.46 This resistance is attributed to the mecA gene product (PBP2a), which is a class B D,D‐transpeptidase containing an approximately 100 residue N‐terminal extension domain, and that has remarkably low reactivity toward β‐lactams.47 Expression of mecA is induced in the presence of β‐lactams by the mec pathway (or in its absence by the analogous bla pathway48, 49), which consists of a transmembrane sensor/transducer protein located in the cytoplasmic membrane (MecR1) that binds β‐lactam, resulting in de‐repression of the mecA gene by cleavage of the mecI repressor (reviewed in Ref. 50). Staphylococcus aureus has four PBPs that are expressed under normal growth conditions in the absence of β‐lactam (PBP1‐4). In the presence of β‐lactams, the four native PBPs become irreversibly inactivated, and the resistant PBP2a is responsible for all cellular PG transpeptidation. Importantly, the expression level of the native PBPs is unaltered under these conditions, and the GTase activity of the β‐lactam bound bi‐functional PBP2 is required for PG synthesis.45 Furthermore, there are greater than 30 other accessory genes that are essential for the methicillin resistance phenotype in MRSA, many of which are involved in cell wall metabolism (e.g., aux and fem genes).51 Interestingly, glycosylated wall teichoic acids are also required for the methicillin resistance phenotype via an unresolved mechanism.52
A kinetic comparison of PBP2a with the β‐lactam sensitive R61 PBP and R61 PBP2x reveals that the β‐lactam resistance of PBP2a is predominantly due to a slow acylation rate (k 2 is 3 orders of magnitude lower than susceptible PBPs) rather than low affinity for the Michaelis‐complex or fast hydrolysis of the acyl‐enzyme (i.e., the PBP2a K D and k 3 values are similar to non‐resistant PBPs).53, 54 A structural comparison of the penicillin susceptible PBP2, with PBP2a from MRSA reveals a distorted active site cleft with conformational changes at the β3 strand and α2 helix, which contains the nucleophilic motif i S403. This rearrangement, while still promoting the initial binding event, provides a misalignment of the lactam‐peptide bond with respect to the catalytic serine. A twisting of the β3 strand with a concomitant conformational change in the α2 helix are therefore required for acylation, a process that presumably provides an energetic penalty resulting in significantly reduced acylation rates.47
Much effort has been put forth to develop novel anti‐MRSA β‐lactams. Ceftobiprole is the active component from the parenteral pro‐drug (ceftobiprole medocaril), a late generation cephalosporin that has activity against MRSA via inhibition of PBP2a. Ceftobiprole contains an oxyimino aminothiadiazolyl (R1 substituent), and a vinylpyrrolidine moiety (R2 group) linked at the 7‐amino and C3 carbon to the cephalosporin dihydro‐thiazine nucleus [Fig. 3(B)].55 In vitro, the activity of ceftobiprole encompasses a broad range of Gram‐negative pathogens including P. aeruginosa, and Gram‐positive pathogens including MRSA, E. faecalis, S. pneumoniae, and methicillin resistant Staphylococcus epidermidis (MRSE).56 Ceftobiprole is impervious to β‐lactamase resistance for many class A and C SBLs,57 and its broad‐spectrum activity makes it a viable treatment option for MRSA among other prominent pathogens.
Kinetically, the effectiveness of ceftobiprole against S. aureus PBP2a is attributed to a combination of a lower K D, increased k 2 and a decreased k 3 when compared with susceptible PBPs.58 The 2.9 Å resolution crystal structure of acyl ceftobiprole bound S. aureus PBP2a reveals that the compound behaves like a traditional β‐lactam in that it is irreversibly acylated by the catalytic S403 [Fig. 3(C)].59 Furthermore, a similar displacement of the β3 strand and the N‐terminal portion of the α2 helix are observed in the ceftobiprole bound structure as was seen previously in other β‐lactam bound PBP2a acyl‐enzyme complexes [Fig. 3(D)].47 The ceftobiprole R1 group occupies two distinct conformations (A and B), within the same enzyme active site [Fig. 3(C,D)]. Both conformations of the oxyimino aminothiadiazolyl side chain had been previously observed in studies involving the similar R1 group of cefotaxime, but never before in the same structure.59 The observed conformational variability likely presents an entropic gain for the bound ceftobiprole, a common affect observed for ligands that bind in multiple conformations.60 Importantly, the ceftobiprole R2 group is sandwiched in a hydrophobic pocket adjacent to the active site, and the R2 pyrrolidine rings form favorable pi–pi interactions with M641, T600, and Y446 [Fig. 3(C)]. It is thought that the extended, planar character of the ceftobiprole R2 group provides the shape complementarity required for binding the narrow PBP2a active site cleft. Furthermore, the non‐specific hydrophobic nature of its interactions within the active site may permit the conformational plasticity required to facilitate the necessary rearrangements during acylation.59 Anti‐MRSA β‐lactams are typically cephalosporins like ceftobiprole, an observation that is thought to be due to increased van der Waals contacts with the R2 of cephalosporins, a functionality that is absent in other β‐lactam classes.61 The motif iii residue T600 (located on strand β3), forms part of the oxyanion hole and interacts with the ceftobiprole R2 functional group. Therefore, this residue seems important to link structural rearrangements in the strand β3 with acylation. However, the lack of a Michaelis complex makes it difficult to decipher the exact role of R1 and R2 during the conformational rearrangements leading to acylation.
A S. aureus PBP2a allosteric binding site has been inferred from kinetic studies,50, 62 and was recently supported by X‐ray crystallography to be an 60 Å from the PBP2a TPase active site.63 The allosteric site is located at the intersection of Lobe 1 (residues 166–240), Lobe 2 (residues 258–277), Lobe 3 (residues 364–390), and the N‐terminal extension domain (residues 27–138). Crystallographic data shows that when the allosteric site is occupied by a PG fragment, a multiresidue conformational change culminates in the “opening” of the active site, presumably to permit substrate entry.63
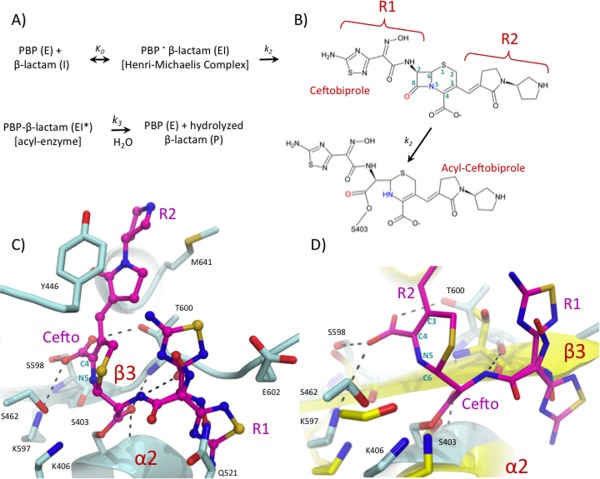
Ceftaroline, the active metabolite of the phosphono prodrug (fosfamil) and late generation parenteral cephalosporin, received approval from the FDA for the treatment of acute skin structure infections and community acquired pneumonia [Fig. 4(A)].64 Ceftaroline has antimicrobial activity against MDR Gram‐positive bacteria including S. aureus and S. pneumoniae and has up to 256‐fold higher affinity for PBP2a than traditional cephalosporins.65 Interestingly, unhydrolyzed ceftaroline was found to mimic the allosteric activation of PBP2a by PG and promote acylation by a second ceftaroline molecule in the active site [Fig. 4(B)].63 Allosteric regulation of PBPs is unprecedented, and it will be interesting to see if related enzymes such as PBP5fm from MDR Enterococcus faecium contain an analogous allosteric site. This work has illuminated the potential for design of allosteric activators as an approach to potentiate the activity of pre‐existing β‐lactams and has shed light on the molecular basis of ceftaroline‐mediated inhibition.
Figure 4.
Inhibition of the S. aureus PBP2a by ceftaroline. (A) General model for ceftaroline mediated PBP acylation. (B) Structural details of ceftaroline mediated PBP2a inhibition. On the left, the overall ceftaroline bound PBP2a protein structure (PDB ID: 3ZG0) is shown in surface representation with the N‐terminal extension, allosteric domain, and TPase domain colored green, yellow, and cyan. On the right side are close‐up views of the ceftaroline bound allosteric and TPase domains with the protein chain depicted in cartoon representation and key residues shown as sticks with non‐carbon atoms colored by atom type. Ceftaroline is shown as pink sticks.
Future prospects for anti MRSA drugs include the late generation carbapenem razupenem, which has high affinity for MRSA PBP2a as well as pbp5R from ampicillin resistant E. faecium.66 The C‐2 thiazole side chain of razupenem is likely responsible for the observed inhibition of these low‐affinity PBPs. A novel class of non‐β‐lactam based PBP2a inhibitors (oxadiazoles) have displayed excellent bactericidal antibiotic properties against MRSA in vivo.67 However, the structural basis for inhibition, and whether or not these compounds bind the PBP2a allosteric site awaits elucidation. Additionally, vancomycin, a glycopeptide used in first‐line treatment of MRSA infection, has shown synergistic activity against vancomycin resistant S. aureus (VRSA) when co‐administered with β‐lactams such as oxacillin. Vancomycin inhibits the synthesis of PG by binding to the D‐ala–D‐ala terminal residues of PG precursors. Susceptible strains can develop resistance by substituting these terminal residues with D‐ala–D‐lac, which is no longer recognized by vancomycin. PBP2a, however, does not recognize this new precursor such that co‐administration of the two drugs can be used to treat affected patients.68, 69 TD‐1792, a hybrid glycopeptide–cephalosporin antibiotic developed by Theravance, has completed a phase II clinical trial for the treatment of complicated Gram‐positive skin and skin structure infections.70 This compound is highly potent against MRSA and VRSA in that it simultaneously inhibits the activity of PBPs and interferes with PG precursor binding.71
Other examples
The genus Enterococci includes some of the most therapeutically challenging opportunistic nosocomial MDR pathogens to date.42 Penicillins such as ampicillin, either alone or in combination with aminoglycosides were the mainstay for treatment of enterococcal infections for more than half a century. Enterococci have an intrinsically low susceptibility to β‐lactams in general, and today ampicillin resistance occurs in approximately 90% of E. faecium clinical isolates.42 The observed resistance is largely due to the presence of a high‐molecular‐weight (HMW) class B PBP5 variant (encoded by pbp5R), which assumes the transpeptidase activity of all other susceptible TPases in the presence of β‐lactam (analogous to PBP2a of MRSA).42 The PBP5 resistant variant belongs to the same subgroup of class B PBPs as the staphylococcal PBP2a.42, 45 In the laboratory, resistant PBP5 is readily disseminated via horizontal gene transfer between microbial populations and in some cases, along with a transposon (Tn5382) that carries vancomycin resistance genes.72 The crystal structure of the penicillin acylated form of resistant PBP5 from E. faecium was solved to 2.4 Å resolution.73 The apo structure however has not yet been solved such that it remains to be seen whether active site rearrangements are an important feature governing acylation. Ceftobiprole shows great potential for the treatment of E. faecium infections and was found to efficiently acylate resistant PBP5.74
Of the six PBPs from Streptococcus pneumoniae PBP1a, PBP2b, PBP2x, and sometimes PBP2a were found to contain resistance mutations in clinical isolates. PBP1a, PBP2b, and PBP2x were found to be mosaic genes in the majority of resistant isolates.75, 76 Mosaic genes are the product of recombination events between different alleles within a single species, or orthologous genes from related species. The M339F and T338A double mutant for the R6 PBP2x significantly reduces the penicillin acylation rate, and the crystal structure of this double mutant was solved to 2.4 Å resolution (PDB ID: 1PYY).77 The motif i catalytic S337 faces away from the active site center and is hydrogen bonded to the main chain nitrogen of T550 rather than K340, and this shift likely results in reduced acylation due to conformational rearrangement required to produce a catalytically competent state. Also, the mutant Q552E crystal structure was solved to 3 Å and reveals that the β3 strand is shifted approximately 0.5 Å closer to the catalytic serine than in the native enzyme.77 This shift narrows the active site cleft and is reminiscent of the closed conformation of PBP2a from S. aureus.
Resistant PBPs have also been found in Neisseria meningitides and Neisseria gonorrhea; (PBP2), Haemophilus influenzae (PBP3), Helicobacter pylori (PBP1a), and numerous other examples.78 Recently, antimicrobial resistance to ceftazidime in Burkholderia pseudomallei has been correlated with a deletion of PBP3, which is the primary target of ceftazidime in other Gram‐negative bacilli including P. aeruginosa and E. coli.79 Furthermore, the growing identification of microbes harboring both β‐lactamases and low affinity PBPs presents a double resistance threat that is particularly worrisome.
Barriers to β‐Lactam Permeability
Due to the accessibility of their periplasmic PBP targets, β‐lactams are among the few antibiotics that are effective against both Gram‐positive and Gram‐negative bacteria. The cell‐wall architecture of Gram‐negatives, however, presents unique challenges for drug entry even for β‐lactams, and is key to the notorious intrinsic resistance of strains such as P. aeruginosa and A. baumannii. Indeed, Gram‐negative bacteria can modify the permeability of their cell walls to effectively limit intracellular antibiotic concentrations. This is commonly achieved by restricting the entry of antibiotics via loss or downregulation of aqueous outer membrane channels known as porins, as well as by the active expulsion of drugs from the cell through efflux pumps.
Porins
β‐lactams are hydrophilic compounds that along with tetracyclines, chloramphenicols, and fluoroquinolones, largely enter Gram‐negative cells using porins. These aqueous channels are the most abundant proteins in the outer membrane of bacteria and act as molecular sieves, allowing only the entry of hydrophilic molecules below a specific exclusion limit, as determined by the channel diameter. According to high‐resolution structures, porins share high structural similarity with small variations in loop topology and surface charge, and generally exist as trimers (Fig. 5). Porins are recognized by their β‐barrel architectures where antiparallel β‐strands are aligned such that alternating hydrophobic and hydrophilic residues respectively line the membrane and water exposed surfaces. These channels are classified into the substrate specific porins that generally consist of 18 β‐strands, and the typically less substrate stringent general porins consisting of 16 β‐strands (reviewed in Ref. 80). It is the general porins that are largely involved in resistance mechanisms, where the degree of conferred resistance depends on the number and type of porins possessed by the bacteria.80 In fact, bacteria often rely on adaptive or mutational changes to their repertoire of porins as their first line of defense against antibiotics. Since porins act not only as selective outer membrane barriers but also facilitate the uptake of nutrients, their expression is organized by responsive regulatory elements that can be coordinated with stress pathways.81 The bacterial ability to alter its surface permeability in response to its environment therefore carries a great selective advantage, and one that is readily used in response to antibiotics. For instance, β‐lactam resistance in E. coli can involve the loss of OmpF82 or mutations in OmpC around the point of pore constriction.83 A further example is seen in P. aeruginosa carbapenem resistant isolates where the facilitating porin, OprD, is lost due to various disruptions including deletion, nonsense mutations, gene insertion, or decreased transcription due to mutation in regulatory elements.84, 85, 86 Although resistance is often accompanied by a fitness cost, a recent study has challenged this paradigm by demonstrating the association of OprD loss and concurrent carbapenem resistance with enhanced in vivo fitness and virulence. Using a P. aeruginosa transposon insertion library, the authors identified an enrichment of disrupted OprD strains with enhanced mucosal colonization and spleen dissemination.87 Although accompanying phenotypes such as increased survival against host serum‐mediated killing, immune response, and acidification were observed, the role of OprD in these complex processes remains unclear.
Figure 5.
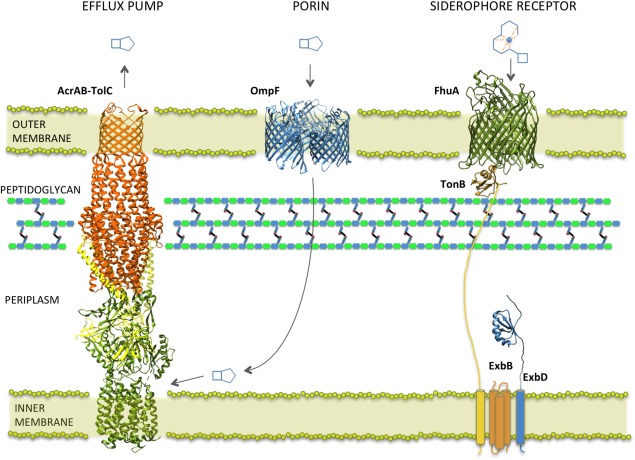
Representative β‐lactam uptake and efflux systems in Gram‐negative E. coli. β‐Lactams enter the periplasm through outer membrane porins such as OmpF (PDB ID 2ZFG), where they inhibit periplasmic PBP targets. Dianionic β‐lactams such as carbenicillin can be expelled from the cell via RND efflux pumps, represented by the AcrAB‐TolC complex (PDB ID 2F1M, 1OYE, 1EK9), where the drug is captured from the periplasmic or periplasmic‐cytoplasmic interface. Siderophore conjugated β‐lactam drugs can use cognate siderophore receptors for entry as represented here by FhuA complexed with TonB/ExbB/ExbD (FhuA and C‐terminus of TonB: PDB ID 2GRX; ExbD: PDB ID 2PFU). The TonB complex couples the proton motive force across the inner membrane to facilitate the active transport mechanism of FhuA across the outer membrane.
Siderophore conjugated β‐lactams
As adaptive and mutational changes that affect porins can substantially limit the diffusion rate of antibiotics, efforts have been made to explore other methods of drug delivery. One such avenue receiving much attention is the exploitation of the bacterial iron uptake system. Iron is an essential nutrient for microorganisms, and although abundant in nature, it has a fairly low bioavailability under aerobic conditions. Due to the low solubility and toxicity of free ferric ions, the human iron transport protein, transferrin, maintains the free ferric ion serum concentration at approximately 10−24 M, whereas a bacterial cell typically requires 105 to 106 ferric ions per generation to maintain a required internal concentration of 10−6 M (reviewed in Ref. 88). As a result, bacteria have evolved aggressive and efficient iron acquisition systems involving the secretion of small iron chelating siderophores, whose main structural forms include the catecholates, hydroxamates, and citrate based polycarboxylates (reviewed in Refs. 89, 90). In Gram‐negative bacteria, these siderophores are taken up by high affinity outer‐membrane receptors that are often specific for individual siderophores (e.g., FepA, Cir, and Fiu catecholate receptors).91 A universal feature required for all siderophore uptake systems is the TonB complex (consisting of TonB, ExbB, and ExbD) that couples the proton motive force across the cytoplasmic membrane to facilitate receptor‐mediated active transport of substrates across the outer membrane92 (Fig. 5). Siderophore receptors typically consist of a 22‐stranded transmembrane β‐barrel that encloses a globular plug domain, where ligand‐binding sites are formed by residues on the extracellular face of the plug as well as on extracellular loops and barrel walls. The periplasmic‐facing N‐terminus of the plug domain contains a “TonB box” motif that interacts (via β‐strand pairing) with the periplasmic C‐terminal domain of TonB93 (Fig. 5). It is proposed that upon siderophore binding, the plug domain of the receptor undergoes a conformational change that allows the transport of its substrate, which is supported by observations of high‐level solvation and loose packing of the plug inside the β‐barrel.94
Along with the advantage of iron scavenging, antibacterial compounds produced by other competing bacteria can also exploit siderophore receptors. These include siderophore‐conjugated antibiotics such as the Actinomyces subtropicus albomycin,95 the Streptomyces griseoflavus ferrimycin,96 and the Streptomyces violaceus salmycins.97 These “Trojan Horse” strategies were adopted by scientists in the 1980s using natural siderophores or synthetic analogue conjugates of nucleosides, glycopeptides, macrolides, fluroquinolones, and most importantly, β‐lactams. Early studies on non‐ β‐lactam siderophore conjugates for the most part resulted in compounds with lower activities than the parent antibiotic, often due to problems with solubility, receptor uptake, or possibly, lack of release of the antibiotic from its siderophore partner upon entry (as needed for proper function) (reviewed in Ref. 98). The intensely investigated β‐lactam based siderophore conjugates have had somewhat better success due to certain inherent advantages. Firstly, the PBP targets of these drugs reside in the periplasm such that only the outer‐membrane needs to be crossed for drug delivery. Secondly, the β‐lactam conjugates can become active as a whole without having to be cleaved from the siderophore since the site of siderophore linkage is remote from the β‐lactam active site.89 A number of catechol and mixed‐ligand‐catechol conjugated β‐lactams have been developed with enhanced activity (compared with antibiotics considered as “gold standards” in treatment) against problematic Gram‐negative bacteria such as P. aeruginosa (reviewed in Refs. 89 and 99). Nevertheless, activity is often drastically decreased in mutants lacking functional TonB or siderophore receptors such as Cir and Fiu. In this way, resistance may be readily acquired by loss of components in siderophore transport systems, as is often observed with P. aeruginosa (reviewed in Ref. 98). Recently, Basilea Pharmaceutica has investigated a series of siderophore monosulfactams, leading to the promising BAL30072 that is currently in phase I clinical development.100 BAL30072 is a monobactam derivative with reduced susceptibility to inactivation by various β‐lactamases, conjugated to a hydroxypyridone moiety that allows easy uptake through the siderophore transport system.34, 101 Resistance to BAL30072 was found to evolve relatively slowly compared with preceding compounds, with infrequent mutations in TonB and siderophore receptors.34 BAL30072 furthermore displayed in vitro antimicrobial activity against a range of Gram‐negative bacteria, including strains of MDR P. aeruginosa, A. baumannii, and Burkholderia pseudomallei.34, 102, 103, 104 In addition, in vitro and in vivo combinations of BAL30072 and carbapenems resulted in additive and synergistic antimicrobial activity, particularly against Enterobacteriaceae and P. aeruginosa.101
Efflux pumps
In Gram‐negative bacteria, efflux pumps are major contributors to antibiotic resistance, in that they form a near impenetrable barrier against antimicrobials and are largely responsible for MDR phenotypes. Furthermore, in limiting the periplasmic and cytoplasmic concentration of nearly all antimicrobial classes, they encourage the acquisition of additional resistance mechanisms, including the alteration of intended antimicrobial targets such as the modified PBPs, as well as production of drug inactivating enzymes such as the β‐lactamases (reviewed in Refs. 105 and 106). In Gram‐negative bacteria, the MDR phenotype is largely conferred by the resistance‐nodulation‐cell division superfamily (RND). RND efflux complexes are assembled as tripartite membrane machineries consisting of a plasma membrane‐located RND pump, an adaptor unit of the membrane fusion protein (MFP) family, and an outer membrane channel belonging to the outer membrane factor (OMF) family107 (Fig. 5). The resulting complex is capable of expelling substrates across the entire cellular envelope, and can possibly capture substrates from the periplasm or from either the periplasmic or cytoplasmic face of the inner membrane, as demonstrated with dianionic β‐lactams such as carbenicillin.107, 108 Despite lacking the structure of the native intact tripartite complex, high resolution structures of each of the individual components are available along with proposed composite models.109, 110 Furthermore, recent efforts by Du et al. have yielded a pseudo‐atomic structure of a fully assembled AcrAB‐TolC MDR efflux pump, using a clever approach involving protein fusion.111 The features summarized here stem from a wealth of recently reviewed structural data.112, 113 The RND pump typically exists as a trimer composed of a total of 12 transmembrane helices and two larger periplasmic loops.114 This unit is believed to function in a rotatory manner based on the alternate protonation of individual subunits, resulting in successive substrate capture and release.115, 116, 117 The MFP adaptor protein, consisting of an extended β‐barrel connected to a long periplasmic α‐helical hairpin by a lipoyl domain, is proposed to stabilize weak interactions between the RND and OMF.118, 119 The outer membrane OMF channel, consisting of a 12 stranded β‐barrel trimeric arrangement that extends into the periplasm via a long (∼100 Å) coiled‐coil α‐helical domain, is believed to open and close at the base by an iris‐like mechanism.120, 121 As affinity tends to rely on physiochemical characteristics rather than a particular structural chemistry, RND pumps can capture and expel a great variety of structurally diverse compounds, as evident in the frequent MDR phenotypes of their carrier strains.122, 123 In P. aeruginosa, for example, the MexAB‐OprM efflux system, combined with low outer membrane permeability, contributes to increased resistance to penicillins and cephalosporins and is largely responsible for the organisms notorious intrinsic resistance.124 Likewise, in A. baumannii, the AdeABC efflux system contributes to high‐level resistance to the majority of β‐lactams including carbapenems.125 β‐lactam uptake and susceptibility is also diminished by AcrAB‐TolC efflux in H. influenzae 126 and K. pneumoniae.127 Consequently, many efforts have been made to reverse or inhibit the activity of efflux pumps in order to restore susceptibility and increase the intracellular concentration of existing antibiotics in these pathogens. Although there are currently no efflux pump inhibitors (EPIs) that have been approved for clinical purposes, several compounds show promise (reviewed in Refs. 128 and 129). Phenyl‐arginine‐β‐naphthylamide (PAβN) (MC‐207) was the first identified EPI to exhibit relatively broad inhibitory activity and, in combination with fluoroquinolones, is effective against efflux‐mediated resistance in several MDR Gram‐negative strains including P. aeruginosa,130 K. pneumoniae,131 and E. coli.132 PAβN derivatives are also being explored, an example being MC‐04, a compound that displays decreased toxicity, increased stability, and greater activity against P. aeruginosa strains that overexpress efflux pumps, as compared with its parent compound.133 Other inhibitors such as the aryl‐piperazines have also been shown to increase intracellular concentrations of commonly used antibiotics and to reverse MDR phenotypes in a number of ESKAPE pathogens that over‐express RND efflux pumps.134, 135, 136 However, to date, few EPIs have shown synergy with β‐lactam based antibiotics,137 likely due to the large contribution of other resistance mechanisms such as the β‐lactamases. Nevertheless, a recent study has shown that artesunate, a derivate of artemisinin isolated from the plant Artemisia annua and used to treat malaria, enhances the activity of β‐lactams against E. coli by inhibiting the MDR efflux pump, AcrAB‐TolC.138 In addition, PAβN, in combination with sub‐inhibitory concentrations of cloxicillin, was observed to restore susceptibility to several β‐lactam antibiotics in certain K. pneumoniae clinical isolates whose resistance was independent of β‐lactamase acquisition.127
Conclusions
Since their discovery, β‐lactams have remained the most widely prescribed antibiotics due to their safety, efficacy, and availability. The emergence and dissemination of bacterial resistance to this class of drugs has nevertheless become a serious problem in the clinic, often leaving few if any treatment options for infections resulting from MDR superbugs. The gravity of the situation has however prompted the development of novel antimicrobials and clever strategies to counter bacterial resistance. The two non‐β‐lactam families of β‐lactamase inhibitors, the DBOs and boronic acids, in combination with novel or existing β‐lactam antibiotics show particular promise. In addition, the conjugation of β‐lactams to siderophores, which allows drug entry through bacterial iron transport systems, is a lucrative option for the treatment of MDR Gram‐negative bacteria with intrinsic resistance due to restricted porin entry and drug efflux. As resistance is a natural consequence of selective pressures imposed by antibiotic use, the battle against bacterial pathogens is inevitable and ongoing. This struggle requires continued surveillance and understanding of emerging resistance mechanisms along with structural efforts that provide avenues for rational drug design.
Acknowledgments
This work was supported by the Canadian Institutes of Health Research (S.S. (Banting Fellow) and D.T.K), Michael Smith Foundation for Health Research (S.S), and Canadian Institute of Health Research operating grant (N.C.J.S). N.C.J.S is a Tier 1 Canada Research Chair.
References
- 1. Lewis K (2013) Platforms for antibiotic discovery. Nat Rev Drug Discov 12:371–387. [DOI] [PubMed] [Google Scholar]
- 2. Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J (2009) Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 48:1–12. [DOI] [PubMed] [Google Scholar]
- 3. Davies J, Davies D (2010) Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev 74:417–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Abraham EP, Chain E (1988) An enzyme from bacteria able to destroy penicillin. 1940. Rev Infect Dis 10:677–678. [PubMed] [Google Scholar]
- 5. Bush K, Fisher JF (2011) Epidemiological expansion, structural studies, and clinical challenges of new β‐lactamases from Gram‐negative bacteria. Annu Rev Microbiol 65:455–478. [DOI] [PubMed] [Google Scholar]
- 6. Bush K (2010) Alarming β‐lactamase‐mediated resistance in multidrug‐resistant Enterobacteriaceae. Curr Opin Microbiol 13:558–564. [DOI] [PubMed] [Google Scholar]
- 7. Baroud M, Dandache I, Araj GF, Wakim R, Kanj S, Kanafani Z, Khairallah M, Sabra A, Shehab M, Dbaibo G, Matar GM (2013) Underlying mechanisms of carbapenem resistance in extended‐spectrum β‐lactamase‐producing Klebsiella pneumoniae and Escherichia coli isolates at a tertiary care centre in Lebanon: role of OXA‐48 and NDM‐1 carbapenemases. Int J Antimicrob Agents 41:75–79. [DOI] [PubMed] [Google Scholar]
- 8. Rawat D, Nair D (2010) Extended‐spectrum β‐lactamases in Gram negative bacteria. J Glob Infect Dis 2:263–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cole ST (2014) Who will develop new antibacterial agents? Philos Trans R Soc London B Biol Sci 369:20130430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tomanicek SJ, Standaert RF, Weiss KL, Ostermann A, Schrader TE, Ng JD, Coates L (2013) Neutron and X‐ray crystal structures of a perdeuterated enzyme inhibitor complex reveal the catalytic proton network of the Toho‐1 β‐lactamase for the acylation reaction. J Biol Chem 288:4715–4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. King DT, Strynadka NC (2013) Targeting metallo‐β‐lactamase enzymes in antibiotic resistance. Future Med Chem 5:1243–1263. [DOI] [PubMed] [Google Scholar]
- 12. Bush K (2013) The ABCD's of β‐lactamase nomenclature. J Infect Chemother 19:549–559. [DOI] [PubMed] [Google Scholar]
- 13. Delmas J, Chen Y, Prati F, Robin F, Shoichet BK, Bonnet R (2008) Structure and dynamics of CTX‐M enzymes reveal insights into substrate accommodation by extended‐spectrum beta‐lactamases. J Mol Biol 375:192–201. [DOI] [PubMed] [Google Scholar]
- 14. Strynadka NC, Adachi H, Jensen SE, Johns K, Sielecki A, Betzel C, Sutoh K, James MN (1992) Molecular structure of the acyl‐enzyme intermediate in beta‐lactam hydrolysis at 1.7 A resolution. Nature 359:700–705. [DOI] [PubMed] [Google Scholar]
- 15. Golemi D, Maveyraud L, Vakulenko S, Samama JP, Mobashery S (2001) Critical involvement of a carbamylated lysine in catalytic function of class D beta‐lactamases. Proc Natl Acad Sci USA 98:14280–14285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen Y, McReynolds A, Shoichet BK (2009) Re‐examining the role of Lys67 in class C beta‐lactamase catalysis. Protein Sci 18:662–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Papp‐Wallace KM, Bethel CR, Gootz TD, Shang W, Stroh J, Lau W, McLeod D, Price L, Marfat A, Distler A, Drawz SM, Chen H, Harry E, Nottingham M, Carey PR, Buynak JD, Bonomo RA (2012) Inactivation of a class A and a class C β‐lactamase by 6β‐(hydroxymethyl)penicillanic acid sulfone. Biochem Pharmacol 83:462–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Drawz SM, Papp‐Wallace KM, Bonomo RA (2014) New β‐lactamase inhibitors: a therapeutic renaissance in an MDR world. Antimicrob Agents Chemother 58:1835–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bebrone C, Lassaux P, Vercheval L, Sohier JS, Jehaes A, Sauvage E, Galleni M (2010) Current challenges in antimicrobial chemotherapy: focus on ß‐lactamase inhibition. Drugs 70:651–679. [DOI] [PubMed] [Google Scholar]
- 20. Shlaes DM (2013) New β‐lactam‐β‐lactamase inhibitor combinations in clinical development. Ann N Y Acad Sci 1277:105–114. [DOI] [PubMed] [Google Scholar]
- 21. Livermore DM, Mushtaq S (2013) Activity of biapenem (RPX2003) combined with the boronate β‐lactamase inhibitor RPX7009 against carbapenem‐resistant Enterobacteriaceae. J Antimicrob Chemother 68:1825–1831. [DOI] [PubMed] [Google Scholar]
- 22. Coleman K (2011) Diazabicyclooctanes (DBOs): a potent new class of non‐β‐lactam β‐lactamase inhibitors. Curr Opin Microbiol 14:550–555. [DOI] [PubMed] [Google Scholar]
- 23. Ehmann DE, Jahic H, Ross PL, Gu RF, Hu J, Durand‐Réville TF, Lahiri S, Thresher J, Livchak S, Gao N, Palmer T, Walkup GK, Fisher SL (2013) Kinetics of avibactam inhibition against Class A, C, and D β‐lactamases. J Biol Chem 288:27960–27971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lagacé‐Wiens P, Walkty A, Karlowsky JA (2014) Ceftazidime‐avibactam: an evidence‐based review of its pharmacology and potential use in the treatment of Gram‐negative bacterial infections. Core Evid 9:13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lahiri SD, Mangani S, Durand‐Reville T, Benvenuti M, De Luca F, Sanyal G, Docquier JD (2013) Structural insight into potent broad‐spectrum inhibition with reversible recyclization mechanism: avibactam in complex with CTX‐M‐15 and Pseudomonas aeruginosa AmpC β‐lactamases. Antimicrob Agents Chemother 57:2496–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ehmann DE, Jahić H, Ross PL, Gu RF, Hu J, Kern G, Walkup GK, Fisher SL (2012) Avibactam is a covalent, reversible, non‐β‐lactam β‐lactamase inhibitor. Proc Natl Acad Sci USA 109:11663–11668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. King DT, King AM, Lal SM, Wright GD, Strynadka NCJ (2015) Molecular mechanism of avibactam‐mediated β‐lactamase inhibition. ACS Infect Dis 1:175–184. [DOI] [PubMed] [Google Scholar]
- 28. Smoum R, Rubinstein A, Dembitsky VM, Srebnik M (2012) Boron containing compounds as protease inhibitors. Chem Rev 112:4156–4220. [DOI] [PubMed] [Google Scholar]
- 29. Baker SJ, Tomsho JW, Benkovic SJ (2011) Boron‐containing inhibitors of synthetases. Chem Soc Rev 40:4279–4285. [DOI] [PubMed] [Google Scholar]
- 30. Eidam O, Romagnoli C, Caselli E, Babaoglu K, Pohlhaus DT, Karpiak J, Bonnet R, Shoichet BK, Prati F (2010) Design, synthesis, crystal structures, and antimicrobial activity of sulfonamide boronic acids as β‐lactamase inhibitors. J Med Chem 53:7852–7863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Strynadka NC, Martin R, Jensen SE, Gold M, Jones JB (1996) Structure‐based design of a potent transition state analogue for TEM‐1 beta‐lactamase. Nat Struct Biol 3:688–695. [DOI] [PubMed] [Google Scholar]
- 32. Yang W, Gao X, Wang B (2003) Boronic acid compounds as potential pharmaceutical agents. Med Res Rev 23:346–368. [DOI] [PubMed] [Google Scholar]
- 33. Livermore DM, Mushtaq S, Warner M (2010) Activity of BAL30376 (monobactam BAL19764 + BAL29880 + clavulanate) versus Gram‐negative bacteria with characterized resistance mechanisms. J Antimicrob Chemother 65:2382–2395. [DOI] [PubMed] [Google Scholar]
- 34. Page MG, Dantier C, Desarbre E (2010) In vitro properties of BAL30072, a novel siderophore sulfactam with activity against multiresistant gram‐negative bacilli. Antimicrob Agents Chemother 54:2291–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Crandon JL, Nicolau DP (2013) Human simulated studies of aztreonam and aztreonam‐avibactam to evaluate activity against challenging Gram‐negative organisms, including metallo‐β‐lactamase producers. Antimicrob Agents Chemother 57:3299–3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. King AM, Reid‐Yu SA, Wang W, King DT, Gianfranco DP, Strynadka NC, Walsh TR, Coombes BK, Wright GD (2014) Aspergillomarasmine A overcomes metallo‐beta‐lactamase antibiotic resistance. Nature510: 503–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tan Q, Ogawa AM, Raghoobar SL, Wisniewski D, Colwell L, Park YW, Young K, Hermes JD, Dininno FP, Hammond ML (2011) Thiophenyl oxime‐derived phosphonates as nano‐molar class C beta‐lactamase inhibitors reducing MIC of imipenem against Pseudomonas aeruginosa and Acinetobacter baumannii . Bioorg Med Chem Lett 21:4363–4365. [DOI] [PubMed] [Google Scholar]
- 38. Brem J, van Berkel SS, Aik W, Rydzik AM, Avison MB, Pettinati I, Umland KD, Kawamura A, Spencer J, Claridge TD, McDonough MA, Schofield CJ (2014) Rhodanine hydrolysis leads to potent thioenolate mediated metallo‐β‐lactamase inhibition. Nat Chem 6:1084–1090. [DOI] [PubMed] [Google Scholar]
- 39. Drawz SM, Bethel CR, Doppalapudi VR, Sheri A, Pagadala SR, Hujer AM, Skalweit MJ, Anderson VE, Chen SG, Buynak JD, Bonomo RA (2010) Penicillin sulfone inhibitors of class D beta‐lactamases. Antimicrob Agents Chemother 54:1414–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lambert PA (2005) Bacterial resistance to antibiotics: modified target sites. Adv Drug Deliv Rev 57:1471–1485. [DOI] [PubMed] [Google Scholar]
- 41. Chambers HF, Deleo FR (2009) Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol 7:629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Arias CA, Murray BE (2012) The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol 10:266–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P (2008) The penicillin‐binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev 32:234–258. [DOI] [PubMed] [Google Scholar]
- 44. Typas A, Banzhaf M, Gross CA, Vollmer W (2012) From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol 10:123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zapun A, Contreras‐Martel C, Vernet T (2008) Penicillin‐binding proteins and beta‐lactam resistance. FEMS Microbiol Rev 32:361–385. [DOI] [PubMed] [Google Scholar]
- 46. Klein E, Smith DL, Laxminarayan R (2007) Hospitalizations and deaths caused by methicillin‐resistant Staphylococcus aureus, United States, 1999‐2005. Emerg Infect Dis 13:1840–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lim D, Strynadka NC (2002) Structural basis for the beta lactam resistance of PBP2a from methicillin‐resistant Staphylococcus aureus . Nat Struct Biol 9:870–876. [DOI] [PubMed] [Google Scholar]
- 48. Zhang HZ, Hackbarth CJ, Chansky KM, Chambers HF (2001) A proteolytic transmembrane signaling pathway and resistance to beta‐lactams in staphylococci. Science 291:1962–1965. [DOI] [PubMed] [Google Scholar]
- 49. Archer GL, Niemeyer DM, Thanassi JA, Pucci MJ (1994) Dissemination among staphylococci of DNA sequences associated with methicillin resistance. Antimicrob Agents Chemother 38:447–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fuda C, Hesek D, Lee M, Morio K, Nowak T, Mobashery S (2005) Activation for catalysis of penicillin‐binding protein 2a from methicillin‐resistant Staphylococcus aureus by bacterial cell wall. J Am Chem Soc 127:2056–2057. [DOI] [PubMed] [Google Scholar]
- 51. De Lencastre H, Wu SW, Pinho MG, Ludovice AM, Filipe S, Gardete S, Sobral R, Gill S, Chung M, Tomasz A (1999) Antibiotic resistance as a stress response: complete sequencing of a large number of chromosomal loci in Staphylococcus aureus strain COL that impact on the expression of resistance to methicillin. Microb Drug Resist 5:163–175. [DOI] [PubMed] [Google Scholar]
- 52. Brown S, Xia G, Luhachack LG, Campbell J, Meredith TC, Chen C, Winstel V, Gekeler C, Irazoqui JE, Peschel A, Walker S (2012) Methicillin resistance in Staphylococcus aureus requires glycosylated wall teichoic acids. Proc Natl Acad Sci USA 109:18909–18914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lu WP, Kincaid E, Sun Y, Bauer MD (2001) Kinetics of beta‐lactam interactions with penicillin‐susceptible and ‐resistant penicillin‐binding protein 2x proteins from Streptococcus pneumoniae. Involvement of acylation and deacylation in beta‐lactam resistance. J Biol Chem 276:31494–31501. [DOI] [PubMed] [Google Scholar]
- 54. Lu WP, Sun Y, Bauer MD, Paule S, Koenigs PM, Kraft WG (1999) Penicillin‐binding protein 2a from methicillin‐resistant Staphylococcus aureus: kinetic characterization of its interactions with beta‐lactams using electrospray mass spectrometry. Biochemistry 38:6537–6546. [DOI] [PubMed] [Google Scholar]
- 55. Bush K, Heep M, Macielag MJ, Noel GJ (2007) Anti‐MRSA beta‐lactams in development, with a focus on ceftobiprole: the first anti‐MRSA beta‐lactam to demonstrate clinical efficacy. Expert Opin Investig Drugs 16:419–429. [DOI] [PubMed] [Google Scholar]
- 56. Murthy B, Schmitt‐Hoffmann A (2008) Pharmacokinetics and pharmacodynamics of ceftobiprole, an anti‐MRSA cephalosporin with broad‐spectrum activity. Clin Pharmacokinet 47:21–33. [DOI] [PubMed] [Google Scholar]
- 57. Queenan AM, Shang W, Kania M, Page MG, Bush K (2007) Interactions of ceftobiprole with beta‐lactamases from molecular classes A to D. Antimicrob Agents Chemother 51:3089–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Page MG (2004) Cephalosporins in clinical development. Expert Opin Investig Drugs 13:973–985. [DOI] [PubMed] [Google Scholar]
- 59. Lovering AL, Gretes MC, Safadi SS, Danel F, de Castro L, Page MG, Strynadka NC (2012) Structural insights into the anti‐methicillin‐resistant Staphylococcus aureus (MRSA) activity of ceftobiprole. J Biol Chem 287:32096–32102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mobley DL, Dill KA (2009) Binding of small‐molecule ligands to proteins: “what you see” is not always “what you get”. Structure 17:489–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Graves‐Woodward K, Pratt RF (1998) Reaction of soluble penicillin‐binding protein 2a of methicillin‐resistant Staphylococcus aureus with beta‐lactams and acyclic substrates: kinetics in homogeneous solution. Biochem J 332:755–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Villegas‐Estrada A, Lee M, Hesek D, Vakulenko SB, Mobashery S (2008) Co‐opting the cell wall in fighting methicillin‐resistant Staphylococcus aureus: potent inhibition of PBP 2a by two anti‐MRSA beta‐lactam antibiotics. J Am Chem Soc 130:9212–9213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Otero LH, Rojas‐Altuve A, Llarrull LI, Carrasco‐López C, Kumarasiri M, Lastochkin E, Fishovitz J, Dawley M, Hesek D, Lee M, Johnson JW, Fisher JF, Chang M, Mobashery S, Hermoso JA (2013) How allosteric control of Staphylococcus aureus penicillin binding protein 2a enables methicillin resistance and physiological function. Proc Natl Acad Sci USA 110:16808–16813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Saravolatz LD, Stein GE, Johnson LB (2011) Ceftaroline: a novel cephalosporin with activity against methicillin‐resistant Staphylococcus aureus . Clin Infect Dis 52:1156–1163. [DOI] [PubMed] [Google Scholar]
- 65. Moisan H, Pruneau M, Malouin F (2010) Binding of ceftaroline to penicillin‐binding proteins of Staphylococcus aureus and Streptococcus pneumoniae . J Antimicrob Chemother 65:713–716. [DOI] [PubMed] [Google Scholar]
- 66. Ueda Y, Kanazawa K, Eguchi K, Takemoto K, Eriguchi Y, Sunagawa M (2005) In vitro and in vivo antibacterial activities of SM‐216601, a new broad‐spectrum parenteral carbapenem. Antimicrob Agents Chemother 49:4185–4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. O'Daniel PI, Peng Z, Pi H, Testero SA, Ding D, Spink E, Leemans E, Boudreau MA, Yamaguchi T, Schroeder VA, Wolter WR, Llarrull LI, Song W, Lastochkin E, Kumarasiri M, Antunes NT, Espahbodi M, Lichtenwalter K, Suckow MA, Vakulenko S, Mobashery S, Chang M (2014) Discovery of a new class of non‐β‐lactam inhibitors of penicillin‐binding proteins with Gram‐positive antibacterial activity. J Am Chem Soc 136:3664–3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Fox PM, Lampen RJ, Stumpf KS, Archer GL, Climo MW (2006) Successful therapy of experimental endocarditis caused by vancomycin‐resistant Staphylococcus aureus with a combination of vancomycin and beta‐lactam antibiotics. Antimicrob Agents Chemother 50:2951–2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dilworth TJ, Ibrahim O, Hall P, Sliwinski J, Walraven C, Mercier RC (2013) beta‐Lactams enhance vancomycin activity against methicillin‐resistant Staphylococcus aureus bacteremia compared to vancomycin alone. Antimicrob Agents Chemother 58:102–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Stryjewski ME, Potgieter PD, Li YP, Barriere SL, Churukian A, Kingsley J, Corey GR, Group T‐I (2012) TD‐1792 versus vancomycin for treatment of complicated skin and skin structure infections. Antimicrob Agents Chemother 56:5476–5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Blais J, Lewis SR, Krause KM, Benton BM (2011) Antistaphylococcal activity of TD‐1792, a multivalent glycopeptide‐cephalosporin antibiotic. Antimicrob Agents Chemother 56:1584–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rice LB, Carias LL, Rudin S, Lakticová V, Wood A, Hutton‐Thomas R (2005) Enterococcus faecium low‐affinity pbp5 is a transferable determinant. Antimicrob Agents Chemother 49:5007–5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sauvage E, Kerff F, Fonzé E, Herman R, Schoot B, Marquette JP, Taburet Y, Prevost D, Dumas J, Leonard G, Stefanic P, Coyette J, Charlier P (2002) The 2.4‐A crystal structure of the penicillin‐resistant penicillin‐binding protein PBP5fm from Enterococcus faecium in complex with benzylpenicillin. Cell Mol Life Sci 59:1223–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Henry X, Amoroso A, Coyette J, Joris B (2010) Interaction of ceftobiprole with the low‐affinity PBP 5 of Enterococcus faecium . Antimicrob Agents Chemother 54:953–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Cornick JE, Bentley SD (2012) Streptococcus pneumoniae: the evolution of antimicrobial resistance to beta‐lactams, fluoroquinolones and macrolides. Microbes Infect 14:573–583. [DOI] [PubMed] [Google Scholar]
- 76. Hakenbeck R (1998) Mosaic genes and their role in penicillin‐resistant Streptococcus pneumoniae . Electrophoresis 19:597–601. [DOI] [PubMed] [Google Scholar]
- 77. Chesnel L, Pernot L, Lemaire D, Champelovier D, Croizé J, Dideberg O, Vernet T, Zapun A (2003) The structural modifications induced by the M339F substitution in PBP2x from Streptococcus pneumoniae further decreases the susceptibility to beta‐lactams of resistant strains. J Biol Chem 278:44448–44456. [DOI] [PubMed] [Google Scholar]
- 78. Ohnishi M, Golparian D, Shimuta K, Saika T, Hoshina S, Iwasaku K, Nakayama S, Kitawaki J, Unemo M (2011) Is Neisseria gonorrhoeae initiating a future era of untreatable gonorrhea?: detailed characterization of the first strain with high‐level resistance to ceftriaxone. Antimicrob Agents Chemother 55:3538–3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Chantratita N, Rholl DA, Sim B, Wuthiekanun V, Limmathurotsakul D, Amornchai P, Thanwisai A, Chua HH, Ooi WF, Holden MT, Day NP, Tan P, Schweizer HP, Peacock SJ (2011) Antimicrobial resistance to ceftazidime involving loss of penicillin‐binding protein 3 in Burkholderia pseudomallei . Proc Natl Acad Sci U S A 108:17165–17170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Galdiero S, Falanga A, Cantisani M, Tarallo R, Della Pepa ME, D'Oriano V, Galdiero M (2013) Microbe‐host interactions: structure and role of Gram‐negative bacterial porins. Curr Protein Pept Sci 13:843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Fernandez L, Hancock RE (2012) Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin Microbiol Rev 25:661–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Harder KJ, Nikaido H, Matsuhashi M (1981) Mutants of Escherichia coli that are resistant to certain beta‐lactam compounds lack the ompF porin. Antimicrob Agents Chemother 20:549–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lou H, Chen M, Black SS, Bushell SR, Ceccarelli M, Mach T, Beis K, Low AS, Bamford VA, Booth IR, Bayley H, Naismith JH (2011) Altered antibiotic transport in OmpC mutants isolated from a series of clinical strains of multi‐drug resistant E. coli . PLoS One 6:e25825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wolter DJ, Hanson ND, Lister PD (2004) Insertional inactivation of oprD in clinical isolates of Pseudomonas aeruginosa leading to carbapenem resistance. FEMS Microbiol Lett 236:137–143. [DOI] [PubMed] [Google Scholar]
- 85. Ochs MM, McCusker MP, Bains M, Hancock RE (1999) Negative regulation of the Pseudomonas aeruginosa outer membrane porin OprD selective for imipenem and basic amino acids. Antimicrob Agents Chemother 43:1085–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Huang H, Hancock RE (1996) The role of specific surface loop regions in determining the function of the imipenem‐specific pore protein OprD of Pseudomonas aeruginosa . J Bacteriol 178:3085–3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Skurnik D, Roux D, Cattoir V, Danilchanka O, Lu X, Yoder‐Himes DR, Han K, Guillard T, Jiang D, Gaultier C, Guerin Fo Aschard H, Leclercq R, Mekalanos JJ, Lory S Pier GB (2013) Enhanced in vivo fitness of carbapenem‐resistant oprD mutants of Pseudomonas aeruginosa revealed through high‐throughput sequencing. Proc Natl Acad Sci USA 110:20747–20752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Raymond KN, Dertz EA, Kim SS (2003) Enterobactin: an archetype for microbial iron transport. Proc Natl Acad Sci USA 100:3584–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Mollmann U, Heinisch L, Bauernfeind A, Kohler T, Ankel‐Fuchs D (2009) Siderophores as drug delivery agents: application of the “Trojan Horse” strategy. Biometals 22:615–624. [DOI] [PubMed] [Google Scholar]
- 90. Holden VI, Bachman MA (2015) Diverging roles of bacterial siderophores during infection. Metallomics 7:986–995. [DOI] [PubMed] [Google Scholar]
- 91. Schalk IJ, Mislin GL, Brillet K (2012) Structure, function and binding selectivity and stereoselectivity of siderophore‐iron outer membrane transporters. Curr Top Membr 69:37–66. [DOI] [PubMed] [Google Scholar]
- 92. Noinaj N, Guillier M, Barnard TJ, Buchanan SK (2010) TonB‐dependent transporters: regulation, structure, and function. Annu Rev Microbiol 64:43–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Pawelek PD, Croteau N, Ng‐Thow‐Hing C, Khursigara CM, Moiseeva N, Allaire M, Coulton JW (2006) Structure of TonB in complex with FhuA, E. coli outer membrane receptor. Science 312:1399–1402. [DOI] [PubMed] [Google Scholar]
- 94. Chimento DP, Kadner RJ, Wiener MC (2005) Comparative structural analysis of TonB‐dependent outer membrane transporters: implications for the transport cycle. Proteins 59:240–251. [DOI] [PubMed] [Google Scholar]
- 95. Gause GF (1955) Recent studies on albomycin, a new antibiotic. Br Med J 2:1177–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Bickel H, Mertens P, Prelog V, Seibl J, Walser A (1965) Constitution of ferrimycin A1. Antimicrob Agents Chemother 5:951–957. [PubMed] [Google Scholar]
- 97. Vértesy L, Aretz W, Fehlhaber H‐W, Kogler H (1995) Salmycin A–D, Antibiotika aus Streptomyces violaceus, DSM 8286, mit Siderophor‐Aminoglycosid‐Struktur. Helvetica Chim Acta 78:46–60. [Google Scholar]
- 98. Page MG (2013) Siderophore conjugates. Ann N Y Acad Sci 1277:115–126. [DOI] [PubMed] [Google Scholar]
- 99. Mislin GL, Schalk IJ (2014) Siderophore‐dependent iron uptake systems as gates for antibiotic Trojan horse strategies against Pseudomonas aeruginosa . Metallomics 6:408–420. [DOI] [PubMed] [Google Scholar]
- 100. Butler MS, Blaskovich MA, Cooper MA (2013) Antibiotics in the clinical pipeline in 2013. J Antibiot 66:571–591. [DOI] [PubMed] [Google Scholar]
- 101. Hofer B, Dantier C, Gebhardt K, Desarbre E, Schmitt‐Hoffmann A, Page MG (2013) Combined effects of the siderophore monosulfactam BAL30072 and carbapenems on multidrug‐resistant Gram‐negative bacilli. J Antimicrob Chemother 68:1120–1129. [DOI] [PubMed] [Google Scholar]
- 102. Higgins PG, Stefanik D, Page MG, Hackel M, Seifert H (2012) In vitro activity of the siderophore monosulfactam BAL30072 against meropenem‐non‐susceptible Acinetobacter baumannii . J Antimicrob Chemother 67:1167–1169. [DOI] [PubMed] [Google Scholar]
- 103. Mushtaq S, Warner M, Livermore D (2010) Activity of the siderophore monobactam BAL30072 against multiresistant non‐fermenters. J Antimicrob Chemother 65:266–270. [DOI] [PubMed] [Google Scholar]
- 104. Mima T, Kvitko BH, Rholl DA, Page MG, Desarbre E, Schweizer HP (2011) In vitro activity of BAL30072 against Burkholderia pseudomallei . Int J Antimicrob Agents 38:157–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Nikaido H, Pages JM (2012) Broad‐specificity efflux pumps and their role in multidrug resistance of Gram‐negative bacteria. FEMS Microbiol Rev 36:340–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Martins A, Spengler G, Molnar J, Amaral L (2012) Sequential responses of bacteria to noxious agents (antibiotics) leading to accumulation of mutations and permanent resistance. Biochem Pharmacol 1:104. [Google Scholar]
- 107. Nikaido H, Takatsuka Y (2009) Mechanisms of RND multidrug efflux pumps. Biochim Biophys Acta 1794:769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Li XZ, Livermore DM, Nikaido H (1994) Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: resistance to tetracycline, chloramphenicol, and norfloxacin. Antimicrob Agents Chemother 38:1732–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Su CC, Long F, Zimmermann MT, Rajashankar KR, Jernigan RL, Yu EW (2011) Crystal structure of the CusBA heavy‐metal efflux complex of Escherichia coli. Nature 470:558–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Symmons MF, Bokma E, Koronakis E, Hughes C, Koronakis V (2009) The assembled structure of a complete tripartite bacterial multidrug efflux pump. Proc Natl Acad Sci USA 106:7173–7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Du D, Wang Z, James NR, Voss JE, Klimont E, Ohene‐Agyei T, Venter H, Chiu W, Luisi BF (2014) Structure of the AcrAB‐TolC multidrug efflux pump. Nature 509:512–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Hinchliffe P, Symmons MF, Hughes C, Koronakis V (2013) Structure and operation of bacterial tripartite pumps. Annu Rev Microbiol 67:221–242. [DOI] [PubMed] [Google Scholar]
- 113. Du D, van Veen HW, Luisi BF (2015) Assembly and operation of bacterial tripartite multidrug efflux pumps. Trends Microbiol 23:311–319. [DOI] [PubMed] [Google Scholar]
- 114. Murakami S, Nakashima R, Yamashita E, Yamaguchi A (2002) Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419:587–593. [DOI] [PubMed] [Google Scholar]
- 115. Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A (2006) Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature 443:173–179. [DOI] [PubMed] [Google Scholar]
- 116. Pos KM (2009) Drug transport mechanism of the AcrB efflux pump. Biochim Biophys Acta 1794:782–793. [DOI] [PubMed] [Google Scholar]
- 117. Seeger MA, Schiefner A, Eicher T, Verrey F, Diederichs K, Pos KM (2006) Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science 313:1295–1298. [DOI] [PubMed] [Google Scholar]
- 118. Higgins MK, Bokma E, Koronakis E, Hughes C, Koronakis V (2004) Structure of the periplasmic component of a bacterial drug efflux pump. Proc Natl Acad Sci USA 101:9994–9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Mikolosko J, Bobyk K, Zgurskaya HI, Ghosh P (2006) Conformational flexibility in the multidrug efflux system protein AcrA. Structure 14:577–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Andersen C, Koronakis E, Bokma E, Eswaran J, Humphreys D, Hughes C, Koronakis V (2002) Transition to the open state of the TolC periplasmic tunnel entrance. Proc Natl Acad Sci U S A 99:11103–11108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Bavro VN, Pietras Z, Furnham N, Perez‐Cano L, Fernandez‐Recio J, Pei XY, Misra R, Luisi B (2008) Assembly and channel opening in a bacterial drug efflux machine. Mol Cell 30:114–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Lewis K (1994) Multidrug resistance pumps in bacteria: variations on a theme. Trends Biochem Sci 19:119–123. [DOI] [PubMed] [Google Scholar]
- 123. Lomovskaya O, Zgurskaya HI, Totrov M, Watkins WJ (2007) Waltzing transporters and 'the dance macabre' between humans and bacteria. Nat Rev Drug Discov 6:56–65. [DOI] [PubMed] [Google Scholar]
- 124. Li XZ, Ma D, Livermore DM, Nikaido H (1994) Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: active efflux as a contributing factor to beta‐lactam resistance. Antimicrob Agents Chemother 38:1742–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Heritier C, Poirel L, Lambert T, Nordmann P (2005) Contribution of acquired carbapenem‐hydrolyzing oxacillinases to carbapenem resistance in Acinetobacter baumannii . Antimicrob Agents Chemother 49:3198–3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Kaczmarek FS, Gootz TD, Dib‐Hajj F, Shang W, Hallowell S, Cronan M (2004) Genetic and molecular characterization of beta‐lactamase‐negative ampicillin‐resistant Haemophilus influenzae with unusually high resistance to ampicillin. Antimicrob Agents Chemother 48:1630–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Pages JM, Lavigne JP, Leflon‐Guibout V, Marcon E, Bert F, Noussair L, Nicolas‐Chanoine MH (2009) Efflux pump, the masked side of beta‐lactam resistance in Klebsiella pneumoniae clinical isolates. PLoS One 4:e4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Sun J, Deng Z, Yan A (2014) Bacterial multidrug efflux pumps: mechanisms, physiology and pharmacological exploitations. Biochem Biophys Res Commun 453:254–267. [DOI] [PubMed] [Google Scholar]
- 129. Venter H, Mowla R, Ohene‐Agyei T, Ma S (2015) RND‐type drug efflux pumps from Gram‐negative bacteria: molecular mechanism and inhibition. Front Microbiol 6:377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Lomovskaya O, Warren MS, Lee A, Galazzo J, Fronko R, Lee M, Blais J, Cho D, Chamberland S, Renau T, Leger R, Hecker S, Watkins W, Hoshino K, Ishida H, Lee VJ (2001) Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob Agents Chemother 45:105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Hasdemir UO, Chevalier J, Nordmann P, Pages JM (2004) Detection and prevalence of active drug efflux mechanism in various multidrug‐resistant Klebsiella pneumoniae strains from Turkey. J Clin Microbiol 42:2701–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Mazzariol A, Tokue Y, Kanegawa TM, Cornaglia G, Nikaido H (2000) High‐level fluoroquinolone‐resistant clinical isolates of Escherichia coli overproduce multidrug efflux protein AcrA. Antimicrob Agents Chemother 44:3441–3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Watkins WJ, Landaverry Y, Leger R, Litman R, Renau TE, Williams N, Yen R, Zhang JZ, Chamberland S, Madsen D, Griffith D, Tembe V, Huie K, Dudley MN (2003) The relationship between physicochemical properties, in vitro activity and pharmacokinetic profiles of analogues of diamine‐containing efflux pump inhibitors. Bioorg Med Chem Lett 13:4241–4244. [DOI] [PubMed] [Google Scholar]
- 134. Coban AY, Guney AK, Tanriverdi Cayci Y, Durupinar B (2011) Effect of 1‐(1‐Naphtylmethyl)‐piperazine, an efflux pump inhibitor, on antimicrobial drug susceptibilities of clinical Acinetobacter baumannii isolates. Curr Microbiol 62:508–511. [DOI] [PubMed] [Google Scholar]
- 135. Bohnert JA, Kern WV (2005) Selected arylpiperazines are capable of reversing multidrug resistance in Escherichia coli overexpressing RND efflux pumps. Antimicrob Agents Chemother 49:849–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Schumacher A, Steinke P, Bohnert JA, Akova M, Jonas D, Kern WV (2006) Effect of 1‐(1‐naphthylmethyl)‐piperazine, a novel putative efflux pump inhibitor, on antimicrobial drug susceptibility in clinical isolates of Enterobacteriaceae other than Escherichia coli . J Antimicrob Chemother 57:344–348. [DOI] [PubMed] [Google Scholar]
- 137. Kourtesi C, Ball AR, Huang YY, Jachak SM, Vera DM, Khondkar P, Gibbons S, Hamblin MR, Tegos GP (2013) Microbial efflux systems and inhibitors: approaches to drug discovery and the challenge of clinical implementation. Open Microbiol J 7:34–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Li B, Yao Q, Pan X‐C, Wang N, Zhang R, Li J, Ding G, Liu X, Wu C, Ran D, Zheng J, Zhou H (2011) Artesunate enhances the antibacterial effect of Î2‐lactam antibiotics against Escherichia coli by increasing antibiotic accumulation via inhibition of the multidrug efflux pump system AcrAB‐TolC. J Antimicrob Chemother 66:769–777. [DOI] [PubMed] [Google Scholar]


