Abstract
Clostridium acetobutylicum is an industrially important Gram-positive organism, which is capable of producing economically important chemicals in the ABE (Acetone, Butanol and Ethanol) fermentation process. Renewed interests in the ABE process necessitate the availability of additional genetics tools to facilitate the derivation of a greater understanding of the underlying metabolic and regulatory control processes in operation through forward genetic strategies. In this study, a xylose inducible, mariner-based, transposon system was developed and shown to allow high-efficient random mutagenesis in the model strain ATCC 824. Of the thiamphenicol resistant colonies obtained, 91.9% were shown to be due to successful transposition of the catP-based mini-transposon element. Phenotypic screening of 200 transposon clones revealed a sporulation-defective clone with an insertion in spo0A, thereby demonstrating that this inducible transposon system can be used for forward genetic studies in C. acetobutylicum.
Keywords: Clostridium, transposon, random mutagenesis, xylose-inducible promoters
A useful method for forward genetic studies in Clostridium acetobutylicum.
INTRODUCTION
Heightened concerns over global warming and fossil fuel supply, security and prices have led to a resurgence of interest in the sustainable production of chemicals and fuels. In this regard, saccharolytic Clostridium species are of particular interest given their former use on a commercial scale in the so-called Acetone-Butanol-Ethanol (ABE) fermentation process, producing the solvents acetone, butanol and ethanol. While several different saccharolytic Clostridium species have been deployed in the ABE process, C. acetobutylicum is widely regarded as the model organism. Not surprisingly, therefore, the genome of C. acetobutylicum ATCC 824 was the first clostridial genome sequence to be described (Nolling et al. 2001).
Renewed interest in reviving the ABE process has led to concerted efforts to derive genetic tools that may be deployed to both better understand the underlying metabolic and regulatory control processes, and to bring about improvements in productivity. Accordingly, genetic tools for directed gene disruption have been widely described. These include gene knock-down methods based on antisense RNA (Desai and Papoutsakis 1999; Tummala, Junne and Papoutsakis 2003), insertional gene disruption methods based on the bacterial mobile group II intron from the ltrB gene of Lactococcus lactis (Heap et al. 2007, 2010a,b; Shao et al. 2007), gene knock-out methods reliant on homologous recombination and the use of replicative and non-replicative vectors (Al-Hinai, Fast and Papoutsakis 2012; Heap et al. 2012; Leang et al. 2013; Ehsaan et al. 2016), and recently developed CRISPR-Cas9 system (Wang et al. 2015; Xu et al. 2015). Such tools have provided the basis for reverse genetic strategies that have focused on the key solvent producing pathways and have led to a number of successful rational metabolic engineering strategies for altering solvent productivity in C. acetobutylicum (Scotcher, Rudolph and Bennett 2005; Jang et al. 2012).
By way of contrast, scant attention has been paid to forward genetic approaches reliant on random mutagens typified by transposon elements. Two conjugative transposons Tn916 and Tn1514 have been studied in C. acetobutylicum ATCC 824, C. saccharobutylicum P262 and C. beijerinckii NCIMB 8052 (Woolley et al. 1989; Bertram, Kuhn and Durre 1990; Mattsson and Rogers 1994). These were used to isolate a number of interesting mutants, including one deficient in ‘degeneration’ (the loss in prolonged culture of the ability to produce solvents) (Kashket and Cao 1993) and others exhibiting increased butanol tolerance (Liyanage, Young and Kashket 2000). However, a number of factors mean these tools are far from ideal. Principles among these are their large size and low efficiency of transposition, in the case of Tn916 the existence of ‘hot spots’ in the chromosome where the majority of the insertions locate and, in the case of Tn1545, the predilection to insert in multiple copies (probably attributed to a high rate of vector retention). This latter property significantly complicates the association of genotype with phenotype, hence, requiring further experimentation to understand the basis of the observed phenotypes.
A number of non-conjugative transposon mutagenesis systems have been described for pathogenic clostridia recently, including two mariner-based transposon systems, in C. perfringens (Liu et al. 2013) and C. difficile (Cartman and Minton 2010). The later was recently adapted to be used in C. acetobutylicum ATCC 824, by equipping the host cell with the ability to produce a foreign sigma factor, TcdR (Zhang, Grosse-Honebrink and Minton 2015). Although the new system proved to be highly effective, it is reliant on prior addition of the TcdR-encoding gene and is, therefore, not applicable if a wild-type clostridial host is the desired target. In addition, embracing the fast developed next generation DNA sequencing technology, transposon-directed insertion site sequencing was recently used in C. difficile to identify a core set of 404 essential genes and 798 genes that are likely to impact spore production (Dembek et al. 2015).
Here, we reported the development of a xylose inducible mariner-based transposon system and its exemplification in wild-type C. acetobutylicum ATCC 824. Moreover, we described the isolation of a sporulation mutant that contains an insertion within the gene spo0A from 200 mutants generated by this transposon system.
MATERIALS AND METHODS
Bacterial strains and media
Bacterial strains utilized in this study are listed in Table 1. Escherichia coli strains were grown in Luria-Bertani medium at 37°C. Clostridium spp. were cultured under anaerobic condition in an anaerobic cabinet (MG1000 Anaerobic Work Station, Don Whitley Scientific Ltd) containing an atmosphere of 80% nitrogen, 10% hydrogen and 10% carbon dioxide. Antibiotics were used at the following concentrations: erythromycin (Em), 20 μg ml−1, thiamphenicol (Tm), 15 μg ml−1 for Clostridium spp.; Em, 500 μg ml−1, chloramphenicol (Cm), 25 μg ml−1, tetracycline (Tc), 10 μg ml−1 for E. coli. Clostridium acetobutylicum ATCC 824 was grown in Clostridium Growth Medium (CGM) (Hartmanis and Gatenbeck 1984) for routine manipulations, or P2 medium (Baer, Blaschek and Smith 1987) with 20 g l−1 glucose for auxotrophic mutant screening.
Table 1.
Bacterial strains and plasmids.
| Strains or plasmids | Relevant characteristicsa | Reference or sourceb |
|---|---|---|
| Strains | ||
| C. acetobutylicum ATCC 824 | Wild type | ATCC |
| E.coli ER2275 | hsdR mcr recA1 endA1 | NEB |
| E.coli DH5α | General cloning host strain | Takara |
| Plasmids | ||
| pAN1 | Φ3TI, p15a origin, Sper | Mermelstein and Papoutsakis (1993) |
| pMTL82254 | Clostridium modular plasmid with catP reporter, pBP1 (Gram+ origin), ColE1+tra (Gram– origin), Emr | Heap et al. (2009) |
| pMTL82254-Pfdx | Clostridium modular plasmid with catP reporter expressed by the fdx promoter, pBP1 (Gram+ origin), ColE1+tra (Gram– origin), Emr | Zhang, Grosse-Honebrink and Minton (2015) |
| pMTL82254-Pcac1339 | Clostridium modular plasmid with catP reporter expressed by promoter of cac1339, pBP1 (Gram+ origin), ColE1+tra (Gram– origin), Emr | This study |
| pMTL82254-Pcac1344 | Clostridium modular plasmid with catP reporter expressed by promoter of cac1344, pBP1 (Gram+ origin), ColE1+tra (Gram– origin), Emr | This study |
| pMTL82254-Pcac2612 | Clostridium modular plasmid with catP reporter expressed by promoter of cac2612, pBP1 (Gram+ origin), ColE1+tra (Gram– origin), Emr | This study |
| pMTL83151 | Clostridium modular plasmid used for construction of IPTG inducible promoter system, pCB102 (Gram+ origin), ColE1+tra (Gram– origin), Tmr | Heap et al. (2009) |
| pMTL-SC0 | Transposon plasmid with pBP1 replicon, Tmr | Cartman and Minton (2010) |
| pMTL-YG0 | Derived from pMTL-SC0 by replacing pBP1 with pCB102 replicon | This study |
| pMTL-YG3 | Derived from pMTL-YG0 by introducing the promoter of cac1339 to express the transposase Himar1 C9 | This study |
a hsdR, host-specific restriction deficient; mcr, methylcytosine-specific restriction abolished; recA1, homologous recombination abolished; endA1, endonucleases abolished; Sper, spectinomycin resistance; Emr, erythromycin resistance; Tmr: thiamphenicol resistance; pBP1, Gram-positive origin of replication; pCB102, Gram-positive origin of replication, which was unstable in C. acetobutylicum
bATCC, American Type Culture Collection; NEB, New England Biolabs.
Plasmids, primers, DNA techniques
Plasmids and primers used in this study are listed in Tables 1 and 2. Chromosomal DNA preparation, plasmid isolation and purification of DNA fragments from agarose gels were carried out using the DNeasy Tissue kit, the QIAprep Miniprep kit and the QIAquick Gel Extraction kit, respectively (Qiagen, Manchester, UK). Restriction enzymes were supplied by New England Biolabs and were used according to the manufacturer's instructions. Escherichia coli strains were transformed by electroporation using a Gene-Pulser (Bio-Rad), as recommended by the manufacturer. PCR amplifications were carried out using the KOD Hot Start Master Mix (Merck, Darmstadt, Germany). Oligonucleotides used in this study are detailed in Table 2, which were synthesized by Eurofins MWG Operon, Germany.
Table 2.
Oligonucleotide primers used in this study.
| Primer name | Sequence (5′-3′) | Description |
|---|---|---|
| 1339-F1 | CATCATATGGAAAACTCCTCCTTAAGATTTATAT | Amplify Cac1339 promoter |
| 1339-R1 | ACCGCGGCCGCTTTATATTTAGTCCCTTGCCTTGCC | Amplify Cac1339 promoter |
| 1344-F1 | CATCATATGACATTAATAAATTAACTGTTATACT | Amplify Cac1344 promoter |
| 1344-R1 | ACCGCGGCCGCTTTTAAAACCCCTTCCCGAAATATT | Amplify Cac1344 promoter |
| 2612-F1 | CATCATATGAATCAAACCCCCTTAATTTTAAATA | Amplify Cac2612 promoter |
| 2612-R1 | TACCGCGGCCGCAATTATATATTTTATGTGTAGAAT | Amplify Cac2612 promoter |
| pCB102-F | TCCGGCGCGCCGATAATTTACAGAAAAGAAAATTA | Amplify pCB102 replicon |
| pCB102-R | TTCGGCCGGCCTGCAGCACATTAAGTATATACTATT | Amplify pCB102 replicon |
| catP-INV-F1 | TATTGTATAGCTTGGTATCATCTCATCATATATCCCCAATTCACC | For inverse PCR |
| catP-INV-R1 | TATTTGTGTGATATCCACTTTAACGGTCATGCTGTAGGTACAAGG | For inverse PCR |
| catP-Sou-F1 | GATTGTTTCCATACCGTTGC | For southern probe synthesis |
| catP-Sou-R1 | AGTTATTAAGTCGGGAGTGC | For southern probe synthesis |
To confirm whether transposition had occurred, inverse (INV) PCR was performed according to the previously reported procedure (Cartman and Minton 2010). All DNA Sanger sequencing was carried out by Source BioScience, UK.
To identify the genomic location of transposon insertions, sequence data were analyzed using DNASTAR (www.dnastar.com) and compared to the published genome sequences of C. acetobutylicum ATCC 824 (Refseq number NC_003030.1 and NC_001988.2; GenBank accession number AE001437 and AE001438) using Artemis (www.sanger.ac.uk/resources/software/artemis/).
Plasmid transfer in C. acetobutylicum
Clostridium acetobutylicum was transformed as described previously (Mermelstein et al. 1992). Prior to transformation, plasmid DNA was purified from E. coli ER2275 cells containing plasmid pAN1. This plasmid contains the φ3TI methyltransferase gene of Bacillus subtilis phage φ3tI, which protects DNA from Cac824I restriction activity in C. acetobutylicum (Mermelstein and Papoutsakis 1993).
Construction of plasmids
The promoters of CAC1339 (putative sugar-proton symporter, araE), CAC1344 (sugar kinase, possible xylulose kinase, xylB) and CAC2612 (xylulose kinase, xylB) were amplified using primers 1339-F1/1339-R1, 1344-F1/1344-R1 and 2612-F1/2612-R1 and then cloned into plasmid pMTL82254 via NdeI/NotI. The final plasmids were sequence verified and named pMTL82254-Pcac1339, pMTL82254-Pcac1344 and pMTL82254-Pcac2612 accordingly.
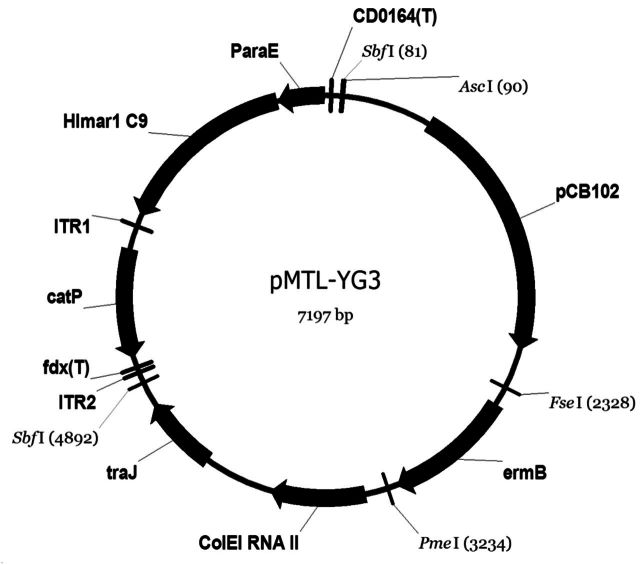
To swap the existing Gram-positive pBP1 replicon in plasmid pMTL-SC0 (Cartman and Minton 2010) with pCB102, a 2403 bp AscI/FseI fragment was excised from pMTL-SC0 and then replaced with a 1625 bp AscI/FseI fragment from pMTL83151, resulting plasmid pMTL-YG0. To generate the final transposon plasmid, the 269 bp NotI-NdeI fragment encompassing the promoter of the CAC1339 gene was cloned into plasmid pMTL-YG0 to express Himar1 C9 transposase, thus, giving rise to plasmid pMTL-YG3 (Fig. 2).
Figure 2.
Vector map of plasmid pMTL-YG3. Expression of the hyperactive mariner transposase gene Himar1 C9 was under the control of the ParaE promoter of the C. acetobutylicum CAC1339 gene (possible sugar-proton symporter, araE). The plasmid backbone consisted of the pCB102 replicon of C. butyricum (Minton and Morris 1981), the macrolide-lincosamide-streptogramin B antibiotic resistance gene ermB, the Gram-negative replicon ColE1, and the conjugal transfer function traJ. The whole mariner element (i.e. transposase gene and catP mini-transposon) can be excised as an SbfI fragment. The transcriptional terminators (T) are identical in sequence to those found immediately downstream of the fdx gene of C. pasteurianum and the CD0164 open reading frame of C. difficile 630. This vector conforms to the pMTL80000 modular system for Clostridium shuttle plasmids (Heap et al. 2009).
Chloramphenicol acetyltransferase assay
Chloramphenicol acetyltransferase (CAT) activity was determined according to the method of Shaw (1975). A quartz cuvette was prepared containing 540 μl of 100 mM Tris buffer (pH 7.8), 200 μl of 2.5 mM DTNB (5,5′-dithiobis-2-nitrobenzoic acid) solution in 100 mM Tris buffer (pH 7.8), 200 μl of 5.0 mM freshly prepared acetyl Coenzyme A solution in deionized water and 10 μl of cell lysate. The cuvette was pre-warmed to 25°C, and the reaction initiated by adding 10 μl of 0.3% w/v Cm solution in deionized water. The initial rate of increase of absorption at 412 nm was measured using an Analytik Jena SPECORD® 250 PLUS spectrophotometer.
Isolation of transposon mutants
The mariner transposon plasmid pMTL-YG3 was in vivo methylated and transformed into C. acetobutylicum ATCC 824 by electroporation as described (Heap et al. 2009), and transformants were selected on CGM agar plates with 2% glucose (Hartmanis and Gatenbeck 1984), supplemented with 20 μg ml−1 erythromycin, for 48 h. All transformants were harvested by flushing the whole plate with CGM broth, then spread onto CGM agar plates with 2% xylose supplemented with 15 μg ml−1 thiamphenicol, to induce Himar1 C9 transposase expression and select transposon mutants. After 48 h of induction, all growth on agar plates were harvested by flushing the whole plate with CGM broth. For further characterization, glycerol was added to the final concentration of 10% and stored at −80°C.
To examine the transposon insertion rate and eliminate plasmids, the harvested cell was subcultured into liquid CGM medium using glucose as the sole carbon source, for three passages with 5% inoculum (12 h every passage). And serial dilutions were made and plated onto CGM agars in triplicate: CGM agar plates without any antibiotics, CGM agar plates supplemented with thiamphenicol and CGM agar plates supplemented with erythromycin.
INV PCR and DNA sequence analysis
INV PCRs were performed as described previously (Cartman and Minton 2010). Briefly, genomic DNA was isolated from mutants and digested overnight with HindIII (200 ng μl−1). The HindIII restriction endonuclease was heat inactivated at 65°C for 30 min. The DNA was diluted to 5 ng μl−1, and then T4 DNA ligase was added to favor self-ligation of the DNA fragments. Ligation reaction was performed at 16°C overnight. Then, the T4 ligase was heat inactivated (65°C for 30 min). INV PCRs were carried out in 50 μl volumes using primers catP-INV-F1 and catP-INV-R1.
Southern blot
Southern blot analysis was carried out using a DIG (digoxigenin) High Prime DNA Labeling and Detection Starter kit I (Roche) as instructed by the manufacturer.
RESULTS AND DISCUSSION
Identification of a xylose inducible promoter
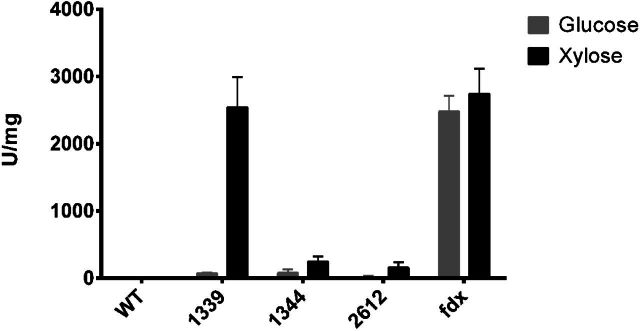
Potential xylose inducible promoters were identified from DNA microarray data generated during growth of C. acetobutylicum ATCC 824 on a mixture of glucose and xylose (Grimmler et al. 2010). In the described experiments, RNA samples were taken from cells grown in a phosphate-limited continuous culture in which either glucose or xylose was the sole carbon source. By comparing the expression profiles of the two cultures, it was possible to identify three genes that were significantly up regulated when growing on xylose compared to glucose. These were CAC1339 (Putative sugar-proton symporter, araE), CAC1344 (Sugar kinase, possible xylulose kinase, xylB) and CAC2612 (Xylulose kinase, xylB). Accordingly, the DNA fragments encompassing the promoter regions of these three genes were cloned upstream of the promoter-less catP gene of pMTL82254 to generate the plasmids pMTL82254-Pcac1339, pMTL82254-Pcac1344 and pMTL82254-Pcac2612. Each plasmid was transformed into C. acetobutylicum as described, along with the positive control of plasmid pMTL82254-Pfdx (catP reporter gene expressed from the promoter of the C. pasteurianum ferredoxin gene, (Zhang, Grosse-Honebrink and Minton 2015) and the negative control of plasmid pMTL82254 that contained a catP reporter gene without a promoter (Heap et al. 2009). The erythromycin resistant transformants obtained were purified by restreaking on CGM agar supplemented with erythromycin and then used in comparative experiments, growing either on glucose or xylose, and measuring production of CAT. Briefly, cells carrying each of the five plasmids were grown up overnight in 5 ml of CGM broth containing erythromycin (20 μg ml−1) and a 1.5 ml aliquot was used to inoculate two batches of 30 ml of P2 medium (Baer, Blaschek and Smith 1987) containing either 5% (w/v) glucose or 5% (w/v) xylose as the carbon source. The cultures were then incubated until their OD600 reached 1.1, at which point (7 h for glucose grown cells and 17 h for xylose grown cells) the cells were harvested by centrifugation (13 000 rpm at 4°C for 10 min) and the pellets obtained were stored at −80°C. Lysates were then prepared and the levels of CAT present were determined by enzyme assay (as described in Materials and Methods). The data demonstrated (Fig. 1) that the cells carrying the plasmid (pMTL82254-Pcac1339) which incorporated the promoter of CAC1339 (Pcac1339) showed the highest CAT activity in the presence of xylose with a 33-fold increase compared to cells grown on glucose. This xylose-induced promoter activity is almost equivalent to the fdx promoter (Pfdx), one of the strongest promoters in Clostridium (Takamizawa et al. 2004). Production of CAT by cells carrying plasmids which containing the other two promoters was also induced when growing on xylose compared to glucose. However, the level of CAT activity in cells in which catP was under the transcriptional control of the Pcac1344 promoter was only one-tenth of that with the Pfdx promoter, while the Pcac2612 activity was apparently even lower (Fig. 1). Given the level of xylose induction achieved, and the relative promoter strength, it was concluded that the promoter of CAC1339 (araE) was the most appropriate transcriptional system for the controlled expression of the Himar1 C9 transposase gene expression in the planned transposon mutagenesis system.
Figure 1.
CAT activities showing the strength of the promoters of the CAC1339, CAC1344, CAC2612 genes of C. acetobutylicum compared to the promoter of the fdx gene of C. pasteurianum in the presence of either glucose (grey) or xylose (black) as the sole carbon source. Data are representative of three replicates.
Construction of a xylose inducible mariner-based transposon system for C. acetobutylicum
To construct the transposon delivery vector, an appropriate Clostridium replicon needs to be selected to facilitate rapid plasmid loss after the transposition events. Segregational stability studies showed that the replicon of pCB102 is the most segregationally unstable in C. acetobutylicum ATCC 824, suggesting that it can be used as a ‘pseudo-suicide vector’ (Cartman and Minton 2010) in the envisaged xylose inducible transposon system. After swapping the existing Gram-positive pBP1 replicon in plasmid pMTL-SC0 (Cartman and Minton 2010) with pCB102, and the cloning of the Pcac1339 promoter proximal to the Himar1 C9 transposase gene, the desired xylose inducible mariner transposon plasmid was generated and designated pMTL-YG3 (Fig. 2).
Isolation and analysis of transposon mutants
To test its effectiveness as a transposon delivery system, the plasmid pMTL-YG3 was transformed into C. acetobutylicum and transformants were selected by plating on CGM agar plates containing glucose as the sole carbon source and supplemented with erythromycin. After incubated at 37°C for 48 h, all transformant colonies were harvested from the agar surface and resuspended in 6 ml of CGM broth and replated as 30 × 0.2 ml aliquots on 30 individual CGM agar plates containing xylose as the sole carbon source. In this case, the agar media was also supplemented with thiamphenicol to select for transposon events. After 48 h, approximately 1000 colonies were visible on each agar plate. All of the colonies were scraped from each of the 30 plates (roughly 30 000) and resuspended in CGM broth containing 10% (v/v) glycerol for storage purposes. To eliminate the transposon delivery vector, the harvested pools of transposon mutants were passaged a total of three times through liquid CGM medium containing glucose as the sole carbon source but lacking any antibiotic supplementation (see Materials and Methods). Serial dilutions of the cell suspension obtained were plated onto CGM agar plates in triplicate. To determine the transposon insertion rate and percentage of plasmid loss, CFU ml−1 were estimated on CGM agar plates containing no supplementation or agar media supplemented with either thiamphenicol (Tm) or erythromycin (Em). After 48 h of incubation, 358, 329 and 7 colonies were visible on the agar plate without any antibiotics, supplemented with Tm or Em, respectively. These data demonstrated over 90% of the total colonies contained a transposon insertion, and only a small fraction (about 2%) of those still harbored the transposon delivery vehicle at the final plating stage. A total of 69 colonies from plates supplemented with Tm were picked and patch plated onto appropriate solidified rich media containing either Tm or Em to confirm the percentage of cells that had lost the plasmid. All 69 colonies were Tm resistant and Em sensitive, indicating that they carry transposon insertions in the chromosome and not the transposon delivery plasmid.
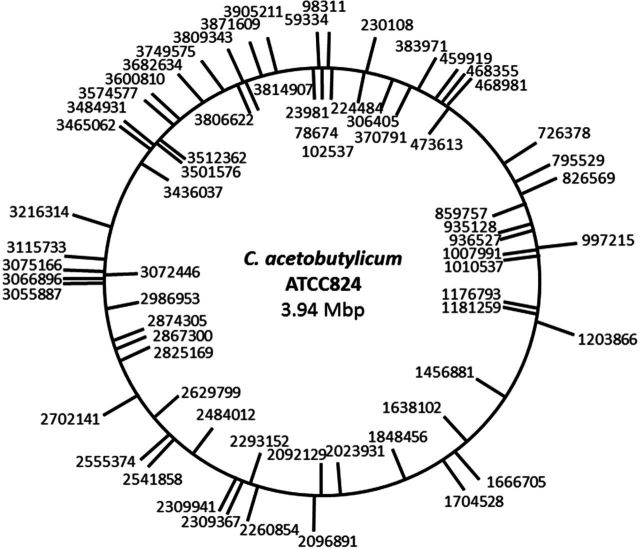
To determine the insertion sites and establish the randomness of the system, genomic DNA was isolated from the above 69 clones and subjected to INV PCR as described (Cartman and Minton 2010). Nucleotide sequencing of the amplified DNA fragments demonstrated that all 69 clones had a single transposon insertion distributed at random around the genome (Fig. 3), indicating that there was no preferred target site within the genome of C. acetobutylicum ATCC 824. Overall, there were 36 insertions in the plus strand and 33 in the minus strand. Moreover, according to the sequencing results, 58 of the 69 insertions sequenced (84%) were located within encoding regions.
Figure 3.
Genetic map of mariner transposon insertions. A total of 69 independent transposon insertions were sequenced. Insertions in the plus orientation are marked on the circle exterior. Insertions in the minus orientation are marked on the circle interior. Numbers indicate the precise point of insertion according to genome sequence data for C. acetobutylicum ATCC 824 (NCBI Ref Seq number NC_ 003030.1; GenBank accession number AE001437) (Nolling et al. 2001).
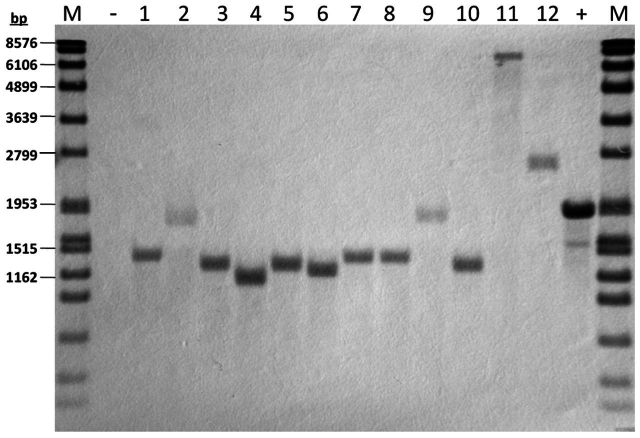
To further examine the independence of each transposition event, Southern blot analysis was performed, in which 12 Tmr colonies from above mentioned mutants library, a wild-type strain (negative control) and the plasmid pMTL-YG3 (positive control) were adopted. As shown in Fig. 4, all 12 colonies had a single transposon insertion, thus, indicating that all transposition events here were a single transposon insertion in the genome.
Figure 4.
Southern blot analysis of mariner transposon insertions in the genome of C. acetobutylicum. M: marker; ‘−’: wild-type strain; ‘+’: plasmid pMTL-YG3. Lane 1–12: colonies harboring transposon insertions.
Phenotype screens and identification of transposon insertions
To demonstrate that the developed xylose inducible mariner-based transposon system can be used for the isolation of mutants with a desired phenotype, over 200 mutants were tested for defects in germination/sporulation. Briefly, the mutant library was plated onto CGM agar plates (supplemented with Tm). When colonies were visible on the plates after incubation at 37°C for 48–72 h, replica cultures of over 200 mutant colonies were inoculated into fresh CBM liquid broth (O'Brien and Morris 1971) in 96-well microtitre plates and anaerobically incubated for two weeks to form spores. The cultures in one 96-well microtitre plate were subjected to heat treatment (80°C for 10 min) and then plated onto CGM agar plate supplemented with Tm, while the cultures in the replicated 96-well microtitre plate were directly plated onto CGM agar plate. Fortunately, we obtained a mutant which was defective in its ability to form colonies after heat shock but that was still able to generate colonies in the absence of a heat treatment. By sequencing the INV PCR products of this spo_ mutant, we were able to show that it contained an insertion in the spo0A gene. This gene encodes the master regulator of sporulation, Spo0A, and has been found to be essential for both sporulation and solvent production in C. acetobutylicum (Harris, Welker and Papoutsakis 2002). Although no novel genes affecting sporulation were identified, a consequence of the low number of mutants screened, the successful isolation of the spo0A mutant did demonstrate that in principle the transposon mutagenesis system developed can be used for forward genetic studies.
In summary, we have constructed a novel random mutagenesis system in which the production of the mariner transposase, and thereby transposition events, is controlled through the use of a xylose inducible promoter. Induction is, therefore, simply controlled through the temporal presence of xylose in the agar medium employed. The use of an inducible promoter to control transposase production may overcome the drawbacks of using constitutive promoters, i.e. early-stage transposition events immediately after electro-transformation which would result in a lack of randomness of the mutant library.
FUNDING
This work was supported by the China Partnering Award of the (), and and . NPM and YZ would also like to acknowledge the financial support of the grant numbers and .
Conflict of interest. None declared.
REFERENCES
- Al-Hinai MA, Fast AG, Papoutsakis ET. Novel system for efficient isolation of Clostridium double-crossover allelic exchange mutants enabling markerless chromosomal gene deletions and DNA integration. Appl Environ Microb. 2012;78:8112–21. doi: 10.1128/AEM.02214-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer SH, Blaschek HP, Smith TL. Effect of butanol challenge and temperature on lipid composition and membrane fluidity of butanol-tolerant Clostridium acetobutylicum. Appl Environ Microb. 1987;53:2854–61. doi: 10.1128/aem.53.12.2854-2861.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram J, Kuhn A, Durre P. Tn916-induced mutants of Clostridium acetobutylicum defective in regulation of solvent formation. Arch Microbiol. 1990;153:373–7. [Google Scholar]
- Cartman ST, Minton NP. A mariner-based transposon system for in vivo random mutagenesis of Clostridium difficile. Appl Environ Microb. 2010;76:1103–9. doi: 10.1128/AEM.02525-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembek M, Barquist L, Boinett CJ, et al. High-throughput analysis of gene essentiality and sporulation in Clostridium difficile. MBio. 2015;6:e02383. doi: 10.1128/mBio.02383-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai RP, Papoutsakis ET. Antisense RNA strategies for metabolic engineering of Clostridium acetobutylicum. Appl Environ Microb. 1999;65:936–45. doi: 10.1128/aem.65.3.936-945.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehsaan M, Kuit W, Zhang Y, et al. Mutant generation by allelic exchange and genome resequencing of the biobutanol organism Clostridium acetobutylicum ATCC 824. Biotechnol Biofuels. 2016;9:4–23. doi: 10.1186/s13068-015-0410-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimmler C, Held C, Liebl W, et al. Transcriptional analysis of catabolite repression in Clostridium acetobutylicum growing on mixtures of D-glucose and D-xylose. J Biotechnol. 2010;150:315–23. doi: 10.1016/j.jbiotec.2010.09.938. [DOI] [PubMed] [Google Scholar]
- Harris LM, Welker NE, Papoutsakis ET. Northern, morphological, and fermentation analysis of spo0A inactivation and overexpression in Clostridium acetobutylicum ATCC 824. J Bacteriol. 2002;184:3586–97. doi: 10.1128/JB.184.13.3586-3597.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmanis MG, Gatenbeck S. Intermediary metabolism in Clostridium acetobutylicum: levels of enzymes involved in the formation of acetate and butyrate. Appl Environ Microb. 1984;47:1277–83. doi: 10.1128/aem.47.6.1277-1283.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heap JT, Cartman ST, Kuehne SA, et al. ClosTron-targeted mutagenesis. Methods Mol Biol. 2010a;646:165–82. doi: 10.1007/978-1-60327-365-7_11. [DOI] [PubMed] [Google Scholar]
- Heap JT, Ehsaan M, Cooksley CM, et al. Integration of DNA into bacterial chromosomes from plasmids without a counter-selection marker. Nucleic Acids Res. 2012;40:e59. doi: 10.1093/nar/gkr1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heap JT, Kuehne SA, Ehsaan M, et al. The ClosTron: mutagenesis in Clostridium refined and streamlined. J Microbiol Meth. 2010b;80:49–55. doi: 10.1016/j.mimet.2009.10.018. [DOI] [PubMed] [Google Scholar]
- Heap JT, Pennington OJ, Cartman ST, et al. The ClosTron: a universal gene knock-out system for the genus Clostridium. J Microbiol Meth. 2007;70:452–64. doi: 10.1016/j.mimet.2007.05.021. [DOI] [PubMed] [Google Scholar]
- Heap JT, Pennington OJ, Cartman ST, et al. A modular system for Clostridium shuttle plasmids. J Microbiol Meth. 2009;78:79–85. doi: 10.1016/j.mimet.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Jang YS, Lee JY, Lee J, et al. Enhanced butanol production obtained by reinforcing the direct butanol-forming route in Clostridium acetobutylicum. MBio. 2012;3:2150–7511. doi: 10.1128/mBio.00314-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashket ER, Cao ZY. Isolation of a degeneration-resistant mutant of Clostridium acetobutylicum NCIMB 8052. Appl Environ Microb. 1993;59:4198–202. doi: 10.1128/aem.59.12.4198-4202.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leang C, Ueki T, Nevin KP, et al. A genetic system for Clostridium ljungdahlii: a chassis for autotrophic production of biocommodities and a model homoacetogen. Appl Environ Microb. 2013;79:1102–9. doi: 10.1128/AEM.02891-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Bouillaut L, Sonenshein AL, et al. Use of a mariner-based transposon mutagenesis system to isolate Clostridium perfringens mutants deficient in gliding motility. J Bacteriol. 2013;195:629–36. doi: 10.1128/JB.01288-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liyanage H, Young M, Kashket ER. Butanol tolerance of Clostridium beijerinckii NCIMB 8052 associated with down-regulation of gldA by antisense RNA. J Mol Microb Biotech. 2000;2:87–93. [PubMed] [Google Scholar]
- Mattsson DM, Rogers P. Analysis of Tn916-induced mutants of Clostridium acetobutylicum altered in solventogenesis and sporulation. J Ind Microbiol. 1994;13:258–68. doi: 10.1007/BF01569758. [DOI] [PubMed] [Google Scholar]
- Mermelstein LD, Papoutsakis ET. In vivo methylation in Escherichia coli by the Bacillus subtilis phage Φ3T I methyltransferase to protect plasmids from restriction upon transformation of Clostridium acetobutylicum ATCC 824. Appl Environ Microb. 1993;59:1077–81. doi: 10.1128/aem.59.4.1077-1081.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermelstein LD, Welker NE, Bennett GN, et al. Expression of cloned homologous fermentative genes in Clostridium acetobutylicum ATCC 824. Biotechnology (N Y) 1992;10:190–5. doi: 10.1038/nbt0292-190. [DOI] [PubMed] [Google Scholar]
- Minton NP, Morris JG. Isolation and partial characterization of 3 cryptic plasmids from strains of Clostridium butyricum. J Gen Microbiol. 1981;127:325–31. [Google Scholar]
- Nolling J, Breton G, Omelchenko MV, et al. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J Bacteriol. 2001;183:4823–38. doi: 10.1128/JB.183.16.4823-4838.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien RW, Morris JG. Oxygen and the growth and metabolism of Clostridium acetobutylicum. J Gen Microbiol. 1971;68:307–18. doi: 10.1099/00221287-68-3-307. [DOI] [PubMed] [Google Scholar]
- Scotcher MC, Rudolph FB, Bennett GN. Expression of abrB310 and sinR, and effects of decreased abrB310 expression on the transition from acidogenesis to solventogenesis, in Clostridium acetobutylicum ATCC 824. Appl Environ Microb. 2005;71:1987–95. doi: 10.1128/AEM.71.4.1987-1995.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao L, Hu S, Yang Y, et al. Targeted gene disruption by use of a group II intron (targetron) vector in Clostridium acetobutylicum. Cell Res. 2007;17:963–5. doi: 10.1038/cr.2007.91. [DOI] [PubMed] [Google Scholar]
- Shaw WV. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Method Enzymol. 1975;43:737–55. doi: 10.1016/0076-6879(75)43141-x. [DOI] [PubMed] [Google Scholar]
- Takamizawa A, Miyata S, Matsushita O, et al. High-level expression of clostridial sialidase using a ferredoxin gene promoter-based plasmid. Protein Expres Purif. 2004;36:70–5. doi: 10.1016/j.pep.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Tummala SB, Junne SG, Papoutsakis ET. Antisense RNA downregulation of coenzyme A transferase combined with alcohol-aldehyde dehydrogenase overexpression leads to predominantly alcohologenic Clostridium acetobutylicum fermentations. J Bacteriol. 2003;185:3644–53. doi: 10.1128/JB.185.12.3644-3653.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang ZT, Seo SO, et al. Markerless chromosomal gene deletion in Clostridium beijerinckii using CRISPR/Cas9 system. J Biotechnol. 2015;200:1–5. doi: 10.1016/j.jbiotec.2015.02.005. [DOI] [PubMed] [Google Scholar]
- Woolley RC, Pennock A, Ashton RJ, et al. Transfer of Tn1545 and Tn916 to Clostridium acetobutylicum. Plasmid. 1989;22:169–74. doi: 10.1016/0147-619x(89)90027-9. [DOI] [PubMed] [Google Scholar]
- Xu T, Li Y, Shi Z, et al. Efficient genome editing in Clostridium cellulolyticum via CRISPR-Cas9 nickase. Appl Environ Microb. 2015;81:4423–31. doi: 10.1128/AEM.00873-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Grosse-Honebrink A, Minton NP. A universal mariner transposon system for forward genetic studies in the genus Clostridium. PLoS One. 2015;10:e0122411. doi: 10.1371/journal.pone.0122411. [DOI] [PMC free article] [PubMed] [Google Scholar]